Abbreviations
- CI
confidence interval
- DLBCL
diffuse large B‐cell lymphoma
- ESI
extranodal site involvement
- FFPE
formalin‐fixed, paraffin‐embedded
- HR
hazard ratio
- IPI
international prognostic index
- ISH
in situ hybridization
- lncRNA
long non‐coding RNA
- MYC
myelocytomatosis oncogene
- NCCN
national comprehensive cancer network
- NHL
non‐Hodgkin lymphoma
- OS
overall survival
- PFS
progression‐free survival
- PVT1
plasmacytoma variant translocation 1
- R‐CHOP
rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone
- RR
relative risk
- RT‐PCR
real‐time PCR
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
Dear Editor,
Diffuse large B‐cell lymphoma (DLBCL) is the most common and aggressive subtype of non‐Hodgkin lymphoma (NHL), accounting for about 40% of all NHL cases [1]. Lacking symptoms at early time and efficient therapeutic methods made DLBCL one of the most life‐threatening types of hematopoietic malignancy [2, 3]. Therefore, identifying novel therapeutic biomarker for early detection and prognosis prediction is urgently needed.
Thanks to large‐scale gene expression profiling between DLBCL and normal B cells, vast groups of genes have been found deregulated. Moreover, various long non‐coding RNAs (lncRNAs) have been reported aberrantly expressed in DLBCL [4, 5]. Previous studies showed that lncRNA plasmacytoma variant translocation 1 (PVT1) is an important epigenetic regulator with critical roles in human tumors [6]. Indeed, by searching the online database (starBase pan‐cancer project database, version 3.0; http://starbase.sysu.edu.cn/), we found that PVT1 was elevated in most kinds of tumors (Supplementary Figure S1A). However, the overall biological role and clinical significance of PVT1 in DLBCL remains largely unknown.
In order to determine the possible involvement of PVT1 in DLBCL, we retrieved 286 formalin‐fixed, paraffin‐embedded (FFPE) DLBCL tissues and 62 normal lymph node FFPE tissues from DLBCL patients treated at Renji hospital (Shanghai, China) between January 1st, 2012 and December 31st, 2017. Furthermore, we also collected 46 paired fresh DLBCL tissues and normal lymph node tissues. The clinical information of the enrolled DLBCL patients was recapitulated in Supplementary Table S1. The materials and methods used in this study were detailed in Supplementary information. We firstly assessed PVT1 expression via real‐time PCR (RT‐PCR) in 46 paired fresh DLBCL tissues and normal lymph node tissues. The expression of PVT1 was found to be significantly increased in DLBCL tissues as compared to their normal counterparts (P < 0.001; Supplementary Figure S1B). PVT1 upregulation in tumor tissues was detected in 39 (84.8%) patients with DLBCL (Supplementary Figure S1C). Furthermore, in situ hybridization (ISH) staining on 286 DLBCL and 62 normal lymph node FFPE tissues confirmed that PVT1 expression was remarkably increased in DLBCL FFPE tissues (P < 0.001; Supplementary Figure S1D). These results suggested that the PVT1 was significantly upregulated in DLBCL tissues.
In order to explore the clinical significance of PVT1 in DLBCL patients, we then analyzed the correlation between PVT1 expression and clinicopathological features in DLBCL patients. We found that the expression of PVT1 was positively correlated with the expression of myelocytomatosis oncogene (MYC) in 46 fresh DLBCL tissues (P < 0.001; Supplementary Figure S2A). Additionally, the ISH score of PVT1 was positively correlated with tumor size and Ki67+ cell rate in those 286 DLBCL (both P < 0.001; Supplementary Figure S2B and C). Both univariate (Chi‐squared test) and multivariate (Logistic regression) analyses confirmed that high PVT1 expression was associated with multiple extranodal site involvement (ESI) in DLBCL patients (P = 0.007; Supplementary Table S1). DLBCL patients were then divided into low and high PVT1 expression groups according to the ISH score for survival analysis (Supplementary Figure S2D). Kaplan‐Meier analysis was performed to assess the prognostic significance of PVT1 expression in 286 DLBCL patients. The results showed that patients with low PVT1 expression had increased progression‐free survival (PFS; hazard ratio [HR] = 2.10; 95% confidence interval [CI] = 1.42‐3.12; P < 0.001) and overall survival (OS) rate (HR = 2.29; 95% CI = 1.45‐3.62; P < 0.001; Figure 1A). The 5‐year PFS and OS rates of high and low PVT1 expression groups were 51.0% vs. 72.8% and 61.4% vs. 82.5%, respectively. Multivariate Cox proportional hazards model showed that high PVT1 expression was associated with reduced PFS (relative risk [RR] = 1.85; 95% CI = 1.19‐2.89; P = 0.006) and OS rates (RR = 1.91, 95% CI = 1.13‐3.23; P = 0.015; Supplementary Table S2). Moreover, we analyzed the PFS and OS by combine PVT1 expression and national comprehensive cancer network international prognostic index (NCCN IPI) to assess the prognosis prediction ability. We found that this combination could further divide the DLBCL patients into four groups, with the group of both low PVT1 expression and low NCCN IPI possess the best PFS and OS while both high PVT1 expression and high NCCN IPI group possess the worst (Figure 1B). Taken together, our results provide evidence showing the potential of PVT1 as an important prognostic factor for DLBCL patients.
FIGURE 1.
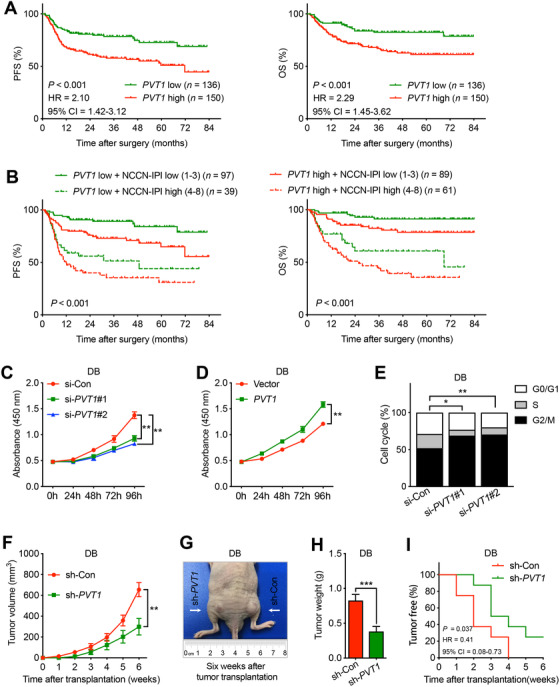
PVT1 is a prognostic factor for DLBCL patients and plays an important oncogenic role in DLBCL in vitro and in vivo.
A. Kaplan‐Meier analysis of the correlation between PVT1 expression and the PFS (left panel) or OS (right panel) in 286 DLBCL patients. B. The significance of combination of PVT1 expression and NCCN IPI score in DLBCL survival (PFS and OS). C. Cell proliferation analysis of DB cells transiently transfected with PVT1 siRNA (si‐PVT1#1 and si‐PVT1#2). DB cells transfected with control siRNA (si‐Con) were served as control. D. Cell proliferation analysis of DB cells transiently transfected with PVT1overexpression plasmids (PVT1). DB cells transfected with empty vector (Vector) served as control. E. Cell cycle distributions of DB cells transfected with PVT1 siRNA (si‐PVT1#1 and si‐PVT1#2). The cell cycle distributions were assessed by measuring DNA content of PI‐stained cells using flow cytometry. DB cells transfected with control siRNA (si‐Con) were served as control. F. Tumor growth curves of DB cells transfected with PVT1 shRNA in vivo. DB cells transfected with control shRNA (sh‐Con) were served as control. G. Representative images of mice at the 6th week after separately subcutaneous transplantation of DB cells transfected with PVT1 shRNA. The mice transplanted with DB cells treated with control shRNA (sh‐Con) were served as control. H. The mean tumor weight of mice received DB cells with or without PVT1 knockdown. I. The tumor‐free percentages of mice received DB cells with or without PVT1 knockdown. Bar, SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Abbreviations: PVT1, plasmacytoma variant translocation 1; DLBCL, diffuse large B‐cell lymphoma; SEM, standard error of mean; PFS, progression‐free survival; OS, overall survival; shRNA, short hairpin RNA; siRNA, small interfering RNA; PI, propidium iodide
For further exploring the biological role of PVT1 in DLBCL, we carried out both in vitro and in vivo analysis. Reducing PVT1 expression by two small interfering RNAs (siRNAs) in four DLBCL cell lines (DB, OCI‐Ly10, BJAB, and OCI‐Ly1) all significantly reduced their proliferation rates (Figure 1C and Supplementary Figure S3A). Contrarily, elevating PVT1 expression by transfecting PVT1 overexpression plasmid promoted DLBCL cells proliferation (Figure 1D and Supplementary Figure S3B). Further cell cycle analysis showed that reduced PVT1 expression in DB and OCI‐Ly1 induced G0/G1 arrest (Figure 1E and Supplementary Figure S3C). Finally, reducing PVT1 expression by short hairpin RNA (shRNA) in both DB and OCI‐Ly1 impeded tumor growth on mouse xenograft models, as shown by reduced tumor volume, tumor weight, and tumor formation rate (Figure 1F–I and Supplementary Figure S4). Taken together, these results indicate that PVT1 influenced DLBCL cells’ proliferation capacity, possibly through regulating cell cycle progression.
Although the current standard treatment with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R‐CHOP) has improved the outcome of DLBCL patients remarkably, there still existed over 30% of patients encounter relapse or refractory [2]. Recent years, the high throughput sequencing technologies have helped define the molecular and biological events with DLBCL development comprehensively [4]. Cumulative evidences suggested the importance of lncRNA in cancer development and progression [5, 6, 7, 8]. A recent study into analyzing the lncRNA expression profile of DLBCL patients has suggested a six‐lncRNA signature could efficiently predict the outcome of DLBCL patients [4]. Therefore, it is meaningful to find novel lncRNAs for pre‐assessment of DLBCL patients’ prognosis.
In the present study, we provided evidences that elevated PVT1 expression was found in DLBCL tissues compared with normal lymph node tissues, and was positively associated with adverse clinicopathological outcomes. Moreover, the PVT1 expression was also a potent independent poor prognostic factor in DLBCL. Consistently, genome‐wide association study confirmed the association between PVT1 variants with DLBCL development [9]. PVT1 has been studied in a variety of pathological processes about tumorigenesis and tumor progression. However, the role of PVT1 played in DLBCL remained to be clarified. Similarly, our results showed that silencing PVT1 expression significantly inhibit DLBCL cell proliferation both in vitro and in vivo. Opposite effect was observed with PVT1 overexpression. Uncontrolled cell proliferation is the hallmark of cancer cells, most of which are fulfilled through directly regulating cell cycle progression [10]. Our flow cytometric results indicated that reducing PVT1 in DB and OCI‐Ly1 cells induced G1 phase arrest and suppressed cell proliferation. However, similar effects were not observed among OCI‐Ly10 and BJAB cells, suggesting other mechanisms might be used by PVT1 in regulating DLBCL tumorigenesis.
In conclusion, our investigations first elucidated the dysregulation of PVT1 expression in DLBCL, and determined the prognostic value of PVT1 for DLBCL patients. Furthermore, we confirmed the oncogenic role of PVT1 in DLBCL development both in vitro and in vivo. These would possibly provide rationale for future targeting PVT1 in DLBCL.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The protocol of this study was approved by the Shanghai Ninth People's Hospital and Renji Hospital Affiliated to School of Medicine, Shanghai Jiao Tong University. The need for informed consent was waived.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The datasets are available from the corresponding authors on reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS' CONTRIBUTIONS
MZ and LY designed and conducted the study. RY, TS, ML, and YS collected clinical samples and data. RY, ML, YS and QL performed the experiments. MZ and LY wrote the manuscript and prepared the figures and tables. All authors reviewed and approved the final manuscript.
Supporting information
FigureS1
FigureS2
FigureS3
FigureS4
Supporting Information
TableS1
TableS2
ACKNOWLEDGEMENTS
Not applicable.
Funding information
This work was supported by the National Natural Science Foundation of China (81802424).
Contributor Information
Linhua Yang, Email: yanglinhua1981@126.com.
Ming Zhan, Email: linyaruo@163.com.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA CANCER. 2019;69(1):7‐34. 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2. Kubuschok B, Held G, Pfreundschuh M. Management of diffuse large B‐cell lymphoma (DLBCL). Cancer Treat Res. 2015;165:271‐88. 10.1007/978-3-319-13150-4_11. [DOI] [PubMed] [Google Scholar]
- 3. Qin Y, Song Y, Shen Z, Du X, Ji W, Hsu W et al. Safety and efficacy of obinutuzumab in Chinese patients with B‐cell lymphomas: a secondary analysis of the GERSHWIN trial. Cancer Commun. 2018;38(1):31 10.1186/s40880-018-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sun J, Cheng L, Shi H, Zhang Z, Zhao H, Wang Z et al. A potential panel of six‐long non‐coding RNA signature to improve survival prediction of diffuse large‐B‐cell lymphoma. Sci Rep. 2016;6:27842 10.1038/srep27842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452‐63. 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiao M, Feng Y, Liu C, Zhang Z. Prognostic values of long noncoding RNA PVT1 in various carcinomas: An updated systematic review and meta‐analysis. Cell Prolif. 2018;51(6):e12519 10.1111/cpr.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luo J, Langer LF, Liu J. A novel role of LncRNA in regulating tumor metabolism and angiogenesis under hypoxia. Cancer Commun. 2019;39(1):2 10.1186/s40880-019-0348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lian Y, Yang J, Lian Y, Xiao C, Hu X, Xu H. DUXAP8, a pseudogene derived lncRNA, promotes growth of pancreatic carcinoma cells by epigenetically silencing CDKN1A and KLF2. Cancer Commun. 2018;38(1):64 10.1186/s40880-018-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cerhan JR, Berndt SI, Vijai J, Ghesquieres H, McKay J, Wang SS et al. Genome‐wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat Genet. 2014;46(11):1233‐8. 10.1038/ng.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672‐7. 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FigureS1
FigureS2
FigureS3
FigureS4
Supporting Information
TableS1
TableS2
Data Availability Statement
The datasets are available from the corresponding authors on reasonable request.


