Abstract
Coronavirus disease 2019 (COVID-19), which is caused by SARS-CoV-2, has been spreading throughout the world. To date, there are still no approved human vaccines for this disease. To develop an effective vaccine, the establishment of animal models for evaluating post-vaccination immune responses is necessary. In this study, we have identified a CTL epitope in the SARS-CoV-2 spike (S) protein that could be used to measure the cellular immune response against this protein. Potential predicted CTL epitopes of the SARS-CoV-2 S protein were investigated by immunizing BALB/c mice with a recombinant of the receptor-binding domain (RBD) of the S protein. Then, CD8+ T cells specific for S-RBD were detected by stimulating with potential epitope peptides and then measuring the interferon-gamma production. Truncation of this peptide revealed that S-RBD-specific CD8+ T cells recognized a H2-Dd-restricted S526–533 peptide. In conclusion, this animal model is suitable for evaluating the immunogenicity of SARS-CoV-2 vaccines.
Keywords: COVID-19, SARS-CoV-2, Spike protein, CTL epitope
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causal agent of coronavirus disease 2019 (COVID-19), was identified in China in late 2019 [1], [2]. As of July 19, 2020, more than 14,307,865 individuals from multiple countries have been infected by this virus, with related deaths totaling 602,525 (https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6). The COVID-19 pandemic has inflicted unprecedented losses both in terms of human lives and global economies. Currently, there are still no approved vaccines available for controlling the rapid and global spread of this disease. Truly effective vaccines for the prevention of COVID-19 are an urgent need worldwide. Various virus-, DNA-, mRNA-, and protein-based vaccines are being developed throughout the world. Almost all of these vaccines contain the whole spike (S) protein or its receptor-binding domain (RBD) as the antigen. SARS-CoV-2 contains four main structural proteins: the envelope (E), membrane (M), nucleocapsid (N), and S proteins. The RBD of the S protein of SARS-CoV-2 (hereinafter S-RBD) binds specifically and efficiently to angiotensin-converting enzyme 2 (ACE2) and thereby supports the entry of the virus into the host cell. It has been shown that neutralizing antibodies recognizing the RBD or other portions of the S protein block the binding of the protein to ACE2, thus preventing the infection of SARS-CoV-2 and other related coronaviruses [3], [4], [5], [6]. Therefore, many of the SARS-CoV-2 vaccines in development aim to induce S-RBD- or S protein-specific neutralizing antibodies by using the RBD or other parts of the S protein as an immunogen.
However, it was recently revealed that the durability of the humoral immune response to SARS-Cov2 is short [7] (unpublished data at https://doi.org/10.1101/2020.07.09.20148429). Furthermore, researchers have reported that plasma levels of the S1 protein-specific neutralizing antibody were under the detectable level in a number of patients who had recovered from COVID-19 (Wu et al. unpublished data at https://doi.org/10.1101/2020.03.30.20047365). T cells specific to SARS-CoV-2 epitopes have been detected in non-exposed healthy individuals [8]. Moreover, it has been reported that SARS-CoV-2-specific T-cell immune memory is induced in convalescent COVID-19 patients [9]. Furthermore, it has been demonstrated that the percentage of activated and proliferating virus-specific CD8+ T cells, such as spike-specific CD8+ T cells, is higher in mild patients, as compared to that in severe COVID-19 patients [10]. Furthermore, studies of SARS-CoV indicate that cellular immune responses induced by SARS-CoV infections have persisted for 11 years and that memory cytotoxic T lymphocytes (CTLs) contributed to protection from SARS-CoV re-challenge in mice [11], [12]. On the basis of these data, it is considered that the induction of a T-cell immune response against SARS-CoV-2 is important for protecting against COVID-19 development. These data also suggest that the capability of a SARS-CoV-2 vaccine to induce not only a humoral but also a cellular response should be taken into account in the development of vaccines and in their preclinical and clinical evaluations. Some animal models, including macaques and mice, were reported to be useful for the evaluation of the immunogenicity and/or efficacy of SARS-CoV-2 vaccines [6], [13], [14]. In particular, mouse models would be useful for the early research of vaccines. However, methods to measure CD8+ CTL responses to specific SARS-CoV-2-derived antigens in mice have not been established. In particular, methods for evaluating the cellular immune responses against spike protein are essential because SARS-CoV-2 vaccines are being developed using this protein as an immunogen. In this study, we found that an epitope peptide (S526–533) that is located at the C-terminus of S-RBD is recognized by CD8+ CTLs in BALB/c mice in a H2-Dd-dependent manner. This is the first identification of a SARS-CoV-2-derived CTL epitope in mice that allows for the precise evaluation of the capability of candidate vaccines to induce a cellular immune response against S-RBD.
2. Materials and methods
2.1. Mice
Female BALB/c mice were purchased from SLC Japan (Shizuoka, Japan) and used at 7–11 weeks of age. They were maintained at the Animal Center of Nagasaki University Graduate School of Medicine (Nagasaki, Japan). The experimental protocol was approved by the Ethics Review Committee for Animal Experimentation of Nagasaki University Graduate School of Medicine.
2.2. Antibodies and reagents
The S-RBD protein of SARS-CoV-2 (amino acid residues (aa) 319–541) was purchased from RayBiotech (Peachtree Corners, GA, USA). Anti-H2-Kd (SF1-1.1.1, mouse Ig2a) and anti-H2-Kd/Dd (34–1-2S, mouse IgG2a) antibodies were purchased from Invitrogen (Carlsbad, CA, USA). Phycoerythrin (PE)-conjugated anti-CD8 mAb (53–6.7, rat IgG2a) was purchased from Biolegend (San Diego, CA, USA). Allophycocyanin (APC)-conjugated anti-interferon-gamma (anti-IFN-γ; clone XMG1.2) was purchased from eBioscience (San Diego, CA, USA). Synthetic S-RBD peptides (summarized in Table 1 ) and 9 m peptide (QYIHSANVL) were obtained from Genscript (Tokyo, Japan). A synthetic oligonucleotide containing an unmethylated CpG dinucleotide (CpG ODN 1668; 5ʹ-TCCATGACGTTCCTGATGCT-3ʹ) was synthesized by Hokkaido System Science (Sapporo, Japan).
Table 1.
List of tested peptide.
| Peptide ID | start | end | length | sequence |
|---|---|---|---|---|
| #1 | 327 | 335 | 9 | VRFPNITNL |
| #2 | 350 | 358 | 9 | VYAWNRKRI |
| #3 | 369 | 390 | 22 | YNSASFSTFKCYGVSPTKLNDL |
| #4 | 395 | 407 | 13 | VYADSFVIRGDEV |
| #5 | 410 | 418 | 9 | IAPGQTGKI |
| #6 | 444 | 452 | 9 | KVGGNYNYL |
| #7 | 472 | 480 | 9 | IYQAGSTPC |
| #8 | 481 | 505 | 25 | NGVEGFNCYFPLQSYGFQPTNGVGY |
| #9 | 497 | 521 | 25 | FQPTNGVGYQPYRVVVLSFELLHAP |
| #10 | 513 | 538 | 26 | LSFELLHAPATVCGPKKSTNLVKNKC |
| #10–1 | 526 | 533 | 8 | GPKKSTNL |
| #10–2 | 525 | 533 | 9 | CGPKKSTNL |
| #10–3 | 524 | 533 | 10 | VCGPKKSTNL |
| #10–4 | 523 | 533 | 11 | TVCGPKKSTNL |
| #10–5 | 522 | 533 | 12 | ATVCGPKKSTNL |
2.3. Immunization
The BALB/c mice were injected subcutaneously with 20 μg of the S-RBD protein and 40 μg of CpG ODN 1668 in 100 μl PBS in the right flank at weekly intervals.
2.4. Flow cytometry and intracellular cytokine staining
For intracellular cytokine staining, splenocytes were first incubated with the synthetic peptides or the control 9 m peptide for 30 min at 37 °C and then incubated for an additional 16 h with GolgiPlug (BD Biosciences, Franklin Lakes, NJ, USA). These cells were then stained with PE-conjugated anti-CD8 mAb. After permeabilization and fixation using a Cytofix/Cytoperm kit according to the manufacturer’s instructions (BD Bioscience), the cells were stained intracellularly with APC-conjugated anti-IFN-γ mAb. After washing, the cells were analyzed on the FACSCanto II system and the results analyzed with FlowJo software (TOMY Digital Biology, Tokyo, Japan).
2.5. Statistical analysis
Values are expressed as the mean ± standard deviation. Differences between groups were examined for statistical significance using the Student t-test. P values of < 0.05 were considered statistically significant.
3. Results and discussion
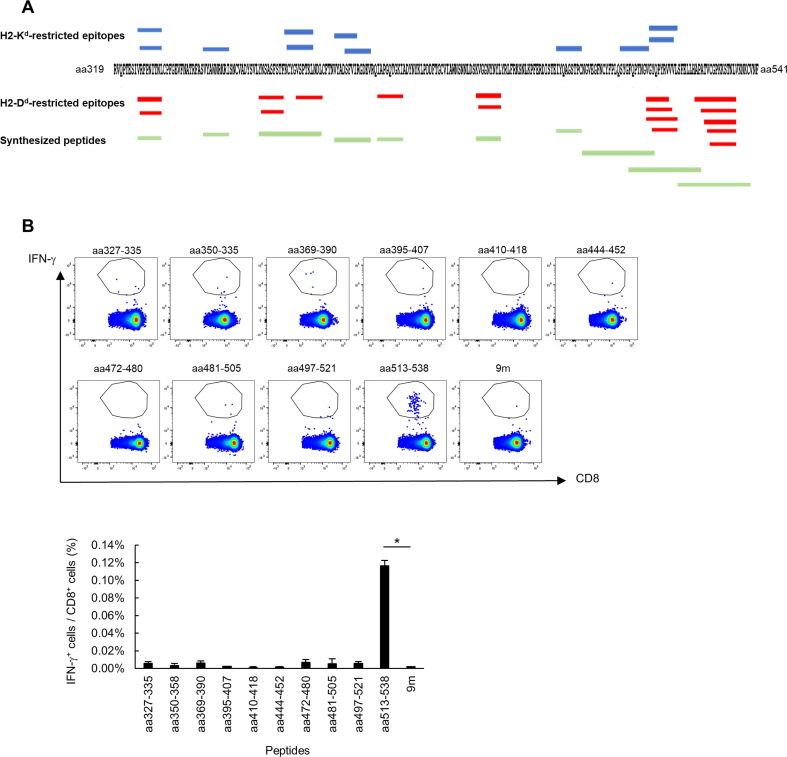
In this study, we attempted to identify minimal epitope(s) in the S-RBD of SARS-CoV-2 that would be recognized by CD8+ T cells in mice immunized with the protein. A recombinant S-RBD protein (aa 319–541) produced in mammalian cells was subcutaneously injected together with CpG ODN 1668 into naive BALB/c mice, twice weekly. At 1 week after the last immunization, the mice were sacrificed and their spleens were removed. Specific CD8+ T-cell induction in the splenocytes was then analyzed by intracellular staining for IFN-γ after stimulation of the cells with short and long peptides in vitro. These peptides included potential S-RBD-derived epitopes that were predicted by the Immune Epitope Database (IEDB) analysis resource (http://www.iedb.org/) to bind to murine major histocompatibility complex (MHC) class I (Table 2 ). We prepared 10 peptides (peptide #1 to #10) that covered all predicted epitopes with a score of greater than 0.15 and a percentile rank of < 0.5 (Table 1 and Fig. 1 A). As shown in Fig. 1B, the CD8+ T cells in splenocytes from the immunized mice produced a significant amount of IFN-γ when stimulated with peptide #10 (sequence: LSFELLHAPATVCGPKKSTNLVKNKC), suggesting the presence of minimal epitopes in the C-terminus region of S-RBD.
Table 2.
Predicted epitopes for spike protein of SARS-CoV-2 (aa319-541).
| Allele | start | end | length | Sequence | score | rank |
|---|---|---|---|---|---|---|
| H2-Kd | 350 | 358 | 9 | VYAWNRKRI | 0.745883 | 0.04 |
| H2-Kd | 504 | 512 | 9 | GYQPYRVVV | 0.740992 | 0.04 |
| H2-Dd | 525 | 533 | 9 | CGPKKSTNL | 0.537362 | 0.02 |
| H2-Dd | 382 | 390 | 9 | VSPTKLNDL | 0.499642 | 0.02 |
| H2-Kd | 379 | 387 | 9 | CYGVSPTKL | 0.498969 | 0.14 |
| H2-Kd | 494 | 503 | 10 | SYGFQPTNGV | 0.457492 | 0.17 |
| H2-Dd | 524 | 533 | 10 | VCGPKKSTNL | 0.432674 | 0.03 |
| H2-Kd | 327 | 335 | 9 | VRFPNITNL | 0.3807 | 0.22 |
| H2-Kd | 395 | 402 | 8 | VYADSFVI | 0.372275 | 0.22 |
| H2-Dd | 505 | 513 | 9 | YQPYRVVVL | 0.35273 | 0.06 |
| H2-Kd | 504 | 513 | 10 | GYQPYRVVVL | 0.339094 | 0.26 |
| H2-Kd | 328 | 335 | 8 | RFPNITNL | 0.319452 | 0.28 |
| H2-Kd | 399 | 407 | 9 | SFVIRGDEV | 0.319166 | 0.28 |
| H2-Dd | 503 | 511 | 9 | VGYQPYRVV | 0.313391 | 0.1 |
| H2-Dd | 503 | 513 | 11 | VGYQPYRVVVL | 0.303071 | 0.11 |
| H2-Kd | 378 | 387 | 10 | KCYGVSPTKL | 0.265013 | 0.36 |
| H2-Dd | 369 | 377 | 9 | YNSASFSTF | 0.245941 | 0.17 |
| H2-Dd | 410 | 418 | 9 | IAPGQTGKI | 0.242132 | 0.18 |
| H2-Dd | 445 | 452 | 8 | VGGNYNYL | 0.233027 | 0.19 |
| H2-Dd | 523 | 533 | 11 | TVCGPKKSTNL | 0.213929 | 0.24 |
| H2-Kd | 472 | 480 | 9 | IYQAGSTPC | 0.204669 | 0.48 |
| H2-Dd | 327 | 335 | 9 | VRFPNITNL | 0.20042 | 0.28 |
| H2-Dd | 522 | 533 | 12 | ATVCGPKKSTNL | 0.188822 | 0.32 |
| H2-Dd | 328 | 335 | 8 | RFPNITNL | 0.156347 | 0.45 |
| H2-Dd | 520 | 533 | 14 | APATVCGPKKSTNL | 0.155495 | 0.46 |
| H2-Dd | 444 | 452 | 9 | KVGGNYNYL | 0.153646 | 0.47 |
| H2-Dd | 370 | 377 | 8 | NSASFSTF | 0.152266 | 0.48 |
| H2-Dd | 503 | 510 | 8 | VGYQPYRV | 0.151477 | 0.49 |
Fig. 1.
S-RBD-specific CD8+ T cells recognize the C-terminus region of the S-RBD protein. (A) Mapping of the predicted epitopes and synthesized peptides. The red and blue lines show the predicted epitopes (H2-Kd and H2-Dd, respectively) and the green lines show the synthesized peptides. (B) BALB/c mice were immunized with the S1 protein twice weekly. Splenocytes were obtained and specific T-cell induction was analyzed with intracellular cytokine staining assays, using stimulation with the indicated peptides. These experiments were repeated two times with similar results. Data are the mean ± SD. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
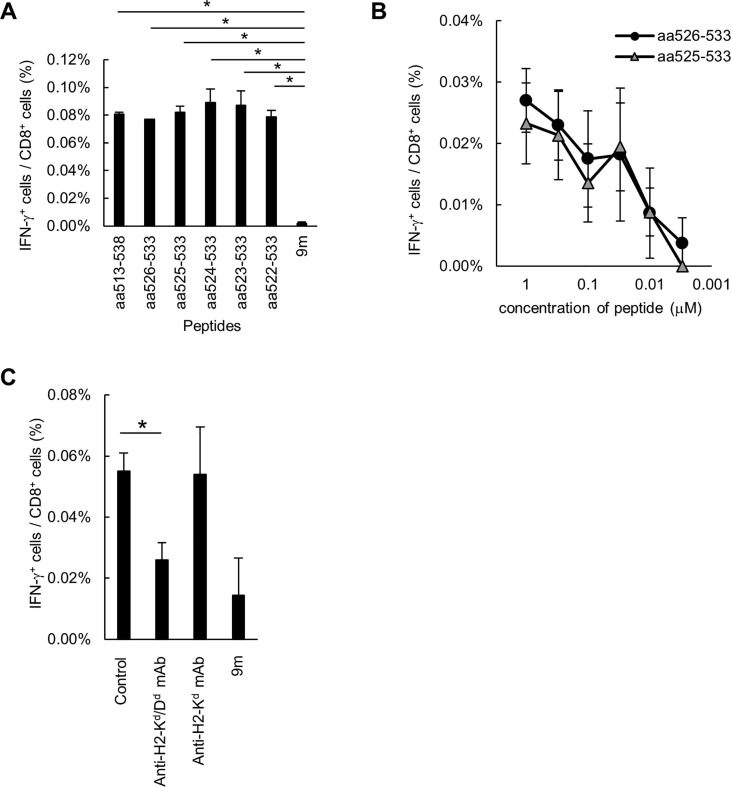
In a study by Zhi et al. [15], the SARS-CoV S peptide CYGVSATKL (aa 366–374) was recognized by CTLs in mice vaccinated with an adenoviral vector encoding the wild-type S protein of the virus. By contrast, a peptide of the homologous region on the SARS-CoV-2 S protein (CYGVSPTKL; aa 379–387) was not recognized by CTLs in our study. The replacement of residue A371 by K or D in this epitope of SARS-CoV led to decreased recognition of the peptide by CTLs, suggesting that residue A371 is critical for epitope recognition by CTLs [16]. To determine the minimal epitope(s) required, we synthesized five truncated peptides of peptide #10 (shown in Table 1) and used them to stimulate CTLs from the immunized mice in vitro. Although peptide #10–1 had an IEDB score of 0.0994 and a percentile rank of 0.97, all the short peptides tested were recognized at a similar degree by CTLs (Fig. 2 A). We next tested lower concentrations of the short peptides, whereupon peptides #10–1 (S526–533, 8-mer) and #10–2 (S525–533, 9-mer) were found to be well recognized by CTLs in concentrations as low as 0.03 nM (Fig. 2B). Because peptide #10–1 was shorter, it was considered the minimal epitope for CTLs. We then assessed the MHC restriction of this recognition using MHC blocking antibodies. The specific CD8+ T-cell response to peptide #10–1 was suppressed by the addition of the anti-H2-Kd/Dd mAb, but not the anti-H2-Kd mAb (Fig. 2C), indicating that this peptide was a H2-Dd-restricted epitope. Taken together, the results indicated that we had succeeded in identifying a H2-Dd-restricted S526–533 peptide as an epitope for SARS-CoV-2 S-RBD-specific CD8+ T cells in mice.
Fig. 2.
Peptide S1526–533 is identified as the H2-Dd-restricted CD8+ CTL epitope. (A, B) BALB/c mice were immunized with the S1 protein twice weekly. Splenocytes were obtained and specific T-cell induction was analyzed with intracellular cytokine staining assays, using stimulation with the indicated peptides. (B) The avidity of the induced S-RBD-specific CD8+ T cells was analyzed with intracellular cytokine staining assays, using stimulation by graded doses of the peptides (ranging from 1 to 0.003 µM). (C) BALB/c mice were immunized with the S1 protein twice weekly. Splenocytes were obtained and specific T-cell induction was stimulated with S1526–533 peptides. MHC restriction of the induced S-RBD-specific CD8+ T cells was analyzed with the intracellular cytokine staining assay after the addition of anti-H2-Kd/Dd or anti-H2-Kd mAbs. These experiments were repeated two times with similar results. Data are the mean ± SD.
In conclusion, we have established a BALB/c mouse model for the evaluation of CTL responses to the S-RBD of SARS-CoV-2. This model could be used to confirm the immunogenicity of candidate vaccines in terms of CTL induction, which has become more recognized as an important mechanism for SARS-CoV-2 eradication. Although a mouse model of SARS-CoV-2 infection has already been established [6], [17], it does not reveal whether the virus-specific CTLs influence the pathology of COVID-19 or contribute to the elimination of the virus, as none of the currently available methods can monitor virus-specific CD8+ T cells. Our mouse model would be useful for addressing these issues by enabling analysis of the distribution, function, and phenotype of specific CD8+ T cells in SARS-CoV-2-infected mice. This would be facilitated by the generation of a peptide:MHC tetramer using the epitope peptide identified in this study. In future, the strategy of epitope identification used in this study would also be useful for the determination of human CTL epitopes in SARS-CoV-2 when used in combination with HLA transgenic mice [18]. Together, these strategies could help toward a more efficient and precise development of SARS-CoV-2 vaccines.
Author contributions
DM and NH designed the experiments; DM and DS performed the experiments; DM analyzed the data; NH, SS, KA and HI contributed reagents, materials, and analytical tools; DM and NH wrote the original draft of the paper; all authors approved the final draft for journal submission.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: [NH is CEO and CTO of United Immunity, Co., Ltd. The other authors have no conflicts of interest related to this study to declare.].
Acknowledgements
We thank YS for excellent technical assistance. This study was supported by Grants-in-Aid for Scientific Research (C) (DM).
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. 579:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727–33. http://doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed]
- 3.Sui J., et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. PNAS. 2004 doi: 10.1073/pnas.0307140101. 101:2536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y., Jiang S., Du L. Prospects for a MERS-CoV spike vaccine. Expert Rev Vaccines. 2018 doi: 10.1080/14760584.2018.1506702. 17:677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Structure Veesler D. Function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(281–2) doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassan AO, et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 2020 [in press]. http://doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed]
- 7.Long QX, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020 [in press]. http://doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed]
- 8.Grifoni A., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(1489–501):e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Fan, et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct Target Ther. 2020;5:156. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swadling Leo, Maini Mala K. T cells in COVID-19 — united in diversity. Nat Immunol. 2020 doi: 10.1038/s41590-020-0798-y. [DOI] [PubMed] [Google Scholar]
- 11.Li C.K., et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008 doi: 10.4049/jimmunol.181.8.5490. 181:5490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng O.W., et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016 doi: 10.1016/j.vaccine.2016.02.063. 34:2008–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrashekar A, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 2020 [in press]. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed]
- 14.Yu J, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020 [in press]. http://doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed]
- 15.Zhi Y., et al. Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology. 2005 doi: 10.1016/j.virol.2005.01.050. 335:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J., Cao Y., Bu X., Wu C. Residue analysis of a CTL epitope of SARS-CoV spike protein by IFN-gamma production and bioinformatics prediction. BMC Immunol. 2012 doi: 10.1186/1471-2172-13-50. 13:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020 [in press]. http://doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed]
- 18.Miyahara Y., et al. Determination of cellularly processed HLA-A2402-restricted novel CTL epitopes derived from two cancer germ line genes, MAGE-A4 and SAGE. Clin Cancer Res. 2005 doi: 10.1158/1078-0432.CCR-04-2585. 11:5581–9. [DOI] [PubMed] [Google Scholar]