Abstract
Background
Chromosomal instability is associated with earlier progression in isocitrate dehydrogenase (IDH)–mutated astrocytomas. Here we evaluated the prognostic significance of polysomy in gliomas tested for 1p/19q status.
Methods
We analyzed 412 histologic oligodendroglial tumors with use of 1p/19q testing at 8 institutions from 1996 to 2013; fluorescence in situ hybridization (FISH) for 1p/19q was performed. Polysomy was defined as more than two 1q and 19p signals in cells. Tumors were divided into groups on the basis of their 1p/19q status and polysomy and were compared for progression-free survival (PFS) and overall survival (OS).
Results
In our cohort, 333 tumors (81%) had 1p/19q loss; of these, 195 (59%) had concurrent polysomy and 138 (41%) lacked polysomy, 79 (19%) had 1p/19q maintenance; of these, 30 (38%) had concurrent polysomy and 49 (62%) lacked polysomy. In agreement with prior studies, the group with 1p/19q loss had significantly better PFS and OS than did the group with 1p/19q maintenance (P < 0.0001 each). Patients with 1p/19q loss and polysomy showed significantly shorter PFS survival than patients with 1p/19q codeletion only (P < 0.0001), but longer PFS and OS than patients with 1p/19q maintenance (P < 0.01 and P < 0.0001). There was no difference in survival between tumors with >30% polysomic cells and those with <30% polysomic cells. Polysomy had no prognostic significance on PFS or OS in patients with 1p/19q maintenance.
Conclusions
The presence of polysomy in oligodendroglial tumors with codeletion of 1p/19q predicts early recurrence and short survival in patients with 1p/19q codeleted tumors.
Keywords: 1p/19q codeletion, glioma, oligodendroglioma, polysomy
Key Points.
1. Some oligodendroglial tumors with 1p/19q codeletion have concurrent polysomy.
2. Polysomy predicts earlier recurrence in patients with 1p/19q codeleted gliomas.
3. Polysomy is predictive regardless of the fraction of polysomic cells.
Importance of the Study.
Patients with oligodendroglial tumors benefit from molecular testing for the detection of 1p/19q codeletion and IDH1/2 mutation. Polysomy has been suggested to correlate with adverse outcome in small cohorts; however, the impact on long-term prognosis remains unclear. Fluorescence in situ hybridization assay detects polysomy in addition to codeletion of 1p and 19q in individual tumor cells. In our cohort of 412 tumors tested, tumors with 1p/19q loss and no polysomy had significantly better PFS than the group with 1p/19q loss and polysomy (P < 0.0001). The percentage of polysomic cells did not have additional prognostic value. Polysomy did not affect the clinical outcome in patients with 1p/19q maintenance. While the tumor diagnosis and contemporary tumor mutation signature have characterized prognosis, the presence of polysomy, which further substratifies 1p/19q codeleted tumors, needs to be considered for prognosis, follow-up, and treatment strategies.
Testing for 1p/19q codeletion status is the oldest biomarker test as well as diagnostic molecular test in neuropathology. Whereas the previous literature reported allelic losses of chromosomes 1p and/or 19q in up to 80% of oligodendroglioma and in approximately 50% of anaplastic oligodendroglial tumors,1–3 our current understanding of molecular pathology makes 1p/19q loss a defining molecular hallmark of oligodendroglioma. The most recent reiteration of the World Health Organization (WHO) classification requires 1p/19q codeletion status for the diagnosis of oligodendroglioma.4 The allelic loss of 1p and 19q in oligodendroglial tumors is mediated by balanced translocation involving chromosomes 1 and 19 at their centromeres with maintenance of the der (1;19)(q10; p10) and subsequent loss of the derivative chromosome der (1;19)(p10;q10).5,6 The loss of chromosomal arms 1p and 19q is invariably accompanied by heterozygous mutations in isocitrate dehydrogenase 1 or 2 genes (IDH1, IDH2) as well as mutations in the telomerase reverse transcriptase (TERT) promoter and the capicua transcriptional repressor (CIC) gene.7–10
Overall, oligodendrogliomas with some diffuse astrocytomas form a larger molecular entity of IDH1/2 mutant gliomas, which are characterized by slow growth and relatively good response to adjuvant therapy relative to their IDH1/2 wild-type counterparts.6,11–13 However, despite a relatively indolent behavior of IDH1/2 mutant tumors, some IDH1/2-mutated astrocytomas14 and oligodendrogliomas can behave aggressively with rapid growth, rapid progression, and short survival. We have recently shown that IDH mutated astrocytomas with increased copy number aberrations can show rapid progression into glioblastoma.15,16
Although oligodendrogliomas show relatively few chromosomal aberrations besides the loss of arms 1p and 19q, we have previously described a relative loss, that is, loss of chromosomal arms 1p and 19q with concurrent polysomy, which was originally defined as the presence of more than 2 fluorescence in situ hybridization (FISH) signals of 1q25 and 19p13.17 Several studies have confirmed that polysomy is associated with adverse outcome.17–20 However, in addition to the small number of patients in each study, the numeric criteria (% positive nuclei) for 1p/19q deletion and polysomy varied among the various published reports.
Earlier studies found relative deletion of 1p/19q/polysomy in high-grade and/or recurrent oligodendrogliomas only, suggesting that this pattern reflected a later stage of tumor progression.18 Polysomy was found to be a marker of early recurrence but did not correlate with overall survival (OS) in primary anaplastic oligodendroglioma in a subsequent study.17 Yet another group of investigators who analyzed low- and high-grade oligodendrogliomas showed that polysomy of either one or both chromosomes, regardless of deletion status, was correlated with younger age at disease onset, and co-polysomy correlated with high histologic tumor grade and poor OS. Co-polysomy, however, did not correlate with recurrence-free survival.20
In a recent study of a cohort of 1p/19q codeleted oligodendroglial tumors, co-polysomy was associated with shorter progression-free survival (PFS) and OS in low-grade oligodendroglial tumors, and a similar trend was noted in high-grade oligodendroglial tumors.19 The apparent discrepancy in conclusions drawn from these studies may be attributed to the use of different numeric criteria for polysomy and inclusion of mixed gliomas (oligoastrocytomas) in some instances. The aim of our study was to retrospectively analyze gliomas tested for 1p/19q codeletion status and determine the prognostic significance of polysomy detected by FISH. For this, we collected the largest retrospective cohort of oligodendrogliomas tested for 1p/19q status from 8 institutions. We here show that polysomy detected by FISH is associated with adverse outcome and short survival in tumors with 1p/19q codeletion and recommend to incorporate evaluation and reporting of polysomy into routine 1p/19q FISH testing.
Materials and Methods
Patient Cohort
This retrospective study was conducted in a cohort of 412 patients with oligodendroglial tumors (oligodendroglioma, oligoastrocytoma, anaplastic oligodendroglioma, and anaplastic oligoastrocytoma) tested with FISH for 1p and 19q. The tests were performed at 8 institutions, including Memorial Sloan-Kettering Cancer Center, NYU Langone Health, University of California San Francisco, University of Pittsburgh Medical Center, University of Kentucky, The University of Texas MD Anderson Cancer Center, University of Michigan Medical School, and Massachusetts General Hospital, for which clinical data were available. Testing for 1p/19q was conducted on cases diagnosed at participating institutions from January 1996 to December 2013. We reviewed medical charts for clinical variables including age, sex, site, extent of resection, adjuvant therapy, Karnofsky performance status (KPS) score, as well as histopathologic, immunohistochemical (for IDH1-R132H and Ki67 levels when available), molecular (for IDH1/2 testing results when available), and original FISH scoring results. Table 1 summarizes the patients’ demographic, clinical, and pathological information. Approval from the institutional review boards of these institutions was obtained before initiation of this study. Final data were contributed to the study in a patient de-identified format.
Table 1.
Clinical and pathological characteristics of patients with tumors tested for 1p/19q by FISH
| Characteristics | Total | 1p/19q Loss without Polysomy | 1p/19q Loss with >30% Polysomy | 1p/19q Loss with <30% Polysomy | 1p/19q Maintenance without Polysomy | 1p/19q Maintenance with Polysomy | P-Value |
|---|---|---|---|---|---|---|---|
| N = 412, 100% | N = 138, 33% | N = 150, 36% | N = 45, 11% | N = 49, 12% | N = 30, 7% | ||
| Age, y | |||||||
| Median | 43 | 42 | 45 | 44 | 43 | 44 | 0.82+ |
| Range | 11–86 | 11–86 | 15–83 | 21–82 | 17–72 | 23–79 | |
| Sex, n | |||||||
| Male | 227 (55%) | 68 (49%) | 92 (61%) | 24 (53%) | 24 (49%) | 19 (63%) | 0.21++ |
| Female | 185 (45%) | 70 (51%) | 58 (39%) | 21 (47%) | 25 (51%) | 11 (37%) | |
| Pathology diagnosis, n | |||||||
| Oligodendroglioma | 365 (89%) | 127 (92%) | 137 (91%) | 31 (69%) | 43 (88%) | 27 (90%) | |
| Oligoastrocytoma | 47 (11%) | 11 (8%) | 13 (9%) | 14 (31%) | 6 (12%) | 3 (10%) | <0.001 |
| WHO grade, n | |||||||
| Anaplastic (WHO grade III) | 232 (56%) | 61 (44%) | 110 (73%) | 19 (42%) | 29 (59%) | 13 (43%) | <0.001 |
| Not anaplastic (WHO grade II) | 180 (44%) | 77 (56%) | 40 (27%) | 26 (58%) | 20 (41%) | 17 (57%) | |
| IDH1/2 mutation, n | |||||||
| Positive | 130 (32%) | 42 (30%) | 65 (43%) | 14 (31%) | 6 (12%) | 3 (10%) | <0.01++ |
| Negative | 15 (3%) | 4 (3%) | 4 (3%) | 1 (2%) | 4 (8%) | 2 (7%) | |
| Unknown | 267 (65%) | 92 (67%) | 81 (54%) | 30 (67%) | 39 (80%) | 25 (83%) | |
| Ki67 scores, n | |||||||
| Median | 10 | 8 | 12 | 9.5 | 10 | 9 | 0.20+ |
| Range | 0.3–71.6 | 1–71.6 | 0.3–67.2 | 1–25.3 | 1.24–31 | 4.7–15 | |
| Location, n | |||||||
| Frontal | 257 (62%) | 97 (70%) | 100 (67%) | 29 (64%) | 19 (39%) | 12 (40%) | <0.001++ |
| Temporal | 73 (18%) | 13 (9%) | 27 (18%) | 7 (16%) | 19 (39%) | 7 (23%) | |
| Parietal | 41 (10%) | 11 (8%) | 15 (10%) | 6 (13%) | 5 (10%) | 4 (13%) | |
| Others | 7 (2%) | 4 (3%) | 0 | 0 | 2 (4%) | 1 (3%) | |
| Unknown | 34 (8%) | 13 (9%) | 8 (5%) | 3 (7%) | 4 (8%) | 6 (20%) | |
| Karnofsky Performance Status | |||||||
| Score | |||||||
| Median | 90 | 90 | 90 | 90 | 90 | 90 | |
| Range | 60–100 | 70–100 | 70–100 | 60–100 | 60–100 | 80–100 | |
| KPS Score Distribution, n | |||||||
| 60 | 2 (<1%) | 0 | 0 | 1 (2%) | 1 (2%) | 0 | 0.13++ |
| 70 | 5 (1%) | 1 (1%) | 1 (1%) | 1 (2%) | 2 (4%) | 0 | |
| 80 | 30 (7%) | 11 (8%) | 12 (8%) | 6 (13%) | 6 (12%) | 1 (3%) | |
| 90 | 70 (17%) | 24 (17%) | 34 (23%) | 5 (11%) | 5 (10%) | 1 (3%) | |
| 100 | 64 (16%) | 12 (9%) | 39 (26%) | 7 (33%) | 7 (14%) | 1 (3%) | |
| Unknown | 241 (59%) | 90 (65%) | 64 (43%) | 32 (71%) | 28 (57%) | 27 (90%) | |
| Initial surgery, n | |||||||
| GTR | 156 (38%) | 51 (37%) | 66 (44%) | 14 (31%) | 16 (33%) | 9 (30%) | 0.24++ |
| STR | 166 (40%) | 55 (40%) | 59 (39%) | 22 (49%) | 20 (41%) | 10 (33%) | |
| Biopsy | 62 (15%) | 22 (16%) | 15 (10%) | 7 (16%) | 10 (20%) | 8 (27%) | |
| Unknown | 28 (7%) | 10 (7%) | 10 (7%) | 2 (4%) | 3 (6%) | 3 (10%) | |
| Initial neoadjuvant/adjuvant chemotherapy, n | |||||||
| Yes | 229 (56%) | 78 (57%) | 90 (60%) | 18 (40%) | 29 (59%) | 14 (47%) | 0.07++ |
| No | 152 (37%) | 52 (38%) | 46 (31%) | 24 (53%) | 16 (33%) | 14 (47%) | |
| Unknown | 31 (7%) | 8 (6%) | 14 (9%) | 3 (7%) | 4 (8%) | 2 (6%) | |
| Initial radiotherapy, n | |||||||
| Yes | 174 (42%) | 55 (40%) | 58 (39%) | 14 (31%) | 33 (67%) | 14 (47%) | <0.001 ++ |
| No | 208 (50%) | 74 (54%) | 81 (54%) | 28 (62%) | 11 (22%) | 14 (47%) | |
| Unknown | 30 (7%) | 8 (5%) | 11 (7%) | 3 (7%) | 5 (10%) | 2 (6%) |
GTR: gross total resection; STR: subtotal resection. +One-way ANOVA analysis; ++ chi-square analysis.
We evaluated PFS and OS. Progression was defined by radiographic enlargement of the existing lesion, by development of a new lesion proven by biopsy or excision, or by clinical deterioration attributed to the tumor based on a new lesion by imaging when biopsy or resection was not considered clinically indicated. The start point of PFS was the time of diagnosis. The endpoint was the time of progression or death if death occurred without progression. Overall survival was defined as the time from the initial diagnosis to death from any cause.
Fluorescence In Situ Hybridization
Dual-color FISH was performed on two 4-µm-thick paraffin sections with use of Vysis 1p36/1q25 and 19q13/19p13 probes (Abott Laboratories) according to the manufacturer’s instructions. One section was used to probe for 1p36 (red) and 1q25 (green); the second section was used to probe for 19q13 (red) and 19p13 (green). Briefly, the formalin-fixed paraffin-embedded tissue sections were deparaffinized, permeabilized, and hybridized.18 For each set of probes, at least 100 non-overlapping nuclei were assessed for signal quantitation to generate a 1p to 1q ratio and a 19q to 19p ratio. The total number of signals was counted, and ratios internally validated at each institution for loss or maintenance were applied.17
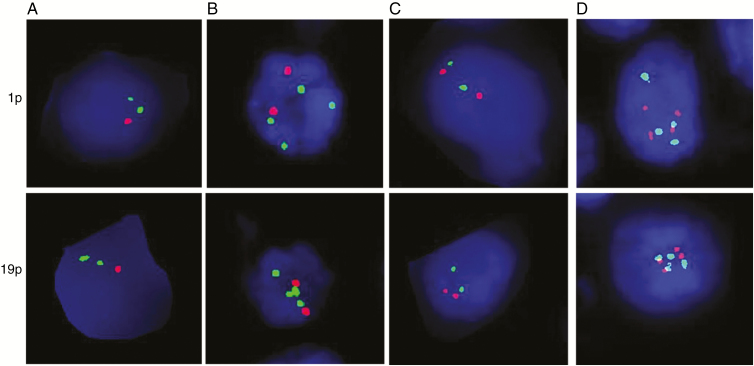
Codeletion of 1p and 19q (1p/19q loss) was defined as loss of both 1p and 19q in tumor cells; 1p/19q maintenance included tumors with maintenance in both 1p and 19q (Fig. 1). Rare cases with either 1p loss only or 19q loss only were excluded from this study. Polysomy was defined as 3 or more signals for both 1q and 19p. Most of the cases showed 2:4 1p:1q and 19q:19p ratio, and while the cases with higher number were present (3:6, 4:8), the subgroups were too small for further analysis. Rare cases with polysomy in either 1q or 19p were excluded from this study. On the basis of our original description, we applied a 30% cutoff for the presence of polysomy.17
Fig. 1.
Representative FISH images illustrate the presence and absence of polysomy. (A) 1p and 19q loss without polysomy, (B) 1p and 19q relative loss with concurrent polysomy, (C) 1p and 19q maintenance without polysomy, and (D) 1p and 19q maintenance with polysomy. Red probe is for 1p or 19q; green probe is for 1q or 19p.
On the basis of 1p/19q loss and polysomy determined by FISH, we divided all cases into 5 groups: 1p/19q loss with >30% polysomy of both chromosomes (n = 150), 1p/19q loss with <30% polysomy (n = 45), 1p/19q loss without polysomy (n = 138), 1p/19q maintenance with polysomy (n = 30), and 1p/19q maintenance without polysomy (n = 49).
Statistical Analysis
Statistical analysis of the associations among categorical variables including FISH patterns, age, sex, pathology diagnosis, WHO grade, IDH1/2 mutation status, Ki67 score, tumor location, KPS score, type of initial surgery, use of neoadjuvant or adjuvant chemotherapy at initial diagnosis, and use of initial radiotherapy were assessed by chi-square analyses. Correlation between age and categorical variable FISH patterns was examined by ANOVA analysis. The Cox proportional hazards model was used to examine the association between the clinical and genetic variables and OS and PFS rates. In particular, predictors were examined in separate models. Then, predictors that were significant at the 0.05 level were included in a single multivariable Cox proportional hazards model, to examine the adjusted effects of all significant predictors. In all cases, Wald chi-square tests were used to determine significance. The Kaplan–Meier survival curves were plotted using GraphPad Prism 6. Data analysis was performed using SPSS (IBM) software. A P-value of less than 0.05 was considered statistically significant.
Results
1p/19q Testing Refines Diagnosis of Oligodendroglioma
In total, 412 oligodendroglial tumors with an original histologic diagnosis of oligodendroglioma or oligoastrocytoma were analyzed. However, 79 (19%) of these tumors did not show 1p/19q loss, making them molecularly incompatible with a diagnosis of oligodendroglioma (Table 1). Interestingly, the vast majority of these misdiagnosed tumors (70 of 79, 89%) were originally diagnosed as oligodendrogliomas, not oligoastrocytomas, from which one might assume that 1p/19q testing would be most informative. IDH1/2 mutation status was tested in 145 cases and mutation identified in 130 tumors, of which 121 also showed 1p/19q loss and would be classified as oligodendroglioma, IDH mutant and 1p/19q codeleted, while oligodendroglial tumors with 1p/19q codeletion detected but unavailable IDH1/2 mutation status would be best classified as oligodendroglioma not otherwise specified, according to the WHO 2016 classification.
Correlation of polysomy and 1p/19q loss with clinical, pathological, and molecular features
Among 333 cases of 1p and 19q codeleted oligodendroglial tumors, 138 cases (41%) showed classic codeletion; 195 cases (59%) showed a relative deletion, that is, codeletion of 1p and 19q with concurrent polysomy including 150 cases with >30% polysomy and 45 cases with <30% polysomy (Table 1). In 79 cases of glial tumors with 1p/19q maintenance, polysomy of chromosomes 1 and 19 was observed in 30 cases (38%) and was absent in 49 cases (62%).
Across our cohort of all patients, the median age at initial diagnosis was 43 years, and the male-to-female ratio was approximately 1.2:1. The distribution of age, sex, Ki67 score, KPS score, type of initial surgery, and use of initial chemotherapy was not different among cases either with or without 1p/19q loss or with or without polysomy. Initial radiotherapy status was available in 382 cases. Radiotherapy was used at first diagnosis more frequently in tumors with 1p/19q maintenance and no polysomy (33/44, 75%) than in those with other FISH patterns (141/338, 42%; P < 0.001).
Histologic diagnosis of oligoastrocytoma was associated more frequently with 1p/19q loss with <30% polysomy (14/45, 31%) than with other FISH patterns (33/367, 9%; P < 0.001).
Similar to previous findings,20 anaplasia was more frequent with 1p/19q loss and a high level polysomy (>30%) (110/150, 73%) than with the other FISH patterns (122/262, 47%; P < 0.001). Mutation analyses for IDH1 and IDH2 were performed in 145 patients from our cohort.
Although a small fraction was tested for IDH1/2 status, similar to previous findings,7 mutations in IDH1 or IDH2 were more frequent in tumors with 1p/19q loss (121/130, 93%) than in tumors with 1p/19q maintenance (9/15, 60%; P < 0.01). IDH1-mutated tumors with 1p/19q maintenance likely represented IDH1/2-mutated diffuse astrocytomas; however, data for alpha thalassemia/mental retardation syndrome X-linked (ATRX) were not available. The majority of tumors were located in the frontal lobe in all groups, and frontal lobe tumors were more frequently associated with 1p/19q loss (226/309, 73%) than with 1p/19q maintenance (31/69, 45%; P < 0.001). Temporal lobe tumors were more frequently associated with 1p/19q maintenance (26/69, 38%) than with 1p/19q loss (47/309, 15%; P < 0.001).
Polysomy in 1p/19q codeleted oligodendroglial tumor is associated with worse OS and PFS
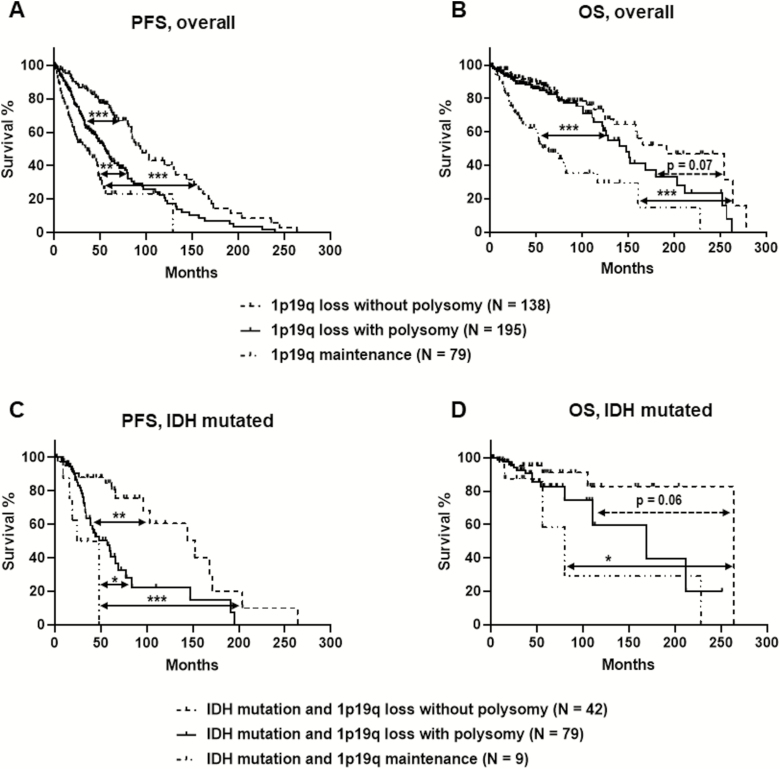
The clinical follow-up time ranged from 1.5 to 278 months, with a median follow-up of 151 months. Oligodendroglial tumors with 1p/19q loss and polysomy, regardless of the degree of polysomy, had worse PFS and increased hazard ratio (median, 55 mo; 95% CI: 46–66 mo) than did tumors with 1p/19q loss and no polysomy (median, 94 mo; 95% CI: 84–132 mo; P < 0.0001) but had better PFS and lower hazard ratio than did tumors with 1p/19q maintenance (median, 36 mo; 95% CI: 22–48 mo; P < 0.0001) (Fig. 2, Table 2). Tumors with 1p/19q loss and polysomy also showed better OS (median, 148 mo; 95% CI: 122–188 mo) than did those with 1p/19q maintenance (median, 64 mo; 95% CI: 39–117 mo; P < 0.0001) and a trend for worse OS than did those with 1p/19q loss without polysomy (median, 192 mo; 95% CI: 134–264 mo; P = 0.07 (Fig. 2, Table 2). Similar findings were observed in a subgroup with available IDH and 1p/19q data (oligodendroglioma, IDH mutant and 1p/19q codeleted). Polysomy in 1p/19q codeleted and IDH mutated tumor were associated with worse PFS (P = 0.0003) and a trend for worse OS (P = 0.06) than those without polysomy (Fig. 2C, D). The IDH mutated and 1p/19q loss tumor with polysomy showed better PFS than did IDH mutated and 1p/19q preserved tumors (P = 0.03); however, no difference was observed in OS between these 2 groups (P = 0.31), which is partially due to the limited case number in the group of IDH mutated and 1p/19q maintenance tumors.
Fig. 2.
Clinical outcomes from the presence and absence of polysomy in all patients with 1p/19q codeleted oligodendroglial tumors (A and B) and in a subset of patients with molecularly defined oligodendroglioma, IDH mutant and 1p/19q codeleted (C and D). (A) Kaplan–Meier estimate shows that tumors with 1p/19q loss and polysomy had worse PFS than did tumors with 1p/19q loss and no polysomy (P < 0.0001) and better PFS than did tumors with 1p/19q maintenance (P < 0.01). (B) Tumors with 1p/19q loss and polysomy had better OS than did tumors with 1p/19q maintenance (P < 0.0001) and a trend for worse OS than tumors with 1p/19q loss and no polysomy (P = 0.07). (C) Kaplan–Meier estimate shows that oligodendroglioma, IDH mutant and 1p/19q codeleted with polysomy has worse PFS than did tumors with 1p/19q loss and no polysomy (P < 0.01) and better PFS than did tumors with 1p/19q maintenance (P < 0.05). (D) Oligodendroglioma, IDH mutant and 1p/19q codeleted with polysomy showed a trend for worse OS compared with IDH mutated tumors with 1p/19q loss without polysomy (P = 0.06). *P < 0.05, **P < 0.01, ***P < 0.0001.
Table 2.
Bivariate (unadjusted) analyses for survival
| Progression-free Survival | Overall Survival | |||
|---|---|---|---|---|
| Covariate | Hazard Ratio (95% CI) | Chi-square P-value | Hazard Ratio (95% CI) | Chi-square P-value |
| 1p/19q status | ||||
| 1p/19q loss without polysomy | – | – | – | – |
| 1p/19q loss with polysomy (all) | 2.08 (1.52, 2.84) | <0.0001 | 1.52 (0.97, 2.38) | 0.07 |
| 1p/19q maintenance (all) | 3.66 (2.47, 5.42) | <0.0001 | 4.26 (2.63, 6.90) | <0.0001 |
| <0.0001 | <0.0001 | |||
| Age | 1.02 (1.01, 1.03) | 0.0003 | 1.04 (1.02, 1.05) | <0.0001 |
| Sex | ||||
| Female | – | – | – | – |
| Male | 1.11 (0.85, 1.45) | 0.43 | 1.10 (0.77, 1.58) | 0.60 |
| Pathology diagnosis | 1.08 (0.72, 1.61) | 0.73 | 1.15 (1.95, 0.68) | 0.60 |
| WHO grade III | 2.09 (1.58, 2.78) | <0.0001 | 2.57 (1.70, 3.87) | <0.0001 |
| Ki67 | 1.01 (0.995, 1.02) | 0.31 | 1.01 (0.995, 1.02) | 0.20 |
| IDH1/2 mutation | 0.76 (0.46, 1.27) | 0.30 | 0.57 (0.24, 1.34) | 0.20 |
| Chemotherapy | 1.12 (0.85, 1.47) | 0.41 | 0.98 (0.68, 1.42) | 0.92 |
| Radiotherapy | 0.92 (0.71, 1.20) | 0.55 | (1.57 (1.09, 2.27) | 0.016 |
|
Karnofsky Performance Status Score |
||||
| 80 | – | – | – | – |
| 90 | 0.77 (0.51, 1.15) | 0.55 (0.32, 0.92) | ||
| 100 | 0.63 (0.43, 0.93) | 0.44 (0.26, 0.77) | ||
| 0.10 | 0.017 | |||
| Initial Surgery | ||||
| GTR | 0.69 (0.49, 0.96) | 0.82 (0.53, 1.28) | ||
| Biopsy only | 1.13 (0.78, 1.64) | 1.35 (0.82, 2.24) | ||
| 0.03 | 0.17 | |||
| Location | ||||
| Temporal | – | – | – | – |
| Frontal | 1.00 (0.72, 1.40) | 0.78 (0.49, 1.26) | ||
| Parietal | 1.26 (0.78, 2.05) | 1.23 (0.72, 2.50) | ||
| Other | 0.87 (0.31, 2.46) | 1.43 (0.42, 4.93) | ||
| 0.66 | 0.16 |
GTR: gross total resection; STR: subtotal resection.
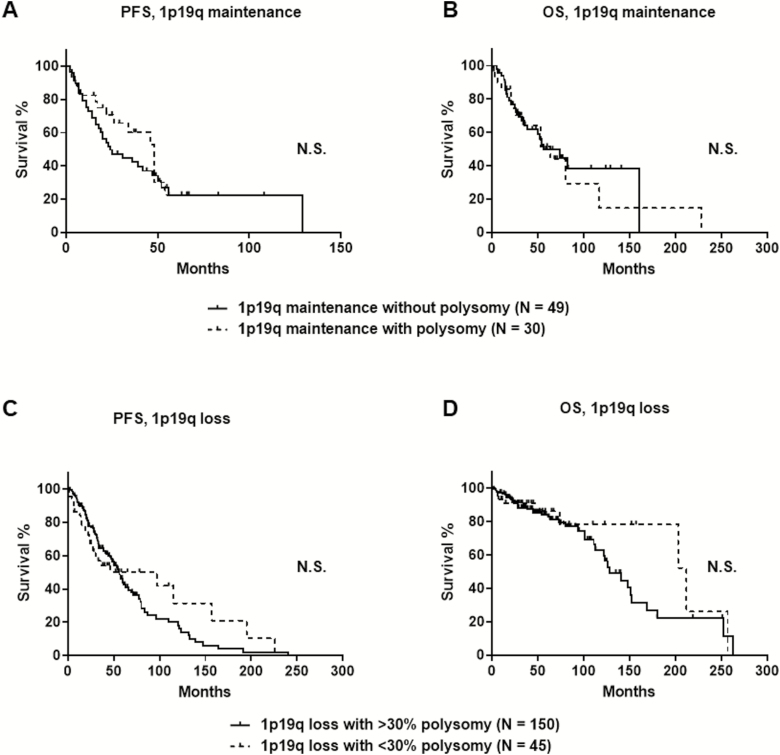
PFS was similar between 1p/19q codeleted tumors with polysomy greater than 30% (median, 55 mo; 95% CI: 46–66 mo) and with polysomy less than 30% (median, 46 mo; 95% CI: 24–157 mo; P = 0.76). Similarly, OS rates were not different between 1p/19q codeleted tumors with polysomy greater than 30% (median, 128 mo; 95% CI: 113–152 mo) and polysomy less than 30% (median, 212 mo; 95% CI: 203–257 mo; P = 0.32) (Fig. 3). This suggests that a 30% cutoff for polysomy is not relevant for laboratory practice, since even a low polysomy was associated with adverse outcome.
Fig. 3.
The presence of polysomy in glial tumors with 1p/19q maintenance did not affect PFS (A, P = 0.36 by log-rank test) or OS (B, P = 0.75 by log- rank test) via Kaplan–Meier estimate. The extent of polysomy with either >30% or <30% polysomy in oligodendroglial tumors with 1p/19q loss did not affect PFS (C, P = 0.76 by log-rank test) or OS (D, P = 0.32 by log-rank test). N.S. = P-value is not significant.
Polysomy in tumors with 1p/19q maintenance has no effect on survival
In contrast, polysomy did not show prognostic value in tumors with 1p/19q maintenance. PFS was similar between tumors with polysomy (median, 48 mo; 95% CI: 22–54 mo) and tumors without polysomy (median, 24 mo; 95% CI: 16–47 mo; P = 0.36). Similarly, OS was not different between tumors with polysomy (median, 64 mo; 95% CI: 26–117 mo) and without polysomy (median, 56 mo; 95% CI: 35–161 mo; P = 0.75) (Fig. 3).
Polysomy is an independent factor of shorter PFS in 1p/19q codeleted tumors
We evaluated the adjusted effects of FISH patterns including polysomy and other clinical and pathological factors on survival. Age and extent of resection were independently prognostic of survival. Multivariate analyses reveal that the PFS and OS in patients with oligodendroglial tumors were independently associated with FISH patterns (P < 0.0001), age (P = 0.001), and WHO grade (P < 0.001). Among 3 FISH patterns, 1p/19q maintenance was independently associated with worse OS and PFS than cases with 1p/19q loss only (P < 0.0001). In tumors with 1p/19q loss, the presence of polysomy was independently associated with worse PFS than cases without polysomy (P < 0.0001). In this study, there were no statistically significant differences in survival between sex, pathology diagnosis, Ki67 score, IDH1/2 mutation status, initial neoadjuvant chemotherapy, or location of tumor (Table 3).
Table 3.
Multivariate analysis for survival
| Covariate | Hazard Ratio (95% CI) | Wald Chi-Square P-Value |
|---|---|---|
| Hazard ratios for PFS | ||
| 1p/19q status | ||
| 1p/19q loss without polysomy | - | |
| 1p/19q loss with polysomy | 1.96 (1.42, 2.71) | |
| 1p/19q maintenance | 3.64 (2.470, 5.46) | |
| <0.0001 | ||
| Age | 1.02 (1.01, 1.03) | 0.001 |
| WHO grade III | 2.88 (1.43, 2.57) | <0.0001 |
| Initial Surgery | ||
| Subtotal resection | – | |
| Gross total resection | 0.64 (0.49, 0.91) | |
| Biopsy only | 1.21 (0.80, 1.70) | |
| 0.007 | ||
| Hazard ratios for OS | ||
| 1p/19q status | ||
| 1p/19q loss without polysomy | – | |
| 1p/19q loss with polysomy | 1.35 (0.84, 2.15) | |
| 1p/19q maintenance | 4.52 (2.70, 7.55) | |
| <0.0001 | ||
| Age | 1.04 (1.02, 1.05) | <0.0001 |
| WHO grade III | 2.24 (1.43, 3.51) | 0.0004 |
| Radiotherapy at initial diagnosis | 0.94 (0.62, 1.41) | 0.81 |
| KPS | ||
| 80 | – | |
| 90 | 0.56 (0.37, 1.10) | |
| 100 | 0.64 (0.28, 1.07) | |
| 0.36 |
Discussion
Recent (re)classification of low-grade gliomas has delineated diagnosis on the basis of genetic alterations rather than on histology. Critical molecular features necessary for accurate classification include 1p/19q codeletion status, IDH1/2 mutation status, and ATRX or TERT promoter mutation status.7,10 Status of 1p/19q in glial tumors is commonly detected via FISH, whereas IDH1/2 and ATRX mutation status and TERT promoter mutation status are assessed via immunohistochemical analysis and/or sequencing. Patients with IDH1/2 mutated tumors, particularly those with concurrent 1p/19q codeletion, have the most favorable clinical outcome, whereas patients with IDH1/2 wild-type tumors typically have poor outcome, regardless of the tumor histologic grade, with a clinical behavior similar to that of glioblastoma.10
The presence of codeletion of 1p/19q in oligodendroglial tumors has clearly shown treatment response and survival advantage.3 However, identifying patients with increased risk of rapid progression remains a challenge for neuropathologists as well as for neuro-oncologists. We have previously observed that polysomy is a negative prognostic marker in anaplastic oligodendrogliomas and that the prognostic role of polysomy has been confirmed in follow-up studies.17,19,20 Polysomy is a phenomenon frequently observed in oligodendroglial tumors—between 20% and 30% across various studies. On the basis of previous studies, polysomy was associated with recurrent and high-grade oligodendroglial tumors.20–22 Studies of high-grade oligodendrogliomas with 1p/19q loss and polysomy showed either worse PFS or a trend for earlier recurrence compared with high-grade tumors with 1p/19q loss and no polysomy; no definitive difference was observed in OS.17,19 Previous studies defined polysomy as 30% or more tumor cells showing 3 or more signals for 1q and 19p.17,19
In the study by Ren et al, worse PFS and OS were seen in low-grade tumors with 1p/19q codeletion and polysomy than in low-grade tumors with 1p/19q codeletion and no polysomy.19 In other studies, higher histologic tumor grade and worse OS were seen in tumors with polysomy than in tumors without polysomy, but differences in PFS were not observed.20 Thus the case selection, small numbers of cases in each study, and slightly different FISH criteria likely contributed to some differences in clinical outcomes from these studies. Therefore, a unified approach to quantification of polysomy is necessary for clinical reporting.
In this study, we defined polysomy as more than 2 signals of 1q and 19p in tumor cells, irrespective of 1p and 19q deletion status; in addition, we used a previously established threshold of 30% to further assess the extent of polysomy on clinical outcome. We demonstrated that polysomy was an independent risk factor and correlated with less favorable PFS and a trend for less favorable OS in 1p/19q codeleted oligodendroglial tumors, regardless of the extent of polysomy. Polysomy in 1p/19q maintenance tumors had no effect on survival outcome. In addition, we observed that 1p/19q codeleted tumors with >30% polysomy correlated with anaplastic oligodendroglioma, whereas the polysomy FISH pattern did not correlate with age or sex. On the basis of these findings, we believe that incorporation of polysomy status in the 1p and 19q FISH report would provide better prognostic assessment of oligodendroglial tumors to delineate patients’ risk of progression than the codeletion status alone.
Polysomy defined by FISH can be used to approximate the copy number status of chromosomes and chromosome arms; however, we were not able to assess the allelic imbalance and copy number of entire chromosomes and chromosome arms. In addition, the molecular mechanism leading to polysomy is not understood, although there has been speculation on the altered cell cycle and cell proliferation as possible mechanisms.
The Ki67 labeling index correlated with tumor grade in oligodendroglial tumors; however, the presence of polysomy did not correlate with the Ki67 labeling index, similarly to findings from previous reports.17,23,24 Further studies using a single-nucleotide polymorphism microarray would help to clarify whether the polysomy FISH pattern stemmed from uniparental disomy versus copy number alterations involving chromosomes, chromosome arms, and/or segments. Identifying the additional genomic instability associated with polysomy FISH pattern at 1 and 19 may help us to understand biologic behavior that dictates the potential malignant transformation.
Due to the retrospective nature of our study and the fact that our cases were diagnosed before 2013, the mutation status of IDH1/2, ATRX, and TERT was not available in most of the cases and only a subset of cases could be molecularly classified as oligodendroglioma, IDH mutant and 1p/19q codeleted, according to the WHO 2016. However, 1p/19q loss is almost invariably associated with IDH1 or IDH2 mutation. Although molecular diagnosis of oligodendroglioma can be reached by mutation testing for IDH1/2, ATRX, and TERT, our study showed that standard FISH testing for 1p and 19q provides additional relevant prognostic information; therefore, diagnosis should not consist of somatic sequencing studies alone, at least not until future deep sequencing studies comparing oligodendrogliomas with and without polysomy reveal the mutational cause of genomic instability.
In summary, we present the largest cohort of 1p/19q tested glioma. We conclusively show that the presence of polysomy can be used to further substratify 1p/19q codeleted oligodendroglial tumors and identify patients with risk of early recurrence and who may benefit from closer follow-up and potentially more intensive regimen. The refined framework could provide precise prediction of early recurrence and short survival in a subset of 1p/19q codeleted glial tumors with polysomy.
Funding
This work was not supported by funding.
Acknowledgments
We thank Kim Vu for graphic editing and Tamara K. Locke in the Department of Scientific Publications at MD Anderson Cancer Center for her editing.
Conflict of interest statement. The authors declare no conflict of interest.
Authorship statement. Experimental design: Hui Chen, Marc Rosenblum, Meera Hameed, Matija Snuderl. Acquisition of data: Hui Chen, Cheddhi Thomas, Sanda Alexandrescu, Craig M. Horbinski, Adriana Olar, Declan McGuone, Sandra Camelo-Piragua, Lu Wang, Elena Pentsova, Joanna Phillips, Kenneth Aldape, A. John Iafrate, Andrew S. Chi, David Zagzag, John G. Golfinos, Dimitris G. Placantonakis, Marc Rosenblum, Meera Hameed, Matija Snuderl. Analysis and interpretation of data: Hui Chen, Cheddhi Thomas, Felipe Andres Munoz, Pamela Ohman-Strickland, Wen Chen, Meera Hameed, Matija Snuderl. Writing, review, and/or revision of the manuscript: Hui Chen, Cheddhi Thomas, Meera Hameed, Matija Snuderl.
References
- 1. Jeuken JW, von Deimling A, Wesseling P. Molecular pathogenesis of oligodendroglial tumors. J Neurooncol. 2004;70(2):161–181. [DOI] [PubMed] [Google Scholar]
- 2. Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108(1):49–56. [DOI] [PubMed] [Google Scholar]
- 3. Reifenberger G, Louis DN. Oligodendroglioma: toward molecular definitions in diagnostic neuro-oncology. J Neuropathol Exp Neurol. 2003;62(2):111–126. [DOI] [PubMed] [Google Scholar]
- 4. Louis D, Ohgaki H, Wiestler O, et al. WHO Classification of Tumors of the Central Nervous System. Revised 4th ed. Lyon, France: International Agency for Research on Cancer (IARC); 2016. [Google Scholar]
- 5. Griffin CA, Burger P, Morsberger L, et al. Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol. 2006;65(10):988–994. [DOI] [PubMed] [Google Scholar]
- 6. Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66(20):9852–9861. [DOI] [PubMed] [Google Scholar]
- 7. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brat DJ, Verhaak RG, Aldape KD, et al. ; Cancer Genome Atlas Research Network Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90(19):1473–1479. [DOI] [PubMed] [Google Scholar]
- 12. Cairncross G, Berkey B, Shaw E, et al. ; Intergroup Radiation Therapy Oncology Group Trial 9402 Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol. 2006;24(18):2707–2714. [DOI] [PubMed] [Google Scholar]
- 13. Perry JR, Louis DN, Cairncross JG. Current treatment of oligodendrogliomas. Arch Neurol. 1999;56(4):434–436. [DOI] [PubMed] [Google Scholar]
- 14. Shirahata M, Ono T, Stichel D, et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018;136(1):153–166. [DOI] [PubMed] [Google Scholar]
- 15. Richardson TE, Snuderl M, Serrano J, et al. Rapid progression to glioblastoma in a subset of IDH-mutated astrocytomas: a genome-wide analysis. J Neurooncol. 2017;133(1):183–192. [DOI] [PubMed] [Google Scholar]
- 16. Richardson TE, Sathe AA, Kanchwala M, et al. Genetic and epigenetic features of rapidly progressing IDH-mutant astrocytomas. J Neuropathol Exp Neurol. 2018;77(7):542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Snuderl M, Eichler AF, Ligon KL, et al. Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clin Cancer Res. 2009;15(20):6430–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perry A, Fuller CE, Banerjee R, Brat DJ, Scheithauer BW. Ancillary FISH analysis for 1p and 19q status: preliminary observations in 287 gliomas and oligodendroglioma mimics. Front Biosci. 2003;8:a1–a9. [DOI] [PubMed] [Google Scholar]
- 19. Ren X, Jiang H, Cui X, et al. Co-polysomy of chromosome 1q and 19p predicts worse prognosis in 1p/19q codeleted oligodendroglial tumors: FISH analysis of 148 consecutive cases. Neuro Oncol. 2013;15(9):1244–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wiens AL, Cheng L, Bertsch EC, Johnson KA, Zhang S, Hattab EM. Polysomy of chromosomes 1 and/or 19 is common and associated with less favorable clinical outcome in oligodendrogliomas: fluorescent in situ hybridization analysis of 84 consecutive cases. J Neuropathol Exp Neurol. 2012;71(7):618–624. [DOI] [PubMed] [Google Scholar]
- 21. Reddy KS. Assessment of 1p/19q deletions by fluorescence in situ hybridization in gliomas. Cancer Genet Cytogenet. 2008;184(2):77–86. [DOI] [PubMed] [Google Scholar]
- 22. Fallon KB, Palmer CA, Roth KA, et al. Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. J Neuropathol Exp Neurol. 2004;63(4):314–322. [DOI] [PubMed] [Google Scholar]
- 23. Hagel C, Krog B, Laas R, Stavrou DK. Prognostic relevance of TP53 mutations, p53 protein, Ki-67 index and conventional histological grading in oligodendrogliomas. J Exp Clin Cancer Res. 1999;18(3):305–309. [PubMed] [Google Scholar]
- 24. Heegaard S, Sommer HM, Broholm H, Broendstrup O. Proliferating cell nuclear antigen and Ki-67 immunohistochemistry of oligodendrogliomas with special reference to prognosis. Cancer. 1995;76(10):1809–1813. [DOI] [PubMed] [Google Scholar]