INTRODUCTION
The clinical and molecular heterogeneity of diffuse large B-cell lymphoma (DLBCL), even beyond the recent WHO reclassification,1 is well recognized. (For simplicity, this review will still use the term DLBCL to cover all the related WHO entities, including high-grade B-cell lymphoma [HGBL].) Yet, efforts to individualize therapy on the basis of this recognition have thus far been met with limited success.2,3 Why this may be, and why this may soon change, is the topic of this review. Recent comprehensive multiplatform genomic analyses have deeply probed and systematically organized the heterogeneity of DLBCL. This new knowledge could facilitate a major paradigm shift in DLBCL, which would finally allow for the successful deployment of precision medicine–based approaches. The goal is clear, and we now have new tools to build a bridge to it. Yet, the chasm that remains is wide, and successfully bridging it will require much foresight and collaboration. Here, we examine the foundation for the traditional classification tools that remain in use today and review the major efforts to personalize therapy on the basis of them. We briefly describe the new genomic categories, highlighting both their potential to profoundly transform the approach to DLBCL treatment and the significant challenges this will entail.
CONTEXT
Key Objective
How can we successfully implement precision medicine strategies in diffuse large B-cell lymphoma (DLBCL)? In this review, we describe the recently proposed genomic categories of DLBCL and their potential use for personalized treatment.
Knowledge Generated
We examine the foundation for the traditional classification tools that remain in use for DLBCL as well as the previous unsuccessful efforts to personalize therapy on the basis of them. We subsequently highlight the recently discovered genomic categories and their potential to transform the management of DLBCL as well as the challenges that this will entail.
Relevance
This review discusses the promises and barriers of novel genomic classifications in DLBCL.
TRADITIONAL CLASSIFICATIONS
While approximately two thirds of patients with DLBCL are cured after frontline chemoimmunotherapy, the remaining patients with relapsed or refractory (RR) disease have poor outcomes.4-6 This variability in sensitivity to standard chemoimmunotherapy could plausibly reflect the underlying molecular heterogeneity of DLBCL, in which case characterizing this heterogeneity could be a critical step in the quest for improved cure rates. A variety of categorization systems are currently available, principally the International Prognostic Index (IPI) and its derivatives, the cell-of-origin (COO) classification, and the various methods that capture double-hit lymphoma (DHL) and related subtypes.
Despite its venerable age and construction before the introduction of rituximab, the IPI has remained the most important and successful clinical risk stratification tool for DLBCL2,7 and has spawned several variants.8,9 Yet despite its powerful and reproducible prognostic value, it has not, with few exceptions,10,11 translated into therapeutic stratification likely because it does not align directly with the molecular heterogeneity of DLBCL. Indeed, in all the major classification studies, old and new, the genomic classification’s prognostic value is complementary, and not substitutable, to that of the IPI.
The first major step toward deciphering the genomic complexity of DLBCL was taken through the inventive application of gene expression profiling (GEP). Using hierarchical clustering algorithms on cDNA microarrays of DLBCL tumors, two principal subtypes of DLBCL were identified: the germinal center B-cell–like (GCB) and the activated B-cell–like (ABC) subtypes.12,13 This COO classification provided important prognostic information.14,15 With the subsequent development of the popular Hans algorithm that is based on a more wieldy immunohistochemistry (IHC) platform, this classification became more broadly used. However, using GEP as the gold standard, the sensitivity of IHC to assign COO is only ∼70% for the GCB group and ∼90% for the non-GCB group.16 Over the past decade, advancements in GEP technologies, especially the ability to use RNA from formalin-fixed paraffin-embedded tissue, has enabled the development of assays that can be more reliably applied to patient samples.17 The Lymph2Cx assay, for example, utilizes a NanoString platform (NanoString Technologies, Seattle, WA) and provides a highly concordant COO classification compared with GEP on fresh tissue, with consistent results across laboratories.18 Thus, RNA-based approaches have become the standard method to assign COO for research purposes.
In addition to the COO classification, another transcriptional profiling classification, known as comprehensive consensus clustering (CCC), has identified distinct variants of DLBCL.19 This classification identified important distinctions in predominant fuel utilization pathways associated with the presence or absence of B-cell receptor signaling and features of the tumor immune/inflammatory infiltrate.19-21 While CCC, like COO, identifies important biologic heterogeneity within DLBCL, it has had a more limited role in clinical practice to date and has not been used to test individualized treatment approaches.
Other biologic features have also emerged as key contributors to prognosis. The presence of MYC translocations has been shown to portend an unfavorable outcome.22,23 Tumors that harbor translocations in MYC and translocations in BCL-2 or BCL-6, the so-called double- (or triple-) hit lymphoma (DHL/THL) have been reproducibly shown to be more chemorefractory.24,25 Intensified regimens, such as dose-adjusted rituximab plus etoposide, prednisone, vincristine, cyclophosphamide, and doxorubicin (R-EPOCH), have been associated with a better outcome (although only retrospectively) in patients with DHL/THL and are preferentially used in many centers.26-28 While such intensified regimens are not targeted therapy, this represents one of the few examples of genomically stratified therapy in DLBCL. The majority of DHL tumors fall within the GCB subtype of DLBCL.29 Efforts to distinguish DHL from the remainder of GCB DLBCLs using RNA sequencing have led to the recognition of a distinct molecular subgroup characterized by a double-hit signature (DHITSig), which in one study, comprised 27% of GCB DLBCLs (only half of which were DHL).30 Similarly, GEP of a large cohort of DLBCL samples identified a distinct subgroup of patients with a molecular high-grade (MHG) profile, which correlated with a significantly worse progression-free survival (PFS).31 Of note, patients with DHL lacking the MHG signature had a similar outcome to patients with GCB DLBCL, which suggests that chromosomal rearrangements are an imperfect way to identify this more aggressive variant. These studies suggest that more sophisticated tools may better predict the chemorefractoriness exhibited by DHL and may provide a surer way to select patients for intensified chemotherapy today—and for more targeted therapies tomorrow.
In several series, tumors with an increase in both MYC and BCL-2 protein expression determined by IHC, without the rearrangements that define DHL, termed double-expressor lymphomas (DELs), were also associated with inferior outcomes.32,33 DELs usually fall within the non-GCB category, but their adverse prognosis seems to be independent of COO.32 In the Alliance/CALGB 50303 randomized study that compared rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) with R-EPOCH in DLBCL, there was no clear advantage of more intensive therapy in the DEL subgroup (although the trial was not powered to detect such a difference).26 Thus at present, DELs are typically treated like other DLBCLs, and the biologic substructure and clinical relevance of this category remain unclear.
TRIALS INCORPORATING MOLECULAR CLASSIFICATION
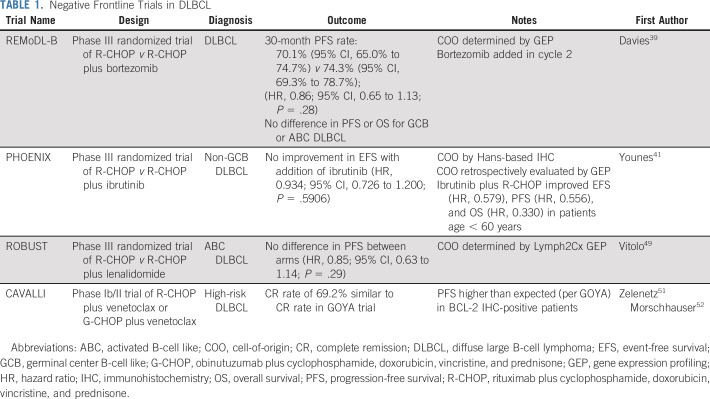
The availability of classifications such as the COO enabled a search for agents with differential activity among different DLBCL subtypes and the incorporation of those agents in frontline therapy.34,35 The main studies are listed in Table 1 and briefly discussed here. Initial studies targeted ABC (or non-GCB) DLBCL, given the generally poorer prognosis associated with this subgroup. Because these tumors often show constitutive activation of nuclear factor-κB (NF-κB), it seemed logical that inhibitors of NF-κB would be selectively more active in ABC DLBCLs. Bortezomib is a proteasome inhibitor that blocks degradation of IκBα, resulting in inhibition of NF-κB activity. Initial studies suggested that bortezomib could overcome the poor prognosis associated with ABC DLBCLs.36,37 However, a randomized phase II trial in patients with non-GCB DLBCL (as determined by IHC) showed no improvement with the addition of bortezomib to chemotherapy.38 More recently, the phase III REMoDL-B trial, which used whole-transcriptome GEP for COO determination, also failed to show an improvement in PFS with the addition of bortezomib, regardless of COO classification.39
TABLE 1.
Negative Frontline Trials in DLBCL
Similarly disappointing results were seen with the Bruton tyrosine kinase (BTK) inhibitor ibrutinib. ABC DLBCLs are also characterized by chronic activation of the B-cell receptor immediately upstream of BTK, which implies possible sensitivity to ibrutinib.40 Here again, preclinical data and a promising phase I/II trial suggested improved responses in ABC DLBCLs.40 This prompted the phase III PHOENIX trial that compared ibrutinib plus R-CHOP with R-CHOP in non-GCB DLBCL.40 Unfortunately, there was no improvement in event-free survival overall.41 Patients were categorized using Hans-based IHC, with retrospective validation of COO by GEP when possible. In exploratory analyses, there seemed to be an advantage of adding ibrutinib in patients < 60 years of age and in patients with DEL, although this benefit was not limited to patients with true ABC tumors.41,42
Lenalidomide, an immunomodulatory agent, was also suggested to have preferential activity in ABC DLBCLs in preclinical models.43 Lenalidomide binds to the E3 ubiquitin ligase complex, modulating its substrate specificity and resulting in the proteasomal degradation of disease-related proteins.44 In ABC DLBCL cells, lenalidomide leads to downregulation of IRF4 and SPIB, transcription factors that together prevent interferon β production and augment NF-κB.45 As with bortezomib, despite an encouraging single-arm phase II study,46 the phase III randomized ROBUST trial in non-GCB DLBCL failed to show an advantage with the addition of lenalidomide to R-CHOP.46-49 These results contrast with those of a randomized phase II study in which the addition of lenalidomide to R-CHOP seemed to confer a PFS benefit, although with a one-sided P value just above the threshold of significance and without clear selectivity for ABC versus GCB.50
In addition to COO, BCL-2 and MYC overexpression and gene rearrangements have been evaluated in studies involving targeted therapy. In the phase Ib/II CAVALLI trial, in which chemoimmunotherapy was combined with the BCL-2 inhibitor venetoclax, the potential predictive impact of COO and DEL was explored.51,52 Compared with historical data from the GOYA trial, outcomes seemed to be improved with the addition of venetoclax in DEL tumors, although not in tumors categorized by COO type. Such cross-trial comparisons must naturally be made with caution, and follow-up randomized studies will be needed to prove the potential therapeutic benefit of venetoclax.53 A trial of venetoclax in combination with R-EPOCH for the treatment of DHL is also ongoing (ClinicalTrials.gov identifier: NCT03036904).
Studies that incorporate targeted therapy in the maintenance setting for patients with DLBCL have also yielded mostly disappointing results to date. The PRELUDE study, for example, found no benefit to adding the PKCβ inhibitor enzastaurin after R-CHOP, regardless of COO subtype.54 The PILLAR-2 trial found no benefit to the addition of everolimus in patients with high-risk DLBCL, as defined by IPI, after completion of frontline therapy.55 While lenalidomide maintenance was associated with an improvement in PFS in elderly patients with DLBCL, the increased toxicity and lack of a documented overall survival benefit have limited the use of this strategy.56,57
In summary, despite robust preclinical justification and considerable investment and enthusiasm, several large and creative phase III trials failed to improve on R-CHOP or provide molecularly stratified therapies in DLBCL, but why? One concern with all studies involving aggressive lymphomas is the biased enrollment of healthier patients into clinical trials. Often, the urgent need to initiate therapy must be weighed against the time required for trial screening, resulting in the exclusion of many patients with clinically aggressive disease and worse expected outcome.58 In addition, there are concerns with regard to the accuracy of COO classification by IHC, which makes it a suboptimal (albeit convenient) method to select patients at trial entry.59 The REMoDL-B study incorporated GEP for COO classification, but given the resulting challenge of incorporating molecular tools in patients in urgent need of therapy, patients did not receive bortezomib until cycle 2 once COO could be determined. One possible solution, adopted in the PHOENIX trial, is to enroll patients on the basis of IHC classification but perform GEP and retroactively analyze the results by GEP classification in prespecified analyses.
While these challenges complicate trial design and interpretation, they are unlikely to entirely explain the failure to develop a successful COO-based frontline treatment selection strategy. Another possible explanation is that the dichotomy of DLBCL into GCB and ABC DLBCL may not capture enough of the underlying molecular heterogeneity of the disease to allow therapeutic targeting. While COO may provide valuable prognostic information, it may not, on an individual level, provide enough granularity to accurately guide treatment selection.
MODERN CLASSIFICATIONS
Technical advances in the past decade have improved the tools available for genomic characterization, allowing for a deeper understanding of recurrent aberrations in DLBCL.60-64 The COO transcriptional framework allowed identification of genetic alterations that are enriched in GCB or ABC tumors.64 More recently, with the inclusion of a broader range of genomic changes in conjunction with new computational tools, gene expression, and functional analyses, our understanding of the genomic underpinnings of DLBCL has considerably deepened.
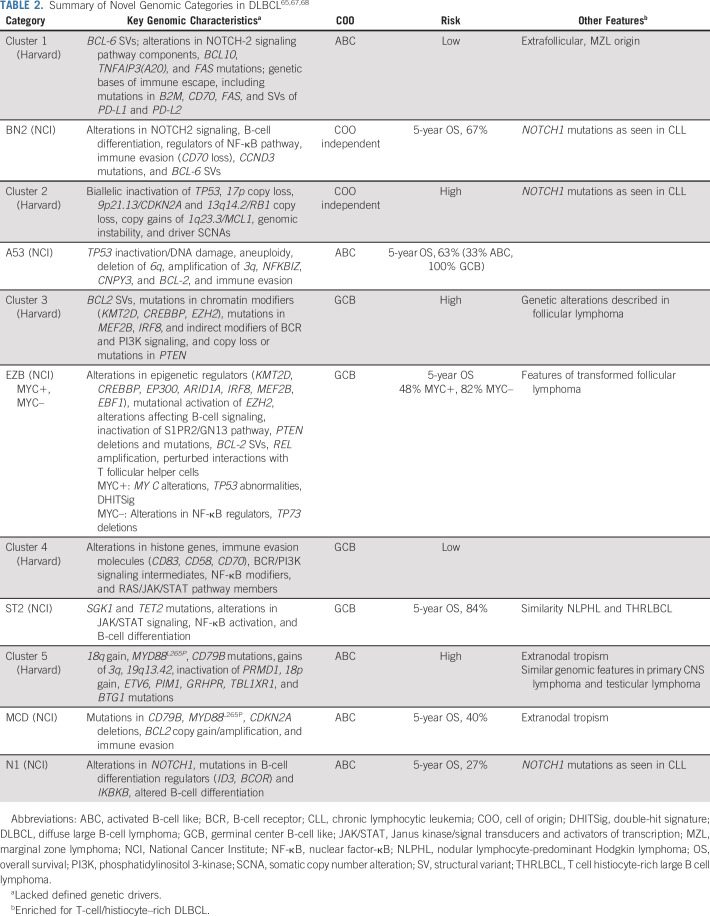
A multiplatform sequencing effort in 304 samples from newly diagnosed patients with DLBCL (herein termed the Harvard cohort) used genomic information to create a novel classification.65 This study encompassed a range of genomic aberrations, including recurrent mutations, somatic copy number alterations (SCNAs), and structural variants (SVs), and classified tumors without a priori consideration of COO grouping. Five genomic clusters were defined, of which the salient features65 are listed in Table 2. Using available RNA-based COO assignments, clusters 1 and 5 were found to be significantly enriched for ABC tumors; in contrast, clusters 3 and 4 were enriched for GCB DLBCLs. In each case, there were significant prognostic differences, with an inferior outcome for patients with cluster 3 and cluster 5 tumors. This study also identified an important group of tumors (cluster 2), comprising approximately one fifth of the original (unselected) tumors, without a clear COO predominance. These results support the view that there is significant genomic variability beyond the COO classification, which may help to explain the variability of the prognostic impact of COO classification across studies and, more importantly, perhaps the difficulty of using the COO to select patients for targeted therapies.34,35 Although co-occurring MYC and BCL-2 alterations occurred most commonly in cluster 3, those with MYC and BCL-6 translocations occurred most commonly in cluster 1, which is biologically distinct from cluster 3. This strengthens the contention that the current concept of DHL may have prognostic relevance but not biologic homogeneity and may not lend itself as a group to a single therapeutic targeting strategy. Finally, an important implication of this study is the need to capture SCNAs and SVs to fully identify coordinate multigene and alteration signatures.
TABLE 2.
A parallel, large-scale sequencing effort from the National Cancer Institute (NCI cohort) similarly highlighted the important residual genomic complexity embedded within traditional COO classification subgroups.67 Genes with recurrent aberrations were identified among 574 DLBCL samples using exome and transcriptome sequencing, array-based copy number analysis, and targeted amplicon resequencing. This multiplatform analysis was layered onto the existing COO classification to define four distinct subgroups. More recently, the same group performed analyses to more fully characterize the genomic underpinnings of their original four subtypes and to broaden the classification scheme to tumors that were not classified in the original analysis.68 As was done in the Harvard analysis, mutations, SCNAs, and fusions were all evaluated as were gene expression signatures. Among the cases not previously characterized in the initial NCI cohort analysis, TP53 was the most frequently mutated gene. In addition, these mutations were associated with high rates of aneuploidy. They therefore defined a fifth group termed A53 (aneuploid with TP53 inactivation). They also created another seed class, ST2 (SGK1 and TET2 mutated). The previously described DHITSig was also evaluated in the context of genomic subgroups, which further stratified the subtype defined by EZH2 and BCL-2 aberrations into MYC+ and MYC− categories. Ultimately, this led the NCI group to define seven genomic subgroups,67,68 as listed in Table 2. These categories more closely aligned with the previously described genomic clusters.65 Using these classifications, the authors created an algorithm, LymphGen, to provide a probabilistic classification of a tumor from an individual patient into a genetic subgroup.
In another study, Lacy et al69 applied a targeted 293-gene panel to a large population-based cohort (n = 928) of patients with DLBCL. This analysis, unlike the two previously described, did not incorporate SCNA or SV information to classify tumors. Despite this, it identified five distinct molecular subgroups, termed MYD88, BCL2, TET2/SGK1, SOCS1/SGK1, and NOTCH2, on the basis of the genetic features enriched in each cluster (Table 2). Approximately one quarter of tumors did not fall within a category and were termed not elsewhere classified. The subgroups generally resembled the previously described NCI and Harvard subgroups.65,67 This analysis did not identify a specific subgroup enriched for TP53 mutations, as was seen in the prior analyses, likely because the analysis did not specifically include SCNAs, which were the most prominent feature of cluster 2 cases in the Harvard analysis.69 Of note, this study identified two different subgroups in which tumors resembled cluster 4 tumors: the TET2/SGK1 subgroup (akin to NCI’s ST2 subgroup)65,69 and the SOCS1/SGK1 subgroup.65,69
In summary, multiple groups have now independently mapped the genomic heterogeneity of DLBCL (Table 2). The resulting classifications have important distinctions and nuances, as briefly mentioned here and listed in Table 2. Altogether, they provide a broad and deep understanding of the biology of DLBCL, much of it beyond the scope of this review. Yet, one remarkable feature is that despite differences in sequencing platforms, types of genomic aberrations evaluated, variant calling algorithms, and methods of statistical analysis, their results are broadly similar, and several subgroups or clusters identified across studies share unmistakable resemblances. In fact, the public availability of sequencing data has already allowed cross-analyses. For example, the Harvard group could recapitulate the 5 clusters using the NCI data set without reference to its original data.70 Similarly, the NCI group found that each LymphGen subtype was drawn predominantly from a single genetic cluster, as defined by the Harvard group, with 75% overall agreement between the analytic methologies.68 The overlapping conclusions of these seminal studies provide convincing support for the presence of previously unrecognized distinct genomic subgroups with coordinate biology.
BRIDGING THE CHASM
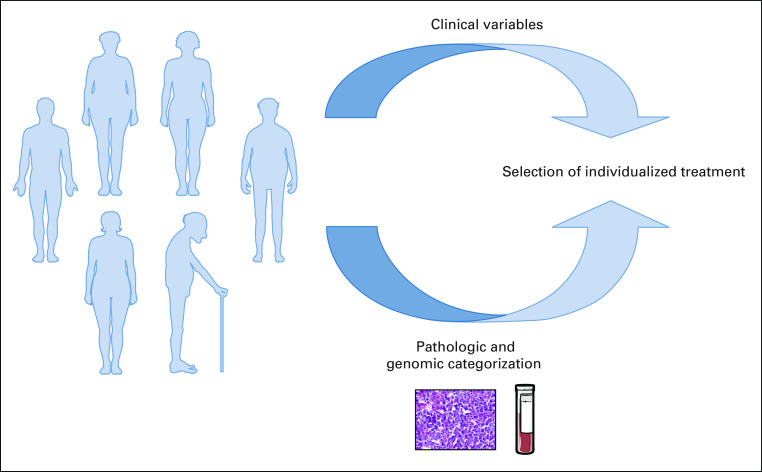
It seems inevitable, on the basis of the results of the aforementioned classification studies, that a more detailed genetic subclassification is poised to supersede the COO framework. At the very least, this should provide a finer way to assess prognosis. But naturally, the greater promise is in its potential to allow for successfully individualized therapy. Indeed, the novel classifications readily provide testable predictions for specific therapeutic vulnerabilities within some of the newly identified groups, which can be tested in preclinical models.71 The ultimate goal can be simply envisioned (Fig 1): On the basis of clinical variables and the genomic classification of an individual patient’s tumor, select a treatment of which at least a component selectively targets the vulnerabilities implied by the genomic abnormalities (eg, R-CHOP plus drug X), and through the optimization of therapy across all subgroups, increase the overall cure rate for all patients.
FIG 1.
Conceptual framework for personalized medicine in diffuse large B-cell lymphoma.
The goal is clear, and with the newfound understanding and classification systems, the tools for building the bridge to it seem to be at hand. Yet before we build, it may be useful to consider the chasm below.
The first two challenges relate to the ease of classification for an individual tumor. First is the challenge of harmonization: While there is clear overlap among the novel genomic classifications, robust clinical implementation will require the development of a common accepted platform for use across studies. The recent NCI analysis, which results in a categorization more similar to that of the Harvard group, may help the field to move toward the implementation of a uniform framework. Without such harmonization, lack of coordination among groups could lead to a profusion of retrospective re-analyses of trial data using different classifiers that only provide exploratory hypotheses that require more trials to definitively confirm. This would ultimately sap a significant amount of research efficiency, especially given the time required at present to design, conduct, and analyze prospective clinical trials. Second is the technical challenge: Both the Harvard and the NCI analyses relied on whole-exome sequencing (WES). This method takes time to result, and time is of the essence in an aggressive disease like DLBCL. As mentioned earlier, the ability to rapidly classify patients who start frontline therapy is likely to be a key ingredient of successful precision approaches in this disease. The most recent study using a targeted next-generation sequencing (NGS) panel instead of WES would allow for a more rapid turnaround time. However, absent SCNA and SV data, such panels do not capture all of the features necessary for accurate classification.69 Similarly, the novel genomic classifications use complex clustering algorithms that are currently limited to research settings. The development of parsimonious classifiers, which recapitulate the power of WES analyses from a smaller number of targeted abnormalities, will allow for more broad applicability of these tools in prospective studies.70 Finally, another practical challenge stems from the difficulty of obtaining biopsy specimens, particularly in a serial fashion, that could capture dynamic changes in a tumor’s genomic profile. The development and availability of liquid biopsies (ie, NGS-based assays of circulating tumor DNA [ctDNA]) that can capture a tumor’s salient genomic characteristics through sampling of peripheral blood will likely be an important contributor to our ability to rapidly and dynamically characterize a tumor’s genomic profile and design more-efficient clinical trials.72-74 Ultimately, the ability to use assays that have a short turnaround time and manageable cost, yet still capture all the essential genomic elements to accurately classify tumors, will be a sine qua non condition of success.
An additional concern is that the genomic subtypes discussed here have all been developed in de novo DLBCL. It remains unknown whether the acquisition of additional genomic aberrations over time and treatment courses alters genomic subgroup classification. If it does, trials in patients with multiply RR disease may not predict well the effectiveness of the new therapies in earlier lines of treatment. This may require a greater willingness to start frontline treatments without demonstrated subgroup-specific benefits in RR disease and the obvious risks this entails. Efforts to sequence and classify samples from patients with RR DLBCL and the use of ctDNA for easier collection and monitoring of clonal evolution will be important to better understand this phenomenon.
Other salient challenges await at the “splitting” end of the granularity spectrum. Launching a phase III clinical trial in DLBCL is relatively straightforward; testing a novel combination in a specific cluster/subgroup, which may represent only ≤ 20% of all patients, less so. Some of this work can be done retrospectively. Indeed, it should be instructive to evaluate the prognostic and possible predictive value of the new classification systems by retrospectively analyzing tumor samples from recent randomized trials. However, the treatments used in these trials were not designed to optimally target any of the new clusters or subgroups, and ultimately, new prospective randomized studies will be required. Those trials, like prior ones, will always fall prey to selection bias and may not faithfully reflect the entire spectrum of patients with DLBCL.
Additional issues will further challenge our ability to leverage the new classifications in prospective attempts to design cluster-specific therapies. Other important aspects of tumors not captured in genomic studies, such as the epigenetic profile and tumor microenvironment (which both may also be altered by prior therapy) may yet create finer subgroups and complicate the task of targeting genomic vulnerabilities.75 Furthermore, it may be naïve to imagine that clinical variables will not remain relevant. Older patients may not tolerate additional therapy as easily; indeed, this is one hypothesis behind the apparent selective benefit of targeted therapy in younger patients in the PHOENIX trial and absence of benefit in the overall population.41 Patients with early-stage disease, with a better prognosis in general, may also not demonstrably benefit from the addition of targeted therapies; combining more selective targeting with chemotherapy de-intensification in this subgroup will be challenging to implement in adequately powered trials. Even factors such as sex and genetic polymorphisms that have an impact on drug metabolism may differentially affect the effectiveness of therapy.76-80 Finally, because R-EPOCH is currently considered by many to be a preferred regimen for patients with DHL, it may be challenging to enroll patients who meet criteria for DHL in a study using an R-CHOP backbone. In fine, splitting clinical trial candidates to the level of our biologic understanding is likely impossible to reconcile with the sample sizes required to power definitive clinical trials. Solving this problem will require some “lumping” that will betray our scientific knowledge. More broadly, it will require improved collaborative structures for running large, multi-arm, flexible, adaptive trials that can seamlessly move from early to late phase, keep up with rapid scientific and technical advances, and recruit from broad populations to ensure adequate power across subgroups. Furthermore, the optimal treatment of a given subgroup/cluster will likely require a combination of targeted therapies, given their coordinate biology (eg, the association of BCL-2 translocation with EZH2 mutations). This will require working with multiple pharmaceutical partners within single trials without sacrificing speed and efficiency and with a clear regulatory path for the approval of such combinations when successful.
Ultimately, the recent genomic studies of DLBCL have dramatically improved our understanding of the complex genomic landscape of this disease. They have revealed the genomic substructure that not only underlies but also extends beyond the COO classification, with clear biologic and prognostic differences among the clusters/subgroups. The insights gained provide a plausible explanation for the negative phase III trial results so far, a new way to retrospectively analyze their findings according to the new classifications, and a platform for designing the next generation of preclinical studies and subsequent prospective clinical trials. But they will only provide the pillars of a bridge to successful personalized therapy, and the chasm is wide. To build a successful bridge across it will require much more from all of us scientifically, technically, and clinically, including, possibly, a deep rethinking of our clinical research methods and research infrastructure in this disease.
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Diffuse Large B-Cell Lymphoma’s New Genomics: The Bridge and the Chasm
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jennifer L. Crombie
Research Funding: Bayer Health (Inst), AbbVie (Inst)
Philippe Armand
Honoraria: Merck, Bristol Myers Squibb
Consulting or Advisory Role: Bristol Myers Squibb, Merck Sharp & Dohme, Adaptive Biotechnologies, Affimed Therapeutics, ADC Therapeutics, Celgene, Daiichi Sankyo, C4 Therapeutics, GenMab, Miltenyi Biotec, Enterome, Tessa Therapeutics, MorphoSys
Research Funding: Merck Sharp & Dohme (Inst), Bristol Myers Squibb (Inst), Tensha Therapeutics (Inst), Roche (Inst), Adaptive Biotechnologies (Inst), Affimed Therapeutics (Inst), Genentech (Inst), IGM Biosciences (Inst), Kite Pharma (Inst)
Travel, Accommodations, Expenses: Bristol Myers Squibb, Merck Sharp & Dohme
No other potential conflicts of interest were reported.
REFERENCES
- 1.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Non-Hodgkin’s Lymphoma Prognostic Factors Project A predictive model for aggressive non-Hodgkin’s lymphoma. N Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 3.Green TM, Young KH, Visco C, et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3460–3467. doi: 10.1200/JCO.2011.41.4342. [DOI] [PubMed] [Google Scholar]
- 4.Gisselbrecht C, Glass B, Mounier N, et al. Salvage regimens with autologous transplantation for relapsed large B-cell lymphoma in the rituximab era. J Clin Oncol. 2010;28:4184–4190. doi: 10.1200/JCO.2010.28.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. doi: 10.1182/blood-2017-03-769620. Crump M, Neelapu SS, Farooq U, et al: Outcomes in refractory diffuse large B-cell lymphoma: Results from the international SCHOLAR-1 study. Blood 130:1800-1808, 2017 [Erratum: Blood 131:587-588, 2018] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neelapu SS, Locke FL, Bartlett NL, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziepert M, Hasenclever D, Kuhnt E, et al. Standard International prognostic index remains a valid predictor of outcome for patients with aggressive CD20+ B-cell lymphoma in the rituximab era. J Clin Oncol. 2010;28:2373–2380. doi: 10.1200/JCO.2009.26.2493. [DOI] [PubMed] [Google Scholar]
- 8.Sehn LH, Berry B, Chhanabhai M, et al. The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2007;109:1857–1861. doi: 10.1182/blood-2006-08-038257. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Z, Sehn LH, Rademaker AW, et al. An enhanced International Prognostic Index (NCCN-IPI) for patients with diffuse large B-cell lymphoma treated in the rituximab era. Blood. 2014;123:837–842. doi: 10.1182/blood-2013-09-524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poeschel V, Held G, Ziepert M, et al. Four versus six cycles of CHOP chemotherapy in combination with six applications of rituximab in patients with aggressive B-cell lymphoma with favourable prognosis (FLYER): A randomised, phase 3, non-inferiority trial. Lancet. 2020;394:2271–2281. doi: 10.1016/S0140-6736(19)33008-9. [DOI] [PubMed] [Google Scholar]
- 11.Récher C, Coiffier B, Haioun C, et al. Intensified chemotherapy with ACVBP plus rituximab versus standard CHOP plus rituximab for the treatment of diffuse large B-cell lymphoma (LNH03-2B): An open-label randomised phase 3 trial. Lancet. 2011;378:1858–1867. doi: 10.1016/S0140-6736(11)61040-4. [DOI] [PubMed] [Google Scholar]
- 12.Scott DW. Cell-of-origin in diffuse large B-cell lymphoma: Are the assays ready for the clinic? Am Soc Clin Oncol Educ Book. 2015:e458–e466. doi: 10.14694/EdBook_AM.2015.35.e458. [DOI] [PubMed] [Google Scholar]
- 13.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 14.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 15.Sehn LH, Gascoyne RD. Diffuse large B-cell lymphoma: Optimizing outcome in the context of clinical and biologic heterogeneity. Blood. 2015;125:22–32. doi: 10.1182/blood-2014-05-577189. [DOI] [PubMed] [Google Scholar]
- 16.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 17.Scott DW, Wright GW, Williams PM, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott DW, Mottok A, Ennishi D, et al. Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol. 2015;33:2848–2856. doi: 10.1200/JCO.2014.60.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Monti S, Juszczynski P, et al. SYK inhibition modulates distinct PI3K/AKT-dependent survival pathways and cholesterol biosynthesis in diffuse large B cell lymphomas. Cancer Cell. 2013;23:826–838. doi: 10.1016/j.ccr.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caro P, Kishan AU, Norberg E, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell. 2012;22:547–560. doi: 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrans S, Crouch S, Smith A, et al. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. J Clin Oncol. 2010;28:3360–3365. doi: 10.1200/JCO.2009.26.3947. [DOI] [PubMed] [Google Scholar]
- 23.Savage KJ, Johnson NA, Ben-Neriah S, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114:3533–3537. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 24.Johnson NA, Savage KJ, Ludkovski O, et al. Lymphomas with concurrent BCL2 and MYC translocations: The critical factors associated with survival. Blood. 2009;114:2273–2279. doi: 10.1182/blood-2009-03-212191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Lin P, Fayad LE, et al. B-cell lymphomas with MYC/8q24 rearrangements and IGH@BCL2/t(14;18)(q32;q21): An aggressive disease with heterogeneous histology, germinal center B-cell immunophenotype and poor outcome. Mod Pathol. 2012;25:145–156. doi: 10.1038/modpathol.2011.147. [DOI] [PubMed] [Google Scholar]
- 26.Bartlett NL, Wilson WH, Jung SH, et al. Dose-Adjusted EPOCH-R compared with R-CHOP as frontline therapy for diffuse large B-cell lymphoma: Clinical outcomes of the phase III Intergroup Trial Alliance/CALGB 50303. J Clin Oncol. 2019;37:1790–1799. doi: 10.1200/JCO.18.01994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oki Y, Noorani M, Lin P, et al. Double hit lymphoma: The MD Anderson Cancer Center clinical experience. Br J Haematol. 2014;166:891–901. doi: 10.1111/bjh.12982. [DOI] [PubMed] [Google Scholar]
- 28.Petrich AM, Gandhi M, Jovanovic B, et al. Impact of induction regimen and stem cell transplantation on outcomes in double-hit lymphoma: A multicenter retrospective analysis. Blood. 2014;124:2354–2361. doi: 10.1182/blood-2014-05-578963. [DOI] [PubMed] [Google Scholar]
- 29.Ennishi D, Mottok A, Ben-Neriah S, et al. Genetic profiling of MYC and BCL2 in diffuse large B-cell lymphoma determines cell-of-origin-specific clinical impact. Blood. 2017;129:2760–2770. doi: 10.1182/blood-2016-11-747022. [DOI] [PubMed] [Google Scholar]
- 30.Ennishi D, Jiang A, Boyle M, et al. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol. 2019;37:190–201. doi: 10.1200/JCO.18.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sha C, Barrans S, Cucco F, et al. Molecular high-grade B-cell lymphoma: Defining a poor-risk group that requires different approaches to therapy. J Clin Oncol. 2019;37:202–212. doi: 10.1200/JCO.18.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson NA, Slack GW, Savage KJ, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staiger AM, Ziepert M, Horn H, et al. Clinical Impact of the Cell-of-Origin Classification and the MYC/ BCL2 Dual Expresser Status in Diffuse Large B-Cell Lymphoma Treated Within Prospective Clinical Trials of the German High-Grade Non-Hodgkin’s Lymphoma Study Group. J Clin Oncol. 2017;35:2515–2526. doi: 10.1200/JCO.2016.70.3660. [DOI] [PubMed] [Google Scholar]
- 34.Goy A. Succeeding in Breaking the R-CHOP Ceiling in DLBCL: Learning from negative trials. J Clin Oncol. 2017;35:3519–3522. doi: 10.1200/JCO.2017.74.7360. [DOI] [PubMed] [Google Scholar]
- 35.Jardin F. Improving R-CHOP in diffuse large B-cell lymphoma is still a challenge. Lancet Oncol. 2019;20:605–606. doi: 10.1016/S1470-2045(19)30021-X. [DOI] [PubMed] [Google Scholar]
- 36.Dunleavy K, Pittaluga S, Czuczman MS, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood. 2009;113:6069–6076. doi: 10.1182/blood-2009-01-199679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruan J, Martin P, Furman RR, et al. Bortezomib plus CHOP-rituximab for previously untreated diffuse large B-cell lymphoma and mantle cell lymphoma. J Clin Oncol. 2011;29:690–697. doi: 10.1200/JCO.2010.31.1142. [DOI] [PubMed] [Google Scholar]
- 38.Leonard JP, Kolibaba KS, Reeves JA, et al. Randomized phase II study of R-CHOP with or without bortezomib in previously untreated patients with non-germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol. 2017;35:3538–3546. doi: 10.1200/JCO.2017.73.2784. [DOI] [PubMed] [Google Scholar]
- 39.Davies A, Cummin TE, Barrans S, et al. Gene-expression profiling of bortezomib added to standard chemoimmunotherapy for diffuse large B-cell lymphoma (REMoDL-B): An open-label, randomised, phase 3 trial. Lancet Oncol. 2019;20:649–662. doi: 10.1016/S1470-2045(18)30935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson WH, Young RM, Schmitz R, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21:922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Younes A, Sehn LH, Johnson P, et al. Randomized phase III trial of ibrutinib and rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in non-germinal center B-cell diffuse large B-cell lymphoma. J Clin Oncol. 2019;37:1285–1295. doi: 10.1200/JCO.18.02403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. doi: 10.1182/bloodadvances.2022009389. Johnson P, Balasubramanian S, Hodkinson B, et al. Clinical Impact of ibrutinib with R-CHOP in untreated non-GCB DLBCL co-expressing BCL2 and MYC genes in the phase 3 PHOENIX trial. Blood 134, 2019 (suppl; abstr 354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang LH, Kosek J, Wang M, et al. Lenalidomide efficacy in activated B-cell-like subtype diffuse large B-cell lymphoma is dependent upon IRF4 and cereblon expression. Br J Haematol. 2013;160:487–502. doi: 10.1111/bjh.12172. [DOI] [PubMed] [Google Scholar]
- 44.Fink EC, Ebert BL. The novel mechanism of lenalidomide activity. Blood. 2015;126:2366–2369. doi: 10.1182/blood-2015-07-567958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y, Shaffer AL, III, Emre NC, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012;21:723–737. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nowakowski GS, LaPlant B, Macon WR, et al. Lenalidomide combined with R-CHOP overcomes negative prognostic impact of non-germinal center B-cell phenotype in newly diagnosed diffuse large B-cell lymphoma: A phase II study. J Clin Oncol. 2015;33:251–257. doi: 10.1200/JCO.2014.55.5714. [DOI] [PubMed] [Google Scholar]
- 47.Czuczman MS, Trněný M, Davies A, et al. A phase 2/3 multicenter, randomized, open-label study to compare the efficacy and safety of lenalidomide versus investigator’s choice in patients with relapsed or refractory diffuse large B-cell lymphoma. Clin Cancer Res. 2017;23:4127–4137. doi: 10.1158/1078-0432.CCR-16-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castellino A, Chiappella A, LaPlant BR, et al. Lenalidomide plus R-CHOP21 in newly diagnosed diffuse large B-cell lymphoma (DLBCL): Long-term follow-up results from a combined analysis from two phase 2 trials. Blood Cancer J. 2018;8:108. doi: 10.1038/s41408-018-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitolo U, Witzig TE, Gascoyne RD, et al. ROBUST: First report of phase III randomized study of lenalidomide/R-CHOP (R2-CHOP) vs placebo/R-CHOP in previously untreated ABC-type diffuse large B-cell lymphoma. Hematol Oncol. 2019;37:36–37. [Google Scholar]
- 50.Nowakowski GS, Hong F, Scott DW, et al. Addition of lenalidomide to R-CHOP (R2CHOP) improves outcomes in newly diagnosed diffuse large B-cell lymphoma (DLBCL): First report of ECOG-ACRIN1412 a randomized phase 2 US Intergroup Study of R2CHOP vs R-CHOP. Hematol Oncol. 2019;37:37–38. [Google Scholar]
- 51.Zelenetz AD, Salles G, Mason KD, et al. Venetoclax plus R- or G-CHOP in non-Hodgkin lymphoma: Results from the CAVALLI phase 1b trial. Blood. 2019;133:1964–1976. doi: 10.1182/blood-2018-11-880526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morschhauser F, Feugier P, Flinn IW, et al: Venetoclax plus rituximab, cyclophosphamide, doxorubicin, vincristine and prednisolone (R-CHOP) improves outcomes in BCL2-positive first-line diffuse large B-cell lymphoma (DLBCL): First safety, efficacy and biomarker analyses from the phase II CAVALLI study. Blood 132, 2018 (suppl; abstr 782) [Google Scholar]
- 53.Vitolo U, Trněný M, Belada D, et al. Obinutuzumab or rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in previously untreated diffuse large B-cell lymphoma. J Clin Oncol. 2017;35:3529–3537. doi: 10.1200/JCO.2017.73.3402. [DOI] [PubMed] [Google Scholar]
- 54.Crump M, Leppä S, Fayad L, et al. Randomized, double-blind, phase III trial of enzastaurin versus placebo in patients achieving remission after first-line therapy for high-risk diffuse large B-cell lymphoma. J Clin Oncol. 2016;34:2484–2492. doi: 10.1200/JCO.2015.65.7171. [DOI] [PubMed] [Google Scholar]
- 55.Witzig TE, Tobinai K, Rigacci L, et al. Adjuvant everolimus in high-risk diffuse large B-cell lymphoma: Final results from the PILLAR-2 randomized phase III trial. Ann Oncol. 2018;29:707–714. doi: 10.1093/annonc/mdx764. [DOI] [PubMed] [Google Scholar]
- 56.Thieblemont C, Howlett S, Casasnovas RO, et al. Lenalidomide maintenance for diffuse large B-cell lymphoma patients responding to R-CHOP: Quality of life, dosing, and safety results from the randomised controlled REMARC study. Br J Haematol. 2020;189:84–96. doi: 10.1111/bjh.16300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thieblemont C, Tilly H, Gomes da Silva M, et al. Lenalidomide maintenance compared with placebo in responding elderly patients with diffuse large B-cell lymphoma treated with first-line rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2017;35:2473–2481. doi: 10.1200/JCO.2017.72.6984. [DOI] [PubMed] [Google Scholar]
- 58.Maurer MJ, Ghesquières H, Link BK, et al. Diagnosis-to-treatment interval is an important clinical factor in newly diagnosed diffuse large B-cell lymphoma and has implication for bias in clinical trials. J Clin Oncol. 2018;36:1603–1610. doi: 10.1200/JCO.2017.76.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutiérrez-García G, Cardesa-Salzmann T, Climent F, et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood. 2011;117:4836–4843. doi: 10.1182/blood-2010-12-322362. [DOI] [PubMed] [Google Scholar]
- 60.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morin RD, Mungall K, Pleasance E, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122:1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. doi: 10.1016/j.cell.2017.09.027. Reddy A, Zhang J, Davis NS, et al: Genetic and functional drivers of diffuse large B cell lymphoma. Cell 171:481-494.e415, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. doi: 10.1038/s41591-018-0016-8. Chapuy B, Stewart C, Dunford AJ, et al: Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 24:679-690, 2018 [Erratum: Nat Med 24:1290-1292, 2018] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reference deleted. [Google Scholar]
- 67.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378:1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. doi: 10.1016/j.ccell.2020.03.015. Wright GW, Huang DW, Phelan JD, et al: A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 37(4):551-568.e514, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lacy SE, Barrans SL, Beer PA, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: A Haematological Malignancy Research Network report. Blood. 2020;135:1759–1771. doi: 10.1182/blood.2019003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chapuy B, Stewart C, Wood T, et al: Validation of the genetically-defined DLBCL subtypes and generation of a parsimonious probabilistic classifier. Blood 134, 2019 (suppl; abstr 920) [Google Scholar]
- 71.Bojarczuk K, Wienand K, Ryan JA, et al. Targeted inhibition of PI3Kα/δ is synergistic with BCL-2 blockade in genetically defined subtypes of DLBCL. Blood. 2019;133:70–80. doi: 10.1182/blood-2018-08-872465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crombie J, Armand P. The emerging role of liquid biopsies in lymphoproliferative disorders. Curr Hematol Malig Rep. 2019;14:11–21. doi: 10.1007/s11899-019-0493-y. [DOI] [PubMed] [Google Scholar]
- 73.Scherer F, Kurtz DM, Newman AM, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med. 2016;8:364ra155. doi: 10.1126/scitranslmed.aai8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scherer F, Kurtz DM, Diehn M, et al. High-throughput sequencing for noninvasive disease detection in hematologic malignancies. Blood. 2017;130:440–452. doi: 10.1182/blood-2017-03-735639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fowler NH, Cheah CY, Gascoyne RD, et al. Role of the tumor microenvironment in mature B-cell lymphoid malignancies. Haematologica. 2016;101:531–540. doi: 10.3324/haematol.2015.139493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ngo L, Hee SW, Lim LC, et al. Prognostic factors in patients with diffuse large B cell lymphoma: Before and after the introduction of rituximab. Leuk Lymphoma. 2008;49:462–469. doi: 10.1080/10428190701809156. [DOI] [PubMed] [Google Scholar]
- 77.Müller C, Murawski N, Wiesen MH, et al. The role of sex and weight on rituximab clearance and serum elimination half-life in elderly patients with DLBCL. Blood. 2012;119:3276–3284. doi: 10.1182/blood-2011-09-380949. [DOI] [PubMed] [Google Scholar]
- 78.Pfreundschuh M, Poeschel V, Zeynalova S, et al. Optimization of rituximab for the treatment of diffuse large B-cell lymphoma (II): Extended rituximab exposure time in the SMARTE-R-CHOP-14 trial of the German High-Grade Non-Hodgkin Lymphoma Study Group. J Clin Oncol. 2014;32:4127–4133. doi: 10.1200/JCO.2013.54.6861. [DOI] [PubMed] [Google Scholar]
- 79.Ghesquieres H, Slager SL, Jardin F, et al. Genome-wide association study of event-free survival in diffuse large B-cell lymphoma treated with immunochemotherapy. J Clin Oncol. 2015;33:3930–3937. doi: 10.1200/JCO.2014.60.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bashash M, Connors JM, Gascoyne RD, et al. Genetic polymorphism at BCL2 as a predictor for rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone efficacy in patients with diffuse large B-cell lymphoma. Haematologica. 2017;102:e199–e202. doi: 10.3324/haematol.2016.159087. [DOI] [PMC free article] [PubMed] [Google Scholar]