Abstract
Alzheimer's disease (AD) is a common worldwide progressive neurodegenerative disease. The dysregulation of miRNA is crucial in neurodegenerative diseases and neuron apoptosis during AD and is closely associated with the MAPK pathway. By bioinformatic website, we found that there was target inhibiting relationship between microRNA (miR)-132 and MAPK1. Therefore, the current study speculated that miR-132 could improve the cognitive function of rats with AD by inhibiting MAPK1 expression. To verify our hypothesis, 10 normal rats and 60 rats with AD were selected and divided into model, Ad-miR-132 negative control (NC), Ad-miR-132, Ad-small interfering (si)MAPK1 NC, Ad-siMAPK1 and Ad-miR-132 + Ad-MAPK1 groups. Rats were evaluated for learning by performing morris water maze tests and pathological changes of the hippocampus were assessed via HE staining. Additionally, hippocampus cell apoptosis was determined using a TUNEL assay and levels of acetylcholinesterase (AChE), reactive oxygen species (ROS), malondialdehyde (MDA), superoxide dismutase (SOD) and glutathione peroxidase (GSH-Px) were evaluated in sera via ELISA. The mRNA and protein expression of miR-132, iNOS, MAPK1 and phosphorylated (p)-MAPK1 was determined in hippocampus tissues via reverse transcription-quantitative PCR and western blotting, respectively. Compared with normal mice, rats with AD had significantly decreased learning abilities, increased cell apoptosis rates, increased levels of AChE, iNOS, ROS, MDA, MAPK1 and p-MAPK1 and decreased levels of SOD, GSH-Px and miR-132. Upregulation of miR-132 group improved the above indictors and silencing MAKP1 worsened the condition of rats. miR-132 upregulation therefore reversed the negative effects caused by MAPK1 silencing in rats with AD. In conclusion, miR-132 inhibited hippocampal iNOS expression and oxidative stress by inhibiting MAPK1expression to improve the cognitive function of rats with AD.
Keywords: miR-132, MAPK1, iNOS, oxidative stress, cognitive function
Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disease that impacts ~50 million people per year, worldwide. In addition, AD it is the most common form (60-80%) of dementia (1). Two major risk factors for AD are traumatic brain injury and cerebrovascular diseases (2). Decreased function of the central cholinergic system leads to impaired cognitive ability that is positively correlated with acetylcholinesterase (AchE) activity, which reflects the state of cellular metabolism and the activity of cholinergic neurons (3). Currently, there is no cure for Alzheimer's disease (AD). Acetylcholinesterase inhibitors and N-methyl-D-aspartate receptor antagonists are the main drugs used for the clinical treatment of AD (4). The mean age of AD patients was over 70 years old, and they would die without effective treatment within 10 years after diagnosis (5,6).
Eukaryotes have an evolutionary defense system against the destructive effect of reactive oxygen species (ROS) overproduction, which is considered crucial for brain health (7,8). Malondialdehyde (MDA), superoxide dismutase (SOD) and glutathione peroxidase (GSH-Px) are common indicators of oxidative Stress (9). Nitric oxide synthase (iNOS) is a gas intercellular signaling carrier. Jiang et al found that inhibiting iNOS could reduce the risk of Alzheimer's disease in rat (10). In the last few decades, great progresses have been made in understanding early onset familial and delayed sporadic AD at molecular levels (11-14).
Numerous biological processes and >60% human genes are regulated by microRNAs (miR) than can be found in most body tissues, including brain tissues, cerebrospinal fluid and serum (15). Dysregulation of miRNA is therefore crucial in neurodegenerative diseases (16). Previous in vitro and in vivo studies have explored the role of miRNAs in the pathogenesis of AD, and demonstrated that miR-92a-3p, miR-181c-5p and miR-210-3p, miRNA-132, miRNA-107 are in abnormal high or low expression level in the brains of patients with AD (17-19). miRNA-22 over-expression may be neuroprotective for neurodegenerative diseases as well as neurodevelopmental disorders by inhibiting apoptosis (20). For instance, miR-132 contributes to dendritic growth of newborn neurons in the hippocampus of adult mice (21). Furthermore, deletion of miR-132 and miR-212 results in induction of tau aggregation and impairment of cognitive skills in mice (22). In addition, expressions of miR-212 and miR-23a are upregulated in post-mortem frontal cortex tissues of patients with AD or with mild cognitive impairment (23).
Neuron apoptosis during AD is closely associated with MAPK pathway (24). MAPKs are serine-threonine kinases that are naturally highly expressed in the central nervous system. Cellular activity includes proliferation, differentiation, survival, death, and transformation (25). The mammalian MAPK family consists of p38 MAPK, ERK and c-Jun NH2-terminal kinases. The p38 signaling pathway is involved in the increase of inflammation and apoptosis subsequent to the overproduction of ROS during oxidative stress, and MAPK1 is one of the important members of the family (26).
Therefore, we hypothesized that miR-132 may be able to downregulate the expression of MAPK1, thus inhibiting the p38 signaling pathway and protecting the brain tissues of rats with AD from inflammatory injury and apoptosis.
Materials and methods
Animals
A total of 70 SPF Sprague-Dawley rats (weight, 235±5 g) were purchased from the Experimental Animal Center of Capital Medical University. A rat model of AD was established by intracerebroventricular administration of 20 µg Aβ25-35 (4 µg/µl, 5 µl; purity ≥97%, Sigma) as previously described (27). Briefly, rats were anesthetized with intraperitoneal injection of 3% pentobarbital sodium (30 mg/kg) and their head was fixed in the table by stereotaxic device (Ruiwode Life Technology Co., Ltd, China) for later injection. Subsequently, a hole was drilled on the right parietal bone (anteroposteriorly, 1mm; laterally right, 1.5 mm; dorsoventrally, 4 mm). Model rats were received intracerebroventricular injection of 5 µl of Aβ25-35 solution (4 µg/µl) at the speed of 1 µl/min and the normal rats were injected with normal saline by same method. The Aβ25-35 solution was incubated at 37˚C for 96 h to induce aggregation before usage. The rats were free to move and eat after surgery.
Rats were separated into seven groups of 10 rats as follows: Normal group (untreated rats), model group (rat model of AD), Ad-miR-132 negative control (NC) group (rats injected with negative control of miR-132 adenovirus vector in the hippocampal CA1 region), Ad-miR-132 group (model rats injected with miR-132 adenovirus vector in the hippocampal CA1 region), Ad-small interfering (si)MAPK1 NC (model rats injected with negative control of siRNA adenovirus vector of MAPK1 gene in the hippocampal CA1 region), Ad-siMAPK1 (model rats injected with siRNA adenovirus vector of MAPK1 in the hippocampal CA1 region), and Ad-miR-132 + Ad-MAPK1 group (model rats injected with miR-132 adenovirus vector and overexpressing adenovirus vector of MAPK1 in the hippocampal CA1 region). The adenoviral vectors used in this experiment were purchased from Tianjin Saierbio Biotechnology Co., Ltd.. After one week, rats were anesthetized with intraperitoneal injection of 3% pentobarbital sodium (30 mg/kg) and, after removal of eyeballs, 0.5 ml of blood samples were collected from the ophthalmic vein, and brains were harvested. Brain tissues and venous blood of five rats per group were used for detection of AChE, ROS, MDA, SOD and GSH-Px in serum. A part of hippocampus tissue was fixed with 10% neutral formalin solution at room temperature for 24 h, dehydrated by gradient alcohol (30-100%), and then embedded in paraffin. The remaining part of brain tissue was stored in liquid nitrogen for further experiments.
Rats were euthanized in the following situations: i) During the study, a rat showed weight loss (rapid loss of 20% of the original weight), loss of appetite (complete loss of appetite for 24 h or 50% loss of appetite for 3 days), would not voluntary eat or drink, or failed to or were reluctant in standing; ii) euthanasia was performed for tissue collection and subsequent experiments. Rats were sacrificed by rapid cervical dislocation following anesthesia with intraperitoneal injection of 3% pentobarbital sodium (30 mg/kg). The death of rats was confirmed by the absence of breath and heartbeat and when rats showed pupil dilation. The experiment was conducted in accordance with the 3R principles and approved by the Animal Ethics Committee of Beijing Tiantan Hospital, Capital Medical University.
Dual-luciferase reporter assay
The binding site of miR-132 to the 3'-UTR of the MAPK1 gene was analyzed via the biological prediction website microRNA.org (http://www.microrna.org/microrna/home.do). The screening of 3'-UTR on this website was mainly analyzed from three aspects: The sequence matching, the thermal stability of the double strand of miRNA and the mRNA and the conservation of target sites. Subsequently, the targeting relationship between miR-132 and MAPK1 was verified by dual-luciferase reporter assay. The 3'-UTR of the MAPK1 gene and the 3'-UTR of the mutated MAPK1 gene were inserted into the luciferase reporter gene vector pGL3-Basic and named PGL3-MAPK1 wild-type (WT) and PGL3-MAPK1 mutant (MUT) respectively (Shanghai GenePharma Co., Ltd.). The Renila Luciferase internal reference plasmid and the two reporter vectors were co-transfected into HEK 293T cells (American Type Culture Collection) with Ad-miR-132 and Ad-miR-132 NC by Lipofectamine 3000 (Thermo Fisher Scientific, USA). After 24 h transfection, dual luciferase assay was performed. Protein of cells in each group was extracted using RIPA (cat. no. R0010; Beijing Solarbio Science & Technology Co., Ltd.) containing PMSF (0.1 mM). The detection of luciferase activity was performed using a kit from Promega Corporation according to the manufacturers' instructions. The relative luciferase activity=firefly luciferase activity/Renilla luciferase activity (15).
Reverse transcription quantitative (RT-q) PCR
Total RNA was extracted from brain tissues using TRIzol (Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into cDNA using TaqMan MicroRNA Assays Reverse Transcription Primer (Thermo Fisher Scientific, Inc.). Quantitative PCR was performed using SYBR® Premix Ex Taq™ II Kit (Takara). The following components were added in the mixture: 25 µl of SYBR® Premix Ex Taq™ II (2x), 2 µl of PCR upstream and downstream primers, 1 µl of ROX Reference Dye (50x), 4 µl of DNA template and 16 µl of ddH2O. Fluorescence quantitative PCR was performed with ABI PRISM® 7300 (Kunke Equipment Co., Ltd., Shanghai, China). The reaction conditions were as follows: 10 min pre-denaturation at 95˚C, followed by 32 cycles at 95˚C for 15 sec and 60˚C for 30 sec, and 72˚C for 1 min. The relative expression levels were normalized to endogenous control U6 and were expressed as 2-ΔΔCq and calculated as follows (28): ΔCt=Ct (target gene)-Ct (U6) and ΔΔCt=ΔCt (experimental group)-ΔCt (control group). The sequences of the primers used are present in Table I.
Table I.
Sequences of the primers used for reverse transcription quantitative PCR.
| Name | Sequences |
|---|---|
| miR-132 | |
| Forward | 5'-TGGATCCCCCCCAGTCCCCGTC CCTCAG-3' |
| Reverse | 5'-TGAATTCGGATACCTTGGCCGG GAGGAC-3' |
| U6 | |
| Forward | 5'-CTCGCTTCGGCAGCACA-3' |
| Reverse | 5'-AACGCTTCACGAATTTGCGT-3' |
Western blotting
Total protein was extracted from brain tissues using RIPA (cat. no. R0010; Beijing Solarbio Science & Technology Co., Ltd.) containing PMSF (0.1 mM). The protein concentration was determined using BCA kit (Thermo Fisher Scientific, Inc.). The sample was mixed with the loading buffer and heated at 100˚C in a water bath for 10 min. Proteins (50 µg) were separated by 10% SDS-PAGE and transferred onto a PVDF membrane (cat. no. ISEQ00010; EMD Millipore). Membranes were blocked using 5% skim milk at 4˚C for 2 h and washed with 0.1% TBST. Membranes were incubated with primary antibodies against phosphorylated (p) p38MAPK (ab31828; 1:1,000; Abcam), MAPK1 (ab31828; 1:5,000; Abcam), iNOS (ab213987; 1:5,000; Abcam) and GAPDH (ab22555; 1:2,000; Abcam) overnight at 4˚C. After three washes with TBST for 6 min, membranes were incubated with the secondary HRP-labeled goat anti-rabbit IgG antibody (TA140003; 1:5,000; OriGene Technologies, Inc.) at room temperature for 2 h. Membranes were washed three times with TBST for 6 min and placed into TBS. Enhanced chemiluminescence reagent (BB-3501; Cytiva) was used to detect the signal on the membrane. Images were acquired on a Bio-Rad image analysis system (Bio-Rad Laboratories, Inc.). The data were analyzed via densitometry using ImageJ software V2.1.4.7 (National Institutes of Health) and normalized to expression of the internal control GAPDH.
Morris water maze
The water maze is a circular pool (150 cm in diameter and 60 cm in height) filled with water at the temperature of 20-25˚C. The pool was divided into four quadrants as follows: Lower right, upper right, lower left and upper left, with a platform installed in the lower right quadrant. After 5 days of navigation training, the rats were placed in water and the time to find the platform within 2 min (escape latency) was recorded. If the rat failed to locate the platform within 2 min, they would be guided to the platform and allowed to stay on it for 10 sec (recorded as 2 min). On the 5th day, the platform in the water was removed to conduct the space exploration experiment. Each rat was placed in the same place of the pool and the time spent in the lower right quadrant, along with the number of times the rat swum by the original platform position were recorded, which corresponds to the number of times passing through the platform.
Hematoxylin and eosin (H&E) staining
The brain tissues were fixed in 10% neutral formalin solution at room temperature for 24 h, dehydrated with gradient alcohol (30-100%), dewaxed with xylene, embedded in a wax bath and sliced (4-6 µm). Sections were dewaxed with xylene, hydrated with gradient alcohol (100-70%), washed with distilled water for 1 min, stained with hematoxylin for 3 min and then rinsed with tap water. Sections were immersed in 0.5% hydrochloric acid for 10 sec, immersed in water for 10 min and stained in eosin solution for 5 min. Slices were conventionally dehydrated and dewaxed again, and mounted by neutral gum. Sections were were observed under an optical microscope (XP-330; Shanghai Bingyu Industry Co., Ltd.). Five fields per section were randomly selected and the number of pyknotic nerve cells was determined.
TUNEL assay
Paraffin embedded sections were dewaxed, rehydrated by 100-70% gradient alcohol, immersed in 3% H2O2 for 12 min and incubated with proteinase K (20 µg/ml in Tris/HCl) for 30 min at room temperature. Sections were washed three times with PBS for 6 min and sections were incubated with 20 µg/ml proteinase K without DNase (ST533; Beyotime Institute of Biotechnology) at 37˚C for 15 min. Then, TUNEL reaction mixture was added and incubated at 37˚C for 1 h in a wet box (C1088; Beyotime Institute of Biotechnology). Samples were washed three times with PBS for 6 min and observed under a fluorescence microscope (ECLIPSE Ti; Nikon Corporation) and cells exhibiting green fluorescent were TUNEL-positive cells. The apoptotic index was calculated as follows: Apoptotic rate=number of TUNEL-positive cells/total number of cells x100.
Detection of AChE and iNOS in brain tissues and ROS, MDA, SOD and GSH-Px in serum
The levels of iNOS and AChE in brain tissues were determined using ELISA kits (cat. nos. 69-98762 and 69-30132, respectively; Merck KGaA). The blood sample were stand at room temperature for 2 h and centrifuged at 10,000 x g for 10 min to obtain serum. The levels of MDA and SOD in serum were measured with the use of ROS (cat. no. E004-1-1), MDA (cat. no. A003-1-2), GSH-Px (cat. no. A006-2-1), and SOD (cat. no. A001-3-3) kits from Nanjing Jiancheng Bioengineering Institute.
Statistical analyses
All data were analyzed using SPSS v21.0 statistical software (IBM Corp.). Data were expressed as the means ± standard deviation. The comparison among multiple groups was performed using one-way analysis of variance followed by Tukey post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-132 inhibits MAPK1 gene expression
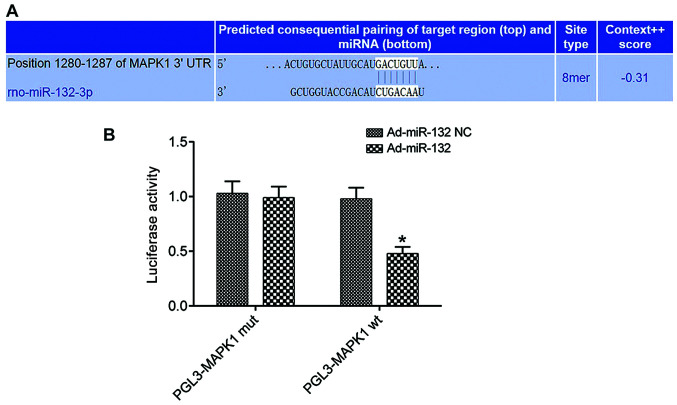
The biological prediction site microrna.org (http://www.microrna.org/microrna/home.do) predicted that miR-132 and MAPK1 had specific binding sites (Fig. 1A). The results from the dual-luciferase reporter assay demonstrated that the luciferase activity in the subgroup PGL3-MAPK1 WT of Ad-miR-132 group was significantly lower compared with that in the Ad-miR-132 NC group (P<0.05). However, the luciferase activity in the subgroup PGL3-MAPK1 MUT did not change significantly (P>0.05; Fig. 1B). miR-132 could inhibit the expression of MAPK1.
Figure 1.
miR-132 targeting negative regulation of MAPK1 gene in HEK 293T cells (n=3). (A) Sequence of the 3'-UTR segment of miR-132 binding to MAPK1. (B) Luciferase activity detected by dual luciferase assay. *P<0.05 vs. Ad-miR-132 NC group. wt, wild-type; mut, mutant; miR, microRNA; NC, negative control.
Expression of miR-132, MAPK1, p-MAPK1 and iNOS
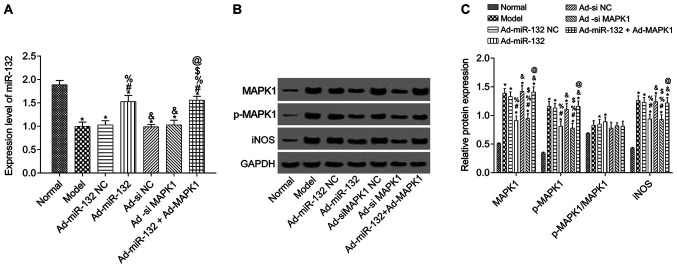
To further verify the results of dual-luciferase reporter assay, miR-132 expression level was detected by RT-qPCR and the expression of MAPK1, p-MAPK1 and iNOS was evaluated by western blotting in hippocampus tissue of rats with AD (Fig. 2). Compared with the Normal group, miR-132 expression level was significantly downregulated in model group, and expression of MAPK1, p-MAPK1 and iNOS was significantly upregulated (P<0.05). Compared with the Model group, miR-132 expression was significantly increased in the Ad-miR-132 group and the Ad-miR-132 + Ad-MAPK1 group (P<0.05). The other groups had similar levels of miR-132 expression (Fig. 2A). Furthermore, the expression of MAPK1, p-MAPK1 and iNOS in Ad-miR-132 NC group, Ad-siMAPK1 NC group and Ad-miR-132 Ad-MAPK1 group was similar to that of the Model group (P>0.05). However, the expression of MAPK1, p-MAPK1 and iNOS in Ad-miR-132 group and Ad-siMAPK1 group were significantly decreased compared with the Model group (all P<0.05). In addition, compared with Ad-miR-132 group, the expression of MAPK1, p-MAPK1 and iNOS in Ad-miR-132 + Ad-MAPK1 group was significantly increased (P<0.05, Fig. 2B and C).
Figure 2.
Expression of miR-132, MAPK1, p-MAPK1 and iNOS (n=5). (A) Expression level of miR-132 in rat brain tissues. (B) Protein expression of MAPK1, p-MAPK1 and iNOS in rat brain tissues. (C) Relative protein expression of MAPK1, p-MAPK1 and iNOS from (B). *P<0.05 vs. Normal group; #P<0.05 vs. Model group; %P<0.05 vs. Ad-miR-132 NC group; &P<0.05 vs. Ad-miR-132 group; $P<0.05 vs. Ad-siMAPK1 NC group; @P<0.05 vs. Ad-siMAPK1 group. iNOS, inducible nitric oxide synthase; NC, negative control; miR, microRNA; p, phosphorylated; si, small interfering.
Learning and memory ability
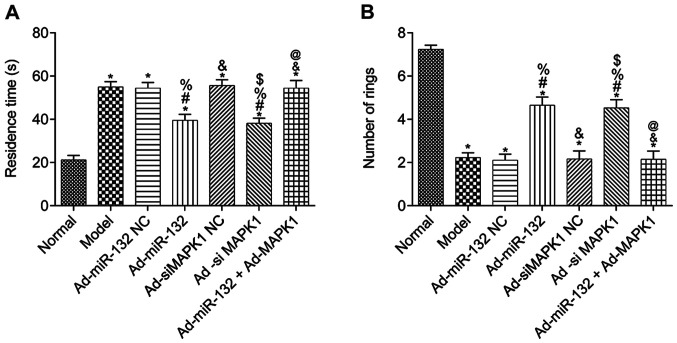
The results from the water maze test demonstrated that compared with the Normal group, the escape latency of rats in all other groups was significantly elevated, and the number of times passing through the rings was significantly decreased (P<0.05; Fig. 3A and B). Furthermore, compared with the Model group, there was no significant difference in Ad-miR-132 NC group, Ad-siMAPK1 NC group and Ad-miR-132 + Ad-MAPK1 group in the two experiments (P>0.05; Fig. 3A and B). However, the Ad-miR-132 group and Ad-siMAPK1 group had significantly decreased escape latency and significantly elevated number of times passing through the rings compared with the Model group (P<0.05; Fig. 3A and B). In addition, compared with the Ad-miR-132 group, the Ad-miR-132 + Ad-MAPK1 group had significantly elevated escape latency and significantly decreased number of times passing through the rings (P<0.05; Fig. 3A and B).
Figure 3.
Learning and memory ability of rats from different groups (n=10). (A) Latency time in water maze test. (B) Number of times passing through the rings. *P<0.05 vs. Normal group; #P<0.05 vs. Model group; %P<0.05 vs. Ad-miR-132 NC group; &P<0.05 vs. Ad-miR-132 group; $P<0.05 vs. Ad-siMAPK1 NC group; @P<0.05 vs. Ad-siMAPK1 group. NC, negative control; miR, microRNA; si, small interfering.
Pathological changes of brain tissues
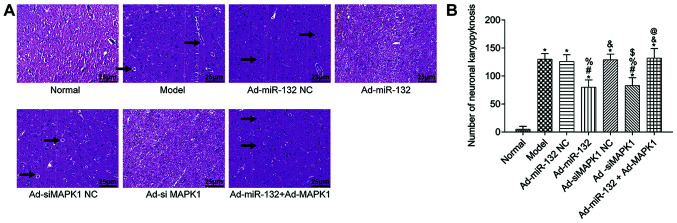
The pathological changes of rat hippocampus tissues were detected by HE staining (Fig. 4). The hippocampus tissue of Normal group showed regular structure and no obvious pathological damage (irregular structure, neuronal shrinkage and deep staining color). However, other groups presented with different degrees of pathological damage, which were less severe in the Ad-miR-132 group and the Ad-siMAPK1 group. The quantification results were consist with above description (Fig. 4B).
Figure 4.
Pathological of rat brain tissue from different groups (n=5). (A) Representative hematoxylin and eosin staining images of rat brain tissues in each group. Brain tissues from the Normal group showed regular structures without obvious pathological damage, whereas the other groups demonstrated pathological damage of varying degrees, with pyknosis of nerve cells (indicated by arrows). Damage degree was less severe in Ad-miR-132 group and Ad-siMAPK1 group. Magnification, x200. Scale bar, 25 µm. (B) Quantitative results of pathological damages. *P<0.05 vs. Normal group; #P<0.05 vs. Model group; %P<0.05 vs. Ad-miR-132 NC group; &P<0.05 vs. Ad-miR-132 group; $P<0.05 vs. Ad-siMAPK1 NC group; @P<0.05 vs. Ad-siMAPK1 group. NC, negative control; miR, microRNA; si, small interfering.
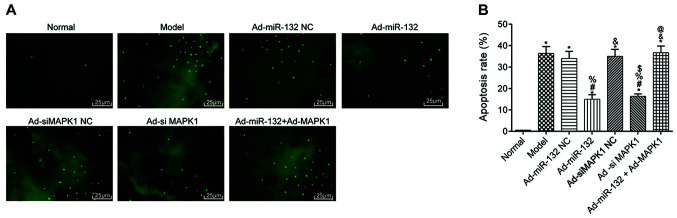
Apoptosis of brain neurons in rats
The apoptosis of hippocampal neurons was detected by TUNEL staining (Fig. 5). The results demonstrated that the apoptosis rate of hippocampus neurons was significantly in all groups compared with the Normal group (P<0.05). Compared with the Model group, there were no statistical difference in the apoptotic rate in the Ad-miR-132 NC group, Ad-siMAPK1 NC group and Ad-miR-132 + Ad-MAPK1 group (P>0.05). However, the Ad-miR-132 group and Ad-siMAPK1 group had significantly decreased apoptosis rate compared with the Model group (P<0.05). In addition, compared with Ad-miR-132 group, the apoptosis rate of hippocampus neurons in the Ad-miR-132 + Ad-MAPK1 group was significantly elevated (P<0.05).
Figure 5.
Apoptosis of hippocampal neurons in each group of rats (n=5; 400X). (A) Pathological changes of brain tissues detected by TUNEL staining. Magnification, x200. (B) Apoptosis rate of rat brain tissues according to (A). *P<0.05 vs. Normal group; #P<0.05 vs. Model group; %P<0.05 vs. Ad-miR-132 NC group; &P<0.05 vs. Ad-miR-132 group; $P<0.05 vs. Ad-siMAPK1 NC group; @P<0.05 vs. Ad-siMAPK1 group. NC, negative control; miR, microRNA; si, small interfering.
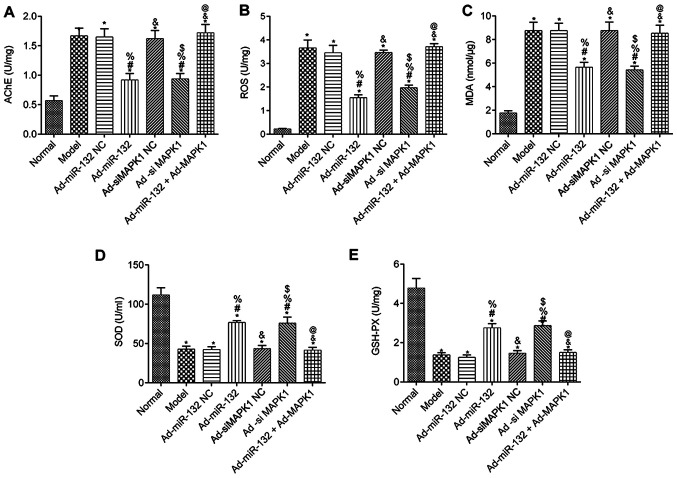
Levels of AChE, ROS, MDA, SOD and GSH-Px in serum of rats
The levels of AChE, ROS, MDA, SOD and GSH-Px in the serum of rats from each group are presented in Fig. 6. Compared with the Normal group, all groups had presented significantly decreased serum levels of SOD and GSH-Px, as well as significantly elevated AChE, ROS and MDA levels (P<0.05). There were no difference in the serum levels of AChE, ROS, MDA, SOD and GSH-Px between the Model group, Ad-miR-132 NC group, Ad-siMAPK1 NC group and Ad-miR-132 + Ad-MAPK1 group (P>0.05). Furthermore, compared with the Model group, the Ad-miR-132 group and Ad-siMAPK1 group presented significantly elevated levels of serum SOD and GSH-Px, and decreased levels of AChE, ROS and MDA (P<0.05). In addition, compared with the Ad-miR-132 group, the Ad-miR-132 + Ad-MAPK1 group had significantly decreased serum levels of SOD and GSH-Px, and elevated serum levels of AChE, ROS and MDA (P<0.05).
Figure 6.
Serums levels of AChE, ROS, MDA, SOD and GSH-PX in rats from different groups (n=5). Serum levels of (A) AChE, (B) ROS, (C) MDA, (D) SOD and (E) GSH-PX. *P<0.05 vs. Normal group; #P<0.05 vs. Model group; %P<0.05 vs. Ad-miR-132 NC group; &P<0.05 vs. Ad-miR-132 group; $P<0.05 vs. Ad-siMAPK1 NC group; @P<0.05 vs. Ad-siMAPK1 group. NC, negative control; miR, microRNA; si, small interfering; GSH-Px, glutathione peroxidase; MDA, malondialdehyde; ROS, reactive oxygen species; SOD, superoxide dismutase; AChE, acetylcholinesterase.
Discussion
AD is a progressive neurodegenerative disease characterized by loss of memory and cognitive function and is considered as a major cause of dementia (27). Recently, numerous potential biomarkers have been used in combination with therapeutic targets for the treatment of AD (29). For example, Lin et al (30) used Osthole to upregualte miRNA-101a-3p. Several miRNAs have been reported to be involved in a variety of diseases (16-19). Because of their stability and endogenous nature, miRNAs may be used as treatment options in AD (31).
Certain miRNAs can control the formation, maturation and function of synapse, and their abnormal expressions might be the basis of synaptic dysfunction (32). Furthermore, a number of specific miRNAs are dysregulated in patients with AD, including the miRNAs of key genes of AD, such as amyloid precursor protein or beta-secretase 1, or of neuronal functions, such as glutamate receptors (33). Che et al (34) demonstrated that miR-132 can regulate the angiogenesis of patients with cerebral ischemia through NF-κB and vascular endothelial growth factor pathways, and reported that overexpression of miR-132 in vitro could decrease iNOS expression. In the present study, specific binding sites of miR-132 and MAPK1 were found through bioinformatics analysis, and dual-luciferase reporter assay confirmed that miR-132 can negatively target MAPK1 gene. This study therefore hypothesized that miR-132 may be involved in the occurrence and development of AD by regulating MAPK signaling pathway, which may provide a promising new pathway for the treatment of AD.
In the present study, rats were treated with miR-132 analogs and siMAPK1, and the results demonstrated that after p38 signaling pathway was disturbed, the learning ability, memory and brain tissue disorder of AD rats were significantly improved, the apoptosis of nerve cells was decreased, and the serum levels of AChE was significantly decreased. Previous studies on neural and non-neuronal cells have demonstrated that p38 regulates apoptosis through a variety of mechanisms, including activation of p53, as well as phosphorylation of c-JUN and c-fos, induction of Bax transposition, and involvement in Fas-FasL-mediated apoptosis, enhancement of c-myc expression and activation of caspase-3 (35,36). In addition, p38 MAPK can enhance the expression of TNF-α, thereby activating p38-induced apoptosis (37). ERK is involved in cell activation and migration and plays an important role in synaptic plasticity and memory in vivo (38). Amyloid beta has been reported to induce JNK activation and cell death (39). Therefore, miR-132 may improve learning ability and memory, improving brain disorder in rats with AD and protecting nerve cells from apoptosis by inhibiting the p38 signaling pathway.
Numerous studies have reported that miRNAs can regulate oxidative stress (40-42). Increased oxidative stress can lead to cell apoptosis and serves a key role in neurodegenerative diseases, such as AD (42). In the present study, rats treated with miR-132 analogs and siMAPK1 presented with decreased serum levels of ROS and MDA, elevated serum levels of SOD and GSH-Px and downregulated iNOS. Overall, these results suggested that oxidative stress was decreased and that the antioxidant defense systems were stimulated. Che et al (34) reported that miR-132 overexpression in vitro inhibited iNOS, which is consistent with the present results. Subsequently, inhibition of p38 signaling pathway may improve brain damage, decrease oxidative stress and improve cognitive dysfunction.
In conclusion, the present study demonstrated that miR-132 inhibit iNOS expression in brain tissue, decreased oxidative stress and ameliorated the cognitive function in AD rats through p38 signaling pathway These findings may help understanding the pathogenesis of AD and may serve the development of novel clinical treatment. However, the association between miR-132 signaling pathway and AD are not fully understood, and the downstream molecules of the p38 signaling pathway need to be further investigated. In addition, since miR-132 and miR-212 are both related to AD, the role of miR-212 in AD requires further investigation.
Acknowledgements
Not applicable.
Funding
This study was supported by the Beijing Municipal Administration of Hospitals' Youth Program (grant no. QML20180506) and the Natural Science Foundation of Beijing (grant no. 17G10258).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LS contributed to manuscript concept. YD, JZ and XS carried out the experiments and analyzed data. GM, GL and ZM prepared the experimental data and performed statistical analysis. LS and YD involved in drafting the manuscript or revising it critically. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Animal Ethics Committee of Beijing Tiantan Hospital, Capital Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Herrera-Espejo S, Santos-Zorrozua B, Álvarez-González P, Lopez-Lopez E, Garcia-Orad Á. A systematic review of microRNA expression as biomarker of late-onset Alzheimer's disease. Mol Neurobiol. 2019;56:8376–8391. doi: 10.1007/s12035-019-01676-9. [DOI] [PubMed] [Google Scholar]
- 2.Pan Y, Liu R, Terpstra E, Wang Y, Qiao F, Wang J, Tong Y, Pan B. Dysregulation and diagnostic potential of microRNA in Alzheimer's disease. J Alzheimers Dis. 2016;49:1–12. doi: 10.3233/JAD-150451. [DOI] [PubMed] [Google Scholar]
- 3.Ossenkoppele R, Mattsson N, Teunissen CE, Barkhof F, Pijnenburg Y, Scheltens P, van der Flier WM, Rabinovici GD. Cerebrospinal fluid biomarkers and cerebral atrophy in distinct clinical variants of probable Alzheimer's disease. Neurobiol Aging. 2015;36:2340–2347. doi: 10.1016/j.neurobiolaging.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X, Cai H, Pan L, Cui G, Qin F, Li Y, Cai Z. Small molecule natural products and Alzheimer's disease. Curr Top Med Chem. 2019;19:187–204. doi: 10.2174/1568026619666190201153257. [DOI] [PubMed] [Google Scholar]
- 5.Panza GA, Taylor BA, MacDonald HV, Johnson BT, Zaleski AL, Livingston J, Thompson PD, Pescatello LS. Can exercise improve cognitive symptoms of Alzheimer's disease? J Am Geriatr Soc. 2018;66:487–495. doi: 10.1111/jgs.15241. [DOI] [PubMed] [Google Scholar]
- 6.Torvinen-Kiiskinen S, Tolppanen AM, Koponen M, Tanskanen A, Tiihonen J, Hartikainen S, Taipale H. Proton pump inhibitor use and risk of hip fractures among community-dwelling persons with Alzheimer's disease-a nested case-control study. Aliment Pharmacol Ther. 2018;47:1135–1142. doi: 10.1111/apt.14589. [DOI] [PubMed] [Google Scholar]
- 7.Nikolova G, Mancheva V. Analysis of the parameters of oxidative stress in patients with Parkinson's disease. Com Clin Pathol. 2012;22:151–155. [Google Scholar]
- 8.Bouchez C, Devin A. mitochondrial biogenesis and mitochondrial reactive oxygen species (ROS): A complex relationship regulated by the cAMP/PKA signaling pathway. Cells. 2019;8(287) doi: 10.3390/cells8040287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gökçe Çokal B, Yurtdaş M, Keskin Güler S, Güneş HN, Ataç Uçar C, Aytaç B, Durak ZE, Yoldaş TK, Durak İ, Çubukçu HC. Serum glutathione peroxidase, xanthine oxidase, and superoxide dismutase activities and malondialdehyde levels in patients with Parkinson's disease. Neurol Sci. 2017;38:425–431. doi: 10.1007/s10072-016-2782-8. [DOI] [PubMed] [Google Scholar]
- 10.Jiang P, Li C, Xiang Z, Jiao B. Tanshinone IIA reduces the risk of Alzheimer's disease by inhibiting iNOS, MMP-2 and NF-κBp65 transcription and translation in the temporal lobes of rat models of Alzheimer's disease. Mol Med Rep. 2014;10:689–694. doi: 10.3892/mmr.2014.2254. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, Reddy AP, Yin X, Reddy PH. Novel MicroRNA-455-3p and its protective effects against abnormal APP processing and amyloid beta toxicity in Alzheimer's disease. Biochim Biophys Acta Mol Basis Dis. 2019;1865:2428–2440. doi: 10.1016/j.bbadis.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson YS, Raby S, Foulds PG, Robinson A, Thompson JC, Sikkink S, Yusuf I, Amin H, DuPlessis D, Troakes C, et al. TDP-43 pathological changes in early onset familial and sporadic Alzheimer's disease, late onset Alzheimer's disease and Down's syndrome: Association with age, hippocampal sclerosis and clinical phenotype. Acta Neuropathol. 2011;122:703–713. doi: 10.1007/s00401-011-0879-y. [DOI] [PubMed] [Google Scholar]
- 13.Cruchaga C, Del-Aguila JL, Saef B, Black K, Fernandez MV, Budde J, Ibanez L, Deming Y, Kapoor M, Tosto G, et al. Polygenic risk score of sporadic late-onset Alzheimer's disease reveals a shared architecture with the familial and early-onset forms. Alzheimers Dement. 2018;14:205–214. doi: 10.1016/j.jalz.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin SC, Pastor P, Cooper B, Cervantes S, Benitez BA, Razquin C, Goate A. Pooled-DNA sequencing identifies novel causative variants in PSEN1, GRN and MAPT in a clinical early-onset and familial Alzheimer's disease Ibero-American cohort. Alzheimers Res Ther. 2012;4(34) doi: 10.1186/alzrt137. Ibero-American Alzheimer Disease Genetics Group Researchers and Cruchaga C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan Q, Si E. MicroRNA-25 aggravates Aβ1-42-induced hippocampal neuron injury in Alzheimer's disease by downregulating KLF2 via the Nrf2 signaling pathway in a mouse model. J Cell Biochem. 2019;120:15891–15905. doi: 10.1002/jcb.28861. [DOI] [PubMed] [Google Scholar]
- 16.Yang K, Feng S, Ren J, Zhou W. Upregulation of microRNA-196a improves cognitive impairment and alleviates neuronal damage in hippocampus tissues of Alzheimer's disease through downregulating LRIG3 expression. J Cell Biochem. 2019;120:17811–17821. doi: 10.1002/jcb.29047. [DOI] [PubMed] [Google Scholar]
- 17.Siedlecki-Wullich D, Català-Solsona J, Fábregas C, Hernández I, Clarimon J, Lleó A, Boada M, Saura CA, Rodríguez-Álvarez J, Miñano-Molina AJ. Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer's disease. Alzheimers Res Ther. 2019;11(46) doi: 10.1186/s13195-019-0501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luikart BW, Bensen AL, Washburn EK, Perederiy JV, Su KG, Li Y, Kernie SG, Parada LF, Westbrook GL. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS One. 2011;6(e19077) doi: 10.1371/journal.pone.0019077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jovicic A, Zaldivar Jolissaint JF, Moser R, Silva Santos Mde F, Luthi-Carter R. MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington's disease-related mechanisms. PLoS One. 2013;8(e54222) doi: 10.1371/journal.pone.0054222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith PY, Hernandez-Rapp J, Jolivette F, Lecours C, Bisht K, Goupil C, Dorval V, Parsi S, Morin F, Planel E, et al. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum Mol Genet. 2015;24:6721–6735. doi: 10.1093/hmg/ddv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen KF, Sakamoto K, Aten S, Snider KH, Loeser J, Hesse AM, Page CE, Pelz C, Arthur JS, Impey S, Obrietan K. Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learn Mem. 2016;23:61–71. doi: 10.1101/lm.039578.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberg RB, Mufson EJ, Counts SE. Evidence for a neuroprotective microRNA pathway in amnestic mild cognitive impairment. Front Neurosci. 2015;9(430) doi: 10.3389/fnins.2015.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Ba M, Du Y, Xia C, Tan S, Ng KP, Ma G. Aβ1-42 increases the expression of neural KATP subunits Kir6.2/SUR1 via the NF-κB, p38 MAPK and PKC signal pathways in rat primary cholinergic neurons. Hum Exp Toxicol. 2019;38:665–674. doi: 10.1177/0960327119833742. [DOI] [PubMed] [Google Scholar]
- 25.Jiang S, Zhao Y, Zhang T, Lan J, Yang J, Yuan L, Zhang Q, Pan K, Zhang K. Galantamine inhibits β-amyloid-induced cytostatic autophagy in PC12 cells through decreasing ROS production. Cell Prolif. 2018;51(e12427) doi: 10.1111/cpr.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng M, Ai D, Sun L, Xu X, Cao X. EGb 761 inhibits Aβ1-42-induced neuroinflammatory response by suppressing P38 MAPK signaling pathway in BV-2 microglial cells. Neuroreport. 2019;30:434–440. doi: 10.1097/WNR.0000000000001223. [DOI] [PubMed] [Google Scholar]
- 27.Galoyan AA, Sarkissian JS, Chavushyan VA, Meliksetyan IB, Avagyan ZE, Poghosyan MV, Vahradyan HG, Mkrtchian HH, Abrahamyan DO. Neuroprotection by hypothalamic peptide proline-rich peptide-1 in Abeta25-35 model of Alzheimer's disease. Alzheimers Dement. 2008;4:332–344. doi: 10.1016/j.jalz.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Brennan S, Keon M, Liu B, Su Z, Saksena NK. Panoramic visualization of circulating microRNAs across neurodegenerative diseases in humans. Mol Neurobiol. 2019;56:7380–7407. doi: 10.1007/s12035-019-1615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y, Liang X, Yao Y, Xiao H, Shi Y, Yang J. Osthole attenuates APP-induced Alzheimer's disease through up-regulating miRNA-101a-3p. Life Sci. 2019;225:117–131. doi: 10.1016/j.lfs.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Gao S, Lin J, Wang T, Shen Y, Li Y, Yang W, Zhou K, Hu H. Qingxin kaiqiao fang ameliorates memory impairment and inhibits apoptosis in APP/PS1 double transgenic mice through the MAPK pathway. Drug Des Devel Ther. 2019;13:459–475. doi: 10.2147/DDDT.S188505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadlon A, Takousis P, Alexopoulos P, Evangelou E, Prokopenko I, Perneczky R. miRNAs identify shared pathways in Alzheimer's and Parkinson's diseases. Trends Mol Med. 2019;25:662–672. doi: 10.1016/j.molmed.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Prakash D, Sudhandiran G. Dietary flavonoid fisetin regulates aluminium chloride-induced neuronal apoptosis in cortex and hippocampus of mice brain. J Nutr Biochem. 2015;26:1527–1539. doi: 10.1016/j.jnutbio.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 34.Che F, Du H, Zhang W, Cheng Z, Tong Y. MicroRNA-132 modifies angiogenesis in patients with ischemic cerebrovascular disease by suppressing the NF-κB and VEGF pathway. Mol Med Rep. 2018;17:2724–2730. doi: 10.3892/mmr.2017.8138. [DOI] [PubMed] [Google Scholar]
- 35.Hwang S, Jeong H, Hong EH, Joo HM, Cho KS, Nam SY. Low-dose ionizing radiation alleviates Aβ42-induced cell death via regulating AKT and p38 pathways in Drosophila Alzheimer's disease models. Biol Open. 2019;8(bio036657) doi: 10.1242/bio.036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SJ, Hwang SG, Shin DY, Kang SS, Chun JS. p38 kinase regulates nitric oxide-induced apoptosis of articular chondrocytes by accumulating p53 via NFkappa B-dependent transcription and stabilization by serine 15 phosphorylation. J Biol Chem. 2002;277:33501–33508. doi: 10.1074/jbc.M202862200. [DOI] [PubMed] [Google Scholar]
- 37.Muraleva NA, Kolosova NG, Stefanova NA. p38 MAPK-dependent alphaB-crystallin phosphorylation in Alzheimer's disease-like pathology in OXYS rats. Exp Gerontol. 2019;119:45–52. doi: 10.1016/j.exger.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Liang Z, Blanchard J, Dai CL, Sun S, Lee MH, Grundke-Iqbal I, Iqbal K, Liu F, Gong CX. A non-transgenic mouse model (icv-STZ mouse) of Alzheimer's disease: Similarities to and differences from the transgenic model (3xTg-AD mouse) Mol Neurobiol. 2013;47:711–725. doi: 10.1007/s12035-012-8375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Z, Jiao R, Wang J, Wang P, Zhu Y, Zhao J, De Jager P, Bennett DA, Jin L, Xiong M. Shared causal paths underlying Alzheimer's dementia and type 2 diabetes. Sci Rep. 2020;10(4107) doi: 10.1038/s41598-020-60682-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishimoto T, Sugihara H, Watanabe M, Sawayama H, Iwatsuki M, Baba Y, Okabe H, Hidaka K, Yokoyama N, Miyake K, et al. Macrophage-derived reactive oxygen species suppress miR-328 targeting CD44 in cancer cells and promote redox adaptation. Carcinogenesis. 2014;35:1003–1011. doi: 10.1093/carcin/bgt402. [DOI] [PubMed] [Google Scholar]
- 41.Ebi H, Sato T, Sugito N, Hosono Y, Yatabe Y, Matsuyama Y, Yamaguchi T, Osada H, Suzuki M, Takahashi T. Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene. 2009;28:3371–3379. doi: 10.1038/onc.2009.201. [DOI] [PubMed] [Google Scholar]
- 42.Taupin P. A dual activity of ROS and oxidative stress on adult neurogenesis and Alzheimer's disease. Cent Nerv Syst Agents Med Chem. 2010;10:16–21. doi: 10.2174/187152410790780172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.