Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is considered as the most significant global public health crisis of the century. Several drug candidates have been suggested as potential therapeutic options for COVID-19, including remdesivir, currently the only authorized drug for use under an Emergency Use Authorization. However, there is only limited information regarding the safety profiles of the proposed drugs, in particular drug-induced cardiotoxicity. Here, we evaluated the antiviral activity and cardiotoxicity of remdesivir using cardiomyocytes-derived from human pluripotent stem cells (hPSC-CMs) as an alternative source of human primary cardiomyocytes (CMs). In this study, remdesivir exhibited up to 60-fold higher antiviral activity in hPSC-CMs compared to Vero E6 cells; however, it also induced moderate cardiotoxicity in these cells. To gain further insight into the drug-induced arrhythmogenic risk, we assessed QT interval prolongation and automaticity of remdesivir-treated hPSC-CMs using a multielectrode array (MEA). As a result, the data indicated a potential risk of QT prolongation when remdesivir is used at concentrations higher than the estimated peak plasma concentration. Therefore, we conclude that close monitoring of the electrocardiographic/QT interval should be advised in SARS-CoV-2-infected patients under remdesivir medication, in particular individuals with pre-existing heart conditions.
Keywords: COVID-19, SARS-CoV-2, Human cardiomyocytes, Pluripotent stem cells, Remdesivir
Graphical abstract
Highlights
-
•
Remdesivir potently inhibits SARS-CoV-2 infection in hPSC-CMs with EC50 at high nanomolar concentrations.
-
•
Remdesivir-induced arrhythmogenic risk in hPSC-CMs was evaluated by multi-electrode array-based assay.
-
•
Remdesivir exhibits potential cardiotoxicity in hPSC-CMs with CC50 values close to the estimated peak plasma concentration.
-
•
Close monitoring of the electrocardiographic/QT interval should be advised in COVID-19 patients under remdesivir medication.
1. Introduction
The world is currently facing unprecedented challenges from the coronavirus disease-19 (COVID-19) pandemic caused by Severe Acute Respiratory Syndrome-coronavirus-2 (SARS-CoV-2). Since its first appearance in December 2019, the highly contagious virus has spread throughout the world with detrimental effects on global socioeconomic and healthcare systems. SARS-CoV-2 infection predominantly manifests respiratory symptoms with a range of disease severity. In some cases, it progresses to multi-organ failure, especially in the elderly and those with other co-morbidities, eventually leading to death (Guan et al., 2020; Tay et al., 2020). In addition, there is growing evidence that SARS-CoV-2 infection causes extrapulmonary manifestations, including cardiovascular complications such as myocardial injury, acute myocardial infarction, arrhythmias, and heart failure that significantly increase the risk of mortality (Long et al., 2020; Shi et al., 2020). Several drug candidates have been suggested as potential therapeutic options for COVID-19, including remdesivir, which is currently the only authorized drug for use under an Emergency Use Authorization (Eastman et al., 2020). However, the proposed drugs for COVID-19 provide only limited information regarding safety profiles, especially drug-induced cardiotoxicity, which is one of the leading causes of post-marketing withdrawal. Therefore, assessment of any potential cardiovascular adverse reactions associated with current COVID-19 pharmacotherapy is of utmost importance to avoid fatal side effects.
Primary human CMs are the ideal platform to examine drug-induced cardiotoxicity, but they are virtually impossible to obtain sufficient quantities for any applications (Mitcheson et al., 1998). Due to many similarities with primary human CMs, CMs-derived from human pluripotent stem cells (hPSC-CMs), which include both human embryonic stem cells (hESCs) and human-induced pluripotent stem cells (hiPSCs), have attracted significant attention as an alternative system for a wide range of applications, including human cardiac disease modeling, cardiac drug toxicity screening, and cell-based cardiac repair (Laflamme et al., 2007). The hPSC-CMs possess a clear cardiac phenotype, exhibiting spontaneous contractile activity, cardiac-type mechanisms of excitation-contraction coupling, and expression of cardiac transcription factors, sarcomeric proteins, and ion channels. In addition, hPSC-CMs are readily generated and purified within weeks using defined protocols and can be maintained over several weeks for further applications. Most importantly, hPSC-CMs are capable of recapitulating cardiac pathophysiology, rendering them ideal for assessing cardiac safety in preclinical drug screening (Moreno and Pearson, 2013; Sharma et al., 2017). In this context, the Comprehensive in vitro Proarrhythmia Assessment (CiPA), which is a non-clinical Safety Pharmacology paradigm, have proposed the use of hPSC-CMs as a reliable cardiotoxicity assay to overcome the limitations of the existing methodologies used for preclinical safety evaluation of pharmaceutical entities (Goineau and Castagne, 2017; Gintant et al., 2016; Sala et al., 2017).
In this study, we generated hPSC-CMs from human embryonic stem cells (hESCs: H9) and human-induced pluripotent stem cells (hiPSCs: CMC-11) and used them to investigate the therapeutic potential of remdesivir in SARS-CoV-2 infected hPSC-CMs. Furthermore, we evaluated the potential cardiovascular risk associated with remdesivir treatment using different approaches such as drug-induced cytotoxicity, electrophysiology, and automaticity of hPSC-CMs. The results in this study provide new insights to better understanding the potency and cardiotoxicity of remdesivir in human CMs.
2. Materials and methods
2.1. Cells and viruses
Vero E6 (ATCC® CRL-1586) cells were maintained in Dulbecco's modified Eagle's medium (DMEM, HyClone) supplemented with 10% fetal bovine serum (FBS; HyClone) at 37 °C. Patient-derived isolate SARS-CoV-2 (hCoV/Korea/KCDC-03/2020) was kindly provided by the Korea Centers for Disease Control and Prevention (KCDC, Osong, Republic of Korea). The working virus stock was propagated in Vero E6 cells. The virus-containing supernatants were collected, clarified by centrifugation, and aliquots were stored at −80 °C until further use. Virus stocks were titrated by plaque assay using Vero E6 cells, as previously described (Shin et al., 2018). All experiments using infectious SARS-CoV-2 were performed in a biosafety level-3 facility at Korea Research Institute of Chemical Technology (KRICT), Daejeon, Republic of Korea.
2.2. Chemicals
Chloroquine and hydroxychloroquine were purchased from Sigma-Aldrich (USA). Remdesivir and favipiravir were obtained from MedChem Express (USA). Nifedipine (L-type Ca2+ channel blocker), isoprenaline (β2-adrenergic agonist), and the hERG K+ channel blockers, E4031 and dofetilide were purchased from Sigma-Aldrich. All compounds were prepared in 100% dimethyl sulfoxide (DMSO, Sigma-Aldrich).
2.3. Differentiation of human pluripotent stem cells-derived cardiomyocytes (hPSC-CMs)
The hPSC (H9: Wicell® and CMC-hiPSC-011: KNIH) cell lines were maintained with the StemMACS iPS-BREW XF, human (Miltenyi Biotec, Germany) on Matrigel (Corning, USA). For cardiac lineage differentiation, hPSCs were seeded onto a hPSC-qualified Matrigel-coated cell culture dish (Eppendorf, Germany) at 140,000 cells/cm2 dish. A 5 μM of Y-27632 (Tocris, UK) was added for the first 24 h after passaging. The medium was changed daily, and hPSCs were allowed to grow in iPS BREW for 3–4 days until cells were 90% confluent. At day 0, cells were treated with 6 μM/ml of CHIR99021 (Tocris) in cardiomyocyte differentiation medium (CDM; RPMI1640 [ThermoFisher Scientific] supplemented with bovine serum albumin [BSA, Sigma-Aldrich] and ascorbic acid [Sigma-Aldrich]). After 48 h of incubation, the medium was changed to CDM supplemented with 2 μM/ml of C59, a Wnt inhibitor (Stemgent Inc., USA), and further incubated for 48 h. On day 5, the medium was replaced with fresh CDM and subsequently changed with fresh medium every other day. Spontaneously, contracting cells began to appear approximately on day 8 to day 10. From day 10 to day 15, CDM containing L-lactic acid was used to select and purify hPSC-CMs metabolically. All images were analyzed using an Eclipse-Ti2 fluorescence microscope (Nikon, Japan).
2.4. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
Total RNA from hPSC-CMs was isolated using the TRIzol reagent (Invitrogen, USA) according to the manufacturer's instructions. The human heart total RNA (Cat #636532, Takara, Japan) was used as control. One microgram of RNA was used for cDNA synthesis using a SuperScript II reverse transcriptase kit (AccuPowerBioneer, Korea). The resultant cDNAs were used to measure mRNA expression levels of human cardiac sarcomeric markers, cTnT and MYH6, as well as ACE2 and TMPRSS2 by qPCR using gene-specific primers (human TNNT2 [GenBank NM_000364]; sense 5′-GGGTTACATCCAGAAGACAG-3′ and antisense 5′-GTTATAGATGCTCTGCCACA-3′, human MYH6 [GeneBank NM_002471]; sense 5′-AGTATGAGGAGTCGCATCT-3′ and antisense 5′-CACATTCTTTCCTCCTTCTC-3′, human ACE2 [GenBank NG_012575.1]; sense 5′-CATTGGAGCAAGTGTTGGATCTT-3, antisense 5′-GAGCTAATGCATGCCATTCTCA-3′, and human TMPRSS2 [GenBank NM_005656]; sense 5′-AATCGGTGTGTTCGCCTCTAC-3, antisense 5′-CGTAGTTCTCGTTCCAGTCGT-3′) and SYBR Green gene expression assays (Roche, Switzerland). Glyceraldehyde 3-phosphate dehydrogenase was used as an endogenous control gene. The gene expression level was calculated using the ΔΔCT method, as described previously (Livak and Schmittgen, 2001). For intracellular viral RNA quantification, total cellular RNA was purified from cell lysates using an RNeasy Mini Kit (Qiagen, CA, USA) according to the manufacturer's instructions. RT-qPCR was performed using a SuperScript III Platinum® SYBR® Green One-Step RT-PCR kit (Invitrogen, USA) with a primer set targeting SARS-CoV-2 receptor-binding domain (RBD) gene (sense 5′-CAATGGTTTAACAGGCACAGG-3′ and antisense 5′-CTCAAGTGTCTGTGGATCACG-3′). The relative viral RNA expression levels were calculated by the ΔΔCT method, and β-actin was used as an endogenous control.
2.5. Immunofluorescence microscopy
The hPSC-CMs were plated onto a gelatin-coated glass dish and cultured for 5 days. Cells were fixed with 4% (w/v) paraformaldehyde (PFA) for 20 min at 4 °C, permeabilized with 0.1% BSA in 0.03% Triton X-100 for 10 min at room temperature (RT), and blocked with 0.03% Triton X-100 containing 10% normal goat serum (NGS, ThermoFisher Scientific) for 30 min at RT. Subsequently, cells were stained with antibodies for ACE2 or TMPRSS2 (1:100 dilution; ThermoFisher Scientific), cTnT (1:500 dilution; Abcam, USA), and ACTN2 (1:200 dilution; Sigma-Aldrich) diluted in 0.03% Triton X-100 and incubated at 4 °C overnight. Cells were washed three times for 10 min with 0.03% Triton X-100 and incubated with Alexa Fluor (AF)-594 goat anti-rabbit and AF488 goat anti-mouse IgG antibodies (ThermoFisher Scientific) for 1 h at RT. Lastly, cells were washed four times before counterstaining the cell nuclei with 4’,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific). All images were analyzed using a fluorescence microscope, Nikon TE2000-U (Nikon, Japan).
2.6. Flow cytometry analysis
The hPSCs-CMs were resuspended and fixed with 4% PFA/permeabilization solution (BD Bioscience, USA) for 15 min. The hPSC-CMs were stained for cTnT using 1:50 AF647-conjugated anti-TNNT2 antibody (Abcam) for 30 min at 4 °C. Subsequently, cells were analyzed using an SH800S Cell Sorter flow cytometer with Cell sorter software Ver 2.1.5 (Sony Biotechnology, USA).
2.7. Cell viability assay
Various concentrations of each compound were added to hPSC-CMs for 24 h or 48 h, and the cell viability was determined using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS, Promega) according to the manufacturer's instructions. Absorbance at 490 nm (A490) was measured using a Synergy™ H1 multi-mode microplate reader (Biotek, USA). All the cell viability assays were performed at least three times.
2.8. Antiviral assay
A cytopathic effect (CPE)-based assay was used to evaluate antiviral activities of chloroquine, hydroxychloroquine, favipiravir, and remdesivir in Vero E6 cells, as previously described with modifications (Shin et al., 2018). Briefly, 1 × 104 Vero E6 cells/well were infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.1 in the presence of various concentrations of each drug. At 72 h post-infection, cell viability was measured using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS, Promega). An image-based assay was used to further evaluate the antiviral activities of chloroquine and remdesivir in hPSC-CMs. An immunofluorescence assay used to detect SARS-CoV-2 infection was optimized for the COVID-19 high-throughput/high-content analysis. Briefly, 3 × 104 hPSC-CMs were seeded in a 96-well plate and allowed to regain contractility. Subsequently, hPSC-CMs were infected with SARS-CoV-2 at an MOI of 2.5. At 48 h post-infection, cells were fixed with 4% PFA for 30 min at room temperature. Infected cells were detected by probing with SARS-CoV-2 S protein monoclonal antibody (GeneTex, USA) and AF488-conjugated goat anti-mouse IgG (Thermo Fisher Scientific). Cell nuclei were counterstained with DAPI. Digital images were acquired from 4 different fields from each well at 20 × magnification using an automated high-throughput confocal fluorescence imaging system (Operetta High Content Imaging System, PerkinElmer, USA). Dose-response curve analyses were performed for each drug, and the 50% effective concentration (EC50) and 50% cytotoxic concentration (CC50) were determined with GraphPad Prism 8 (GraphPad Software, USA) using the non-linear regression formula: log (inhibitor) vs. response-variable response (four parameters) model.
2.9. Viral plaque assay
Infectious SARS-CoV-2 titers were determined by plaque assay. Briefly, Vero E6 cells were prepared in 48-well plates at a density of 1 × 105 cells/well and cultured overnight 37 °C in a 5% CO2 incubator. Cells were inoculated with ten-fold serially diluted cell culture supernatants for 1 h at 37 °C. After adsorption, cells were washed with PBS and overlaid with DMEM containing 0.75% agarose (Sigma-Aldrich). At 72 h post-infection, plaques were stained with 0.05% crystal violet solution (Sigma-Aldrich). Plaque image capturing and counting analyses were performed using ImmunoSpot (Cellular Technology Limited, USA).
2.10. Electrophysiological cardiotoxicity assay
Conventional human ether-à-go-go-related gene (hERG) assay and hPSC-CM-based multielectrode array (MEA) were used in the study. Inhibition of hERG currents was evaluated in stably overexpressing hERG HEK-293 cell line. The hERG currents were recorded under the conventional whole-cell voltage-clamp configuration (Axon instrument patch-clamp, Molecular Devices, USA) using the intracellular solution containing 120 mM K-gluconate, 20 mM KCl, 10 mM HEPES, 5 mM EGTA, 1 mM MgCl2, and 1.5 mM Mg-ATP (pH 7.3 adjusted with KOH) and the extracellular solution containing 130 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 12.5 mM glucose, and 10 mM HEPES (pH 7.4 adjusted with NaOH). The peak hERG current was activated at −50 mV of repolarization, followed by a 50 mV depolarization from −80 mV of holding voltage. The hERG current inhibition was analyzed by treating respective concentrations of remdesivir and chloroquine for 3 min. hPSC-CMs exhibiting stable contraction were prepared for conventional patch-clamp and MEA analyses. The current-clamp recording was performed for action potential (AP) analysis from hPSC-CMs using the intracellular solution containing 120 mM K-aspartate, 25 mM KCl, 5 mM NaCl, 10 mM HEPES, 0.1 mM EGTA, 1 mM MgCl2, and 3 mM Mg-ATP (pH 7.25 adjusted with KOH) and the extracellular solution containing 145 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM glucose, and 10 mM HEPES (pH 7.4 adjusted with NaOH). For MEA analysis, hPSC-CMs were seeded on 96-well MEA plates (Cytoview MEA plate, Axion BioSystems, USA) prior to drug treatment. Field potential (FP) was recorded from spontaneously contracting hPSC-CMs using the MEA system (Maestro Pro MEA, Axion BioSystems). The changes of FP parameters were compared with two independent 5 min recordings between 30 min exposure of drugs. Each well in the plate was treated with only one concentration with a replicate set of 5 wells for each concentration. The drugs were prepared at 10 × final concentration in culture media and added to the wells by 10% volume replacement. The MEA experimental protocol was adapted from the CiPA study (Millard et al., 2018; Gintant et al., 2019). Data were analyzed with the Cardiac Analysis Tool and the AxIS Metric Plotting Tool software offered via Axion BioSystems.
2.11. Statistical analysis. Data were analyzed using a two-tailed paired Student's t-test. Differences were considered statistically significant when the P-value was <0.05.
3. Results
3.1. Generation and characterization of human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs)
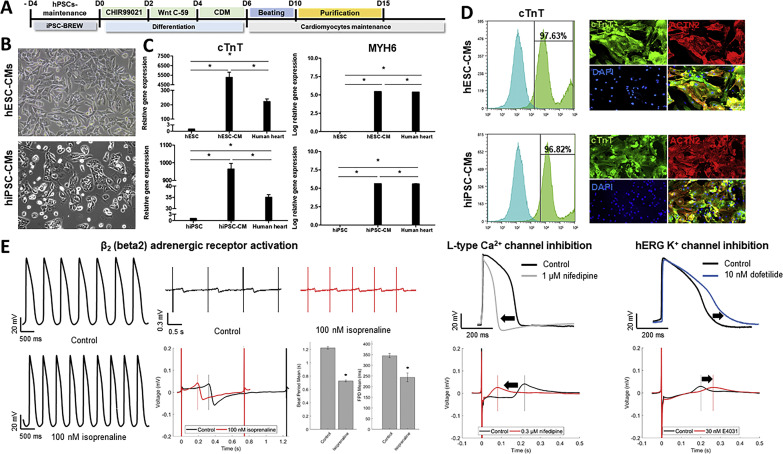
The hPSC-CMs used in the present study were obtained from two independent hPSC lines, including hESCs (H9) and hiPSCs (CMC-11), using a chemically defined differentiation protocol as described previously (Fig. 1 A) (Burridge et al., 2015; Park et al., 2019). After purification, hPSC-CMs exhibited typical human cardiomyocyte morphology and spontaneous beating (Fig. 1B). In addition, qRT-PCR analyses showed a significant expression of cardiomyocyte-specific genes (cTnT and MYH6) in both hESC-CMs and hiPSC-CMs. In contrast, minimal expression of cardiac-specific genes was detected in undifferentiated hPSCs (hESC and hiPSCs). Human heart mRNA was used as a control (Fig. 1C). Furthermore, flow cytometry analysis confirmed that >97% of hESC-CMs and hiPSC-CMs were positive for cTnT, a well-known specific marker for CMs (Fig. 1D, left panels). Similarly, most of the cells showed CM-like morphology and were stained positive for both cardiac sarcomeric markers, cTnT, and α-actinin (ACTN2) by immunofluorescence assay (Fig. 1D, right panels). Collectively, these results clearly indicated the successful generation of highly purified hPSC-CMs. Subsequently, the electrophysiological properties of hPSC-CMs were evaluated by patch-clamp analysis and multielectrode array (MEA) system (Fig. 1E). The patch clamp analysis showed that hPSC-CMs exhibited spontaneous contraction, and regular action potentials (AP) from negative diastolic membrane potential (far left panels). Notably, the duration of AP was relatively long, consistent with the human ventricular-type AP. The MEA analysis also showed a spontaneous and repetitive generation of field potential (FP), reflecting the propagation of AP along with the multicellular layer. In addition to the basal electrophysiological activity, treatment of 100 nM isoprenaline, a β2-adrenergic receptor agonist, substantially increased the rate of AP in hPSC-CMs (Fig. 1E left panels). These results clearly demonstrated that hPSC-CMs used in this study are physiologically functional, displaying the adrenergic responses. Moreover, hPSC-CMs demonstrated drug-dependent repolarization changes (Fig. 1E right panels). Treatment of 1 μM nifedipine, an L-type Ca2+ channel blocker, shortened action potential duration (APD) and repolarization in hPSC-CMs by inhibition of L-type Ca2+ current. In contrast, treatment with anti-arrhythmic agents, dofetilide (10 nM), and E4031 (30 nM) significantly delayed the repolarization through inhibition of hERG K+ currents. Taken altogether, these results clearly indicated that hPSC-CMs are feasible cellular model for further experiments.
Fig. 1.
Characterization of cardiomyocytes-derived from human pluripotent stem cells (hPSC-CMs). (A) Schematic diagram of hPSC differentiation into cardiomyocytes. (B) The morphology of differentiated hPSC-CMs derived from two hPSC cell lines (hESCs: H9 and hiPSCs: CMC-11; 10 × magnification). (C) The RT-qPCR was performed to measure the expression of cardiac sarcomeric markers, cTnT, and MYH6 genes, in hPSC-CMs. The data represent the mean (±SD) of at least two independent experiments performed in triplicate. Statistical analyses were determined using paired student's t-tests, and P < 0.05 was considered as significant (*). (D) Expression of TNNT2 and ACTN2 in hPSC-CMs (hESC-CMs [upper panel] and hiPSC-CMs [lower panel]) at day 15 of differentiation were determined by flow cytometry and immunofluorescence analyses. Scale bar, 100 μm. (E) Beating cardiomyocytes were prepared for action potential (AP) recording using the patch-clamp technique in hESC-CMs and field potential (FP) recording using a multielectrode array (MEA) system in hiPSC-CMs. The hPSC-CMs were treated with 100 nM isoprenaline (β2-adrenergic receptor agonist), 0.3 μM and 1 μM nifedipine (L-type Ca2+ channel blocker), and 10 nM dofetilide and 30 nM E4031 (hERG K+ channel blockers).
3.2. Susceptibility of hPSC-CMs to SARS-CoV-2 infection
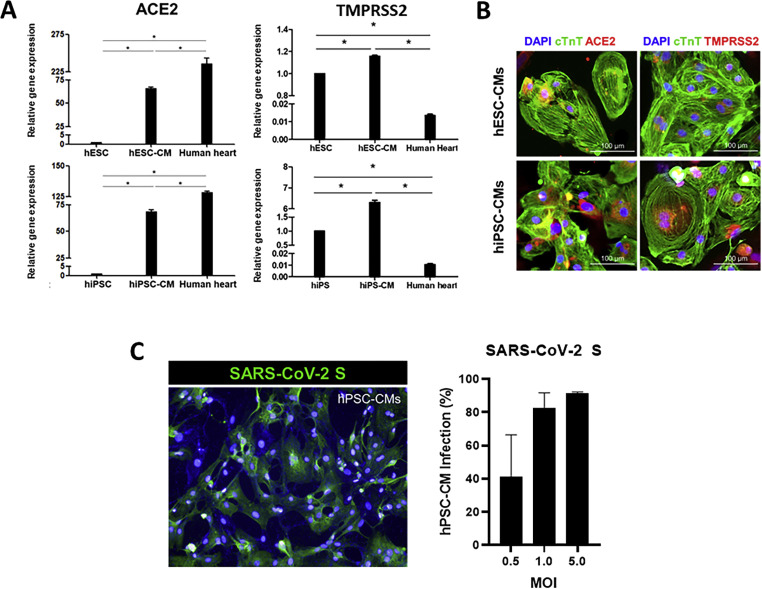
To determine whether human CMs support SARS-CoV-2 infection, we first evaluated the presence of angiotensin-converting enzyme-2 (ACE2), a functional receptor of SARS-CoV-2, and the transmembrane protease serine 2 (TMPRSS2), which mediates priming of the viral S-protein in hPSC-CMs. As a result, high expression levels of ACE2 and TMPRSS2 mRNA were detected in cTnT-positive hPSC-CMs compared to undifferentiated hPSCs (Fig. 2 A). In addition, expression of ACE2 and TMPRSS2 proteins were confirmed in both hESC-CMs and hiPSC-CMs by immunofluorescence assay, suggesting that differentiated hPSC-CMs possess functional receptor and protease necessary for efficient SARS-CoV-2 infection (Fig. 2B). Subsequently, to test the susceptibility of human CMs to SARS-CoV-2 infection, hPSC-CMs were infected with SARS-CoV-2 (hCoV/Korea/KCDC-03/2020) at the indicated multiplicity of infection (MOI). As a result, the expression of the viral spike (S) protein in infected hPSC-CMs was detected in a dose-dependent manner by immunofluorescence assay (Fig. 2C). Maximum viral infection was detected at 48 h in cells infected with MOI 1 or higher, indicating that hPSC-CMs support SARS-CoV-2 infection.
Fig. 2.
SARS-CoV-2 infects hPSC-CMs. (A) The mRNA expression levels of ACE2 and TMPRSS2 genes in hPSC-CMs (hESCs: H9 and hiPSCs: CMC-11) were measured by RT-qPCR. The data represent the mean (±SD) of at least two independent experiments performed in triplicate. Statistical analyses were determined using paired student's t-tests, and P < 0.05 was considered as significant (*). (B) Immunofluorescence assay was performed to evaluate the expression of ACE2 and TMPRSS2 proteins in cTnT-positive hPSC-CMs. ACE2 (red) or TMPRSS2 (red) was co-expressed with cTnT (green) positive hESC-CMs and hiPSC-CMs. (C) SARS-CoV-2 infection of hPSC-CMs. The hiPSC-CMs were infected with SARS-CoV-2 (betaCoV/Korea/KCDC) at the indicated multiplicity of infection (MOI) for 48 h. Immunofluorescence image of SARS-CoV-2 infected hiPSC-CMs positively stained for the viral spike (S) protein (green) is shown. The percentage of SARS-CoV-2 infected hiPSC-CMs at various MOI was determined by the quantification of S protein-positive cells. The data represent the mean (±SD) of at least two independent experiments performed in duplicate.
3.3. In vitro evaluation of the antiviral activity of repurposable drugs against SARS-CoV-2 in Vero E6 cells
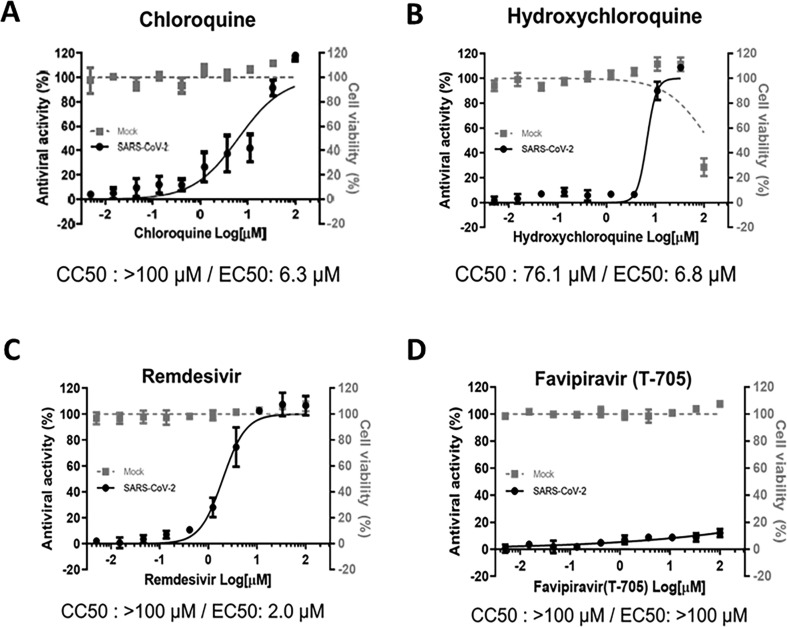
Prior to testing the antiviral activities of selected repurposing drugs in hPSC-CMs, we first determined the in vitro potency of the compounds that are currently undergoing preclinical and/or clinical trials using a well-established Vero E6-based antiviral assay. Briefly, Vero E6 cells infected with SARS-CoV-2 (hCoV/Korea/KCDC-03/2020) were treated with increasing concentrations of chloroquine, hydroxychloroquine, remdesivir, or favipiravir, and the EC50 and CC50 values were determined by dose-response curve analyses. In agreement with previous studies, chloroquine and its derivative, hydroxychloroquine, showed in vitro antiviral activity against SARS-CoV-2 with estimated EC50 values of 6.3 and 6.8 μM and CC50 of >100 and 76.1 μM, respectively (Fig. 3 A and B). In addition, remdesivir exhibited prominent antiviral activity with an EC50 of 2 μM and a CC50 of >100 μM, resulting in a selectivity index (SI) of >50 (Fig. 3C). Similarly, the number of SARS-CoV-2 infectious viral particles and viral RNA expression levels were significantly reduced when infected Vero E6 cells were treated with 3 μM or higher concentration of remdesivir, whereas significant inhibition of viral replication was present only when cells were treated with 10 μM chloroquine (Supplementary Fig. 1). Notably, favipiravir, a pyrazine derivative currently undergoing clinical trial, showed no apparent in vitro antiviral activity against SARS-CoV-2 (Fig. 3D). Therefore, chloroquine and remdesivir were selected to further assess their antiviral potencies and safety profiles in human CMs using human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs).
Fig. 3.
Antiviral efficacy of chloroquine, hydroxychloroquine, remdesivir, and favipiravir in Vero E6 cells. (A to D) Dose-response curve analyses were performed in Vero E6 cells infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.1 in the presence of various concentrations of chloroquine (A), hydroxychloroquine (B), remdesivir (C), or favipiravir (T-705) (D). At 72 h post-infection, cell viability was measured using the CellTiter 96® AQueous One Solution Cell Proliferation Assay. The data represent the mean (±SD) of at least three independent experiments performed in duplicate. Mock-infected (grey square) or infected with SARS-CoV-2 (black circle). CC50, 50% cytotoxic concentrations; EC50, 50% effective concentration.
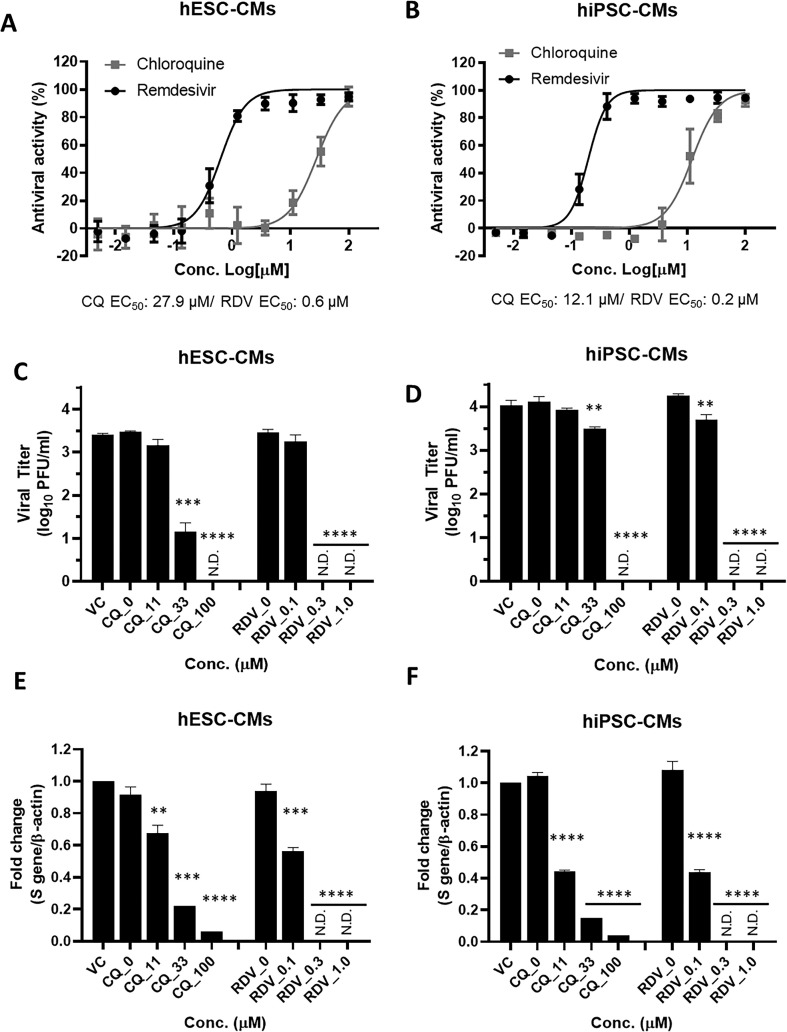
3.4. Remdesivir is a potent antiviral inhibitor of SARS-CoV-2 in hPSC-CMs
To evaluate the antiviral activity of chloroquine and remdesivir against SARS-CoV-2 in human CMs, the drug dose-response curve analyses were performed in hPSC-CMs using the high-content analysis system. The hPSC-CMs were replated at 3 × 104/well in 96-well plates and allowed to regain contractility (Supplementary video 1). Subsequently, hESC-CMs and hiPSC-CMs infected with SARS-CoV-2 at an MOI of 2.5 were treated with increasing concentrations of chloroquine and remdesivir. At 48 h post-infection, the antiviral activity was determined by immunofluorescence detection of S protein for direct imaging and quantification of virus-infected cells. In SARS-CoV-2 infected hESC-CMs and hiPSC-CMs, remdesivir demonstrated potent antiviral activity with EC50 values of 0.6 μM and 0.2 μM, respectively. Notably, remdesivir exhibited 46-fold and 60-fold higher antiviral activity in hESC-CMs and hiPSC-CMs, respectively, compared with those in Vero E6 cells (Fig. 4 A and B; Table 1 ). Similarly, dose-dependent inhibition of viral replication was observed in remdesivir-treated hPSC-CMs as determined by plaque assay and RT-qPCR. Complete suppression of viral particle production occurred when hESC-CMs and hiPSC-CMs were treated with 0.3 μM and 1 μM of remdesivir, whereas there was a minimal reduction of viral titer in 0.1 μM remdesivir-treated hPSC-CMs (Fig. 4C and D). Consistent with a reduction in viral titers, a dose-dependent reduction in the intracellular viral genome was detected by RT-qPCR in hPSC-CMs treated with remdesivir. Treatment of 0.3 μM and 1 μM remdesivir caused complete inhibition of intracellular viral mRNA synthesis (Fig. 4E and F). In contrast to remdesivir, the antiviral activity of chloroquine decreased significantly in SARS-CoV-2-infected hESC-CMs (EC50: 27.9 μM) and hiPSC-CMs (EC50: 12.1 μM) compared to its activity in Vero E6 cells (Fig. 4A and B; Table 1). Similarly, a complete suppression of infectious viral particle production occurred when hESC-CMs and hiPSC-CMs were treated with the highest tested concentration of chloroquine. Treatment with 33 μM chloroquine led to a 2.3-log10 PFU/ml and 0.3-log10 PFU/ml reduction in SARS-CoV-2 titers in hESC-CMs and hiPSC-CMs, respectively, whereas 11 μM chloroquine did not affect the viral titers (Fig. 4C and D). Likewise, quantification of the viral genome in chloroquine-treated hPSC-CMs determined by RT-qPCR showed a similar trend as the viral yield assay (Fig. 4E and F). Taken together, the data indicated that remdesivir potently inhibits SARS-CoV-2 infection in human cardiomyocyte cultures by reducing the production of infectious progeny virus as well as the synthesis of viral RNA at a nanomolar concentration. In contrast, the inhibitory effect of chloroquine was significantly reduced in hPSC-CMs compared to Vero E6 cells (Table 1).
Fig. 4.
Remdesivir potently inhibits SARS-CoV-2 infection in hPSC-CMs. (A and B) Dose-response curve (DRC) analyses of SARS-CoV-2 inhibition by chloroquine and remdesivir in hPSC-CMs. The hESC-CMs (A) and hiPSC-CMs (B) were infected with SARS-CoV-2 at an MOI 2.5 in the presence of various concentrations of chloroquine (grey square) or remdesivir (black circle). At 48 h p.i., the percentage of infected cells was visualized and measured using an automated high-throughput confocal fluorescence imaging system. The data represent the mean (±SD) of at least three independent experiments performed in duplicate. (C to F) Reduction of SARS-CoV-2 replication by chloroquine and remdesivir in hPSC-CMs as determined by infectious viral titer and RT-qPCR. The hESC-CMs (C and E) and hiPSC-CMs (D and F) were infected with SARS-CoV-2 at an MOI of 2.5 in the presence of the indicated concentrations of chloroquine or remdesivir. (C and D) Viral titers from hPSC-CM supernatants were determined with plaque assay in Vero E6 cells. (E and F) Quantification of intracellular SARS-CoV-2 genome RNA by RT-qPCR. Total RNA was isolated from lysates of infected cells for quantification of intracellular SARS-CoV-2 RNA levels (S gene), and results were normalized to β-actin mRNA. Data represent means (±SD) of at least two independent experiments performed in duplicate. Statistical analyses were determined using paired student's t-tests, and significant differences are indicated by **P < 0.01, ***P < 0.005, and ****P < 0.0001. CQ, chloroquine; RDV, remdesivir; N.D, not detected.
Table 1.
Antiviral activities of chloroquine and remdesivir in Vero E6 and hPSC-CMs.
| Chloroquine (μM) |
Remdesivir (μM) |
|||||
|---|---|---|---|---|---|---|
| CC50 | EC50 | EC90 | CC50 | EC50 | EC90 | |
| Vero E6 | >100 | 6.3 | 33 | >100 | 2 | 8.6 |
| hESC-CM | 84.9 | 27.9 | >84.9 | 10.2 | 0.6 | 1.3 |
| hiPSC-CM | >100 | 12.1 | 24.1 | 15.6 | 0.2 | 0.33 |
Supplementary video related to this article can be found at https://doi.org/10.1016/j.antiviral.2020.104955.
The following is the supplementary data related to this article:
3.5. Evaluation of in vitro remdesivir-induced cardiotoxicity using hPSC-CMs
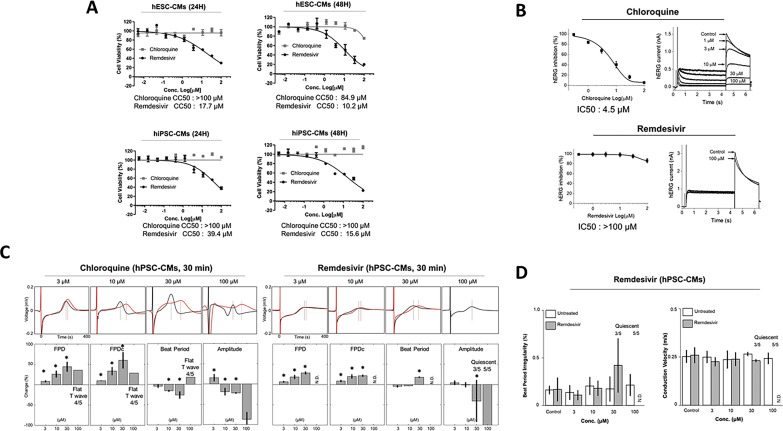
Next, we investigated whether remdesivir could induce cardiotoxic effects in human CMs within the range of effective antiviral concentration (EC50) determined in vitro. Chloroquine, a drug with an increased arrhythmogenic risk, was included as a positive control for toxicity (Traebert et al., 2004). The hPSC-CMs were treated with increasing concentrations of chloroquine and remdesivir, and the cell viability was measured at 24 h and 48 h using cell proliferation assay. As results, remdesivir displayed significant cardiotoxicity with estimated CC50: 39.4 μM and 15.6 μM at 24 and 48 h, respectively, in hiPSC-CMs, while it was slightly more toxic in hESC-CMs with CC50: 17.7 μM and 10.2 μM at 24 and 48 h, respectively (Fig. 5 A). The results suggested that remdesivir induces significant cytotoxic effects in both hESC-CMs and hiPSC-CMs. Moreover, longer remdesivir treatment time (48 h) substantially reduced the cell viability of CMs as compared to 24 h post-treatment suggesting potential accumulative toxicity of the drug. The data together suggested that remdesivir induces cardiotoxicity in hPSC-CMs with CC50 values close to its estimated peak plasma concentration (Cmax: 9 μM) reported in the literature (Agency, 2020). In contrast, chloroquine did not present apparent cardiotoxicity in hPSC-CMs (CC50 > 100 μM) at 24 h, while its cardiotoxicity slightly increased at 48 h post-treatment in hESC-CMs with an estimated CC50: 84.9 μM.
Fig. 5.
Remdesivir induces cardiotoxicity in hPSC-CMs. (A) Cardiomyocyte toxicity of chloroquine and remdesivir in hPSC-CMs. Dose-response curve analyses of drug-induced cardiotoxicity in hPSC-CMs (hESC-CMs and hiPSC-CMs) were performed in the presence of various concentrations of each drug. At 24 h and 48 h post-treatment, cell viability was measured using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS, Promega). The data represent the mean (±SD) of at least two independent experiments performed in triplicate. CC50, 50% cytotoxic concentration. (B) The inhibitory effects of chloroquine and remdesivir on hERG currents in stably overexpressing hERG HEK-293 cell line. The hERG currents were recorded with the conventional whole-cell voltage-clamp configuration. The peak hERG current was activated at −50 mV, followed by a 50 mV depolarization from −80 mV of holding voltage. The data represent the mean (±SD) of at least two independent experiments performed in triplicate. (C, D) Arrhythmogenic effects of chloroquine and remdesivir in hiPSC-CMs. The FP signals were analyzed with FP duration (FPD), corrected FPD (FPDc; field potential duration corrected by Fredericia's formula), beat period, the amplitude of Na+ Peak (amplitude), beat period irregularity (coefficient of variation of the beat period), and conduction velocity using Cardiac Analysis Tool 2.2.7 (Axion BioSystems). All data are expressed as mean ± SD (n = 5, exclusion of quiescent and flat T). Statistical analyses were determined using paired student's t-tests, and P < 0.05 was considered as significant (*). N.D, not detected.
To further gain insight into the risk of drug-induced arrhythmogenic effects, conventional hERG inhibition assay and hiPSC-CM-based MEA were performed (Fig. 5B and C). Not surprisingly, the treatment of chloroquine, a well-described arrhythmogenic drug, significantly inhibited hERG currents with and IC50 4.5 μM (Fig. 5B, upper panel) and increased the field potential duration (FPD) of hiPSC-CMs in a dose-dependent manner. Similarly, chloroquine significantly altered cardiomyocyte electrophysiology, causing a pronounced reduction of Na+ peak amplitude and T-wave flattening (Fig. 5C, left panels). The results demonstrated that the detrimental arrhythmogenic effects of known QT-interval prolonging drugs, such as chloroquine, can be recapitulated using hiPSC-CMs as a model. In contrast to chloroquine, no significant changes in hERG currents were observed with remdesivir at tested concentrations (IC50 > 100 μM), suggesting that remdesivir presents a more favorable safety profile compared to chloroquine (Fig. 5B). However, dose-dependent prolongations of FPD were detected by MEA analysis in hiPSC-CMs treated with remdesivir for 30 min. In addition, 30 μM or higher concentration of remdesivir reduced the Na+ peak amplitudes and spontaneous beating rates in hiPSC-CMs, followed by complete cessation of spontaneous beating (Fig. 5C, right panels). Moreover, the treatment of hiPSC-CMs with 30 μM of remdesivir resulted in a 0.17–0.42% increase of beat period irregularities and 0.266 to 0.230 m/s decrease of conduction velocity (n = 2), suggesting its potential risk of arrhythmogenic effects in human CMs (Fig. 5D). Early afterdepolarizations (EADs) and delayed afterdepolarizations (DADs) were not detected in remdesivir-treated hiPSC-CMs (data not shown). Taken together, our results suggested that hiPSC-CMs subjected to acute remdesivir treatment exhibit arrhythmogenic risk with altered cardiomyocyte electrophysiological properties in a dose-dependent manner, indicating that overdose or drug accumulation may elicit significant cardiac adverse effects, such as QT interval prolongation.
4. Discussion
There is escalating evidence that patients suffering from COVID-19 show profound cardiac complications with elevated cardiac injury biomarkers and deteriorated ventricular heart functions (Bansal, 2020; Aggarwal et al., 2020a; Madjid et al., 2020). At present, there is a shortlist of compounds known for their broad-spectrum antiviral activities, including remdesivir, which is the only authorized drug for use in severe COVID-19 patients under an Emergency Use Authorization (Eastman et al., 2020; Jakhar and Kaur, 2020; Aggarwal et al., 2020b). However, most drug evaluation studies are based on in vitro models using immortalized cell lines that clearly have limitations of recapitulating human physiology and pharmacological responses. As a consequence, there is an urgent need to create human disease-relevant models capable of recapitulating mechanisms responsible for antiviral activity and drug toxicity in human clinical findings. In this context, successful differentiation of human pluripotent stem cells (hPSCs), which include both human embryonic stem cells (hESCs) and human-induced pluripotent stem cells (hiPSCs), into functional human cells or tissues provides valuable tools for modeling human diseases and drug discovery, including infectious diseases. In particular, hPSC-CMs are regarded as one of the most promising sources for various applications, such as cell-based cardiac repair, cardiac drug toxicology screening, and human cardiac disease modeling, mostly due to the unavailability of human primary CMs (Sharma et al., 2014, 2017; Lan et al., 2013; Sun et al., 2012; Burridge et al., 2016). Although the human primary CMs are the ideal cell sources for applications mentioned above, they can only be obtained by isolating from the human heart, which is theoretically impossible.
In this study, hPSC-CMs differentiated from two hPSC lines, such as hESCs (H9) and hiPSCs (CMC-11), were used to investigate the therapeutic potential of anti-SARS-CoV-2 repurposing drug candidates. Consistent with recent publications (Sharma et al., 2020; Bojkova et al., 2020), we confirmed that hPSC-CMs are not only permissive to SARS-CoV-2 entry but also supportive of the production of infectious progeny viruses. In addition, our results showed that remdesivir potently inhibits SARS-CoV-2 replication in both hESC-CMs and hiPSC-CMs with nanomolar EC50 values. Notably, the potency of remdesivir in human CMs was 46-fold–60-fold higher compared to that in Vero E6 cells (EC50: 2 μM). Consistent with our findings, it has been previously noted that remdesivir exhibits weaker anti-SARS-CoV-2 activity in Vero E6 cells compared to that in human primary airway epithelial cultures (Pruijssers et al., 2020). Further studies indicated that the antiviral activity of remdesivir directly correlated with the intracellular concentration of the pharmacologically active triphosphate metabolite (TP) (Pruijssers et al., 2020; Choy et al., 2020; Ko et al., 2020). It has also been described the inefficient metabolism of the nucleotide prodrug sofosbuvir in Vero cells elsewhere (Mumtaz et al., 2017). In this context, we speculate that hPSC-CMs possess a higher capacity to metabolize remdesivir resulting in higher antiviral potency than in Vero E6 cells; however, measurements of active TP form in these cultures were not included in this study. In contrast, the antiviral activity of chloroquine decreased significantly in SARS-CoV-2-infected hPSC-CMs compared to its activity in Vero E6 cells. Chloroquine and hydroxychloroquine are known to affect the acidic pH of intracellular compartments that are needed for the replication of several viruses, including coronaviruses (Savarino et al., 2003). Thus, it is not surprising that the antiviral potency of host-targeting drugs varies between host cells, especially in cells that have unique physiological properties. These discrepancies might in part account for the failure of clinical trials using chloroquine and hydroxychloroquine that did not show any promising results regarding the COVID-19 treatment (Geleris et al., 2020; Borba et al., 2020). The differences in antiviral activity among various cell types, as illustrated here and elsewhere, highlight the importance of using physiologically relevant in vitro human cell/tissue models. Furthermore, the slight differences in EC50 values of the tested compounds observed between hESC-CMs and hiPSC-CMs could be explained by differences in the cardiomyocyte subtypes within the culture or in the phenotype of the initial hPSC (Goineau and Castagne, 2018). Moreover, the phenotypic plasticity of these cells during their in vitro life span cannot be excluded as a factor displaying different phenotypes such as disparity in pharmacological sensitivity (Gintant et al., 2019). Nevertheless, the authenticity of hPSC-CMs used in this study has been thoroughly examined, and we demonstrated that both hESC-CMs and hiPSC-CMs closely recapitulate the human cardiomyocyte phenotypes (Fig. 1).
Drug-induced cardiotoxicity can contribute to life-threatening complications, including heart failure, myocardial ischemia, and cardiac arrhythmias. Thus, preclinical evaluation of cardiovascular adverse reactions associated with new drugs is a primary concern for pharmaceutical industries. Here, we assessed for the first time remdesivir-induced cardiotoxicity and proarrhythmic potential in hPSC-CMs using methodologies adapted from the CiPA study. In agreement with previous reports (Ladipo et al., 1983), we showed that chloroquine significantly alters the CM electrophysiology causing pronounced arrhythmogenic effects with prolongation of QT intervals and T-wave flattening. The results not only corroborated the cardiac adverse effects of an arrhythmogenic drug, such as chloroquine but also indicated the robustness and validity of hPSC-CMs as a reliable cardiotoxicity assay. Surprisingly, the results from cell viability assay showed that while chloroquine treatment did not cause cytotoxic effects in hPSC-CMs, remdesivir induced significantly higher cytotoxicity in hPSC-CMs (CC50, 10.2–39.4 μM) compared to that in Vero E6 cells (>100 μM). In addition, longer remdesivir treatment time (48 h) substantially reduced the cell viability of CMs as compared to 24 h post-treatment suggesting potential accumulative toxicity of the drug. Further researches are needed to investigate the mechanisms of remdesivir-induced cardiotoxicity in human CMs. Moreover, we examined drug-induced cardiotoxicity by measuring electrophysiological abnormalities, contractile dysfunction, and structural toxicity in hPSC-CMs-treated with various concentrations of remdesivir (Fig. 5). The results indicated that remdesivir lengthened FPD in a dose-dependent manner with reduced Na+ peak amplitudes and spontaneous beating rates in hiPSC-CMs that eventually, at higher concentrations, caused a complete cessation of spontaneous beating, suggesting that overdose or drug accumulation may elicit detrimental cardiac adverse effects. Taken together, we speculate that there is a potential risk of QT prolongation if remdesivir is used at concentrations higher than the estimated peak plasma concentration (Cmax: 9 μM). Therefore, close monitoring of the electrocardiographic/QT interval should be advised in patients under remdesivir medication, in particular individuals with pre-existing heart conditions and/or severe COVID-19. Additional study remains to be performed to evaluate remdesivir-associated cardiotoxicity, including arrhythmia inducing potential in SARS-CoV-2 infected hPSC-CMs.
In summary, we generated hPSC-CMs to evaluate anti-SARS-CoV-2 activity as well as the safety of remdesivir in physiologically and disease-relevant human cells. This study provides clinically valuable insight into the risk-benefit assessment of remdesivir for COVID-19 treatment, in particular, its use in patients at risk for cardiac diseases. All these regards, based on the accumulated results from previous reports and our current study, employment of a hPSC-CM model in drug safety screening platforms is a powerful solution that can fill many of the limitations in currently available systems, providing data that will aid in identifying potential cardiotoxicities and allow the development of the safest and most effective drug candidates including new drugs for COVID-19.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
The human pluripotent stem cell line (CMC11-iPSC) and the SARS-CoV-2 (NCCP No. 43326) were provided by the National Stem Cell Bank of Korea and the National Culture Collection for Pathogens (Korea National Institute of Health), respectively. This research was supported by the Ministry of Health and Welfare (grant number HI20C0184 to SWC, Republic of Korea), the Ministry of Trade, Industry & Energy (Technology Innovation Program [grant number 20009748 to SJP], Republic of Korea); the Korea Centers for Disease Control and Prevention (grant number 2020-ER6106-00 to SHM); the Ministry of Science and ICT (National Research Foundation of Korea [grant number NRF-2020M3A9I2081687 to JSS and EJ], Republic of Korea); and KRICT intramural funding (grant number KK-2032-20, Republic of Korea). YYG was supported by a start-up fund from City University of Hong Kong, Hong Kong SAR, China.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104955.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Agency E.M. 2020. Summary on Compassionate Use: Remdesivir Gilead.https://www.ema.europa.eu/en/documents/other/summary-compassionate-use-remdesivir-gilead_en.pdf [Google Scholar]
- Aggarwal G., Cheruiyot I., Aggarwal S., Wong J., Lippi G., Lavie C.J., Henry B.M., Sanchis-Gomar F. Association of cardiovascular disease with coronavirus disease 2019 (COVID-19) severity: a meta-analysis. Curr. Probl. Cardiol. 2020;45:100617. doi: 10.1016/j.cpcardiol.2020.100617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal G., Henry B.M., Aggarwal S., Bangalore S. Cardiovascular safety of potential drugs for the treatment of coronavirus disease 2019. Am. J. Cardiol. 2020;128:147–150. doi: 10.1016/j.amjcard.2020.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M. Cardiovascular disease and COVID-19. Diabetes Metabol. Syndr. 2020;14:247–250. doi: 10.1016/j.dsx.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Wagner J.U.G., Shumliakivska M., Aslan G.S., Saleem U., Hansen A., Luxan G., Gunther S., Pham M.D., Krishnan J., Harter P.N., Ermel U.H., Frangakis A.S., Milting H., Zeiher A.M., Klingel K., Cinatl J., Dendorfer A., Eschenhagen T., Tschope C., Ciesek S., Dimmeler S. SARS-CoV-2 infects and induces cytotoxic effects in human cardiomyocytes. Cardiovasc. Res. 2020 doi: 10.1093/cvr/cvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borba M.G.S., Val F.F.A., Sampaio V.S., Alexandre M.A.A., Melo G.C., Brito M., Mourao M.P.G., Brito-Sousa J.D., Baia-da-Silva D., Guerra M.V.F., Hajjar L.A., Pinto R.C., Balieiro A.A.S., Pacheco A.G.F., Santos J.D.O., Jr., Naveca F.G., Xavier M.S., Siqueira A.M., Schwarzbold A., Croda J., Nogueira M.L., Romero G.A.S., Bassat Q., Fontes C.J., Albuquerque B.C., Daniel-Ribeiro C.T., Monteiro W.M., Lacerda M.V.G., CloroCovid T. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Net. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PubMed] [Google Scholar]
- Burridge P.W., Holmstrom A., Wu J.C. Chemically defined culture and cardiomyocyte differentiation of human pluripotent stem cells. Curr. Protoc. Human Genet. 2015;87:21 3 1–21 3 15. doi: 10.1002/0471142905.hg2103s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Li Y.F., Matsa E., Wu H., Ong S.G., Sharma A., Holmstrom A., Chang A.C., Coronado M.J., Ebert A.D., Knowles J.W., Telli M.L., Witteles R.M., Blau H.M., Bernstein D., Altman R.B., Wu J.C. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 2016;22:547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K.T., Wong A.Y., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent. Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D.K., Kubin C., Barr R.G., Sobieszczyk M.E., Schluger N.W. Observational study of hydroxychloroquine in hospitalized patients with covid-19. N. Engl. J. Med. 2020;382:2411–2418. doi: 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gintant G., Sager P.T., Stockbridge N. Evolution of strategies to improve preclinical cardiac safety testing. Nat. Rev. Drug Discov. 2016;15:457–471. doi: 10.1038/nrd.2015.34. [DOI] [PubMed] [Google Scholar]
- Gintant G., Burridge P., Gepstein L., Harding S., Herron T., Hong C., Jalife J., Wu J.C. Use of human induced pluripotent stem cell-derived cardiomyocytes in preclinical cancer drug cardiotoxicity testing: a scientific statement from the American heart association. Circ. Res. 2019;125:e75–e92. doi: 10.1161/RES.0000000000000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goineau S., Castagne V. Proarrhythmic risk assessment using conventional and new in vitro assays. Regul. Toxicol. Pharmacol. 2017;88:1–11. doi: 10.1016/j.yrtph.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Goineau S., Castagne V. Electrophysiological characteristics and pharmacological sensitivity of two lines of human induced pluripotent stem cell derived cardiomyocytes coming from two different suppliers. J. Pharmacol. Toxicol. Methods. 2018;90:58–66. doi: 10.1016/j.vascn.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S., China Medical Treatment Expert Group for C. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakhar D., Kaur I. Potential of chloroquine and hydroxychloroquine to treat COVID-19 causes fears of shortages among people with systemic lupus erythematosus. Nat. Med. 2020;26:632. doi: 10.1038/s41591-020-0853-0. [DOI] [PubMed] [Google Scholar]
- Ko M., Jeon S., Ryu W.S., Kim S. Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells. J. Med. Virol. 2020 doi: 10.1002/jmv.26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladipo G.O., Essien E.E., Andy J.J. Complete heart block in chronic chloroquine poisoning. Int. J. Cardiol. 1983;4:198–200. doi: 10.1016/0167-5273(83)90136-5. [DOI] [PubMed] [Google Scholar]
- Laflamme M.A., Chen K.Y., Naumova A.V., Muskheli V., Fugate J.A., Dupras S.K., Reinecke H., Xu C., Hassanipour M., Police S., O'Sullivan C., Collins L., Chen Y., Minami E., Gill E.A., Ueno S., Yuan C., Gold J., Murry C.E. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Lan F., Lee A.S., Liang P., Sanchez-Freire V., Nguyen P.K., Wang L., Han L., Yen M., Wang Y., Sun N., Abilez O.J., Hu S., Ebert A.D., Navarrete E.G., Simmons C.S., Wheeler M., Pruitt B., Lewis R., Yamaguchi Y., Ashley E.A., Bers D.M., Robbins R.C., Longaker M.T., Wu J.C. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long B., Brady W.J., Koyfman A., Gottlieb M. Cardiovascular complications in COVID-19. Am. J. Emerg. Med. 2020;38:1504–1507. doi: 10.1016/j.ajem.2020.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madjid M., Safavi-Naeini P., Solomon S.D., Vardeny O. Potential effects of coronaviruses on the cardiovascular system: a review. JAMA Cardiol. 2020;5:831–840. doi: 10.1001/jamacardio.2020.1286. [DOI] [PubMed] [Google Scholar]
- Millard D., Dang Q., Shi H., Zhang X., Strock C., Kraushaar U., Zeng H., Levesque P., Lu H.R., Guillon J.M., Wu J.C., Li Y., Luerman G., Anson B., Guo L., Clements M., Abassi Y.A., Ross J., Pierson J., Gintant G. Cross-site reliability of human induced pluripotent stem cell-derived cardiomyocyte based safety assays using microelectrode arrays: results from a blinded CiPA pilot study. Toxicol. Sci. 2018;164:550–562. doi: 10.1093/toxsci/kfy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitcheson J.S., Hancox J.C., Levi A.J. Cultured adult cardiac myocytes: future applications, culture methods, morphological and electrophysiological properties. Cardiovasc. Res. 1998;39:280–300. doi: 10.1016/s0008-6363(98)00128-x. [DOI] [PubMed] [Google Scholar]
- Moreno L., Pearson A.D. How can attrition rates be reduced in cancer drug discovery? Expet Opin. Drug Discov. 2013;8:363–368. doi: 10.1517/17460441.2013.768984. [DOI] [PubMed] [Google Scholar]
- Mumtaz N., Jimmerson L.C., Bushman L.R., Kiser J.J., Aron G., Reusken C., Koopmans M.P.G., van Kampen J.J.A. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antivir. Res. 2017;146:161–163. doi: 10.1016/j.antiviral.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Park S.J., Kim R.Y., Park B.W., Lee S., Choi S.W., Park J.H., Choi J.J., Kim S.W., Jang J., Cho D.W., Chung H.M., Moon S.H., Ban K., Park H.J. Dual stem cell therapy synergistically improves cardiac function and vascular regeneration following myocardial infarction. Nat. Commun. 2019;10:3123. doi: 10.1038/s41467-019-11091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A.J., George A.S., Schafer A., Leist S.R., Gralinksi L.E., Dinnon K.H., 3rd, Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., Gully K., Martinez D.R., Brown A.J., Graham R.L., Perry J.K., Du Pont V., Pitts J., Ma B., Babusis D., Murakami E., Feng J.Y., Bilello J.P., Porter D.P., Cihlar T., Baric R.S., Denison M.R., Sheahan T.P. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32:107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala L., Bellin M., Mummery C.L. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: has the time come? Br. J. Pharmacol. 2017;174:3749–3765. doi: 10.1111/bph.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases? Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Marceau C., Hamaguchi R., Burridge P.W., Rajarajan K., Churko J.M., Wu H., Sallam K.I., Matsa E., Sturzu A.C., Che Y., Ebert A., Diecke S., Liang P., Red-Horse K., Carette J.E., Wu S.M., Wu J.C. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ. Res. 2014;115:556–566. doi: 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Burridge P.W., McKeithan W.L., Serrano R., Shukla P., Sayed N., Churko J.M., Kitani T., Wu H., Holmstrom A., Matsa E., Zhang Y., Kumar A., Fan A.C., Del Alamo J.C., Wu S.M., Moslehi J.J., Mercola M., Wu J.C. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Garcia G., Arumugaswami V., Svendsen C.N. bioRxiv; 2020. Human iPSC-Derived Cardiomyocytes Are Susceptible to SARS-CoV-2 Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., Gong W., Liu X., Liang J., Zhao Q., Huang H., Yang B., Huang C. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in wuhan, China. JAMA Cardiol. 2020;5(7):802–810. doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.S., Jung E., Kim M., Baric R.S., Go Y.Y. Saracatinib inhibits Middle East respiratory syndrome-coronavirus replication in vitro. Viruses. 2018;10 doi: 10.3390/v10060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Yazawa M., Liu J., Han L., Sanchez-Freire V., Abilez O.J., Navarrete E.G., Hu S., Wang L., Lee A., Pavlovic A., Lin S., Chen R., Hajjar R.J., Snyder M.P., Dolmetsch R.E., Butte M.J., Ashley E.A., Longaker M.T., Robbins R.C., Wu J.C. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traebert M., Dumotier B., Meister L., Hoffmann P., Dominguez-Estevez M., Suter W. Inhibition of hERG K+ currents by antimalarial drugs in stably transfected HEK293 cells. Eur. J. Pharmacol. 2004;484:41–48. doi: 10.1016/j.ejphar.2003.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.