Abstract
Purpose:
To develop a rapid T2 mapping protocol using optimized spiral acquisition, accelerated reconstruction, and model fitting.
Materials and Methods:
A T2-prepared stack-of-spiral gradient echo (GRE) pulse sequence was applied. A model-based approach joined with compressed sensing was compared with the two methods applied separately for accelerated reconstruction and T2 mapping. A 2-parameter-weighted fitting method was compared with 2- or 3-parameter models for accurate T2 estimation under the influences of noise and B1 inhomogeneity. The performance was evaluated using both digital phantoms and healthy volunteers. Mitigating partial voluming with cerebrospinal fluid (CSF) was also tested.
Results:
Simulations demonstrates that the 2-parameter-weighted fitting approach was robust to a large range of B1 scales and SNR levels. With an in-plane acceleration factor of 5, the model-based compressed sensing-incorporated method yielded around 8% normalized errors compared to references. The T2 estimation with and without CSF nulling was consistent with literature values.
Conclusion:
This work demonstrated the feasibility of a T2 quantification technique with 3D high-resolution and whole-brain coverage in 2–3 min. The proposed iterative reconstruction method, which utilized the model consistency, data consistency and spatial sparsity jointly, provided reasonable T2 estimation. The technique also allowed mitigation of CSF partial volume effect.
Keywords: T2 mapping, 3D stack-of-spiral, model-based reconstruction, compressed sensing, cerebrospinal fluid nulling, 2-parameter-weighted fitting
1. Introduction
Quantitative relaxometry is a desired MRI tool for longitudinal or cross-sectional characterization of lesion structures [1,2]. Conventional T2 mapping methods generate and reconstruct T2 weighted images frame-by-frame followed by voxel-wise fitting. The associated long acquisition time hinders its practical utility, especially for applications that require 3D high spatial resolution and broad volume coverage.
Various rapid imaging techniques have been adapted to T2 mapping experiments. Parallel imaging exploits the data redundancy generated by multiple receiver coils to recover missing k-space samples [3,4]. With the development of compressed sensing (CS) [5], several constrained reconstruction methods have been developed for acceleration and applied to parameter mapping [6–14]. Block et al. proposed iterative reconstruction using a total variation (TV) constraint on radial acquisition [6]; Lustig et al. applied sparsifying transform of images acquired with incoherent sampling for acceleration [7]. In addition to the undersampling of spatial characteristics, redundancy in the temporal [8] or parametric [9–12] dimensions of image series has been explored as well. Some methods combined the aforementioned constraints [13,14]. Another promising strategy for fast parameter mapping is model-based reconstruction, which incorporated the underlying signal model as prior knowledge in an iterative reconstruction to estimate parameter maps directly from k-space data [15–21]. Advanced reconstruction imposing subspace constraints has also been demonstrated for parameter mapping [22–24].
Majority of these studies applied 2D multi-slice acquisition with Cartesian trajectories [9,10,13,15,18], which achieved up to 5-fold acceleration. 3D acquisition typically uses the smaller slice thickness without gaps and even isotropic resolution, which allows visualization of small lesions in any reformatted orientation. 3D radial trajectory has been adopted for T1 and/or T2 estimation with undersampling [25,26]. Spiral trajectory offers the great advantages of high acquisition efficiency [27] and accelerated reconstruction [28], as well as robustness to motion artifacts [29]. 3D T2 mapping has been performed using pulse sequences based on gradient echo (GRE) steady state conditions [30], multi-echo fast spin echo (FSE) [25], or T2 magnetization preparation followed by GRE [26,31–37] or FSE [38,39].
During the last decade or so, brain T2 mapping has largely been applied with advanced 2D acquisitions [9,12–16,18,23,40,41] and few with 3D methods [25,30,37]. In the clinical setting, multi-parametric MRI would be straightforward for corregistration when different magnetization preparation modules are appended with the same acquisition readout. In this work, we chose a T2-prepared GRE sequence combined with 3D stack-of-spiral acquisition for T2 mapping with whole-brain coverage and high spatial resolution. Different fitting models, k-space sampling strategies, and reconstruction techniques were evaluated in both numerical simulation and brain scans for optimum performance. Suppression of cerebrospinal fluid (CSF) signal to reduce its partial volume effect was also tested for brain T2 quantification.
2. Materials and Methods
2.1. Simulation of T2 Fitting
All numerical simulations were implemented in MATLAB 2019B (Mathworks, Natick, MA, USA). To investigate the effects of various B1 offsets on the 90⁰/−90⁰ hard pulses used at the beginning and end of the T2 preparation module (Appendix) as well as different noise levels, numerical simulations were conducted to study the performance of three non-linear T2-fitting models: 1) classic 2-paramter fitting to a mono-exponential decay; 2) 2-parameter-weighted fitting with each signal intensity as the weighting; and 3) 3-paramter fitting with an additional constant to account for the effect of incorrect B1 setting.
Typically, the longitudinal magnetization after a T2 preparation module follows an exponential decay with respect to its duration (TEprep):
| (1) |
Where A0 is the longitudinal magnetization prior to the T2 preparation. This is a 2-parameter model where only two parameters, A and T2 are fitted, with several TEprep fixed as a vector , and corresponding signal obtained as a vector . The classic 2-parameter fitting makes use of the least-square-fitting with a cost function of:
| (2) |
Alternatively the 2-parameter-weighted fitting weights the least-square-fitting with signal intensity for different TEprep, resulting in a modified cost function:
| (3) |
where reshapes the vector to a diagonal matrix, which weights obtained signal differently.
Considering B1 inhomogeneity (Eq. (A3) in the Appendix), a constant ε is added to the exponential decay to make up a 3-parameter model:
| (4) |
Therefore, the cost functions for the 3-parameter fitting model can be formulated as:
| (5) |
For all three fitting models, Levenberg–Marquardt algorithm [42] was used to solve the non-linear least squares problems. The voxel-wise fitting iterations were calculated in a matrix form to avoid the slow for-loops in MATLAB. T2 values were set from 40 ms to 200 ms in intervals of 10 ms and the T1 value was set as 1000 ms. Their signal intensities at the end of different TEprep, [20, 40, 80, 120, 160] ms, with B1+ = 1.0 and 0.8, were calculated based on Eq. (A3) in the Appendix. With signal-to-noise ratio (SNR) of the data at TEprep = 20 ms increasing from 10 to 50 in intervals of 5, random noise with normal distribution were added to the signal of various T2 weighting. For each noise level, the data generation was repeated 10000 times through a Monte Carlo simulation in order to compute the mean and standard deviation (std) of the estimated T2. The mean values of the error percentages of the fitted parameters with respect to the input values were analyzed as an indicator of accuracy of each fitting approach. The corresponding coefficient of variation values (CoV = std / mean) were measured as an indicator of precision.
2.2. Simulation of k-space Sampling and Reconstruction
To evaluate the performance of different k-space sampling and reconstruction methods used in the in vivo experiments, a 2D digital phantom (192×192) with four T2 values, [80, 100,150, 200] ms, and a uniform T1 value (1000 ms) was adopted and the T2 weighting was attained with five echo times (TEs), [20, 40, 80, 120, 160] ms, respectively.
2.3. k-Space Sampling
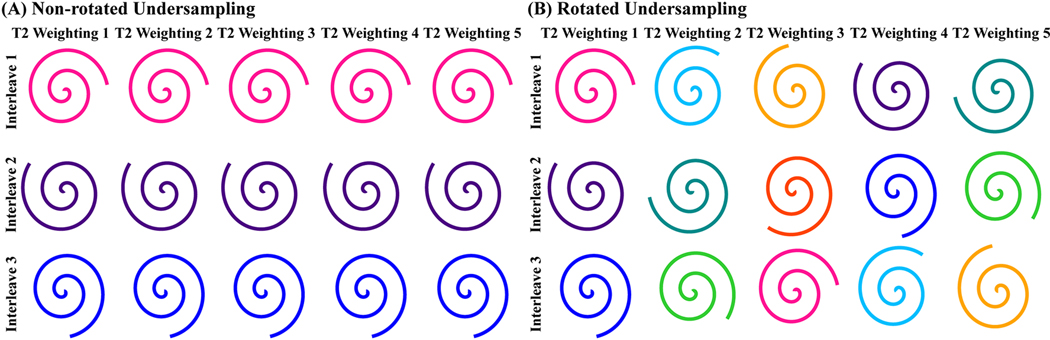
A 2D variable-density spiral trajectory [29,43] was applied to generate k-space data based on the Bloch equation simulation. Specific parameters for the sampling include: the central 15% k-space was fully sampled; the peripheral 15% k-space was sampled with a acceleration factor of 3; the the sampling density reduced linearly between central and peripheral 15% k-space; the readout duration of the spiral trajectory for each phase-encoding interleave was kept 10 ms to mitigate blurring due to off-resonance [44] and the T2* decay [45] during the k-space sampling and the dwell time of the readout sampling was 4.3 μs; number of interleaves was 8 with acceleration factor of 2; 8 simulated coils were uniformly distributed around the digital phantom. Retrospective undersampling was performed for each TE by selecting a subset of three equally spaced interleaves (total acceleration factor of 5). For k-space with different TEprep, both non-rotated and rotated undersampling schemes were tested, with the sampled spiral interleaves identical or being rotated with a fixed angle as between adjacent interleaves (360⁰ / 8 = 45⁰) (Figure 1).
Figure 1:
(A) Non-rotated retrospective undersampling scheme: the same interleaves selected for each T2 preparation; (B) Rotated retrospective undersampling scheme: different interleaves selected for each T2 preparation.
2.4. Reconstruction
For T2 estimation, a model-based reconstruction method incorporating CS in spatial domain was applied, which can be formulated as:
| (6) |
where E is the encoding matrix including the nonuniform Fourier transform and the complex coil sensitivity, is the image series, d is the k-space data, S is the 2-parameter T2 decay model (Eq. (1)), is the adjoint operator mapping image series to T2 maps with 2-parameter-weighted fitting model (Eq. (3)), and TV is the in-plane spatial total variation [6]. ||·||2 stands for the spatial l2norm. ||·||1 is the l1-norm along parameter dimension, which was implemented to eliminate outliers in the T2 fitting process [17]. Parameter λ balances the data consistency and the model consistency. Parameter α trades sparsity with data consistency, and specifically, if α = 0, the objective function degenerates to model-based reconstruction [17]. Fully sampled central k-space was used to obtain a low-resolution coil sensitivity map. The algorithm was implemented using a projected-gradient approach [46], with a projection designed to apply the model-consistency condition [17]. Data consistency, model consistency and spatial sparsity constraint were combined into a joint constrained problem and iteratively optimized with the maximum iteration number set to 60, which is empirically determined to ensure full convergence. The proposed method was compared with regular SENSE [47], CS SENSE [48] and model-based methods without CS constraint [17]. SENSE and CS SENSE were implemented by non-linear conjugate gradient algorithm [7] and the model-based methods was implemented using the projected-gradient approach described above. All reconstruction and fitting process were implemented in MATLAB 2019B (Mathworks, Natick, MA, USA).
2.5. In vivo Experiments
Experiments were performed on a 3T Philips Ingenia scanner (Philips Medical Systems, Best, The Netherlands) with a 32-channel head-only coil for signal reception. Four healthy volunteers (3 females and 1 male, 44–59 years old) were enrolled after providing informed consent in accordance with the Institutional Review Board guidelines.
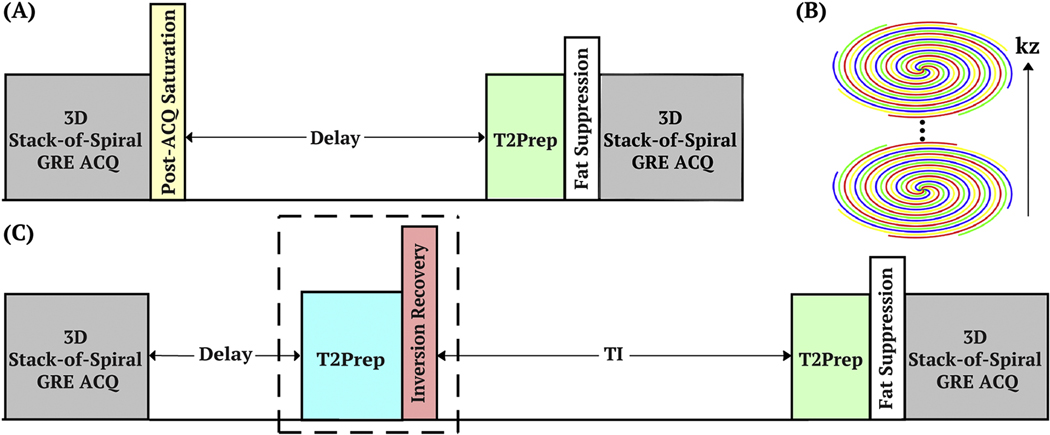
The pulse sequence diagram for T2 mapping is shown in Figure 2A. A post-acquisition spatially selective saturation pulse train was applied with a fixed delay (1500 ms) to ensure the same longitudinal magnetization before each T2 preparation module. The non-selective T2 preparation pulse train starts with a hard pulse excitation (90°x), followed by a series of composite refocusing pulses (90°x180°y90°x) with an MLEV phase-cycling pattern [49] and then a flip-back pulse (90°-x). In order to successively generate different T2 contrasts with TEprep = [20, 40, 80, 120, 160] ms, the number of refocusing pulses was chosen to be [2, 4, 8, 12, 16], respectively, with a constant inter-echo spacing of τCPMG = 10 ms [50]. Immediately following the T2 preparation, a frequency-selective fat-suppression module was inserted before data acquisition.
Figure 2:
(A) Diagram of the pulse sequence with T2-prepared gradient echo (GRE) readout for each T2 weighting. (B) 3D stack-of-spiral trajectory with turbo direction applied along slice direction. (C) Diagram of the pulse sequence with a CSF nulling module (the dashed box) to remove the CSF partial volume effect. Abbreviations: acquisition (ACQ), T2 preparation (T2Prep).
A GRE sequence, turbo-field-echo (TFE), with a train of low-flip-angle excitation pulses was applied with 3D segmented stack-of-spiral trajectory (Figure 2B) in the axial orientation. In addition to the variable-density spiral trajectory described in the simulation section (acceleration factor of 2), a fully sampled readout with a uniform density spiral trajectory (no acceleration) was acquired for reference. Readout duration of the fully sampled uniform density acquisition was identical to the variable-density case (10 ms) and 16 phase encoding interleaves were required to fill the k-space, indicating a net prospective acceleration factor of 2 (16 / 8) using variable-density spiral trajectory and a net acceleration factor of 5.3 (16 / 3) after retrospective undersampling (approximately 5-fold). Acquisition parameters: TR/TE/flip angle = 15 ms/1.2 ms/12°; the acquisition resolution was 1.2 mm3 isotropic and the reconstructed voxel size was 0.6 mm3 isotropic; field of view (FOV) = 220×220×96 mm3, slice oversampling = 1.25, number of acquired slices = 100; low-high profile order with TFE factor = 25 along the slice direction. Thus, each spiral interleave required four shots for the full encoding of the slice direction, and for the variable-density undersampled and uniform density fully sampled spiral trajectories, 8 and 16 spiral interleaves for in-plane encoding or 32 and 64 shots needed to be interleaved, respectively. With 2000 ms TFE shot interval and five separate T2 preparations, total scan duration was 5.5 min and 11 min for the two datasets. Note that the undersampled acquisition with acceleration factor of 5 would only take 2.2 min. The reference T2 map was fitted by the non-linear 2-parameter-weighted T2 decay model using fully sampled data voxel by voxel. Singular value decomposition (SVD) was used to compress 32 channels to 8 virtual channels to mitigate computational burden in the reconstruction. Retrospective undersampling and reconstruction methods were the same as for simulation. And the 2-paramter-weighted fitting model was applied.
When CSF suppression is desired, an SNR-improved inversion recovery method can be implemented [51]. This CSF-suppression module (the dashed box in Figure 2C) utilized a T2 preparation module (TEprep = 300 ms) with double refocused hyperbolic tangent pulses (5.0 ms, tanh/tan, maximum amplitude of 575 Hz and a frequency sweep of 9 kHz). Since CSF has a long T2 value (~1500 ms [52]), thus would be less affected by the prior T2 weighting and inverted and nulled by the following inversion pulse with a delay TI = 1100 ms. The brain tissue with relatively short T2 values were largely saturated after T2 preparation and thus recovered with a higher longitudinal magnetization than experiencing inversion alone. Only variable-density spiral trajectory was used in the CSF-nulled sequence. With a 3000 ms TFE shot-interval, total scan time was 8 min and the acquisition of undersampling with acceleration factor of 5 would take 3.2 min.
2.6. Data Analysis
Quantitative evaluation was performed on simulation and in vivo experiments. To compare the performance of different reconstruction methods and retrospective undersampling schemes, the relative difference of T2 maps to the reference values were computed in each voxel. Normalized root-mean-square-error (nRMSE) was calculated over the entire digital phantom for simulation. For in vivo experiments, CSF and nearby tissue were removed by masking areas of T2 values longer than 200 ms. For results with CSF nulling, the mask was created by thresholding the intensity of the images of TEprep = 20 ms. The nRMSE values were estimated from the segmented brain tissue as well as manually selected regions of interest (ROIs) of white matter and gray matter. To compare the pulse sequences with and without the CSF nulling module, estimated T2 values and corresponding differences were reported in ROIs of frontal gray matter (FGM), frontal white matter (FWM), globus pallidus (GPA), occipital gray matter (OGM), occipital white matter (OWM), splenium, and thalamus.
3. Results
3.1. Simulation of T2 Fitting
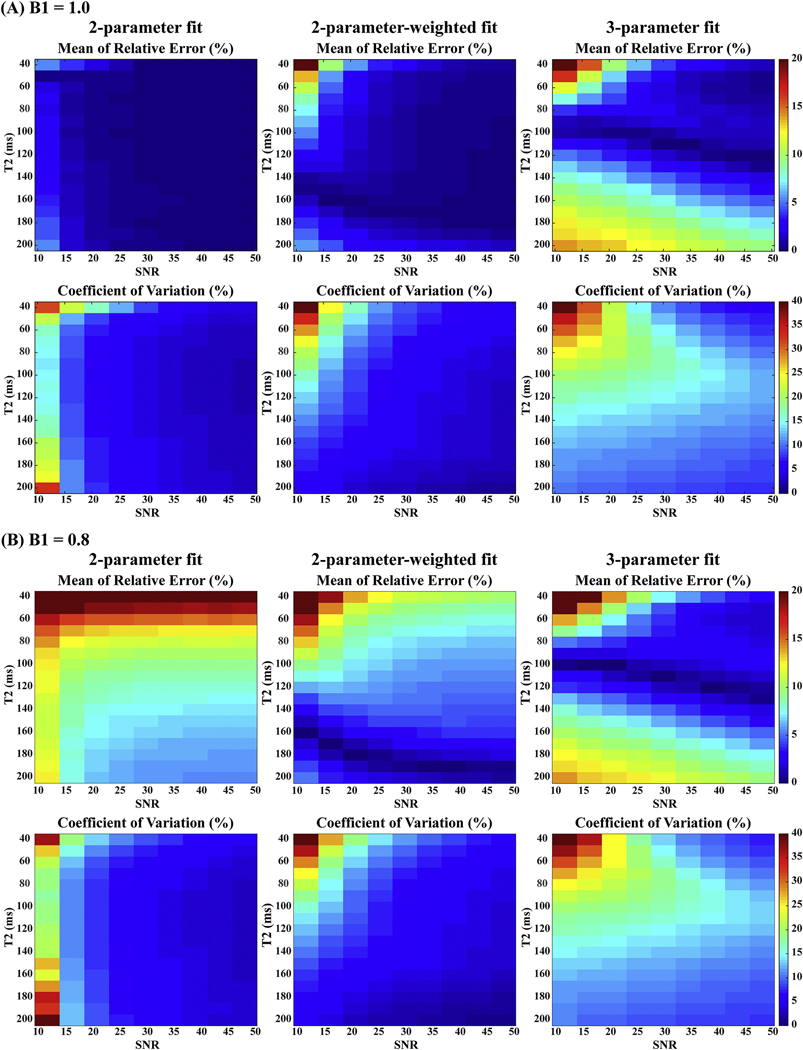
Figure 3 shows the mean of relative errors of estimated T2 and the corresponding CoV (std / mean) using the prescribed T2 preparation modules under different noise levels with three T2 fitting approaches respectively. For each fitting, higher SNR invariably delivered more accurate and precise estimates as expected. When B1 scale was ideal (B1+ = 1.0), at the same SNR levels, the 2-parameter or 2-parameter-weighted fitting ensured distinct robustness than fitting with the 3-parameter model. The relative T2 errors and CoV of the 2-parameter-weighted fitting were slightly higher than the results from the 2-parameter fitting, under 5% and 10% respectively for most cases. 3-parameter fitting was much more sensitive to noise with higher T2 errors and CoV in most cases. When B1 scale was with offset (B1+ = 0.8), 2-parameter fitting was 15–20% overestimated for shorter T2 values (40–80 ms), which indicated that 2-parameter fitting was most sensitive to B1 inhomogeneities. In contrast, 2-parameter-weighted fitting reduced these relative errors largely to under 15%. The 3-parameter fitting yielded minimal sensitivity to B1 inhomogeneities, with relative errors less than 5% when SNR was high, but was much more sensitive to noise. The 2-parameter-weighted fitting was thus chosen for its overall stable performance with various B1 scales and noise levels.
Figure 3:
Simulation results of T2 estimation using 2-parameter, 2-parameter-weighted, and 3parameter fitting models: the mean of relative errors (indicating accuracy) and coefficient of variation (indicating precision) of 10000 repetitions as a function of SNR using Monte Carlo simulation with (A) B1+ = 1.0 and (B) B1+ = 0.8. Compared to the other two fitting approaches, the 2-parameter-weighted fitting demonstrated a balanced robustness to different B1 scales and noise levels.
3.2. Simulation of k-space Sampling and Reconstruction
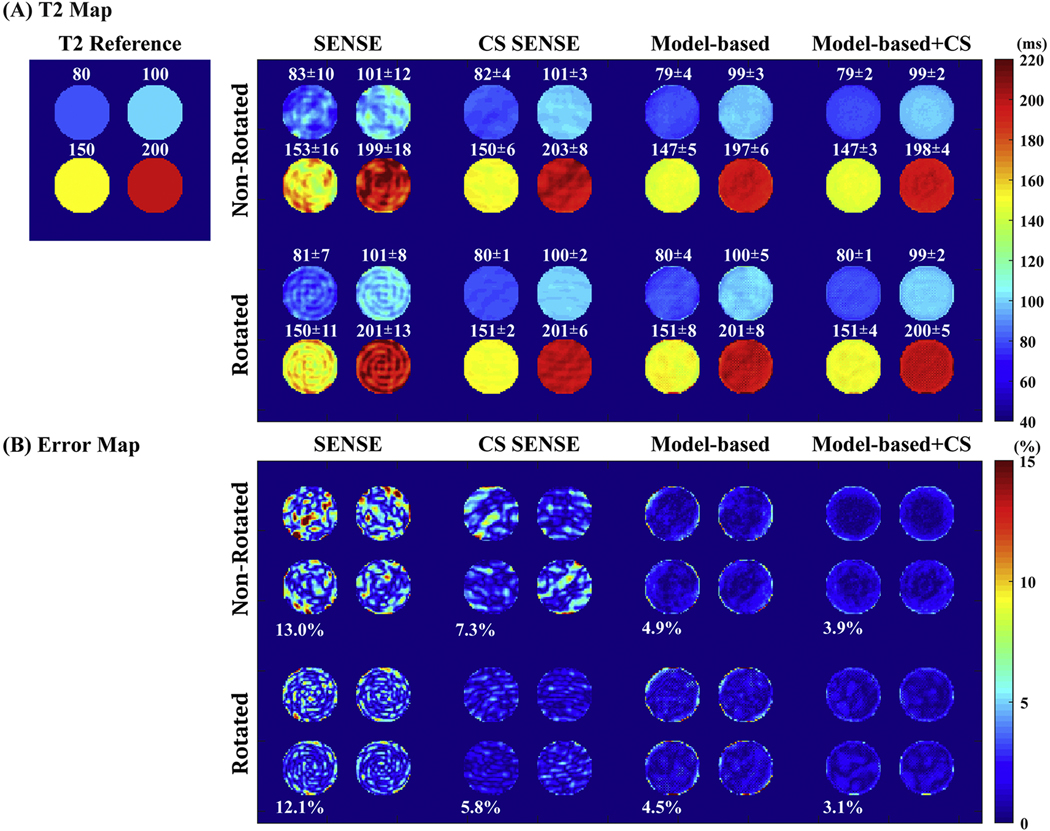
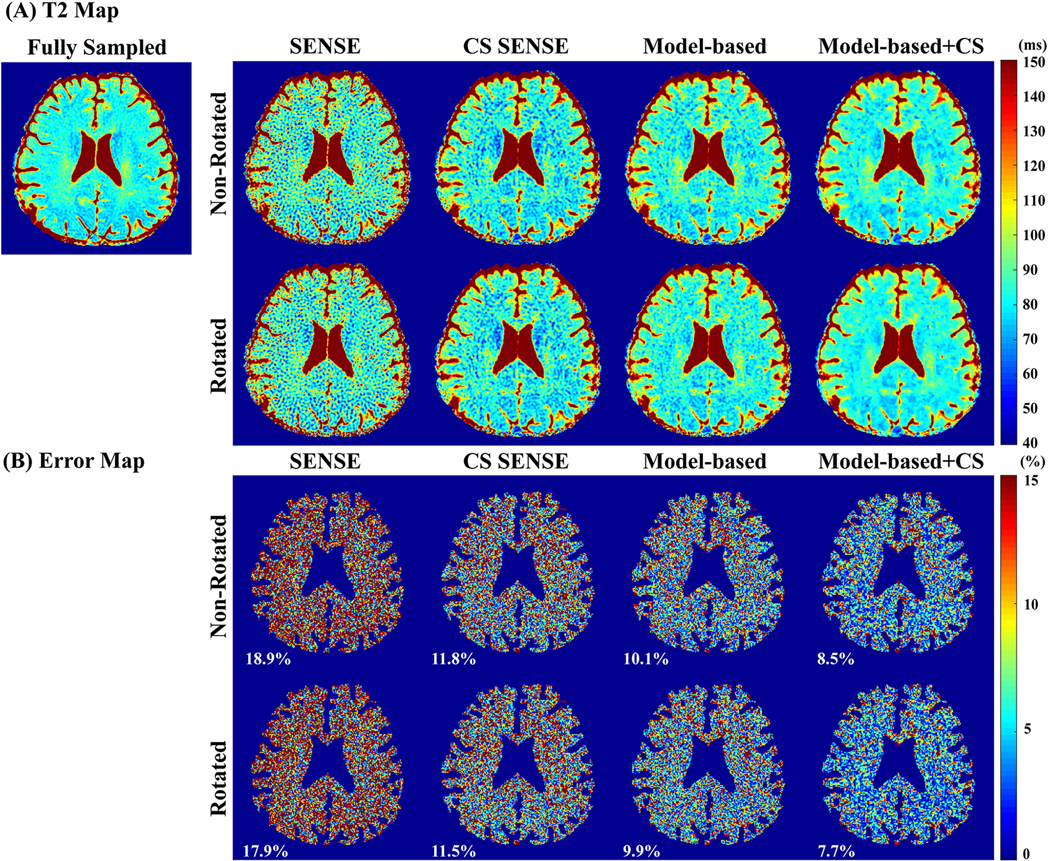
Figure 4A shows the simulation results of T2 maps obtained by different undersampling schemes and reconstruction methods with 5-fold acceleration. CS SENSE reduced the aliasing artifacts which were apparent in the T2 maps obtained using SENSE alone. Model-based methods with joint reconstruction provided more accurate T2 estimation with less artifacts and noises than applying the individual reconstruction followed by 2-parameter-weighted voxel-by-voxel fitting. When incorporating CS, model-based approach further improved the performance with reduced T2 std by reducing artifacts manifested as stripes. Results derived from rotating k-space trajectories through the T2 weighting (bottom panel) show slight improvements than the corresponding ones from the non-rotating methods (top panel). These qualitative observations were confirmed by the absolute normalized errors of the estimated T2 values across the pixels within the digital phantom, as shown in Figure 4B. The nRMSE of SENSE, CS SENSE, model-based, and model-based CS-incorporated were 13.0%, 7.3%, 4.9%, and 3.9% using non-rotated spiral trajectories, slightly higher than 12.1%, 6.8%, 4.1%, and 3.1% using rotated spiral trajectories (P < 0.05 on the corresponding error maps, two-tailed student’s t-test). The nRMSE of model-based methods were all less than 5% with the model-based CS-incorporated delivering the minimal errors. The computation speed to fit a 192×192 T2 map from 5 time points was around 0.07 s and 0.10 s on a 2.3 GHz 4-core CPU with 8 GB memory for 2-parameter and 3-parameter models respectively.
Figure 4:
Simulation results of (A) T2 maps: (left) T2 reference map composed of four regions with values of 80 ms, 100 ms. 150 ms and 200 ms, respectively; (right) T2 maps estimated by different undersampling schemes (non-rotated and rotated trajectories along T2 weighting dimension) and different reconstruction methods (SENSE, CS SENSE, model-based, and model-based CS-incorporated) with an in-plane acceleration factor of 5. Corresponding mean and std of the T2 values of each region is labeled above the region. (B) The absolute normalized error maps in percentage of different undersampling schemes and different reconstruction methods. The corresponding nRMSE of the T2 estimation of the entire digital phantom are labeled on left bottom corner of each cell.
3.3. In vivo Experiments
The T2 maps and corresponding error maps obtained by different sampling and reconstruction methods of one slice from one volunteer are displayed Figure 5. Similar to the simulation result of digital phantom (Figure 4), aliasing artifacts and noises were most notable when using SENSE, and were minimized when applying the model-based CS-incorporated method, for either non-rotated or rotated spiral trajectories. The nRMSE of SENSE, CS SENSE, model-based, and model-based CS-incorporated of brain tissue without CSF partial volume averaged from four volunteers were 16.9%, 11.4%,10.0%, and 8.9% using non-rotated trajectories, vs. 15.9%, 11.2%, 9.6%, and 8.2% using rotated trajectories. The model-based CS-incorporated method generated 47%, 22%, 11% and 48%, 27%, 15% less nRMSE using non-rotated and rotated trajectories than the other three reconstruction approaches, respectively (P < 0.05 on the corresponding error maps, two-tailed student’s t-test). Compared to using non-rotated trajectories, rotating schemes returned slightly improved accuracy, similar to the simulation results.
Figure 5:
In vivo experiment results of a 44-year-old male: (A) T2 maps and (B) corresponding normalized error maps estimated by different undersampling and reconstruction methods with an in-plane 5-fold acceleration. Error maps were masked by an eroded mask of T2 maps to exclude CSF. The corresponding nRMSEs of the masked maps were labeled on left bottom corner of each cell in the error maps.
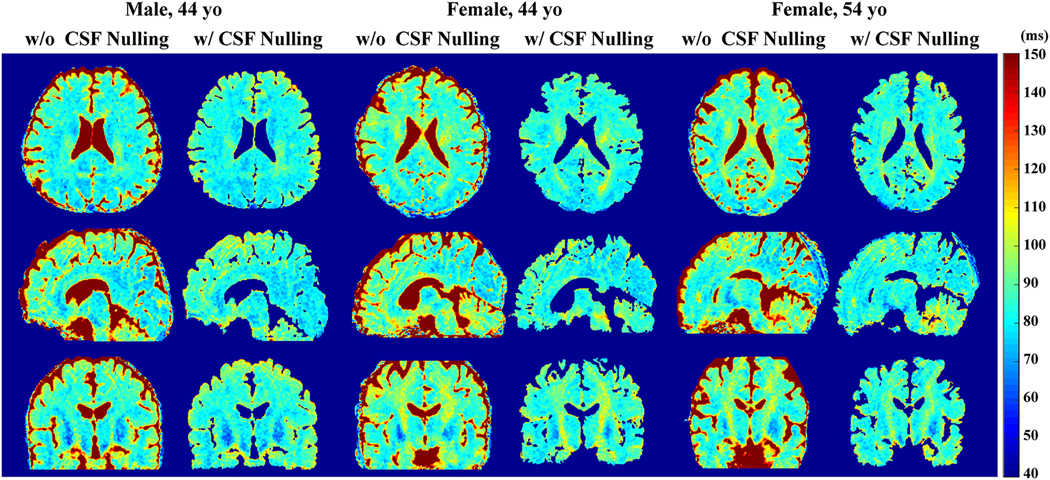
The CSF-nulling sequence (Figure 2C) yielded similar results among different reconstruction techniques (data not shown). The whole-brain T2 maps with and without the CSF nulling module, estimated by the Model-based CS-incorporated method using rotated spiral trajectories, are exhibited for three healthy volunteers (Figure 6) for comparison, showing largely in agreement. The corresponding whole-brain T2-weighted images at TEprep = 80 ms are shown in Figure S2, where the CSF was mostly suppressed with the sequence with the CSF nulling module. The averaged T2 values of several brain tissues (See Supporting Information Figure S3 for ROI selections) are listed in Table 1. The absolute T2 values were close to other literature values [19,53,54], with white matter in the range from 70 to 80 ms, and gray matter in the range from 90 to 110 ms. For all ROIs, the estimated T2 values were found to be slightly shorter by the sequence with CSF nulling. Note that the SNR (mean/std) within these ROIs of the T2-weighted images at TEprep = 20 ms were around 10–50 (data not shown) with 5-fold reconstruction, which correspond to the setting in simulation.
Figure 6:
Whole-brain cross-sectional T2 maps along axial, sagittal and coronal views of three healthy volunteers estimated by pulse sequences with and without CSF nulling, using the rotated spiral trajectories and the model-based CS-incorporated reconstruction method with an in-plane 5-fold acceleration.
Table 1:
The averaged T2 values (ms, mean±std) within ROIs of typical structures, estimated by pulse sequences with and without CSF nulling, using the rotated spiral trajectories and the model-based CS SENSE incorporated reconstruction method. ROI: FWM = frontal white matter; FGM = frontal gray matter; GPA = globus pallidus; OWM = occipital white matter; OGM = occipital gray matter.
| without CSF nulling | with CSF nulling | |
|---|---|---|
| FGM | 92±5 | 90±4 |
| FWM | 80±2 | 75±2 |
| GPA | 68±7 | 64±4 |
| OGM | 81±3 | 76±4 |
| OWM | 78±3 | 74±3 |
| Splenium | 85±3 | 81±2 |
| Thalamus | 75±3 | 70±4 |
4. Discussion
This work demonstrated the feasibility of a rapid (2–3 min with 5-fold in-plane acceleration) T2 quantification technique with 3D high-resolution (1.2 mm3 isotropic) and whole-brain coverage. The method utilized a T2-prepared GRE sequence and the model-based CS-incorporated joint reconstruction. The reliability of its T2 estimation across the brain was achieved with around 8% normalized errors compared to references using fully sampled data.
This work only explored 3D stack-of-spiral trajectory with in-plane variable-density. The acceleration could be further improved with undersampling along slice direction or alternative 3D non-Cartesian trajectories [55,56]. The combination of model-based and CS reconstructions in this study delivered 11–28% less errors than applying them separately. This is expected, as the joint spatial and parametric constraints would improve the performance of the reconstructions.
The non-rotated and rotated undersampling schemes in spiral trajectory along the T2 weighting dimension were implemented, which were similar to the shifting in Cartesian trajectory [18,57] and golden-angle radial trajectory [8,26]. It was presumed that different undersampling trajectories induced different artifacts which might be compensated and reduced when reconstructing the k-space data along the parametric dimension jointly. However, the improvement by rotated trajectory was not as significant as in Cartesian or radial trajectory. One possible reason was that the 45⁰ angle fixed between interleaves within one T2 preparation or rotated between T2 preparations in the acquired spiral trajectories were not the golden angle [58] and thus might not be incoherent for compressed sensing. Further improvements may be achieved by applying golden-angle interleaves along parametric dimension or exploring other trajectories to fulfill incoherent sampling.
In the current implementation, the iterative reconstruction was implemented slice-by-slice, through 2D nonuniform Fast Fourier transform (NUFFT) [59] on CPU and projected-gradient constraint optimization. An optimal implementation is to conduct iteration and 3D NUFFT on GPU. The projected-gradient algorithm converges quickly to obtain a local optimal solution, but there is no guarantee on the convergence of the non-convex problem to a global minimum [16]. Magnetic resonance fingerprinting (MRF) [60–62] and other non-MRF techniques [9,32,63,64] circumvented this problem with dictionary matching reconstruction by generating data with varying sequence parameters and sampling trajectories together.
An essential module of the proposed pulse sequence is T2 preparation, which is commonly adopted for T2 weighting with 3D readout. The effect of the imperfect B1 scales on the prescribed 90° flip down and flip-up hard pulses appends the desired pure T2 weighting with a complexed T1 component (Eq. (A3) in the Appendix). This deviation from the simple mono-exponential decay model can cause fitting bias for T2 prepared approaches, which has been largely overlooked. Although more exact fitting with a 3-parameter model would avert this issue, its accuracy and precision were rather poor with moderate SNR levels (Figure 3). Our simulation results indicated that the 2-parameter-weighted fitting was robust to a wide range of B1 scales and noise levels. The simulation was performed for T1 = 1000 ms. According to Eq. (A3) in the Appendix, longer T1 values, such as of gray matter or at higher field, would induce more bias to 2-parameter fitting model and approximate to the square of cos(B1+ × 90°).
In order to remove the previous T2 weighting history before each T2 preparation module, a post-acquisition saturation pulse was applied with a fixed delay in this work. Another strategy is to add sufficient number of dummy pulses after the GRE readout to ensure the same steady state conditions are reached with each T2 weighting [34]. This could potentially enhance the available signal as it leads to higher longitudinal magnetization before the T2 preparation.
Routine T2-weighted FLAIR imaging enhances the conspicuity of brain lesions with long T2 by suppressing the CSF signal. Similarly, brain T2 mapping would be more readily translated to the clinic if the fluid signal is removed. In this study, a CSF nulling module (Figure 2C) was applied to mitigate CSF partial volume effect. This method was shown to be effective for T2 estimation of typical structures in the brain, which provided small differences with the results obtained by the sequence without CSF nulling (Figure 6, Table 1). This method inevitably suffered some loss of SNR efficiency due to the longer recovery time per acquisition, which is also the challenge faced by the 3D FLAIR sequence [65]. Alternatively, CSF partial voluming could be accounted for with additional sampling of the relaxation curves and fitted by a two-compartment three- or even four-parameter model, which would certainly be more SNR demanding and computationally complex.
The proposed protocol was referenced with the same T2 prepared spiral GRE protocol using fully sampled data. A fully sampled Cartesian spin-echo acquisition would be desired for a more rigorous validation. However, the gold-standard single-echo spin-echo sequence is quite time-consuming even for a 2D scan [32,64] and thus not practical for 3D high-resolution human experiments. Multi-echo spin-echo sequence would be more efficient but its signal is not pure mono-exponential decay as stimulated echoes could be generated from imperfect refocusing [63,66]. The accuracy, precision and reproducibility of our method should be evaluated using system phantoms with known relaxation properties [67,68]. Note that if spatial resolution is lowered than the 1.2 mm3 isotropic chosen for this study, acquisition time would be even faster than 2–3 min, and attained SNR would be higher as well due to the larger voxel size, at the cost of more partial voluming between gray matter and white matter and between normal tissues and lesions. Further investigation among patients with central nervous system disease is needed to explore the clinical value of the proposed technique.
5. Conclusion
In this work, a T2-prepared 3D stack-of-spiral GRE pulse sequence was introduced for T2 mapping with high isotropic resolution, whole-brain coverage, and clinical acceptable scan time. A non-linear 2-parameter-weighted fitting approach was implemented to reduce sensitivity to wide B1 inhomogeneity and high noise level. An iterative model-based reconstruction method, which jointly utilized the model consistency, data consistency, and spatial sparsity, achieved reasonable T2 estimation with an in-plane acceleration factor of 5. This sequence also allowed flexibility of appending with a CSF nulling module to suppress CSF partial volume effect and thus potentially ameliorate lesion detection.
Supplementary Material
Acknowledgments
Grant support from NIH (R01 HL138182, R01 HL144751, R01 HL135500, S10 OD021648) and American Society of Hematology (Scholar Award).
Appendix:
The T2 preparation module (Supplementary Material Figure S1) is typically composed of a hard pulse excitation (90°), followed by paired refocusing pulses (180°) and then a flip-back pulse (−90°). The refocusing pulses are robust to B1 inhomogeneities as they are either composite or adiabatic pulses, so perfect 180° is assumed in the following analytical derivation. In contrast, the flip angles of the hard pulses at the beginning or end of the T2 preparation module, although prescribed as 90⁰, are proportional to the B1 scales:
Assuming the longitudinal and transverse magnetization before T2 preparation (point A) are:
where M0 is the equilibrium magnetization at fully recovery.
After the first pulse, the magnetizations become:
The formula of T1 recovery and T2 decay are:
| (A.1) |
| (A.2) |
where Mz(t) and Mxy(t) are the longitudinal and transverse magnetizations at time = t, respectively.
T2 weighting is set by the duration of the T2 preparation module (T). The longitudinal magnetizations before each of the following pulses are:
And the transverse magnetization before the flip-back pulse is:
Therefore, the longitudinal magnetization at the end of T2 preparation is:
| (A.3) |
In addition to the T2 weighting, the signal after T2 preparation also has dependence on the B1 scale and T1 relaxation. If θ is equal to 90°, it becomes pure T2 dependent as desired:
| (A.4) |
Note that Eq. (A3) does not deviate significantly if the number of refocusing pulses applied in the T2 preparation module is more than 2, as done in the current study (simulation analysis results not shown).
Footnotes
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Deoni SCL. Quantitative Relaxometry of the Brain. Top Magn Reson Imaging 2010;21:101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Margaret Cheng H-L, Stikov N, Ghugre NR, Wright GA. Practical medical applications of quantitative MR relaxometry. J Magn Reson Imaging 2012;36:805–24. [DOI] [PubMed] [Google Scholar]
- [3].Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magn Reson Med 1999;42:952–62. [PubMed] [Google Scholar]
- [4].Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47:1202–10. [DOI] [PubMed] [Google Scholar]
- [5].Donoho DL. Compressed sensing. IEEE Trans Inf Theory 2006;52:1289–306. [Google Scholar]
- [6].Block KT, Uecker M, Frahm J. Undersampled radial MRI with multiple coils. Iterative image reconstruction using a total variation constraint. Magn Reson Med 2007;57:1086–98. [DOI] [PubMed] [Google Scholar]
- [7].Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58:1182–95. [DOI] [PubMed] [Google Scholar]
- [8].Feng L, Grimm R, Block KT, Chandarana H, Kim S, Xu J, et al. Golden-angle radial sparse parallel MRI: Combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn Reson Med 2014;72:707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Doneva M, Börnert P, Eggers H, Stehning C, Sénégas J, Mertins A. Compressed sensing reconstruction for magnetic resonance parameter mapping. Magn Reson Med 2010;64:1114–20. [DOI] [PubMed] [Google Scholar]
- [10].Velikina JV, Alexander AL, Samsonov A. Accelerating MR parameter mapping using sparsity-promoting regularization in parametric dimension. Magn Reson Med 2013;70:1263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Majumdar A, Ward RK. Accelerating multi-echo T2 weighted MR imaging: Analysis prior group-sparse optimization. J Magn Reson 2011;210:90–7. [DOI] [PubMed] [Google Scholar]
- [12].Zhang T, Pauly JM, Levesque IR. Accelerating parameter mapping with a locally low rank constraint. Magn Reson Med 2015;73:655–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhao B, Lu W, Hitchens TK, Lam F, Ho C, Liang Z-P. Accelerated MR parameter mapping with low-rank and sparsity constraints. Magn Reson Med 2015;74:489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang C, Graff CG, Clarkson EW, Bilgin A, Altbach MI. T2 mapping from highly undersampled data by reconstruction of principal component coefficient maps using compressed sensing. Magn Reson Med 2012;67:1355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sumpf TJ, Uecker M, Boretius S, Frahm J. Model-based nonlinear inverse reconstruction for T2 mapping using highly undersampled spin-echo MRI. J Magn Reson Imaging 2011;34:420–8. [DOI] [PubMed] [Google Scholar]
- [16].Block KT, Uecker M, Frahm J. Model-Based Iterative Reconstruction for Radial Fast Spin-Echo MRI. IEEE Trans Med Imaging 2009;28:1759–69. [DOI] [PubMed] [Google Scholar]
- [17].Samsonov A Accelerated MR Parameter Mapping Using Robust Model-Consistency Reconstruction. Proc. 23th Annu. Meet. ISMRM, vol. 3711, Toronto, Canada: 2015. [Google Scholar]
- [18].Hilbert T, Sumpf TJ, Weiland E, Frahm J, Thiran J-P, Meuli R, et al. Accelerated T2 mapping combining parallel MRI and model-based reconstruction: GRAPPATINI. J Magn Reson Imaging 2018;48:359–68. [DOI] [PubMed] [Google Scholar]
- [19].Hilbert T, Schulz J, Marques JP, Thiran J, Krueger G, Norris DG, et al. Fast model‐based T2 mapping using SAR‐reduced simultaneous multislice excitation. Magn Reson Med 2019;82:2090–103. [DOI] [PubMed] [Google Scholar]
- [20].Guo Y, Lingala SG, Zhu Y, Lebel RM, Nayak KS. Direct estimation of tracer-kinetic parameter maps from highly undersampled brain dynamic contrast enhanced MRI. Magn Reson Med 2017;78:1566–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang X, Roeloffs V, Klosowski J, Tan Z, Voit D, Uecker M, et al. Model-based T 1 mapping with sparsity constraints using single-shot inversion-recovery radial FLASH. Magn Reson Med 2018;79:730–40. [DOI] [PubMed] [Google Scholar]
- [22].Tamir JI, Uecker M, Chen W, Lai P, Alley MT, Vasanawala SS, et al. T 2 shuffling: Sharp, multicontrast, volumetric fast spin-echo imaging. Magn Reson Med 2017;77:180–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mandava S, Keerthivasan MB, Li Z, Martin DR, Altbach MI, Bilgin A. Accelerated MR parameter mapping with a union of local subspaces constraint. Magn Reson Med 2018;80:2744–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roeloffs V, Uecker M, Frahm J. Joint T1 and T2 mapping with tiny dictionaries and subspace-constrained reconstruction. IEEE Trans Med Imaging 2019;39:1008–14. [DOI] [PubMed] [Google Scholar]
- [25].Keerthivasan MB, Saranathan M, Johnson K, Fu Z, Weinkauf CC, Martin DR, et al. An efficient 3D stack‐of‐stars turbo spin echo pulse sequence for simultaneous T2‐weighted imaging and T2 mapping. Magn Reson Med 2019;82:326–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Qi H, Sun J, Qiao H, Zhao X, Guo R, Balu N, et al. Simultaneous T1 and T2 mapping of the carotid plaque (SIMPLE) with T2 and inversion recovery prepared 3D radial imaging. Magn Reson Med 2018;80:2598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ahn CB, Kim JH, Cho ZH. High-Speed Spiral-Scan Echo Planar NMR Imaging-I. IEEE Trans Med Imaging 1986;5:2–7. [DOI] [PubMed] [Google Scholar]
- [28].Lustig M, Donoho DL, Santos JM, Pauly JM. Compressed sensing MRI. IEEE Signal Process Mag 2008;25:72–82. [Google Scholar]
- [29].Liao J-R, Pauly JM, Brosnan TJ, Pelc NJ. Reduction of motion artifacts in cine MRI using variable-density spiral trajectories. Magn Reson Med 1997;37:569–75. [DOI] [PubMed] [Google Scholar]
- [30].Deoni SCL, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn Reson Med 2003;49:515–26. [DOI] [PubMed] [Google Scholar]
- [31].Nguyen TD, Wisnieff C, Cooper MA, Kumar D, Raj A, Spincemaille P, et al. T 2 prep three-dimensional spiral imaging with efficient whole brain coverage for myelin water quantification at 1.5 tesla. Magn Reson Med 2012;67:614–21. [DOI] [PubMed] [Google Scholar]
- [32].Roccia E, Vidya Shankar R, Neji R, Cruz G, Munoz C, Botnar R, et al. Accelerated 3D T 2 mapping with dictionary‐based matching for prostate imaging. Magn Reson Med 2019;81:1795–805. [DOI] [PubMed] [Google Scholar]
- [33].Colotti R, Omoumi P, Bonanno G, Ledoux J-B, van Heeswijk RB. Isotropic threedimensional T2 mapping of knee cartilage: Development and validation. J Magn Reson Imaging 2018;47:362–71. [DOI] [PubMed] [Google Scholar]
- [34].Coolen BF, Poot DHJ, Liem MI, Smits LP, Gao S, Kotek G, et al. Three-dimensional quantitative T1 and T2 mapping of the carotid artery: Sequence design and in vivo feasibility. Magn Reson Med 2016;75:1008–17. [DOI] [PubMed] [Google Scholar]
- [35].van Heeswijk RB, Piccini D, Feliciano H, Hullin R, Schwitter J, Stuber M. Self-navigated isotropic three-dimensional cardiac T2 mapping. Magn Reson Med 2015;73:1549–54. [DOI] [PubMed] [Google Scholar]
- [36].Ding H, Fernandez-de-Manuel L, Schär M, Schuleri KH, Halperin H, He L, et al. Three-dimensional whole-heart T2 mapping at 3T. Magn Reson Med 2015;74:803–16. [DOI] [PubMed] [Google Scholar]
- [37].Mussard EPC, Hilbert T, Forman C, Meuli R, Kober T. High-resolution 3D T2 mapping of the Brain Using T2-prepared Cartesian Spiral Phyllotaxis FLASH and Compressed Sensing. Proc. 25th Annu. Meet. ISMRM, Honolulu, USA: 2017. [Google Scholar]
- [38].Weidlich D, Sarah Schlaeger, Kooijman H, Börnert P, Kirschke JS, Rummeny EJ, et al. T2 mapping with magnetization-prepared 3D TSE based on a modified BIR-4 T2 preparation. NMR Biomed 2017;30:e3773. [DOI] [PubMed] [Google Scholar]
- [39].Yuan J, Usman A, Reid SA, King KF, Patterson AJ, Gillard JH, et al. Three-dimensional black-blood T2 mapping with compressed sensing and data-driven parallel imaging in the carotid artery. Magn Reson Imaging 2017;37:62–9. [DOI] [PubMed] [Google Scholar]
- [40].Peng X, Ying L, Liu Y, Yuan J, Liu X, Liang D. Accelerated exponential parameterization of T2 relaxation with model-driven low rank and sparsity priors (MORASA). Magn Reson Med 2016;76:1865–78. [DOI] [PubMed] [Google Scholar]
- [41].Peng X, Liu X, Zheng H, Liang D. Exploiting parameter sparsity in model-based reconstruction to accelerate proton density and T2 mapping. Med Eng Phys 2014;36:1428–35. [DOI] [PubMed] [Google Scholar]
- [42].Moré JJ. The Levenberg-Marquardt algorithm: implementation and theory. Numer Anal 1978:105–16. [Google Scholar]
- [43].Spielman DM, Pauly JM, Meyer CH. Magnetic resonance fluoroscopy using spirals with variable sampling densities. Magn Reson Med 1995;34:388–94. [DOI] [PubMed] [Google Scholar]
- [44].Meyer CH, Hu BS, Nishimura DG, Macovski A. Fast Spiral Coronary Artery Imaging. Magn Reson Med 1992;28:202–13. [DOI] [PubMed] [Google Scholar]
- [45].Qin Q Point spread functions of the T2 decay in k-space trajectories with long echo train. Magn Reson Imaging 2012;30:1134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Samsonov AA, Kholmovski EG, Parker DL, Johnson CR. POCSENSE: POCS-based reconstruction for sensitivity encoded magnetic resonance imaging. Magn Reson Med 2004;52:1397–406. [DOI] [PubMed] [Google Scholar]
- [47].Pruessmann KP, Weiger M, Börnert P, Boesiger P. Advances in sensitivity encoding with arbitrary k -space trajectories. Magn Reson Med 2001;46:638–51. [DOI] [PubMed] [Google Scholar]
- [48].Liang D, Liu B, Wang J, Ying L. Accelerating SENSE using compressed sensing. Magn Reson Med 2009;62:1574–84. [DOI] [PubMed] [Google Scholar]
- [49].Levitt MH, Freeman R, Frenkiel T. Broadband heteronuclear decoupling. J Magn Reson 1982;47:328–30. [Google Scholar]
- [50].Qin Q, Grgac K, van Zijl PCM. Determination of whole-brain oxygen extraction fractions by fast measurement of blood T2 in the jugular vein. Magn Reson Med 2011;65:471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wong EC, Liu TT, Luh W-M, Frank LR, Buxton RB. T1 and T2 selective method for improved SNR in CSF-attenuated imaging: T2-FLAIR. Magn Reson Med 2001;45:529–32. [DOI] [PubMed] [Google Scholar]
- [52].Qin Q A simple approach for three-dimensional mapping of baseline cerebrospinal fluid volume fraction. Magn Reson Med 2011;65:385–91. [DOI] [PubMed] [Google Scholar]
- [53].Lu H, Nagae-Poetscher LM, Golay X, Lin D, Pomper M, van Zijl PCM. Routine clinical brain MRI sequences for use at 3.0 Tesla. J Magn Reson Imaging 2005;22:13–22. [DOI] [PubMed] [Google Scholar]
- [54].Wansapura JP, Holland SK, Dunn RS, Ball WS. NMR relaxation times in the human brain at 3.0 tesla. J Magn Reson Imaging 1999;9:531–8. [DOI] [PubMed] [Google Scholar]
- [55].Pipe JG, Zwart NR, Aboussouan EA, Robison RK, Devaraj A, Johnson KO. A new design and rationale for 3D orthogonally oversampled k-space trajectories. Magn Reson Med 2011;66:1303–11. [DOI] [PubMed] [Google Scholar]
- [56].Gurney PT, Hargreaves BA, Nishimura DG. Design and analysis of a practical 3D cones trajectory. Magn Reson Med 2006;55:575–82. [DOI] [PubMed] [Google Scholar]
- [57].Hu C, Peters DC. SUPER: A blockwise curve-fitting method for accelerating MR parametric mapping with fast reconstruction. Magn Reson Med 2019;81:3515–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An Optimal Radial Profile Order Based on the Golden Ratio for Time-Resolved MRI. IEEE Trans Med Imaging 2007;26:68–76. [DOI] [PubMed] [Google Scholar]
- [59].Fessler JA, Sutton BP. Nonuniform fast fourier transforms using min-max interpolation. IEEE Trans Signal Process 2003;51:560–74. [Google Scholar]
- [60].Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, et al. Magnetic resonance fingerprinting. Nature 2013;495:187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].McGivney DF, Boyac1oğlu R, Jiang Y, Poorman ME, Seiberlich N, Gulani V, et al. Magnetic resonance fingerprinting review part 2: Technique and directions. J Magn Reson Imaging 2020;51:993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Assländer J A Perspective on MR Fingerprinting. J Magn Reson Imaging 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ben-Eliezer N, Sodickson DK, Block KT. Rapid and accurate T2 mapping from multi-spin-echo data using Bloch-simulation-based reconstruction. Magn Reson Med 2015;73:809–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ben-Eliezer N, Sodickson DK, Shepherd T, Wiggins GC, Block KT. Accelerated and motion-robust in vivo T2 mapping from radially undersampled data using bloch-simulation-based iterative reconstruction. Magn Reson Med 2016;75:1346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Visser F, Zwanenburg JJM, Hoogduin JM, Luijten PR. High-resolution magnetization-prepared 3D-FLAIR imaging at 7.0 Tesla. Magn Reson Med 2010;64:194–202. [DOI] [PubMed] [Google Scholar]
- [66].Hennig J Multiecho imaging sequences with low refocusing flip angles. J Magn Reson 1969 1988;78:397–407. [Google Scholar]
- [67].Jiang Y, Ma D, Keenan KE, Stupic KF, Gulani V, Griswold MA. Repeatability of magnetic resonance fingerprinting T 1 and T 2 estimates assessed using the ISMRM/NIST MRI system phantom. Magn Reson Med 2017;78:1452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Keenan KE, Biller JR, Delfino JG, Boss MA, Does MD, Evelhoch JL, et al. Recommendations towards standards for quantitative MRI (qMRI) and outstanding needs. J Magn Reson Imaging 2019;49:e26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.