Highlights
-
•
The molecular characterization and pathogenic mechanisms of SARS-CoV-2 play an important role in the drug design as well as vaccine development.
-
•
Computational methods for vaccine development are more appropriate than conventional methods.
-
•
Toll-like receptors enhance the innate immune system to recognize the pathogen.
-
•
The entry of SARS-CoV-2 is dependent on the receptor-binding domain (RBD) of the virus and angiotensin-converting enzyme (ACE-2) of the host.
Keywords: SARS-CoV-2, COVID-19, Molecular characterization, Pathogen-host interaction, Vaccine development, SARS-CoV-2 neuroinvasion
Abstract
COVID-19 has forsaken the world because of extremely high infection rates and high mortality rates. At present we have neither medicine nor vaccine to prevent this pandemic. Lockdowns, curfews, isolations, quarantines, and social distancing are the only ways to mitigate their infection. This is badly affecting the mental health of people. Hence, there is an urgent need to address this issue. Coronavirus disease 2019 (COVID-19) is caused by a novel Betacorona virus named SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) which has emerged in the city of Wuhan in China and declared a pandemic by WHO since it affected almost all the countries the world, infected 24,182,030 people and caused 825,798 death as per data are compiled from John Hopkins University (JHU). The genome of SARS-CoV-2 has a single-stranded positive (+) sense RNA of ∼30 kb nucleotides. Phylogenetic analysis reveals that SARS-CoV-2 shares the highest nucleotide sequence similarity (∼79 %) with SARS-CoV. Envelope and nucleocapsids are two evolutionary conserved regions of SARS-CoV-2 having a sequence identity of about 96 % and 89.6 %, respectively as compared to SARS-CoV. The characterization of SARS-CoV-2 is based on polymerase chain reaction (PCR) and metagenomic next-generation sequencing. Transmission of this virus in the human occurs through the respiratory tract and decreases the respiration efficiency of lungs. Humans are generally susceptible to SARS-CoV-2 with an incubation period of 2–14 days. The virus first infects the lower airway and bind with angiotensin-converting enzyme 2 (ACE2) of alveolar epithelial cells. Due to the unavailability of drugs or vaccines, it is very urgent to design potential vaccines or drugs for COVID-19. Reverse vaccinology and immunoinformatic play an important role in designing potential vaccines against SARS-CoV-2. The suitable vaccine selects for SARS-CoV-2 based on binding energy between the target protein and the designed vaccine. The stability and activity of the designed vaccine can be estimated by using molecular docking and dynamic simulation approaches. This review mainly focused on the brief up to date information about COVID-19, molecular characterization, pathogen-host interaction pathways involved during COVID-19 infection. It also covers potential vaccine design against COVID-19 by using various computational approaches. SARS-CoV-2 enters brain tissue through the different pathway and harm human's brain and causes severe neurological disruption.
1. Introduction
The origin of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was first reported in Wuhan, China in December 2019 (Singhal, 2020). This virus causes severe pneumonia, a respiratory disease now described as COVID-19 (Coronavirus Disease, 2019) officially declared by the World Health Organization (WHO). International Virus Classification Commission (ICTV) classified novel coronavirus as SARS-CoV-2 (Yang and Wang, 2020). WHO declared COVID-19 as a global pandemic due to uncontrolled transmission and unavailability of the treatment of COVID-19. Coronavirus infected patient shows common symptoms like difficulty in respiration, cough, fever, shortness of breath (CDC, 2020a). The critical condition of this disease is multi-organ damage (lung and kidney) and finally causes the death of COVID-19 suffered person. Generally, lung damage is the main reason for the death of COVID-19 patients (Wu et al., 2020a, b, c, d).
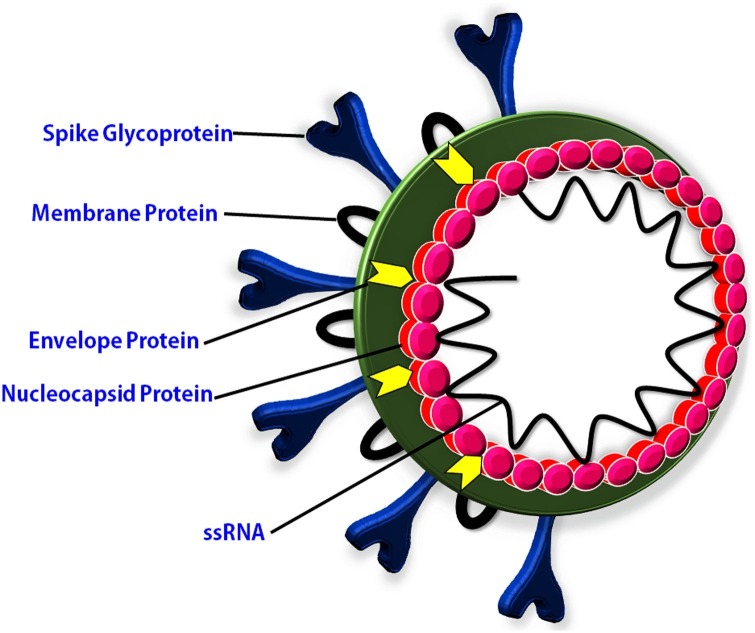
COVID-19 is not the first only disease caused by coronaviruses. In the past two decades, three coronavirus diseases have been reported (Mackenzie and Smith, 2020). COVID-19, Severe Acute Respiratory Syndrome (SARS), and Middle East Respiratory Syndrome (MERS) are the three global epidemics caused by a coronavirus (Memish et al., 2020). The outbreak of SARS-CoV disease was reported in 2002–2003 and caused about 8000 infections in humans (World Health Organization (WHO, 2020e). MERS-CoV disease was emerged in 2012 and caused 2106 infection in humans (Memish et al., 2020). Behind the coronaviruses diseases, other viral diseases such as influenza, polio, ebola, AIDS (acquired immunodeficiency syndrome), and hepatitis also affected the human population Worldwide (Mourya et al., 2019). A major outbreak of viral diseases since the early 20th century and their impacts on the human population have been mentioned in Table 1 . These viral diseases have caused a large number of infections and death during the period of the 20th and 21 st centuries. COVID-19 is one of the highly transmitted among these diseases. The high transmission rate of COVID-19 might be due to a large incubation period and the structure of spike glycoprotein (Li et al., 2020a, b, c). The novel coronavirus was named as SARS-CoV-2 due to similar characteristics to SARS-CoV. It consists of spike glycoprotein (S), membrane protein (M), an envelope protein (E), nucleocapsid protein (N), and linear single-stranded positive (+) sense RNA of ∼30 kb (Fig. 1 ). The genome study of SARS-CoV-2 shows a close relation with SARS-CoV and MERS-CoV. Phylogenetic analysis revealed that SARS-CoV-2 has emerged from Bat SARS-like CoV because lineage B beta-CoV originated from bat and lineage B beta-CoV share 96 % sequence similarity with SARS-like CoV (Chen et al., 2020a, b). The pathogenic mechanism of SARS-CoV-2 is also very similar to the SARS-CoV as well as MERS-CoV. SARS-CoV-2 use angiotensin-converting enzyme 2 (ACE2) receptor during entry into the host cell. Same ACE2 receptor expressed during SARS-CoV infection in unciliated bronchial epithelial cells and type II pneumocytes (Qian et al., 2013). MERS-CoV recognizes to dipeptidyl peptidase 4 (DPP4) receptor during infection in human cells (Raj et al., 2013).
Table 1.
Viral pandemic in the 20th and 21st century.
| Disease | Year | Infection | Mortality (Estimated death) | Country badly Affected | References |
|---|---|---|---|---|---|
| Influenza | 1918−19 | 500 million | 2−3 % (50 million) | Killed > half million people in USA | (CDC, 2020b) |
| Polio | Peak of 1940s & 1950s | 42,173 | 2,720 | US, Canada, UK | (Mehndiratta et al., 2014) |
| Marburg Disease | 1967s | 24 | Up to 88 % | Angola, Congo, Kenya, South Africa, Uganda | (World Health Organization (WHO, 2020d) |
| Ebola hemorrhagic disease | 1970s & 1990s | 602 | 431 | Sudan, Yambuku | (Laupland and Valiquette, 2014) |
| HIV | 1980s & 1990s | 3,064 | 1,292 | United States | (Healthline, 2020) |
| Hepatitis | 1996 | 400,000−600,000 | 300 million | US | (CDC, 2020c) |
| Lassa fever | 1969 | 300,000−500,000 per year | 5000 people | West Africa, Sierra Leone, Guinea | (World Health Organization (WHO, 2020a) |
| West Nile Fever | 2002 | 4,156 | 284 | Canada, Mexico, Ontario, Quebec | (Chancey et al., 2015) |
| SARS | 2003 | 8422 | 11 % | China, Hong Kong, Taiwan | (World Health Organization (WHO, 2020b) |
| MERS | 2012 | 2500 | (35 %) 858 | Saudi Arabia | (World Health Organization (WHO, 2020c) |
| COVID-19 | 2019 | 24,182,030 | 825,798 | >210 counties | (Johns Hopkins University (JHU, 2020) |
Fig. 1.
The structure of COVID-19 which cause SARS-CoV-2.
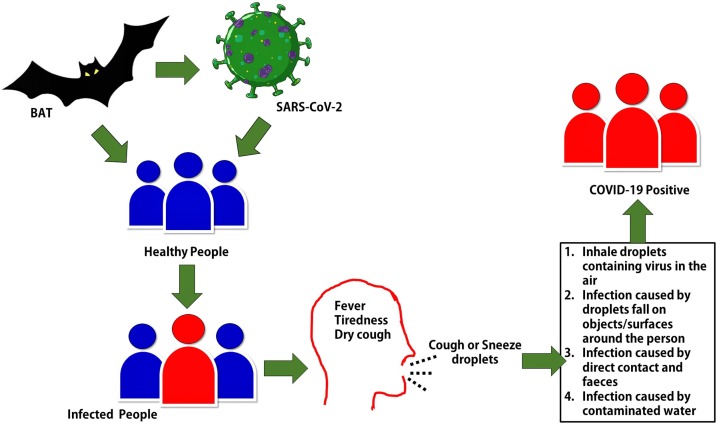
Several studies have reported that transmission of SARS-CoV-2 from bat to human via the food chain. Therefore, bats are considered as the primary host of SARS-CoV-2 due to its similarity with bat coronaviruses (Hampton, 2005; Banerjee et al., 2019; Li et al., 2005). Transmission of SARS-CoV-2 human to human mainly occurs through respiratory droplets and close contacts with an infected person. Fig. 2 depicts the mode of transmission of SARS-CoV-2. The mortality rate of MERS-CoV and SARS-CoV is higher than COVID-19 but the transmission rate of COVID-19 is found several times higher compared with SARS-CoV and MERS-CoV (Petrosillo et al., 2020). The higher transmission rate makes it more transmissible and contagious. COVID-19 caused infection in about 241 million people and 8 million deaths as of August 27, 2020, worldwide (Johns Hopkins University (JHU, 2020). United States of America (USA) is the most affected country in the World. The list of most COVID-19 affected countries and current COVID-19 reports in the respected countries have been mentioned in Table 2 .
Fig. 2.
Transmission and infection of SARS-CoV-2.
Table 2.
Mortality and Recovery by COVID-19 in major affected countries till August 27, 2020.
| Country | Total confirmed cases | Mortality | Recovery | References |
|---|---|---|---|---|
| USA | 5,822,927 | 179,735 | 2,084,465 | (Johns Hopkins University (JHU, 2020) |
| Brazil | 3,717,156 | 117,665 | 3,082,447 | (Johns Hopkins University (JHU, 2020) |
| India | 3,310,234 | 60,472 | 2,523,771 | (Johns Hopkins University (JHU, 2020) |
| Russia | 968,297 | 16,638 | 784,277 | (Johns Hopkins University (JHU, 2020) |
| South Africa | 615,701 | 13,502 | 525,242 | (Johns Hopkins University (JHU, 2020) |
| Mexico | 573,888 | 62,076 | 472,197 | (Johns Hopkins University (JHU, 2020) |
| Peru | 607,701 | 28,001 | 414,577 | (Johns Hopkins University (JHU, 2020) |
| Spain | 419,849 | 28,971 | 150,376 | (Johns Hopkins University (JHU, 2020) |
| Italy | 262,540 | 35,458 | 206,329 | (Johns Hopkins University (JHU, 2020) |
| France | 291,374 | 30,549 | 85,811 | (Johns Hopkins University (JHU, 2020) |
| Germany | 239,014 | 9,290 | 212,464 | (Johns Hopkins University (JHU, 2020) |
| China | 87,363 | 4,658 | 80,594 | (Johns Hopkins University (JHU, 2020) |
| Iran | 365,606 | 16,569 | 261,200 | (Johns Hopkins University (JHU, 2020) |
USA, Brazil, India, Russia, and South Africa are the top COVID-19 affected countries. Across the world, various governing bodies are taking various decisions to stop the spread of the SARS-CoV-2 virus through social distancing and lockdown (Gutiérrez, 2020). The high transmission rates in these countries might be due to late partially or late lockdown and environmental factors such as temperature (Yuki et al., 2020). Early and complete lockdown of the whole country is playing an important role in the reduction of social gatherings and eventually responsible for the low transmission rate in India (Gupta et al., 2020).
There is no specific treatment available for COVID-19. The treatment of COVID-19 symptoms and oxygen supply via mechanical ventilation is the major therapy in the present time (Begley, 2020). Some broad-spectrum antibiotics and plasma therapy are also used for the reduction of viral load but these therapies are unable to eliminate COVID-19 (Mandal, 2020). Hence, a vaccine is a potential option for COVID-19 treatment. The researchers are working to design a potential vaccine for COVID-19. Vaccine development is a very complex process; however, computation biology can design easily potential vaccines (Rahman et al., 2020; Enayatkhani et al., 2020). A designed vaccine based on structural and non-structural viral protein provokes protective immune responses (Dong et al., 2020). Modern bioinformatics approaches for vaccine design are cost-effective, time-saver, and more effective compared to traditional methods (Sieber et al., 2018; Wolf et al., 2010). This review focused on the genomic and molecular characterization of SARS-CoV-2, pathogenic mechanism, and in-silico vaccine design. This review also covered up to date information about COVID-19 transmission Worldwide.
2. Molecular characterization
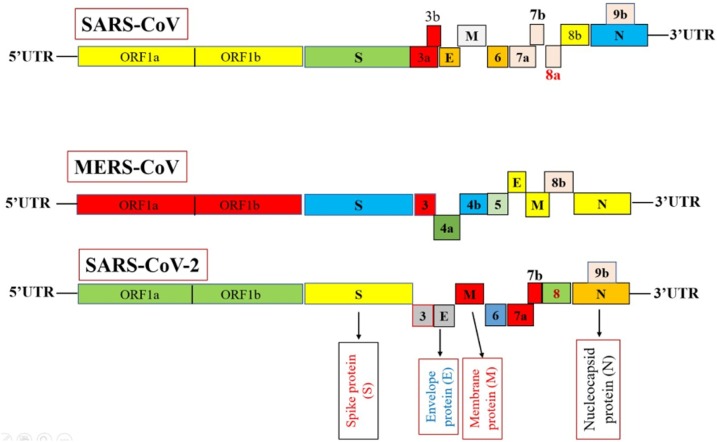
Disease entity of viral infection depends on both molecular and virology techniques including virus isolation and diagnostic test (Ellis and Zambon, 2002). Characterization of SARS-CoV-2 can be accomplished by using molecular approaches. Zhou et al. (2020a, b) reported the characterization and identification of SARS-CoV-2 cause an epidemic of respiratory illness in a human. Zhang et al. (2020) isolated the viral sample from the bronchoalveolar lavage fluid of SARS-CoV-2 infected patients and characterized the viral sample using RT-PCR using degenerate primers and probes designed for detection of SARS-CoV-2. The presence of SARS-CoV-2 in the sample was confirmed by metagenomic sequencing approaches such as high-throughput sequencing (Zhou et al., 2020a, b). The genome size of SARS-CoV-2 was found at about ∼30 kb in length (29,891 bp). Comparative genome analysis with other coronaviruses revealed that the virus genome was closely similar to previously characterized coronaviruses. Fig. 3 shows the genome similarity of SARS-CoV-2 with MERS-CoV and SARS-CoV.
Fig. 3.
Genome similarity and dissimilarity among MERS-CoV, SARS-CoV and SARS-CoV-2.
Therefore, after the identification of SARS-CoV-2, the phylogenetic relationship among the viruses of the same family was studied using bioinformatics approaches (Ching et al., 2003). The genome size of SARS-CoV-2 was 29,891 nucleotides long which contains 8,903 adenosines, 9,574 uracils, 5,852 guanines, and 5,482 cytosines. The G + C content of SARS-CoV-2 is 32 %–43 %. The genome of SARS-CoV-2 encodes 9860 amino acids. Which are arranged in series of 5′UTR, replicase (orf1a/b), Spike(S), Envelope (E), Membrane (M), Nucleocapsid (N), 3′UTR. The genome of SARS-CoV-2 contains 5′UTR, 3′UTR, and six functional open reading frames (ORFs) such as ORF1a/b, S, E, M, and accessory gene including ORF3b and ORF8 (Chan et al., 2020). ORF1a/b translates into a polyproteins pp1a and pp1ab. These polyproteins are further processed by virally encoded chymotrypsin-like protease (3CLpro) or main protease (Mpro) and one or two papain-like protease (PLpro). Papain-like protease cleaved into 16 non-structural proteins which play a role in the replication and transcription of virus infection (Mousavizadeh and Ghasemi, 2020). Serine type protease encoded by nsp5 is the Main protease (Mpro). 3CLpro is dimeric form, and each monomer contains two subunits, the catalytic N-terminal residue, and the C-terminal residue. 3CLpro sequence shares 96 % identity in both SARS-CoV and SARS-CoV-2 (Gil et al., 2020). Various researches it has been reported that the main protease of SARS-CoV-2 plays a significant role in replication, transcription, and translation of viral proteins (Kumar et al., 2020a, b, c). Mpro represents a potential target due to its involvement in essential viral functions (Estrada, 2020). ORF3b encoded a completely novel short protein and ORF8 likely encodes a secreted protein form by alpha-helix followed by beta-sheet contains six strands of the unknown function. S gene of SARS-CoV-2 has a sequence identity of less than 75 % to bat SARS-like-CoV (SL-CoVZXC21 and SL-CoVZC45) and human SARS-CoV. S gene of SARS-CoV-2 encodes spike glycoprotein was longer as compared to SARS-CoV (Zhou et al., 2020a, b; Paraskevis et al., 2020).
From the phylogenetic analysis of coronaviruses, we analyzed that SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-OC43, and HCoV-HKU1 are beta coronavirus and HCoV-NL63, HCoV- 229E are alpha coronaviruses. Nucleotide sequences of SARS-CoV-2 were retrieved from the National Centre for Biotechnology Information database (NCBI). Multiple sequence alignment of SARS-CoV-2 with reference sequences was done using Clustal W software with the default parameter. Phylogenetic tree analysis of the complete genome using maximum likelihood (ML) method-based Tamura-Nei model was done using MEGA software with 100 bootstrap replicates (Zhou et al., 2020a, b). The obtained phylogenetic tree has been shown in Fig. 4 . Multiple sequence alignment is a sequence alignment of more than three biological sequences. It minimizes the number of insertion or deletion (gaps). It is used to reconstruct the phylogenetic tree and show the evolutionary relationship with a common ancestor. Functionally important sites including binding sites, active sites are identified using MSA (Chatzou et al., 2016).
Fig. 4.
Phylogenetic tree of coronaviruses using MEGA software.
Phylogenetic tree constructed using whole genomes of coronaviruses and it represents the evolutionary relationship among coronaviruses. Several branches show the bootstrap value (100) check the reliability of the tree. Based on a phylogenetic tree, SARS-CoV-2 shares a common ancestor with two other bat SARS-like coronaviruses such as bat-SL-CoVZC45 and bat-SL-CoVZXC21 with 88 % sequence similarity. Because both bat viruses are present in the same cluster which shows monophyletic groups but distantly related from SARS-CoV about 79 % and MERS-CoV about 50 % that shows paraphyletic groups (Zhou et al., 2020a, b).
From previous research, homology modeling shows that SARS-CoV-2 has the same receptor-binding domain as SARS-CoV. Therefore, it confirmed that SARS-CoV-2 uses ACE2 as a receptor, despite the presence of difference in amino acid in the SARS-CoV-2 receptor-binding domain (Walls et al., 2020). Based on virus genome sequencing and phylogenetic analysis, the bat is considered as the primary source and origin of SARS-CoV-2 (Zhou et al., 2020a, b).
3. Host-pathogen interaction
The SARS-CoV-2 is first reported from the wild animal market of Wuhan city. The bat is considered as the primary host of SARS-CoV-2 due to close genome similarity with bat SARS-CoV-2 (Huang et al., 2020). The previous study (Cyranoski, 2020) suggested that pangolins are the intermediates host of SARS-CoV-2. Isolated virus from pangolins showed 99 % genome similarity with the human SARS-CoV-2. The COVID-19 transmission in animal to human was due to the consumption of COVID-19 infected animals as food. After entering into the human host, SARS-CoV-2 is transmitted rapidly in the human population. The most probability of transmission of SARS-CoV-2 from human to human is via respiratory droplets and close contacts with respiratory symptoms (Chan et al., 2020). As of August 27, 2020, a total of 24,182,030 confirmed cases were reported worldwide with total death of 825,798 (Johns Hopkins University (JHU, 2020). The high transmission rate of SARS-CoV-2 might be due to the spreading of the virus via asymptomatic-infected person and some modifications in the spike proteins (S) (Rothe et al., 2020). The Spike protein (S) plays an important role in virus attachment, fusion, and entry. The spike glycoproteins (S) are also considered as potential target sites for antiviral drugs and vaccines. Receptor binding domain (RBD) presents in the spike protein of SARS-CoV-2 and is strongly bound to the human ACE-2 (Tai et al., 2020). Hence, SARS-CoV-2 used angiotensin-converting enzymes 2 (ACE2) for entry purposes into the host cell. ACE2 express on the cell surface of a wide range of animals including human. ACE2 could help in the interspecies and human to human transmission of COVID-19 (Hoffmann et al., 2020).
The recent study defined the more accurate COVID-19 transmission situation and outbreak in the future. Researchers use a mathematical model to define reproduction number (R0) which is related to the spreading of COVID-19 from one infected person. If R0 is below 1, it indicates that disease can spread in half of the persons through COVID-19 infected person and disease is considered as in controlled situation. If the value of the R0 is more than 1, it indicates that the disease is very contagious and considered under an uncontrolled situation (Thompson, 2020). Liu et al. (2020) reported 3.28 average R0 of COVID-19 in China from the period between January 1, 2020, to February 7, 2020. This indicates COVID-19 is an uncontrolled and highly infectious disease. SARS-CoV-2 has a high R0 comparison with SARS-CoV (R0 = 1.4) and MERS-CoV (R0 = 2.5), indicates that SAR-CoV-2 is more spreadable than SARS-CoV and MERS-CoV and may cause a global pandemic (Li et al., 2020a, b, c; Wu et al., 2020a, b, c, d). The majority of COVID-19 patients suffered only lung infection because it is only associated with respiratory disease. The asymptotic incubation period of COVID-19 varies from 2-14 days (Wu et al., 2020a, b, c, d; Chen et al., 2020a, b).
3.1. SARS-CoV-2 entry and intracellular replication
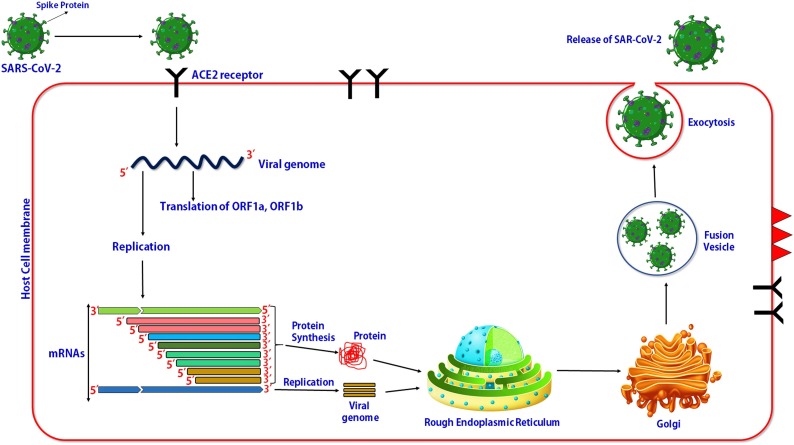
The entry of SARS-CoV-2 into the human host cells through upper body opening like mouth and nasal opening. SARS-CoV-2 recognizes to host cell receptors such as ACE2 present on the outer membrane of the host cell (Zhou et al., 2020a, b; Jia et al., 2005; Wan et al., 2020). Current studies by Lan et al. (2020) reported that the receptor-binding domain (RBD) in the SARS-CoV-2 spike protein strongly binds to ACE2 with higher affinity. The summarized mechanism of viral-host interaction has been shown in Fig. 5 . The figure shows the viral host interaction and multiplication of viruses within the host cell. This figure shows the entry and replication of the viral genome of SRAS-CoV-2 virus and synthesis of the SARS-CoV-2 virus in the endoplasmic reticulum by using viral component and finally release of viral particle (complete viruses) via exocytosis.
Fig. 5.
Viral host interaction and multiplication of viruses within host cell.
Receptor binding domain (RBD) on the surface of SARS-CoV-2 binds to ACE2 on the surface of the host cell receptor (Tortorici and Veesler, 2019). The direct membrane fusion between virus and plasma membrane also reported the second entry mechanism of entry of SARS-CoV into the host cell (Simmons et al., 2004). Belouzard et al. (2009) reported the proteolytic cleavage of ACE2 protein during nitration of viral spike protein and host receptor (ACE2). After entry of the viral RNA genome into the host cell cytoplasm, it starts to synthesize some important viral proteins such as pp1a and pp1ab. The positive-sense viral RNA also enter start to replicate and form multiple viral genomes (Xia et al., 2020; Yu et al., 2020; de Wilde et al., 2018; Perlman and Netland, 2009). Translated viral protein enters into the rough endoplasmic reticulum and Golgi complex for post-translational modification. The newly synthesized viral RNA and protein form complete viral particles assembled into fusion vesicles. These vesicles fused with the plasma membrane and virus particles come out from the host cell and target to next healthy (de Wit et al., 2016).
3.2. Antigen presentation during coronavirus infection
When a virus enters the host cell, its antigen is presented on the antigen-presenting cells (APCs). The antigen presentation of the viral antigen plays an important role in ant-viral immunity. HLA (human leukocyte antigen) and MHC (major histocompatibility complex) are the major antigen-presenting cells for SARS-CoV-2 antigen. The viral antigen is presented by HLA and MHC and is recognized by CTLs (cytotoxic T lymphocytes). Understanding the mechanism of the antigen presentation might be helpful in drug development and vaccine design. The similar receptor expression and antigen presentation also occur in the SARS-CoV (Liu et al., 2010). The gene polymorphism of mannose-binding lectin also correlated with the SARS-CoV-2 infection (Tu et al., 2015). These studies will be provided with important clues for understanding the pathogenic mechanism and treatment for COVID-19.
3.3. Anti-viral immunity and cytokine storm
Viral antigen stimulates viral-specific B and T cells. Two viral-specific antibodies IgM and IgG produced in COVID-19 infection after 1st week of infection. However, IgM antibodies disappeared after 12 weeks of viral infection but IgG remained for a long time, this indicates that IgG is the main antibodies for viral infection (Li et al., 2003). IgG antibody is specific for S and N antigen of SARS-CoV-2 (de Wit et al., 2020). The latest report shows that the portion of CD4+ and CD8+ significantly decreases in the peripheral blood in SARS-CoV-2 patient, whereas its status is excessive activation, as evidence by the high number of HLA-DR (CD3.47) and CD38 (CD8 39.4 %) in double-positive fraction (Xu et al., 2020). CD4+ and CD8+ cells can persist up to four years after SARS-CoV infection and can perform T cell proliferation and IFN-Ƴ production (Fan et al., 2009). The CD8+ cell performs the same mechanisms in the MERS-CoV-2 (Liu et al., 2017, 2006).
The ARDS (acute respiratory distress syndrome) is the main death cause of COVID-19 infection. It is the common immunopathological events in the MERS-CoV, SARS-CoV, and SARS-CoV-2 (Xu et al., 2020). The mechanism of the ARSD is based on cytokine storm in which release of the uncontrolled amount of pro-inflammatory cytokines such as IFN-ɑ, IFN-Ƴ, IL-6, etc (Cameron et al., 2008; Channappanavar and Perlman, 2017). Uncontrolled cytokine is responsible for the violent immune attack and causes damage to multiple organs. The SARS-CoV-2 patient died due to damage to the lungs and in some cases kidney and liver damage (Xu et al., 2020).
4. Treatment of COVID-19
There is no specific anti-viral treatment for COVID-19 and no vaccine available. Currently, the treatment of symptomatic and mechanical ventilation (for oxygen supply) is the major treatment option for patients suffering from COVID-19 infection (Mahmud et al., 2020). Nowadays, some broad-spectrum antibiotics such as favilavir and remdesivir also are used to reduce the viral load. Behind this, few other drugs such as lopinavir, ritonavir, and chloroquine are also considered as an option for COVID-19 treatment. Remdesivir (GS5734) is an RNA polymerase inhibitor and has in vitro activity against RNA viruses, including Ebola and MERS-CoV infection. This drug is also considered an option against COVID-19 infection (de Wit et al., 2020). Plasma therapy is based on recovered COVID-19 patients and has promising applications in COVID-19 treatment. Vaccination is the basic and effective treatment of this global pandemic but vaccine formation is under development and needs furthermore investigation (Kumar et al., 2020a, b, c). Food and Drug Administration (FDA) approved the clinical study of COVID-19 infected patients to test the safety and efficacy of umbilical cord-derived mesenchymal stem cells to stop the lung inflammation (EPR, 2020). MSCs therapy prevents the extra production of the immune system. Through intravenous infusion, after entering the human body in the lung MSCs accumulated, which protects the pulmonary microenvironment, improve alveolar epithelial cells, inhibit pulmonary fibrosis and protects lung function (Leng et al., 2020). current options of COVID-19 treatment are summarized as:
4.1. Anti-viral drug
There are no effective drugs available for the complete treatment of COVID-19. However, some broad-spectrum antivirals are used for COVID-19 symptoms as well as reducing viral load. The drugs which have an effective role against SARS-CoV, MERS-CoV, Ebola, HIV, and malaria parasites are also used against COVID-19 (Wang et al., 2020). Remdesivir and protease inhibitors nucleotide analogue is used as a treatment for SARS-CoV-2 (Agostini et al., 2018). A combination of Remdesivir with chloroquine or interferon beta is highly effective for the prevention of SARS-CoV-2 infection (Wang et al., 2020). This combination has not caused any side effects during treatment (Wang et al., 2020; Sheahan et al., 2020; Holshue et al., 2020). Hydroxychloroquine is the analogue of chloroquine and having some additional safety aspect comparison to chloroquine. Hydroxychloroquine is widely used for the treatment of COVID-19 (Singh et al., 2020). The government of India exports hydroxychloroquine medicine to some COVID-19 affected countries like the United State of America (USA) and Afghanistan (The Economic times, 2020). Nowadays, Favilavir and Remdesivir are the most effective antibiotic comparisons to other anti-viral drugs and are considered as an option for COVID-19 treatment.
4.2. Plasma therapy and monoclonal antibodies
Plasma therapy is the type of passive therapy in which immunity forms against COVID-19 recovered patients directly or indirectly used for the treatment of a patient suffering from COVID-19 infection (Zhao and He, 2020). When the SARS-CoV-2 pathogen enters into the body of a person, the immune system of that person is recognized as foreign and form immunity against COVID-19. The antibodies (IgM and IgG) formation starts during the second week of infection and the patient shows recovery after the second week of COVID-19. The COVID-19 specific antibodies and other immune components such as memory cells remain in the plasma of the recovered patients. The plasma collects from the recovered per and for COVID-19 after several purification stages (Buonaguro et al., 2020). Plasma therapy has been proved beneficial in several critically SARS-CoV-2 infected patients in China and India.
Anti-SARS-CoV-2 therapy includes monoclonal antibodies block viral RNA, the viral protein which involves the replication and translation, and prevents COVID-19 infection (Zumla et al., 2016). S protein contains two subunits S1 and S2. S1 is the amino N-terminal receptor-binding subunit and S2 is the carboxy C-terminal membrane fusion subunit (Gui et al., 2017). The location of the protease cleavage site is S1/S2 junction which required activation of membrane fusion and virus entry (Testa et al., 2020). Targeting the S1 protein by the generation of neutralizing monoclonal antibodies and S2 targeted the fusion inhibitors may be essential therapeutics to kill the coronaviruses (Zumla et al., 2016). In Italy, great investigation on tocilizumab for the treatment of COVID-19. It is generally used in the treatment of rheumatoid arthritis and considered a promising role in COVID-19 treatment. This investigation is performing by the Istituto Nazionale Tumori, Fondazione Pascale di Napoli of Italy.
4.3. Stem cell therapy
In COVID-19 patients, cytokine storm is the production of a large number of cytokines and immune cells. Mesenchymal stem cell therapy stops the production of the storm release of cytokines by the immune system and enhances the endogenous reparative function of the stem cells. (MSCs) treatment for COVID-19 patients has been started recently in several countries such as China, the USA, Iran, and Jordon. In a previous study, mesenchymal stem cell therapy has been widely used in type 2 diabetes, autoimmune disease, spinal cord injury, graft-versus-host disease (GVHD), and several other diseases. MSCs are multipotent cells present in the different locations of the body such as bone marrow, placenta, and umbilical cord. MSCs have powerful immunomodulatory and regenerative properties (Golchin et al., 2020). After MSCs therapy, the number of peripheral lymphocytes was increased, the C-reactive protein decreased, and cytokine-secreting immune cells activated such as CXCR3+CD4+ T cells, CXCR3+CD8+ T cells, and CXCR3 + NK cells disappeared on day 3−6. also, a group of CD14+CD11c + CD11bmid regulatory DC cell population dramatically increased. The level of TNF-α was significantly decreased at the same time while increased the level of IL-10 in the MSCs treatment group compared with conventional therapy. Also, the gene expression profile showed that MSCs were ACE2- and TMPRSS2- which showed that MSCs are free from COVID-19 infection. Thus, the intravenous transplantation of MSCs was safe and effective for the treatment of COVID-19 infected patients, especially for the patients in critically severe conditions. MSCs stop the viral infection due to cytokine’s improved qualities (Leng et al., 2020).
4.4. Vaccine for COVID-19
SARS-CoV-2 which emerged in Wuhan city in December 2019 to date has been transmitted all over the World. The transmission of COVID-19 is uncontrolled due to the absence of effective and complete treatment. Currently, antiviral drugs and other therapies are unable to cure this global pandemic (Chakraborty et al., 2020). Hence, there is an urgent public need to develop a suitable vaccine. The vaccine development of COVID-19 started after the sequencing of SARS-CoV-2 and was published in January 2020. Nowadays, various pharmaceutics and drug companies working with collaboration with the research institute to develop an effective vaccine for COVID-19. It is a time-dependent process and needs to pass several clinical trials (Scavone et al., 2020). The example of some COVID-19 vaccines has been given in Table 3 . These vaccine candidates are underdeveloped in several phases.
Table 3.
Potential vaccine for COVID-19 in clinical trials.
| Vaccine name | Platform | Type of vaccine | Company name | Clinical phase | References |
|---|---|---|---|---|---|
| ChAdOx1 nCoV-19 | Non-replicating viral vector | ChAdOx1 | University of Oxford/AstraZeneca | Phase-III (24ISRCTN8995144) | (World Health Organization (WHO, 2020f) |
| mRNA-1273 | RNA | Lipid nanoparticle dispersion encapsulated mRNA | Moderna therapeutics | Phase-III (NCT04470427) | (World Health Organization (WHO, 2020f) |
| Sinovac | Inactivated viral vaccine | Inactivated viral vaccine | Sinovac | (NCT04456595) | (World Health Organization (WHO, 2020f) |
| Beijing Institute of Biological Products/Sinopharm | Inactivated | Inactivated | Beijing Institute of Biological Products/Sinopharm | (ChiCTR2000034780) | (World Health Organization (WHO, 2020f) |
| Wuhan Institute of Biological Products/Sinopharm | Inactivated viral vaccine | Inactivated vaccine | Wuhan Institute of Biological Products/Sinopharm | (ChiCTR2000034780) | (World Health Organization (WHO, 2020f) |
| BioNTech/Fosun Pharma/Pfizer | RNA | 3 LNP-mRNAs | BioNTech/Fosun Pharma/Pfizer | (NCT04368728) | (World Health Organization (WHO, 2020f) |
| Ad5-nCoV | Non-replicating viral vector | Adenovirus type 5 vector | CanSino biologics | Phase-II (ChiCTR000031781) | (World Health Organization (WHO, 2020f) |
| RBD-Dimer | Protein Subunit | Adjuvanted recombinant protein (RBD-Dimer) | Anhui ZhifeiLongcom Biopharmaceutical/Institute of Microbiology, Chinese Academy of Sciences | (NCT04466085) | (World Health Organization (WHO, 2020f) |
| INO-4800 | DNA | DNA plasmid encoding S protein delivered by electroporation | Inovio Pharmaceuticals, CEPI | Phase-I (NCT04336410) | (National Institutes of Health (NIH, 2020) |
| LV-SMENP-DC | Antigen-specific CTLs | Dendritic cells modified with a lentiviral vector expressing synthetic minigene based on domains of selected viral proteins; administered with antigen-specific CTLs | Shenzhen Geno-Immune Medical Institute | Phase-I Phase-II (NCT04276896) | (National Institutes of Health (NIH, 2020) |
| Covid-19/aAPC | Pathogen-specific aAPC | aAPCs modified with a lentiviral vector expressing synthetic minigene based on domains of selected viral proteins | Shenzhen Geno-Immune Medical Institute | Phase-I (NCT04299724) | (National Institutes of Health (NIH, 2020) |
Moderna therapeutics developed a vaccine and started clinical trials of mRNA-based vaccine (mRNA-1273) just after 2 months of sequence identification (Moderna, 2020). Thanh et al. (2020) described the platforms for vaccine development based on mRNA that provides the flexibility for antigen manipulation and rapid development process. Recombinant protein-based vaccine development may be advantageous owing to typical large-scale production capabilities. The use of an adjuvant can be important in this global pandemic situation. Adjuvants are small molecules that facilitate the immune reaction. Thereby reducing the amount of antigen protein required per dose. This method is more appropriate for COVID-19 vaccine development (Thanh et al., 2020).
These above-mentioned vaccine candidates are under several phases of clinical trials. Their vaccines may be taking more time (several months or years) until complete development. Hence, it is an urgent need to develop a potential vaccine against COVID-19 as soon as possible. In this pandemic situation, modern computation approaches may be more effective and less time-consuming compare to traditional approaches.
5. In silico approaches for vaccine design
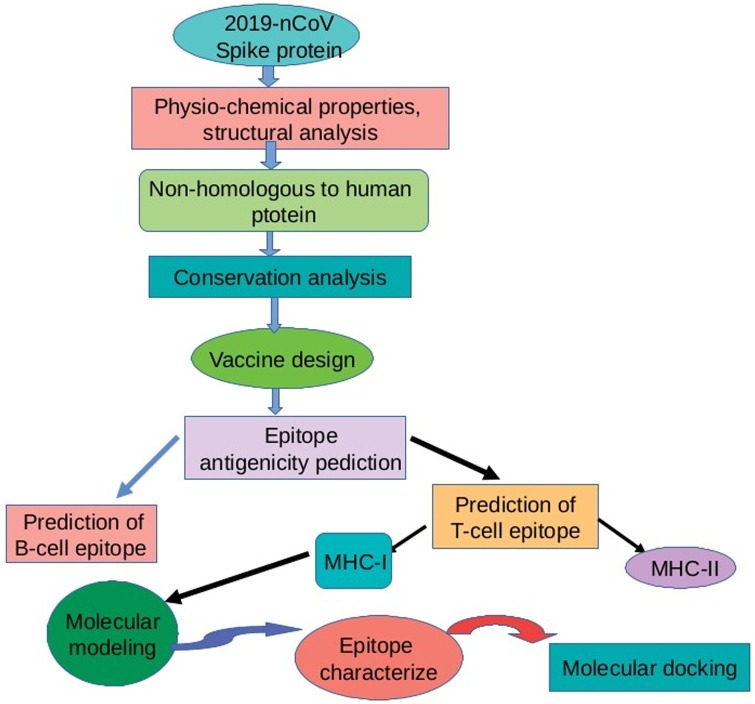
The traditional vaccine takes months or years of trials and also very expensive. Therefore, vaccine design using bioinformatics resources is more advantageous due to cost-effective and time saver. Biological information is well organized present in genetics, biotechnology, and molecular biology and bioinformatics resources stored the biological data and information (Orozco et al., 2013). Bioinformatics approaches including structural analysis, MD simulations, and molecular docking are also used for vaccine design. The overview of vaccine design using bioinformatics approaches are shown in Fig. 6 .
Fig. 6.
Application of computational biology in COVID-19 vaccine development.
In the current situation, vaccine development is an urgent need of the World. The drawback of traditional vaccine development techniques will be overcome by using computational approaches. Immunoinformatic approaches are more useful such as reverse vaccinology, epitope prediction, and structural vaccinology including rational approaches that are required for potential vaccine candidates. Protein scaffolding and epitope prediction play an essential role in vaccine design. Bioinformatics tools are also used for potential vaccine design against new emerging diseases. The vaccine can be designed against COVID-19 based on SARS-CoV, which has a high genetic similarity with the SARS-CoV-2 (Ahmed et al., 2020). Several bioinformatics approaches can be used to design a potential vaccine against COVID-19.
5.1. Reverse vaccinology
Reverse vaccinology (RV) technology is useful for epitope mapping and monovalent peptide vaccine prediction. In reverse vaccinology, a novel antigen of the virus is detected through entered proteome of SARS-CoV-2 using bioinformatics tools. Vaxign reverse vaccinology tool is used for developing a suitable vaccine to fight against SARS-CoV-2 (Sarkar et al., 2020).
5.1.1. Vaccine adjuvants
Peptide-based vaccine adjuvants can be identified by using VaxinPAD. It is used to predict Antigen-presenting cell (APC), epitope, or immuno-modulatory peptides. It made up of aluminum to enhance the host immune response and play a major role in vaccine development (Tom et al., 2019). Adjuvants can be designed by a different component such as TLR4 agonists, T-helper agonists, and flagellin. Toll-like receptors (TLRs) can be used as adjuvant it enhances immune response (Mbow et al., 2010; Kanzler et al., 2007). TLR3 and TLR4 play an important role in pathogen recognition and enhances the innate immune system. Agonists of Toll-like receptors 3 and 4 (TLR3 and TLR4) are potential adjuvant for different vaccines. The mechanism of the adjuvant action of TLR3 and TLR4 agonists is largely associated with their ability to activate APCs
5.2. Epitope prediction tools
An epitope is part of the antigen to which antibody binds and is recognized by the host immune system. Immunoinformatic tools are used to predict B-cell epitopes, T-cell epitopes, MHC binders, and vaccine adjuvant. Immune epitope database (IEDB) is a repository database that contains available antigenic regions or experimentally validated epitopes that tend to regulate the immune system. IEDB is used for the prediction of B-cell and T-cell epitope (Beaver et al., 2007). Epitope prediction tools design potential vaccines as the epitope stimulate immune cells by both B-cell and T-cell (Kreiter et al., 2015). Several bioinformatics approaches such as support vector machine, neural network, hidden Markov models, and QSAR (quantitative structure-activity relationship analysis) approaches are used to analyze the peptide interactions.
5.2.1. B-cell epitopes
The amino acid sequences of B-cell epitopes analyze using the BepiPred server. It is based on both the hidden Markov model and the amino acid propensity scales method. Kumar (2020) performed vaccine design and the author reported that B-cell epitope VLLPLVSSQCVNLTTRTQLPPAYTN has high antigenicity property. This antigenic epitope considers as suitable for vaccine candidates and developed vaccines could be helpful in viral control.
5.2.2. T-cell epitopes
Immunoinformatic tools such as CTLPred predicted Cytotoxic T lymphocytes (CTL) are useful tools. It is evaluated by an artificial neural network and support vector machine. The major histocompatibility complex plays an important role in the human immune response. It expresses on the cell surface and binds to antigenic peptides. ProPred 1 and ProPred tools are used in the identification of MHC-I and MHC-II binders, respectively (Patronov and Doytchinov, 2013).
5.2.3. Multi-epitope prediction
A multi-epitope-based peptide vaccine can be designed against SARS-CoV-2 using immunoinformatic approaches.
AAY linker is used to link the CTL epitope, GPGPG linker used for HTL epitope, and B KK linker used for the B-cell epitope. EAAAK linker is used to increase the vaccine immunogenicity by adjoining the β-defensin (45 mer) amino acid sequence and pan-HLA DR binding epitopes (13aa) to the N-terminal of the vaccine. The β-defensin adjuvant boosts innate immunity cells and recruits naive T cells with the help of chemokine receptor-6 (CCR-6). Different linkers (AYY, KK, and GPGPG) play potential roles in producing flexibility, protein folding, and separation of functional domains, and therefore, make a more stable protein structure (Dong et al., 2020; Kumar et al., 2020a).
5.3. Allergenicity and antigenicity prediction
The allergenicity of selected protein can be predicted using AllerTOP (https://www.ddg-pharmfac.net/AllerTOP/) and AllergenFP (http://ddg-pharmfac.net/AllergenFP/) are two online tools. AllerTOP prediction tool gives more appropriate results because it has better accuracy (88.7 %) compared to AllergenFP (87.9 %). Antigenicity of designed protein can be predicted using VaxiJen server. ToxinPredserver tools are based on SVM and this tool applies to the prediction of toxicity of designed proteins. After antigenicity, toxicity and allergenicity tests the protein found as antigenic, non-allergenic, and non-toxic. This protein considers as the best potential for vaccine design (Dimitrov et al., 2013).
5.4. Physicochemical parameters analysis
The physicochemical parameters are molecular weight, isoelectric point (pI), extinct coefficient, aliphatic index, grand average of hydropathicity, stability, and instability index of designed vaccine evaluated by ProtParam online server (Gasteiger et al., 2005).
5.5. Molecular docking
Molecular docking approaches performed the study of molecular interaction for vaccine development (Kumar et al., 2020a, b, c). Molecular docking assays have been widely applied to study and understand the mechanistic interaction of drugs with target protein receptor (Kumar et al., 2020a, b, c). Molecular docking was performed using Autodock Vina and molecular interaction analysis was done using the Autodock tool. Autodock is an automated docking tool it is used to predict how small molecules such as drugs bind to the 3D structure of the receptor. Molecular docking analyses were executed using the Hex 8.0 module. Hex server is a protein docking server based on a fast Fourier transform. Molecular docking used for prediction of binding affinity of chimeric peptide vaccine with immune receptor including TLR3 and TLR4 (Rahman et al., 2020; Saha and Prasad, 2020). Discovery studio visualizer software is used for the determination of the best result of molecular docking (Ullah et al., 2020).
5.6. Molecular dynamics simulation
Molecular dynamics simulation approaches help in analyzing the stable binding of drugs with the target protein. It evaluates the stability of the receptor-ligand complex. Molecular dynamics simulation studies were performed for a long time and analyzed root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and radius of gyration (RG) plots (Kumar et al., 2020a, b, c). With the help of RMSD, RMSF, RG, and other parameters help in analyzing the atomic fluctuations of the interaction complex system (Kumar et al., 2020a, b, c). RMSD plot was determined to analyze each residue dynamics of the target receptor. (RMSF) was obtained for amino acid side chains to analyze the fluctuations of amino acid in the docked complex (Ojha et al., 2020). Molecular dynamics simulation performed by GROMACS 2018 package and iMODS server. GROMACS is one of the fastest molecular dynamics packages for simulation of lipids, proteins, and nucleic acids. iMODS is an online server, it predicts the deformability, B-factor (mobility profiles) and eigenvalues, variance and co-variance and elastic network of protein. The lower eigenvalue shows the deformability of the complex. This server uses for the analysis of protein flexibility (Kovacs et al., 2004).
5.7. Codon optimization and other available tools
The codon optimization method is used for the enhancement of the expression of vaccine protein. Codon Optimization Online (COOL) tool to synthesize genes. A COOL tool is used for the optimization of various parameters (codon optimization parameters) such as codon adaptation, codon pairing, and codon usage (Chin et al., 2014). Codon adaptation is important to step in silico cloning, which predicts the best codon for a specific amino acid. Java Codon Adaptation Tool uses for codon adaption and plays an important role in the prediction of protein sequence for potential vaccine design (Grote et al., 2005).
6. Role of COVID-19 on brain tissue
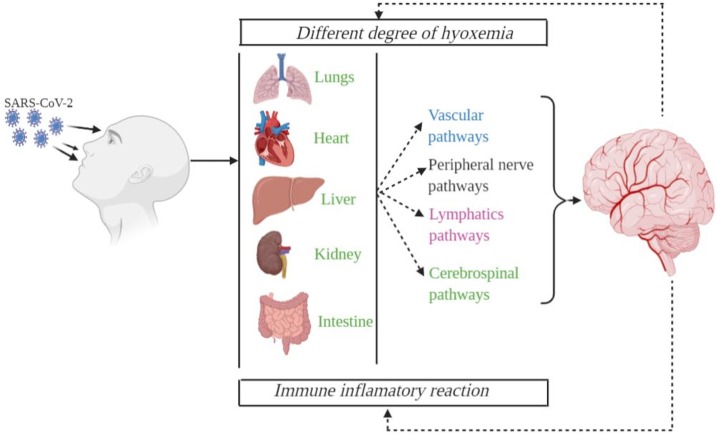
COVID-19 influences the lungs as well as focus on the sensory system. SARS-CoV-2 may arrive at the brain tissue through a neural route from lung chemoreceptors, seriously influencing the cardiorespiratory center (Li et al., 2020a, b, c). SARS-CoV-2 additionally influence the olfactory nerve terminal in the nasal hole. Furthermore, SARS-CoV-2 may come to the cerebral vasculature through the overall flow, breaking the blood-brain barrier and attacking and harming the cerebrum parenchyma. SARS-CoV-2 may bind to its receptor Angiotensin-Converting Enzyme 2 (ACE2) communicated in endothelial cells of cerebral vessels, and inside the brain parenchyma in the two neurons and microglia. Since the blood-brain barrier is disturbed in hypertension and hypertension is continuous comorbidity for COVID-19, these patients may have a higher danger of cerebral complexities. The SARS-CoV-2 infects brain tissue and causes a serious neurological interruption. The main organs and routes of SARS-CoV-2 neuroinvasion are shown in Fig. 7 .
Fig. 7.
Main organs and routes of SARS-CoV-2 neuroinvasion. The SARS-CoV-2 enters into body and binds with ACE-2 receptors presents in the cells of major vital organs such as heart, lungs, kidney, liver. SARS-CoV-2. SARS-CoV-2 activate intrinsic immune response, ARDS as well as damage of peripheral tissue. SARS-CoV-2 invasion in the brain through cerebrospinal fluid pathways, lymphatics pathways, peripheral nerve as well as vascular pathways.
As indicated by ongoing investigations, the coronavirus displays neurotropic qualities and instigates neurological diseases. It likewise announced that the SARS-CoVs-2 infection could found in the human cerebrum. The coronavirus disease modifies the CNS and can lead the host immune response to triggers into a steady issue prompting neurological signs. The infected patients assessed right on time for neurological manifestations with migraines, thinking brokenness, paraesthesia, and other pathologic side effects (Wu et al., 2020a, b, c, d). Because of the pandemic, the antagonistic psychosomatic result is expected to increment among people. The virus can also enter the cerebrum through contaminating endothelial cells lining brain vasculature; an electron-microscopic analysis of the frontal lobe distinguished SARS-COV-2 viral particles in the endothelium with some indications for virus transit to the neuropil. The SARS-COv-2 can enter the CNS utilizing the perivascular spaces of the lymphatic system. Moreover, viruses can attack the brain through different nerves, such as the trigeminal nerve, which ventures nociceptive terminals to nasal cavities. Similarly, tactile filaments of the vagus nerve, that innervate the respiratory tract, can introduce another attack route. Additional proof of the SARS-CoV-2 neuro-infection, oedema, and neuronal degeneration were accounted for in posthumous mind tests, while for a situation of encephalitis genome sequencing affirmed viral nearness in the cerebrospinal fluid (Li et al., 2020a, b, c). Researchers have discovered that organoids (little tissue culture produced using human cells that stimulate entire organs) known as "mini-brains" can be infected by the SARS-CoV-2 that causes COVID-19 (IndiaTV, 2020). Structure and function of spike protein are highly important for cell infection and entering the brain cells
7. Conclusion
COVID-19 has brought the entire planet to its knee challenged the entire mankind in various aspects which include health system, economy, research, and development, etc. SARS-CoV-2 emerged in the wild animal market of Wuhan city, China, and currently transmitted all over the World. WHO declared COVID-19 as a global emergency based on its infectious properties. Human to human transmission of COVID-19 occurs through respiratory droplets and human close contact. The transmission rate of COVID-19 is much higher compared to other diseases such as SARS-CoV and MERS-CoV caused by coronaviruses. WHO and other health organizations suggested the lockdowns, curfews, isolations, quarantines, and social distancing as the ways to mitigate transmission of infection. Based on the genomic study, the SARS-CoV-2 is very similar to SARS-CoV and MERS-CoV viruses. Phylogenetic analysis reveals that SARS-CoV-2 shares the highest nucleotide sequence similarity (79 %) with SARS-CoV. The characterization of the SARS-CoV-2 is based on the PCR and metagenomic next-generation sequencing. The pathogenic mechanism of SARS-CoV-2 is very similar to the SARS-CoV virus. SARS-CoV-2 first infects the lower airway and uses the ACE-2 receptor the same as SARS-CoV. Due to the unavailability of any drug or vaccine, it is needful to design potential vaccines or drugs for COVID-19. Computational methods for design the COVID-19 vaccine are more appropriate compared to traditional methods due to cost-effective, time saver, and more specificity. Immunoinformatics, reverse vaccinology, and molecular docking is major approaches for vaccine development. This review concluded brief information about COVID-19, transmission, genomic and molecular characterization, the pathogenic mechanism of COVID-19, and current treatment option. It also covers potential vaccine design against COVID-19 by using bioinformatics approaches. Infected patients with SARA-CoV-2 should be evaluated early for neurological symptoms, including headache, consciousness disorder, paraesthesia, and other pathological signs
Funding
The authors declare that there is no financial support for this research work.
Declaration of Competing Interest
The authors declare that there is no conflict of interest.
Acknowledgments
Prof. M.P. Singh gratefully acknowledge Prof. A.K. Singh, Ex-Vice-Chancellor, University of Allahabad, India for his unending support and encouragement towards academic excellence. Nidhi Singh achnowledge UGC, New Delhi, India for providing fellowship to pursue Ph.D. Dr. Sachchida Nand Rai acknowledge UGC for awarding Dr. DS Kothari PDF (Ref. No-F.4-2/2006 (BSR)/BL/19-20/0032).
References
- Agostini M.L., Andres E.L., Sims A.C. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9:e00221–18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:E254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Kulcsar K., Misra V. Bats and coronaviruses. Viruses. 2019 doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver J.E., Bourne P.E., Ponomarenko J.V. Epitope viewer:a Java application for the visualization and analysis of immune epitopes in the Immune Epitope Database and Analysis Resource (IEDB) Immunome Res. 2007 doi: 10.1186/1745-7580-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley S. With ventilators running out, doctors say the machines are overused for Covid-19. STAT. 2020 https://www.statnews.com/2020/04/08/doctors-say-ventilators-overused-for-covid-19/ Accessed 8 April 2020. [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaguro F.M., Puzanov I., Ascierto P.A. Anti-IL6R role in treatment of COVID-19-related ARDS. J. Transl. Med. 2020;18:165. doi: 10.1186/s12967-020-02333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Bermejo-Martin J.F., Danesh A. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDCP) 2020. Coronavirus Disease 2019 (COVID-19). Symptoms of Coronavirus.https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html Accessed: April 22, 2020. [Google Scholar]
- Centers for Disease Control and Prevention (CDCP) 2020. Viral Hepatitis.https://www.cdc.gov/hepatitis/outbreaks/index.htm Accessed: April 22, 2020. [Google Scholar]
- Centers for Disease Control and Prevention(CDCP) 2020. Influenza (Flu). 1918 Pandemic (H1N1 virus)https://www.cdc.gov/flu/pandemic-resources/1918-pandemic-h1n1.html Accessed: April 22, 2020. [Google Scholar]
- Chakraborty C., Sharma A.R., Sharma G. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4016–4026. doi: 10.26355/eurrev_202004_20871. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K.H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancey C., Grinev A., Volkov E. The global ecology and epidemiology of west nile virus. Biomed Res. Int. 2015 doi: 10.1155/2015/376230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzou M., Magis C., Chang J.M., Kemena C., Bussotti G., Erb I., Notredame C. Multiple sequence alignment modeling: methods and applications. Brief. Bioinf. 2016;17:1009–1023. doi: 10.1093/bib/bbv099. [DOI] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J.X., Chung B.K.S., Lee D.Y. Codon optimization online (COOL): a web-based multi-objective optimization platform for synthetic gene design. Bioinformatics. 2014;30:2210–2212. doi: 10.1093/bioinformatics/btu192. [DOI] [PubMed] [Google Scholar]
- Ching K.Y., Chow, Hon C.C. Molecular advances in severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Genomics Proteomics Bioinf. 2003;1:247–262. doi: 10.1016/S1672-0229(03)01031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D. Springer Nature; Heidelberg: 2020. Did Pangolins Spread the China Coronavirus to People.https://www.nature.com/articles/d41586-020-00364-2 Accessed 7 February 2020. [DOI] [PubMed] [Google Scholar]
- de Wilde A.H., Snijde r E.J., Kikkert M. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Feldmann F., Cronin J. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U.S.A. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I., Flower D.R., Doytchinova I. AllerTOP-a server for in silico prediction of allergens. BMC Bioinf. 2013 doi: 10.1186/1471-2105-14-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R., Chu Z., Yu F., Zha Y. Contriving multi-epitope subunit of vaccine for COVID-19: immunoinformatics approaches. Front. Immunol. 2020 doi: 10.3389/fimmu.2020.01784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.S., Zambon M.C. Molecular diagnosis of influenza. Rev. Med. Virol. 2002;12:375–389. doi: 10.1002/rmv.370. [DOI] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Gouklani H. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. J Biomol Str Dynami. 2020 doi: 10.1080/07391102.2020.1756411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada E. Topological analysis of SARS CoV-2 main protease. Chaos. 2020 doi: 10.1063/5.0013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Pharmaceutical Review . 2020. US Researchers to Study Stem Cell Therapy in COVID-19 Patients.https://www.europeanpharmaceuticalreview.com/news/116794/us-researchers-to-study-stem-cell-therapy-in-covid-19-patients/ Accessed 27 April 2020. [Google Scholar]
- Fan Y.Y., Huang Z.T., Li L. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch. Virol. 2009;154:1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A. The Proteomics Protocols Handbook. Springer; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- Gil C., Ginex T., Maestro I. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- Golchin A., Seyedjafari E., Ardeshirylajimi A. Mesenchymal stem cell therapy for COVID-19: present or future. Stem Cell Rev and Rep. 2020 doi: 10.1007/s12015-020-09973-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote A., Hiller K., Scheer M. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:526–531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Praharaj I., Bhatnagar T. Severe acute respiratory illness surveillance for coronavirus disease 2019, India, 2020. Indian J. Med. 2020 doi: 10.4103/ijmr.IJMR_1035_20. Res.doi:10.4103/ijmr.IJMR_1035_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez P. Coronavirus map: how COVID-19 is spreading across the world. The Guardian. 2020 Accessed 22 April 2020. [Google Scholar]
- Hampton T. Bats may be SARS reservoir. JAMA. 2005;294:2291. doi: 10.1001/jama.294.18.2291. [DOI] [PubMed] [Google Scholar]
- Healthline . 2020. The History of HIV and AIDS in the United States.https://www.healthline.com/health/hiv-aids/history#1981-1990s Accessed 22 April 2020. [Google Scholar]
- Hoffmann M.K.W.H., Kruger N., Muller M. 2020. The Novel Coronavirus 2019 (2019-nCoV) Uses the SARS-coronavirus Receptor ACE2 and the Cellular Protease TMPRSS2 for Entry Into Target Cells. bioRxiv [preprint] https://www. biorxiv.org/content/10.1101/2020.01.31.929042v1. [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IndiaTV . 2020. How COVID-19 Virus Can Infect Human Brain Cells.https://www.indiatvnews.com/health/how-covid-19-virus-can-infect-human-brain-cells-630696 Accessed 1August 2020. [Google Scholar]
- Jia H.P., Look D.C., Shi L. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins University (JHU) 2020. COVID-19 Dashboard by the Center for System Science and Engineering (CSSE)https://coronavirus.jhu.edu/map.html Accessed 27 August 2020. [Google Scholar]
- Kanzler H., Barrat F.J., Hessel E.M. Therapeutic targeting of innate immunity with toll-like receptor agonists and antagonists. Nat. Med. 2007;13:552–559. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- Kovacs J.A., Chacon P., Abagyan R. Predictions of protein flexibility: first‐order measures. Proteins: Struct, Funct, Bioinf. 2004;56:661–668. doi: 10.1002/prot.20151. [DOI] [PubMed] [Google Scholar]
- Kreiter S., Vormehr M., van de Roemer N. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. 2020. Drug and Vaccine Design Against Novel Coronavirus (2019-nCoV) Spike Protein Through Computational Approach. Preprints 2020020071. [DOI] [Google Scholar]
- Kumar G.V., Jeyanthi V., Ramakrishnan S. A short review on antibody therapy for COVID-19. New Microbes New Infect. 2020 doi: 10.1016/j.nmni.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Sood D., Spek P.J., Sharma H.S., Chandra R. Molecular binding mechanism and pharmacology comparative analysis of noscapine for repurposing against SARS-CoV-2 protease. J. Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00367. [DOI] [PubMed] [Google Scholar]
- Kumar N., Sood D., Sharma N., Chandra R. Multiepitope subunit vaccine to evoke immune response against acute encephalitis. J. Chem. Inf. Model. 2020;60:421–433. doi: 10.1021/acs.jcim.9b01051. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Laupland B.K., Valiquette L. Ebola virus disease. Can. J. Infect. Dis. Med. Microbiol. 2014;25:128–129. doi: 10.1155/2014/527378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Z., Zhu R., Hou W. Transplantation of ACE2- mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020;11:216–228. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li H., Liu S.M., Yu X.H. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu T., Yang N. Neurological manifestations of patients with COVID-19: potential routes of SARS-CoV-2 neuroinvasion from the periphery to the brain. Front. Med. 2020 doi: 10.1007/s11684-020-0786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Fontanet A., Zhang P.H. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis. 2006;193:792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wu P., Gao F. Novel immunodominant peptide presentation strategy: a featured HLA-A∗2402-restricted cytotoxic T-lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Virol. 2010;84:11849–11857. doi: 10.1128/JVI.01464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.J., Zhao M., Liu K. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gayle A.A., Smith A.W. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie S.J., Smith D.W. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don’t. Microbiol. Aust. 2020 doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud E., Dauerman H.L., Welt F.G. Management of acute myocardial infarction during the COVID-19 pandemic. J. Am. Coll. Cardiol. 2020 doi: 10.1016/j.jacc.2020.04.039. S0735-1097:35026-35029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal D. Coronavirus threat to Indian population: risk factors, transmission dynamics and preparedness to prevent the spread of the virus. Virusdisease. 2020;20:1–4. doi: 10.1007/s13337-020-00581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbow M.L., De Gregorio E., Valiante N.M. New adjuvants for human vaccines. CurrOpin Immunol. 2010;22:411–416. doi: 10.1016/j.coi.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Mehndiratta M.M., Mehndiratta P., Pande R. Poliomyelitis historical facts, epidemiology, and current challenges in eradication. Neurohospitalist. 2014;4:223–229. doi: 10.1177/1941874414533352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Perlman S., Kerkhove M.D.V. Middle east respiratory syndrome. Lancet. 2020;395:1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moderna Therapeutics . 2020. Moderna Announces First Participant Dosed in NIH-led Phase 1 Study of mRNA Vaccine (mRNA-1273) Against Novel Coronavirus.https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-first-participant-dosed-nih-led-phase-1-study Accessed 16 March 2020. [Google Scholar]
- Mourya D.T., Yadav P.D., Ullas P.T. Emerging/re-emerging viral diseases & new viruses on the Indian horizon. Indian J. Med. Res. 2019;149:447–467. doi: 10.4103/ijmr.IJMR_1239_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavizadeh L., Ghasemi Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. S1684-1182:30082-30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health (NIH) U.S. Department of Health and Human Services; 2020. NIH Clinical Trial of Investigational Vaccine for COVID-19 Begins.https://www.nih.gov/news-events/news-releases/nih-clinical-trial-investigational-vaccine-covid-19-begins Accessed 25 March 2020. [Google Scholar]
- Ojha R., Gupta N., Naik B. High throughput and comprehensive approach to develop multiepitope vaccine against minacious COVID-19. Eur. J. Pharm. Sci. 2020;151:105375. doi: 10.1016/j.ejps.2020.105375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco A., Morera J., Jimenez S. A review of bioinformatics training applied to research in molecular medicine, agriculture and biodiversity in Costa rica and Central America. Brief Bioinform. 2013;14:661–670. doi: 10.1093/bib/bbt033. [DOI] [PubMed] [Google Scholar]
- Paraskevis D., Kostaki E.G., Magiorkinis G. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020 doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patronov A.I., Doytchinov I. Vol. 3. 2013. (T-Cell Epitope Vaccine Design by Immunoinformatics). 20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonu O. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.03.026. https:// doi.org/10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Travanty E.A., Oko L. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome -coronavirus. Am. J. Respir. Cell Mol. Biol. 2013;48:742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S.M., Hoque M.N., Islam M.R. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2 etiologic agent of global pandemic COVID-19: an in silico approach. BioRxiv. 2020 doi: 10.1101/2020.03.30.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus -EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. [preprint] 2020 doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R., Prasad B.V.S.L. Silico approach for designing of a multi-epitope based vaccine against novel Coronavirus (SARS-COV-2) BioRvix. 2020 doi: 10.1101/2020.03.31.017459. [DOI] [Google Scholar]
- Sarkar B., Ullah M.A., Johora F.T. The essential facts of wuhan novel coronavirus outbreak in china and epitope-based vaccine designing against 2019-nCoV. BioRvix. 2020 doi: 10.1101/2020.02.05.935072. [DOI] [Google Scholar]
- Scavone C., Brusco S., Bertini M. Current pharmacological treatments for COVID-19: what’s next? Br. J. Pharmacol. 2020 doi: 10.1111/bph.15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber C.C., Kiesswetter E., Kwetkat A. Prevention: public healthcare, nutrition, physical activity, vaccination. In: Roller-Wirnsberger R., Singler K., Polidori M.C., editors. Learning Geriatric Medicine: a Study Guide for Medical Students. Springer International Publishing; Cham, Switzerland: 2018. pp. 237–262. [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entryProc. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Singh A., Shaikh A. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: a systematic search and a narrative review with a special reference to India and other developing countries. Diabetes MetabSyndr. 2020;14:241–246. doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus Disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa S., Prandoni P., Paoletti O. Direct oral anticoagulant plasma levels striking increase in severe COVID-19 respiratory syndrome patients treated with antiviral agents. The cremona experiences. J. ThrombHaemost. 2020 doi: 10.1111/jth.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh L.T., Andreadakis Z., Kumar A. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020 doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- The Economic Times . 2020. Hydroxychloroquine Consignment From India Arrives in US.https://economictimes.indiatimes.com/news/politics-and-nation/hydroxychloroquine-consignment-from-india-arrives-in-us/articleshow/75103328.cms?from=mdr [Google Scholar]
- Thompson R. Pandemic potential of 2019-nCoV. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom J.K., Albin T.J., Manna S. Applications of immunomodulatory immune synergiesto adjuvant discovery and vaccine development. Trends Biotechnol. 2019;37:373–388. doi: 10.1016/j.tibtech.2018.10.004. [DOI] [PubMed] [Google Scholar]
- Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X., Chong W.P., Zhai Y. Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. J. Infect. 2015;71:101–109. doi: 10.1016/j.jinf.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah M.A., Sarkar B., Islam S.S. 2020. Exploiting the Reverse Vaccinology Approach to Design Novel Subunit Vaccine Against Ebola Virus. medRxiv preprint. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019 -nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M.C., Freiberg A.N., Zhang T. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3157–3162. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) 2020. Lassa Fever.https://www.who.int/news-room/fact-sheets/detail/lassa-fever Accessed 22 April 2020. [Google Scholar]
- World Health Organization (WHO) 2020. Emergencies Preparedness, Response.https://www.who.int/csr/don/2003_05_28/en/ Accessed 22 April 2020. [Google Scholar]
- World Health Organization (WHO) 2020. Middle East Respiratory Syndrome Coronavirus (MERS-CoV)https://www.who.int/emergencies/mers-cov/en/ Accessed 22 April 2020. [Google Scholar]
- World Health Organization (WHO) 2020. Marburg Virus Disease.https://www.who.int/health-topics/marburg-virus-disease/#tab=tab_1 Accessed 22 April 2020. [Google Scholar]
- World Health Organization (WHO) 2020. International Travel and Health.https://www.who.int/ith/diseases/sars/en/ Accessed 22 March 2020. [Google Scholar]
- World Health Organization (WHO) 2020. DRAFT Landscape of COVID-19 Candidate Vaccines – 31 July 2020. file:///C:/Users/Downloads/novel-coronavirus-landscape-covid-19cc0e97e4ea1b4458a05bbd6f5ac6d3fe.pdf Accessed 31 July 2020. [Google Scholar]
- Wu C., Chen X., Cai Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. https://doi.org/10.1038/ s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30260-9. https://doi.org/10.1016/S0 140-6736(20)30260-30269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xu X., Chen Z. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020;87:18–22. doi: 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Resp Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Wang X. COVID-19: a new challenge for human beings. Cell. Mol. Immunol. 2020;31:1–3. doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Du L., Ojcius D.M. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;20 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B. Molecular and serological investigation of 2019- nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., He Y. Challenges of convalescent plasma therapy on COVID-19. J. Clin. Virol. 2020 doi: 10.1016/j.jcv.2020.104358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J. Network-based drug repurposing for human coronavirus. medRxiv. 2020 doi: 10.1101/2020.02.03.20020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F.W., Azhar E.I. Coronaviruses-drug discoveryand therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]