Abstract
Background:
Octamer binding transcription factor 4 (Oct4) is critically important in the development and progression of cancer, and is considered a potential biomarker for tumor prognosis. However, the prognostic value of Oct4 in patients with solid tumors remains elusive. Herein, we conducted a meta-analysis to assess the prognostic value of Oct4 in patients with solid tumors.
Methods:
We conducted a literature search on PubMed, Embase, and Web of Science databases to retrieve comprehensive and eligible studies published until December 2019. The study was conducted per the Preferred Reporting Items for Systematic Reviews and Meta-Analysis guidelines. The pooled hazard ratios (HRs) with 95% confidence intervals (CIs) of overall survival (OS) and disease-free survival (DFS)/recurrence-free survival (RFS)/progress-free survival (PFS) were used to evaluate the prognostic value of Oct4 in patients with solid tumors via either random or fixed-effects models.
Results:
In total, 36 studies with 5198 patients were included in the meta-analysis. Notably, elevated Oct4 expression was associated with worse OS (pooled HR: 2.02, 95% CI: 1.55–2.62, P < .001) and DFS/RFS/PFS (pooled HR: 2.34, 95% CI: 1.88–2.92, P < .001).
Conclusion:
This work demonstrated that patients with solid tumors show high expression of Oct4 which is linked to worse prognosis in patients with solid tumors including hepatocellular carcinoma (OS, DFS/RFS/PFS), esophageal squamous cell carcinoma (OS), gastric cancer (OS), cervical cancer (OS, DFS/RFS/PFS), and colorectal cancer (OS, DFS/RFS/PFS), this implicated Oct4 as a potential biomarker to predict the prognosis of tumors.
Keywords: meta-analysis, octamer transcription factor 4, prognosis, solid tumor
1. Introduction
Among the several deadly diseases, neoplasm is highly associated with frequent death cases. The GLOBOCAN report released on September 12, 2018 showed that 18.1 million people were diagnosed with cancers, and 9.6 million deaths occurred, this was based on studies conducted from 185 countries.[1] Although researchers have focused on exploring the diagnosis and treatment of cancers, information on the clinical outcome and prognosis of cancer patients remains scanty. Cancer-associated biomolecules participate in proliferation, invasion, and metastasis of tumors and can be utilized as biomarkers, however, only a few have been used clinically. Thus, there is an urgent need to uncover additional valuable biomolecules that can accurately predict the prognosis and biological behavior of tumors at an early stage. The relationship between stem cells and cancers has been elucidated through an in-depth exploration of stem cells. Many researchers previously hold the opinion that cancer is related to stem cells, initially referred to as cancer stem cells (CSCs). Notably, CSCs are rare in cancer tissues,[2] though they have differentiation potential and self-renewal capacity, thus play a role in recurrence, metastasis, heterogeneity, multidrug resistance, and radiation resistance of tumors.[3,4]
Octamer binding transcription factor 4 (Oct4) is encoded by the Pit-Oct-Unc domain, class 5, transcription factor (POU5F1) gene, which is located on chromosome 6p21 and 17B1 in human and mouse genome, respectively.[5] Oct4 has previously been expressed in the embryonic stem cells (ESCs), germline stem cells, and CSCs.[6–8] During embryo development, the expression of Oct4 impacts the differentiation and dedifferentiation of ESCs, thereby maintaining the self-renewal ability of ESCs. Besides, Oct4 is highly expressed in germ cell tumors and embryonic cell tumors thus is considered a potential molecular marker of tumor germ cells.[5] In the recent past, some studies reported that Oct4 was highly expressed in CSCs.[9,10] CSCs could evade the lethality of radiation and chemotherapeutic agents more easily compared to other tumor cells, this was attributed to self-renewal ability, metastasis to distant sites, and infinite proliferation.[11,12] Further, the study inferred Oct4-positive cancer cells likely represent CSCs.[11,12] Other investigations have revealed that Oct4 is expressed abnormally in solid cancers, such as cervical carcinoma,[13] gastric carcinoma,[14] bladder carcinoma[15] among others.
Oct4 drives stemness in CSCs and has a potential role in chemoresistance and clinical prognostic value of cancer patients.[16] Of note, Oct4 expression has been revealed to be significantly correlated with tumor size, histological differentiation, and primary tumor classification.[17,18] However, the prognostic value of Oct4 is unclear. Several literature findings reveal that low overall survival (OS) and disease-free survival (DFS)/recurrence-free survival (RFS)/progress-free survival (PFS) is related to Oct4 overexpression in many types of tumors.[19–21] For instance, reports from 2 studies indicated that Oct4 overexpression was not significantly associated with OS in advanced small cell lung cancer and tongue squamous cell carcinoma (TSCC).[22,23] In hypopharyngeal squamous cell carcinoma and oral squamous cell carcinoma (OSCC), higher expression of Oct4 indicated a better prognosis.[24,25] This prompted us to conduct a meta-analysis aimed at evaluating the prognostic value of Oct4 in solid tumors.
2. Material and methods
2.1. Search strategy and ethical approval
Here, 2 authors (XY Zhao and Y Sun) independently retrieved articles published until December 1, 2019 from electronic databases (Pubmed, Embase, and Web of Science) by conducting a systematic search. The following strategies based on keywords and Mesh terms were used to identify eligible studies: “Pou5f1 OR Oct3 OR Oct4” AND “Neoplasia OR Neoplasias OR Tumor OR Cancer OR Cancers OR Malignant Neoplasms OR Malignant Neoplasm OR Neoplasm OR Neoplasms OR Malignant OR Malignancy OR Malignancies” AND “Prognoses OR Prognostic Factors OR Factor, Prognostic OR Factors, Prognostic OR Prognostic Factor OR outcome OR survival”. Furthermore, the reference lists in eligible researches were carefully scrutinized not to miss out on pertinent studies. In this study, all the materials are based on published articles, and no patients or animals involved, thus, ethics approval is not necessary.
2.2. Inclusion and exclusion criteria
The inclusion criteria were as follows:
-
1.
the research published in English
-
2.
studies involving cohorts;
-
3.
the value of Oct4 expression in solid tumors was shown;
-
4.
the full text of the article can be found;
-
5.
sufficient and effective data, such as Kaplan–Meier (KM) plot, hazard risk (HR) or relative risk (RR), or odds ratio (OR) with 95% confidence interval (CI).
The exclusion criteria were as follows:
-
1.
repeated researches;
-
2.
basic and animal articles;
-
3.
conference abstracts, reviews, case reports;
-
4.
HR/RR/OR and 95% CI could not be obtained via evaluation.
The retrieved articles were screened carefully by 2 authors (XY Zhao and H Lu). A third author (HF Wang) was consulted to solve any conflicting searches.
2.3. Data extraction
Data on the first authors name, the year of publication, region of the population enrolled, cancer type, sample size, tumor stage, the maximum month of follow-up, detection method, Oct4 (−/+), the cut-off value of Oct4 overexpression, multivariate analysis (yes/no), the source of HR/RR/OR, HR/RR/OR and corresponding 95% CIs for OS/DFS/RFS/PFS were collected independently by 3 investigators (XY Zhao, H Lu, and L Liu). When a study provided data on univariate and multivariate analysis, then, the latter would be selected considering the influence of confounding factors which potentially gave inaccurate results. Besides, if a study did not provide survival data, KM curves would be used to obtain consequences of interest in line with methods suggested by Tierney et al.[26]
2.4. Quality assessment
The Newcastle-Ottawa scale (NOS) adopted to evaluate the quality of each included study, 2 investigators (XY Zhao, Y Sun) completed this part. A third author (HF Wang) was consulted to settle any conflicting findings. The NOS scores ranged from 0 to 9 including 3 categories for cohort researches (the selection of study groups, comparability of groups, and ascertainment of outcomes),.[27] Studies with scores ≥6 were treated to be high-quality.
2.5. Statistical analysis
The STATA version 12.0 (Stata Corporation, College Station, TX, USA) was used to analyze all statistical data. We used HRs with 95% CIs extracted from selected studies to evaluate the prognosis value. Since the outcome of the tumor is rare in all populations, differences between the OR, RR, and HR could generally be ignored, the pooled ORs or RRs with 95% CIs were suitable for the assessment thus were treated as HRs for data analysis.[28] The Chi-Squared (evaluating the P value) and I2 tests among studies were used to evaluate heterogeneity results, this indicated significant heterogeneity if the I2 ≥ 50% and P ≤ .05. Then, the random-effects model (the DerSimonian-Laird method) was used to analyze the pooled HRs. Otherwise, the fixed-effects model (Mante-Haenszel method) was selected (I2 < 50% and P > .05). Subgroup analysis and meta-regression analysis were conducted to ascertain the source of heterogeneity. Moreover, we conducted a sensitivity analysis by independently eliminating each study. Publication bias was evaluated using Funnel plots, Eggers and Begg tests. P < .05 was considered statistically significant.
3. Results
3.1. Characteristics
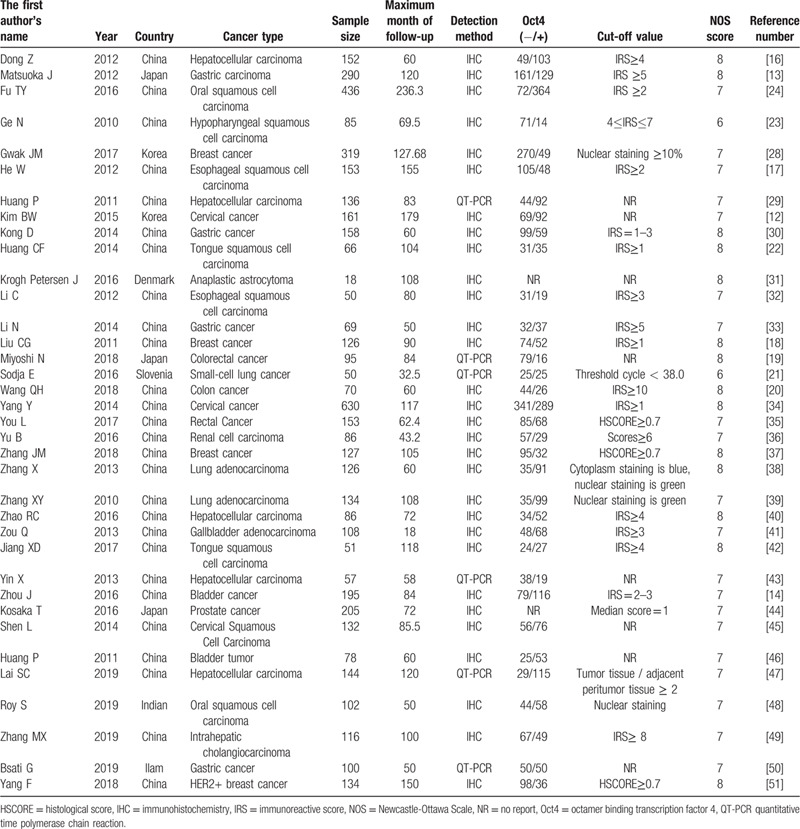
A total of 3222 articles were retrieved from the database via the search strategy. Following the above inclusion and exclusion criteria, 36 articles[14,15,17,18,22–25,29–55]were enrolled in this meta-analysis (Fig. 1) and had 5198 cancer patients from China, Korea, Slovenia, Iran, Denmark, and Japan, who had been diagnosed with all types of solid tumors involving hepatocellular carcinoma (HCC), gastric cancer (GC), OSCC, esophageal squamous cell carcinoma (ESCC), cervical cancer, TSCC, breast carcinoma, colorectal carcinoma, gallbladder carcinoma, lung carcinoma, bladder carcinoma, anaplastic astrocytoma, prostate cancer, renal cell carcinoma, and so on. The key characteristics of the 36 articles are summarized in Table 1. Notably, 31 articles reported that Oct4 expression was associated with OS, whereas 12 articles revealed that Oct4 expression was correlated with DFS/RFS/PFS. The majority of studies reported HRs directly, however, in 2 studies, HRs were indirectly estimated by the KM survival curves. The NOS scores of 36 studies ranged from 6 to 9, an indication that each study adopted a reliable methodology thus was suited for further analyses.
Figure 1.
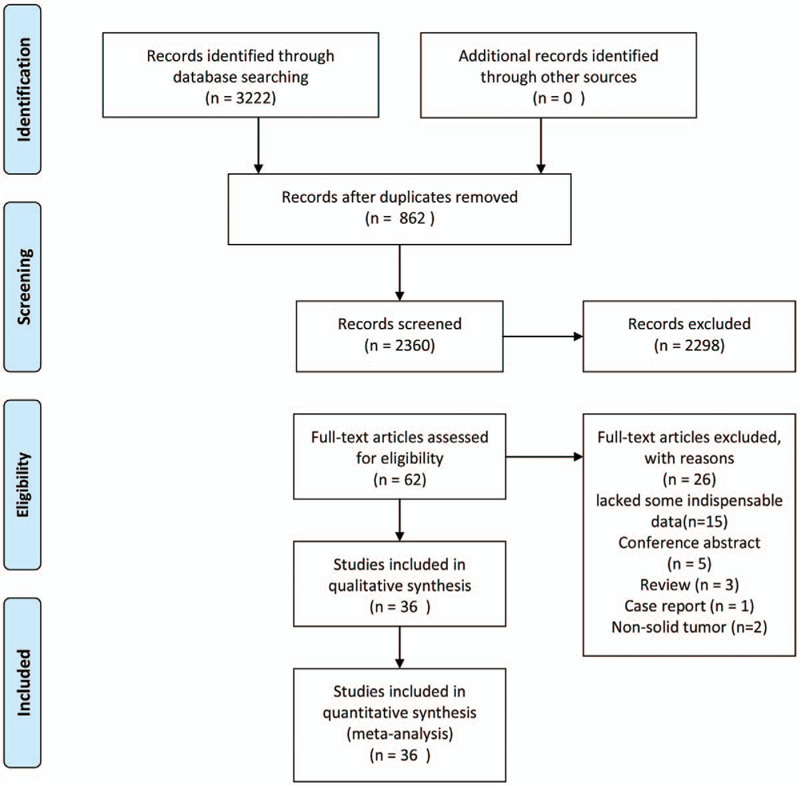
Flow diagram for the selection of studies in the meta-analysis.
Table 1.
Characteristics of the included studies.
3.2. High expression of Oct4 for OS and DFS/RFS/PFS
Upon analysis of data retrieved from 31 articles with a total of 4395 patients, we revealed that Oct4 overexpression was remarkably associated with worse OS in patients with solid tumors. The pooled HR was 2.02 (95% CI: 1.55–2.62, P < .001) (Fig. 2). We adopted a random-effects model to pool HRs because of existing apparent statistical heterogeneity (I2 = 82.3%, P < .001) in these studies. However, due to the small number of studies on the relationship between Oct4 and DFS/RFS/PFS, DFS was combined with RFS/PFS and defined as the “DFS/RFS/PFS” group. Of note, 12 studies with 1569 patients reported on the association of Oct4 overexpression with DFS/RFS/PFS. In these studies, there was no apparent heterogeneity (I2 = 15.60%, P = .291), thus we applied the fixed-effects model, which revealed that high expression of Oct4 was remarkably associated with to poor DFS/RFS/PFS in patients with solid tumors. The pooled HR was 2.34 (95% CI: 1.88–2.92, P < .001), (Fig. 3).
Figure 2.
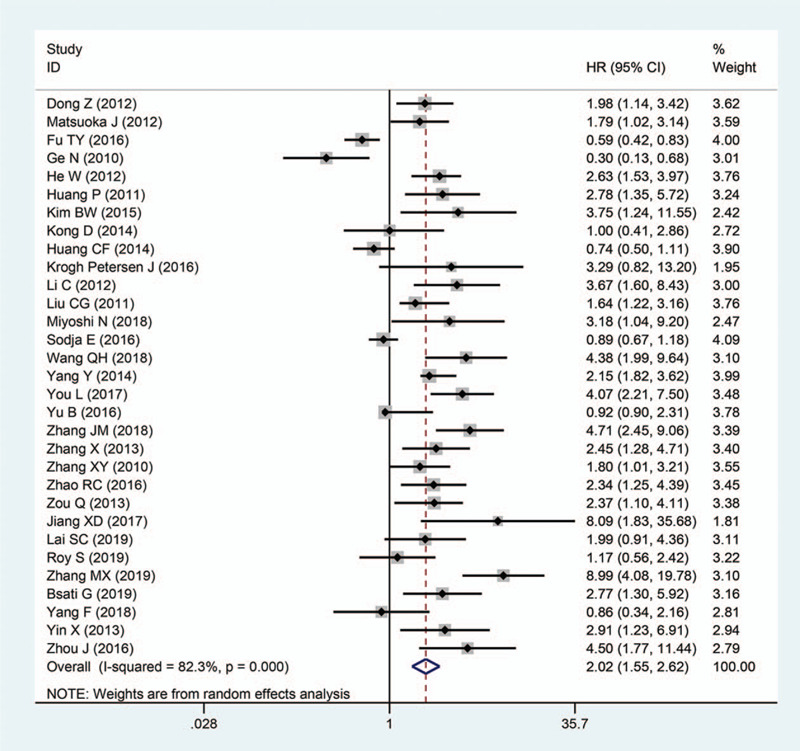
Forest plots showing the relationship between Oct4 and OS.
Figure 3.
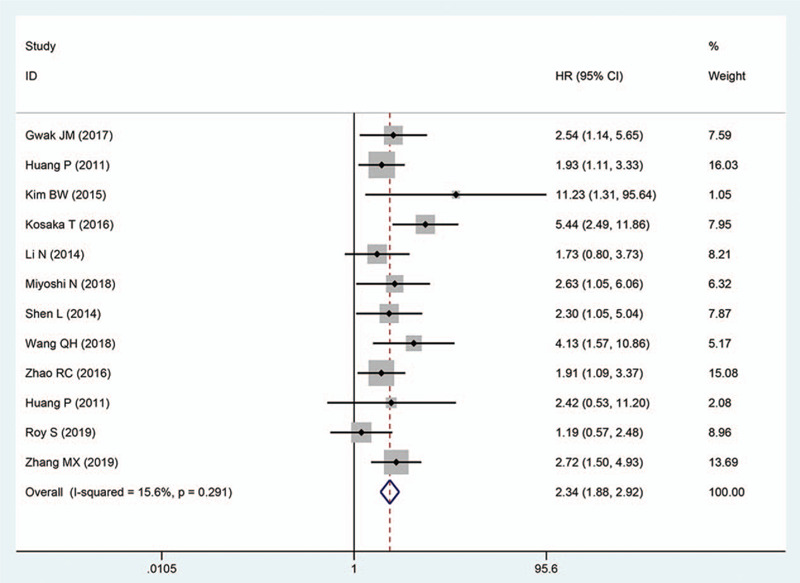
Forest plots showing the relationship between Oct4 and DFS/RFS/PFS.
3.3. Subgroup and meta-regression analyses
On account of the conspicuous heterogeneity in these studies, we conducted subgroup analysis via the random-effects model for OS considering the following parameters: tumor types, digestive system tumor, sample size, the maximum month of follow-up, and the source of HR. In accordance with tumor type, the elevated Oct4 levels demonstrated a worse prognosis in patients with HCC (pooled HR: 2.30; 95% CI: 1.69–3.12; P < .001), GC (pooled HR: 1.81; 95% CI: 1.12–2.95; P = .016), ESCC (pooled HR: 2.85; 95% CI: 1.89–4.32; P < .001), cervical cancer (pooled HR: 2.26; 95% CI: 1.63–3.15; P < .001) and colorectal cancer (pooled HR: 4.00; 95% CI: 2.57–6.22; P < .001) (Fig. 4). Nevertheless, no significant relationship was found between the overexpression of Oct4 and OS in breast cancer, TSCC, OSCC, and lung carcinoma. Other outcomes of subgroup analysis are highlighted in Table 2. All frost plots of different subgroups on the association of Oct4 overexpression with OS are displayed in Fig. 5. Considering the significant heterogeneity, we performed a meta-regression analysis for OS. In general, the differences were not statistically significant in OS as shown in Table 2.
Figure 4.
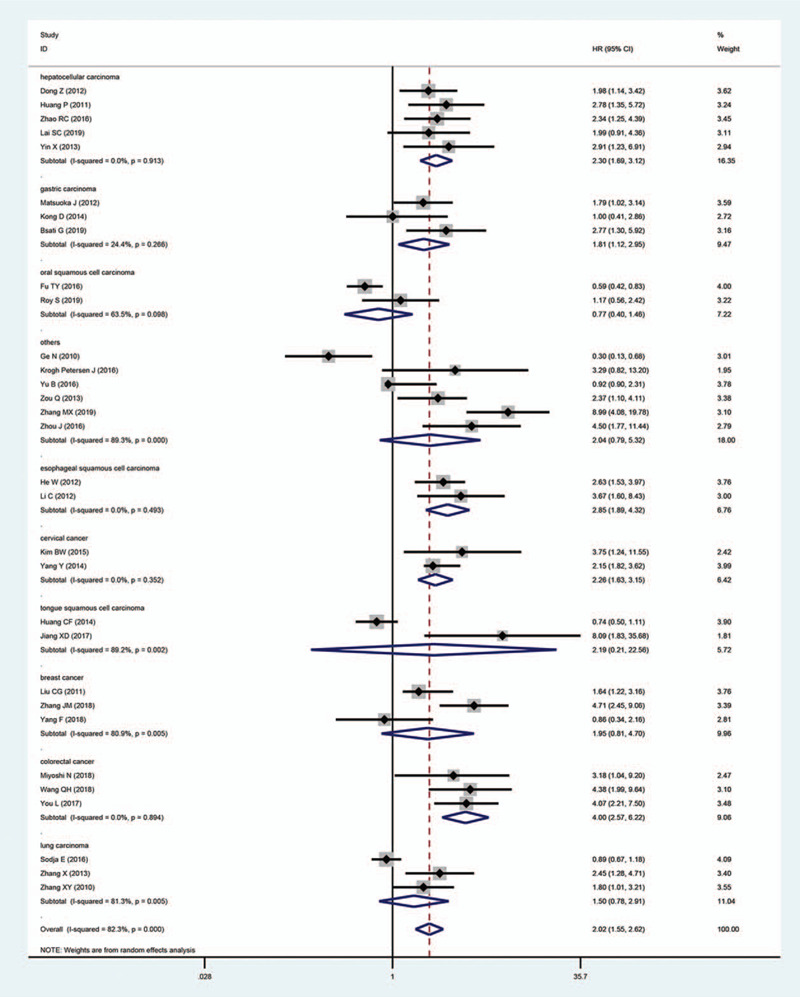
Forest plots to assess the relationship between Oct4 and OS based on the subgroup of tumor type.
Table 2.
Pooled HR for OS based on subgroup analysis.
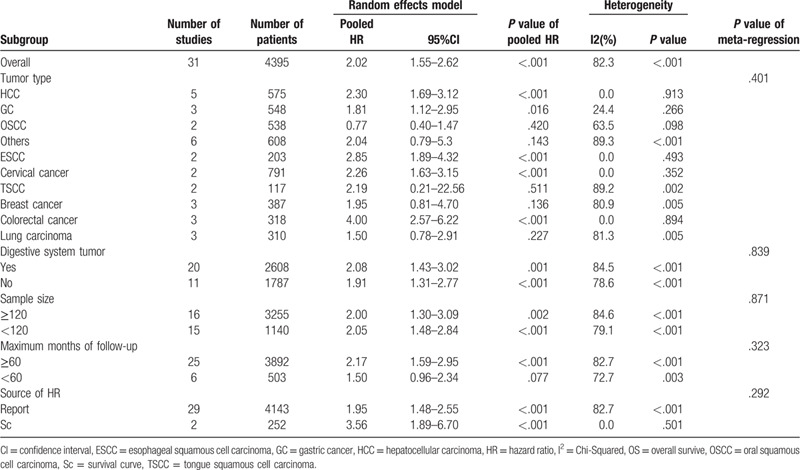
Figure 5.
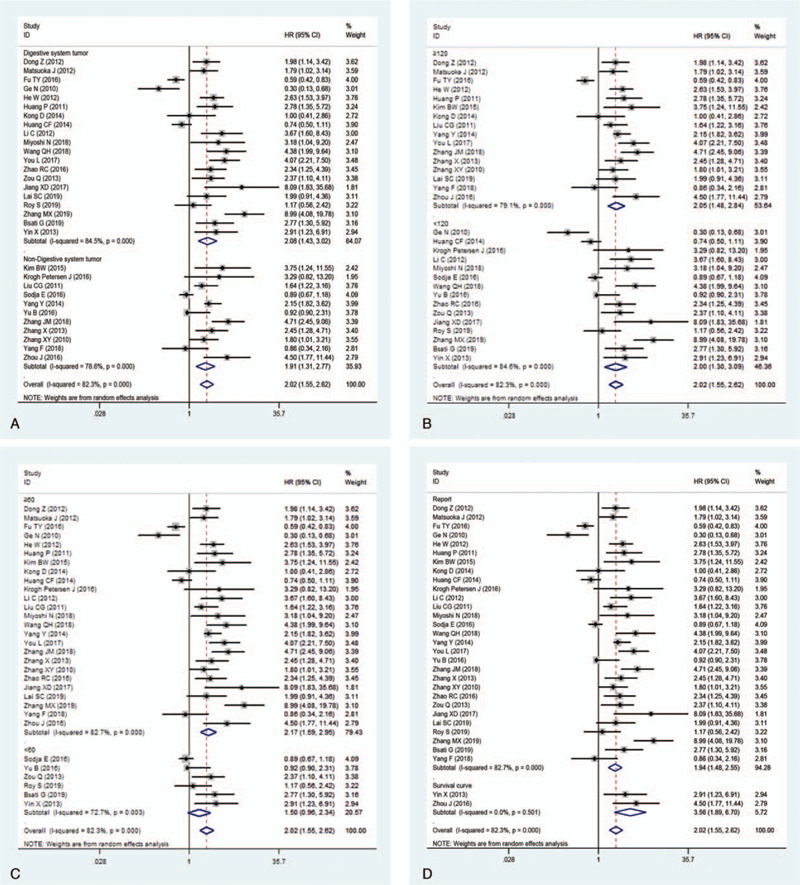
Subgroup analysis of OS. A, digestive system tumor; B, sample size; C, maximum month of follow-up; D, the source of HR.
Despite not observing obvious statistical heterogeneity, we conducted subgroup analysis to assess each subgroup of pooled HRs using the fixed-effects model considering the parameters including tumor type, digestive system tumor, sample size, and maximum follow-up period (months). Results indicated that patients overexpressing Oct4 had poorer DFS/RFS/PFS, including HCC (pooled HR: 1.92; 95% CI: 1.30–2.85; P = .001), cervical cancer (pooled HR: 2.77; 95% CI: 1.33–5.79; P = .007), colorectal cancer (pooled HR: 3.22; 95% CI: 1.68–6.16; P < .001), others (pooled HR: 2.39; 95% CI: 1.74–3.27; P < .001). Detailed results are displayed in Table 3, whereas all frost plots of subgroup analysis for DFS/RFS/PFS are shown in Fig. 6. Moreover, the relationships between Oct4 expression and clinicopathological features were assessed in HCC and GC. The results were shown in Table 4. The level of Oct4 expression was significantly related to the lymph node metastasis and vascular invasion in GC.
Table 3.
Pooled HR for DFS/RFS/PFS based on subgroup analysis.
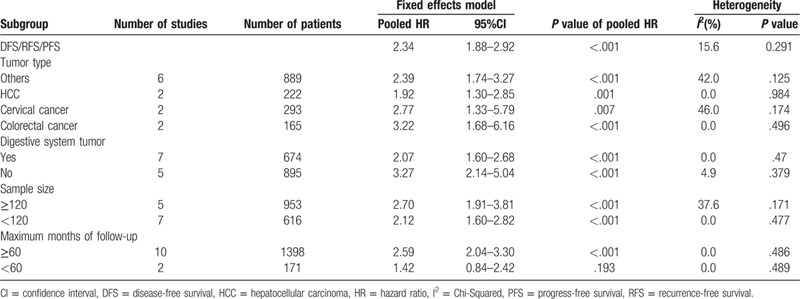
Figure 6.

Subgroup analysis of DFS/RFS/PFS. A, type of tumor; B, digestive system tumor; C, maximum month of follow-up; D, sample size.
Table 4.
Results of meta-analysis of increased Oct4 expression and clinicopathological features of HCC and GC.
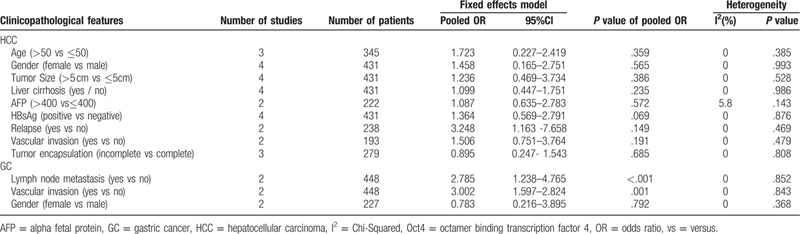
3.4. Sensitivity analysis and publication bias
Here, we conducted a sensitivity analysis by sequentially eliminating studies independently. Any study could not influence the outcomes of the relationship between OS and DFS/RFS/PFS (Fig. 7). The funnel plots for OS and DFS/RFS/PFS (Fig. 8) seemed asymmetric, although the Begg test (OS: P = .139; DFS/RFS/PFS: P = 1.000) and Egger's tests (OS: P = .116; DFS/RFS/PFS: P = .142) were not statistically significant. Consequently, the trim-and-filled model was introduced to neutralize potential bias, notably the correlation of Oct4 with survival was statistically significant (OS, HR: 1.59, 95% CI: 1.24–2.06, P < .0001; DFS/PFS/RFS, HR: 2.301, 95% CI: 1.849–2.864, P < .0001). According to the Cochran manual, the potential cause of funnel plot asymmetry in this study was selection bias, which includes publication bias and selective result reports, the low methodological quality which results in a false exaggeration of efficacy in small sample studies, true heterogeneity, human factors, and opportunities.
Figure 7.
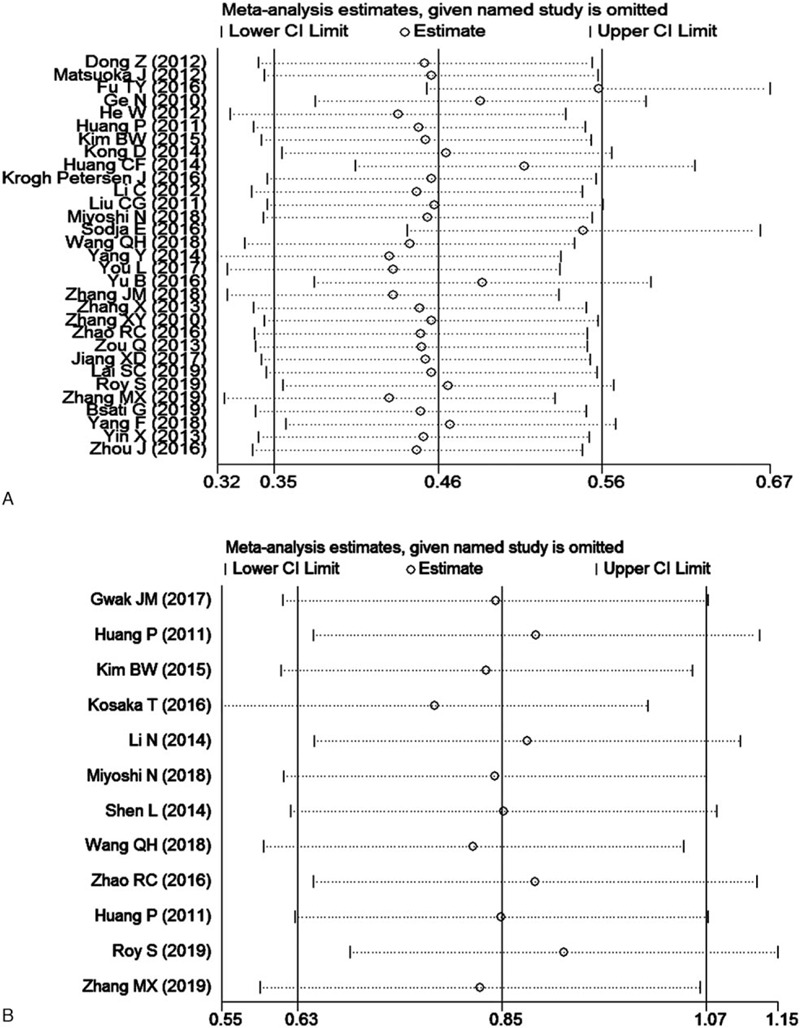
Sensitivity analysis of the meta-analysis. A, OS; B, DFS/RFS/PFS.
Figure 8.
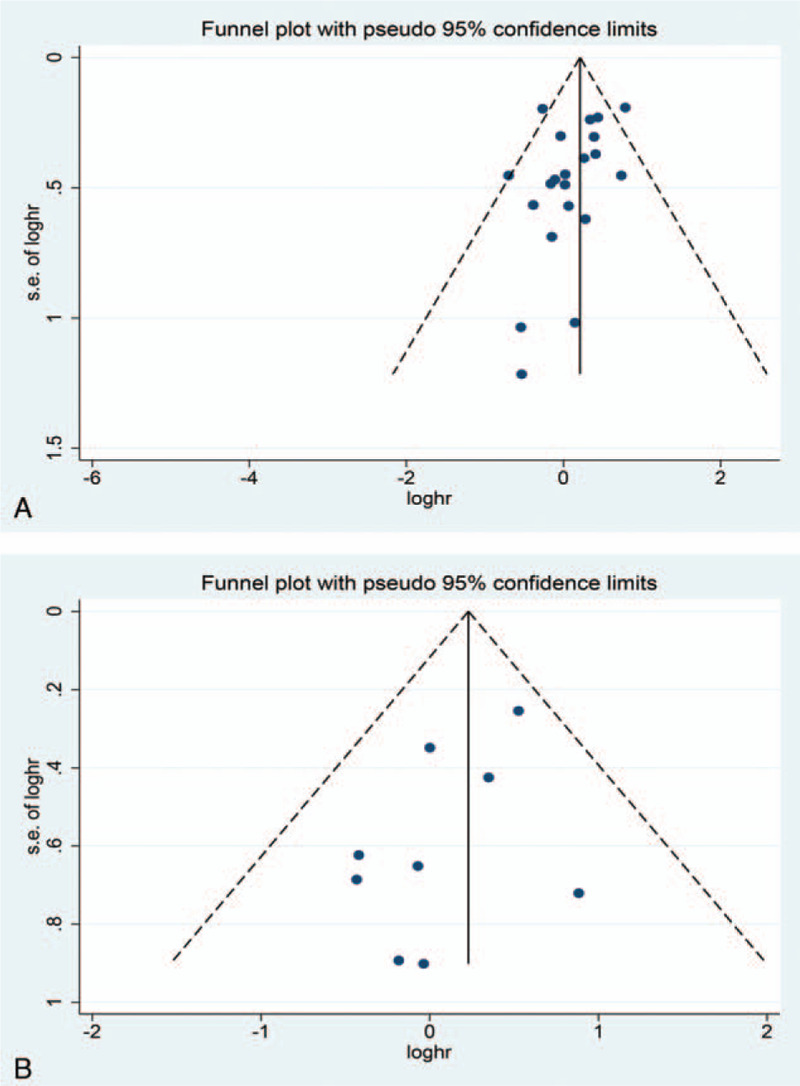
Funnel plot for publication bias assessment. A, OS; B, DFS/RFS/PFS.
4. Discussion
Cancer is lethal and poses a threat to mankind. Exploring more cancer markers is highly crucial for the diagnosis and prognosis of cancer. Notably, CSCs account for only a small fraction of cells in tumors and have self-renewal ability, producing multiple cell progeny thus have been revealed to play a role in metastasis, invasion, therapy resistance, and recurrence.[3,9,56] Additionally, CSCs release a variety of stemness molecules, among them, Oct4, SRY-related HMG-box gene 2, Nanog, among others. Of note, Oct4, in particular, has been reported to induce a variety of CSCs thus exerts potential regulatory roles.[57,58] Therefore, we speculated that CSC surface markers could serve as a biomarker for predicting the prognosis of cancer, this provides molecular targeted therapy for a variety of cancers using the therapeutic antibodies specific to CSC surface markers.
Oct4, as a CSCs marker, exhibits “stemness” characteristic, which is linked to a variety of biological behaviors and can cause cell immortality or account for the self-renewal ability, making cancer cells invasive.[23] Some studies indicated that high expression of Oct4 was observed in multiple human solid tumors. Based on these reports, we speculated that Oct4 could be utilized as a putative prognostic marker for predicting the prognosis of solid tumors. Furthermore, Oct4 had been confirmed to participate in initiation, chemoresistance, radiation resistance, metastasis, and invasion of solid tumors via cancer cell proliferation, migration, epithelial-mesenchymal transition, anti-apoptosis, and dedifferentiation,[59–62] this led to poor prognosis of tumor patients.
In addition, Oct4 function either directly or indirectly in the biological behavior of tumors. For instance, Oct4 can play a role in chemoresistance via multiple signaling pathways such as WNT/Notch-β-catenin-TCF-Oct4, PI3K-AKt-FOXC1-β-catenin-TCF-Oct4, HIF2α-NF-κB-Oct4, among others.[16] Interestingly, in HCC, Oct4 has been revealed to confer chemoresistance on HCC cells through protein kinase B Akt-mediated upregulation of ATP-binding cassette transporter G2, while it promoted cancer cell proliferation and migration via the survivin/STAT3 pathway, leading to poor prognosis.[63,64] Moreover, a study reported that Oct4 played an important role in radiation resistance by promoting the epithelial-mesenchymal transformation process in rectal carcinoma.[65] Elsewhere, Li et al demonstrated that Oct4 was essential in an antiapoptotic behavior of chemo-resistant colorectal cancer cells enriched for CSCs, whose effects were associated with STAT3/Survivin.[66,67] Also, Oct4 promoted tumorigenesis by inhibiting apoptosis in cervical carcinoma, implicating it as a key molecule involved in the inhibition of tumor cell apoptosis.[66,67] Additional findings revealed that greatly induced the transition of epithelial-mesenchymal via VEGF-C/VEGFR-3 signal pathway, thus contributed to metastasis.[68] Conversely, the expression of programmed death-ligand 1 in tumor cells was induced by activating Oct4 signaling to play a role in immune evasion, this suggested that CSCs might participate in tumor metastasis through immune evasion.[69] Another investigation showed that Oct4 could regulate the stability of mitosis and inactivate retinoblastoma tumor suppressor pathway, thus enhancing the aggressiveness of ovarian cancer.[70] Besides, Kumar et al observed that Oct4-mediated tumor cell dedifferentiation and potentially played a key role in tumor progression.[57] On the other hand, Oct4 knockdown could significantly reduce migration and progression in pancreatic cancer and colorectal cancer and cause breast CSC-like cell apoptosis, this strongly suggested that targeting Oct4 might offer vital clinical applications in cancer therapy.[62,71,72] Conclusively, these findings demonstrated that Oct4 remarkably impacts the process of tumor initiation, development, and progression.
Moreover, findings from our study implicated Oct4 as a detrimental prognostic marker for malignant tumors. High expression of Oct4 was dramatically associated with worse OS and DFS/RFS/PFS in patients with solid tumors. Besides, elevated Oct4 levels indicated worse prognosis in patients with HCC, ESCC, GC, cervical cancer, and colorectal cancer. These observations suggested that Oct4 may be utilized as a potential prognostic marker and therapeutic target for most solid tumors. Notably, only 2 studies revealed that patients exhibiting high expression of Oct4 survived longer and had a lower recurrence rate in hypopharyngeal squamous cell carcinoma, and OSCC patients expressing high levels of Oct4 had better cumulative OS, the underlying mechanism is unclear, thus, additional in-depth researches are needed.[24,25] Nevertheless, when the 2 above-mentioned items were excluded from the analysis, the association between Oct4 and OS in solid tumor patients did not change (excluded: pooled HR: 2.23; 95% CI: 1.75–2.84; P < .001, included: pooled HR: 2.02, 95% CI: 1.55–2.62, P < .001).
There were a few limitations that should be addressed in our study. First, the number of published articles on each type of tumor was relatively small, such as for TSCC, bladder cancer, gastric cancer among others, therefore, the included studies were mixed and analyzed in order to assess the relationship between the Oct4 expression and solid tumors, which might be a limitation in our study. In the future, we will also analyze and assess the role of Oct4 in a particular type of cancer as the number of studies increases. Second, survival curves were used to evaluate HRs in 2 of the included studies, this possibly affected the precision of results. Third, in this study, a large number of patients were Asian, thus, the results may not be applicable to other ethnic groups and might not be generalized and be valid globally. More assessments on the relationship between the expression level of Oct4 and prognosis in cancer patients of other ethnic groups are needed to further validate our findings. Fourth, various cut-off values of Oct4 were applied in each study. Fifth, different subtypes of Oct4 might lead to varying prognosis for cancer patients, however, in the included studies, no subtypes were distinguished. Therefore, these data could not be extracted. Finally, the articles included were all in English, which might result in potential language bias.
5. Conclusion
This study provides the first report that sheds light on the prognostic role of elevated Oct4 expression in multiple solid tumors. Oct4 was revealed as a potential novel biomarker and a potential therapeutic target for cancer patients. Notably, overexpression of Oct4 is linked to poor prognosis in patients with solid tumors, among them, HCC (OS, DFS/RFS/PFS), ESCC (OS), GC (OS), cervical cancer (OS, DFS/RFS/PFS), and colorectal cancer (OS, DFS/RFS/PFS). However, additional high-quality studies are needed to explore the relationship between Oct4 expression and prognosis in patients with each type of tumor.
Author contributions
Data curation: Xiaoyan Zhao, Hui Lu.
Funding acquisition: Huafang Wang.
Methodology: Li Liu, Huafang Wang.
Writing – original draft: Xiaoyan Zhao, Hui Lu.
Writing – review & editing: Xiaoyan Zhao, Yan Sun, Li Liu, Huafang Wang.
Footnotes
Abbreviations: AFP = alpha fetal protein, Akt = protein kinase B, ATP = adenosine-triphosphate, CIs = confidence intervals, CSCs = cancer stem cells, DFS = disease-free survival, ESCC = esophageal squamous cell carcinoma, ESCs = embryonic stem cells, FOXC1 = forkhead box protein C1, GC = gastric cancer, HCC = hepatocellular carcinoma, HIF2-α = hypoxia-inducible factor-alpha, HRs = hazard ratios, KM = Kaplan–Meier, NF-κB = nuclear factor kappa-light-chain-en-hancer of activated B, NOS = Newcastle-Ottawa scale, Oct4 = octamer binding transcription factor 4, OR = odds ratio, OS = overall survival, OSCC = oral squamous cell carcinoma, PFS = progress-free survival, PI3K = phosphatidylinositol 3-kinase, POU5F1 = Pit-Oct-Unc domain, class 5, transcription factor 1, RFS = recurrence free survival, RR = relative risk, STAT3 = signal transducing activator of transcription 3, TCF = transcription factor 3, TSCC = tongue squamous cell carcinoma, VEGF-C = vascular endothelial growth factor, VEGFR-3 = vascular endothelial growth factor receptor-3, vs = versus, WNT = wingless/integrated.
How to cite this article: Zhao X, Lu H, Sun Y, Liu L, Wang H. Prognostic value of octamer binding transcription factor 4 for patients with solid tumors: a meta-analysis. Medicine. 2020;99:42(e22804).
XZ and HL contributed equally to this work.
The study was supported by grants from the National Nature Science Foundation of China, No.81770134.
The authors follow field-specific standards for data deposition in publicly available resource. The data analyzed during this study are available from the corresponding author on reasonable request.
The authors report no conflicts of interest.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].The L. GLOBOCAN 2018: counting the toll of cancer. Lancet (London, England) 2018;392:985. [DOI] [PubMed] [Google Scholar]
- [2].Chissoe SL, Bodenteich A, Wang YF, et al. Sequence and analysis of the human ABL gene, the BCR gene, and regions involved in the Philadelphia chromosomal translocation. Genomics 1995;27:67–82. [DOI] [PubMed] [Google Scholar]
- [3].Nassar D, Blanpain C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu Rev Pathol 2016;11:47–76. [DOI] [PubMed] [Google Scholar]
- [4].Toh TB, Lim JJ, Chow EK-H. Epigenetics in cancer stem cells. Mol Cancer 2017;16:29–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998;95:379–91. [DOI] [PubMed] [Google Scholar]
- [6].Niwa H. How is pluripotency determined and maintained? Development (Cambridge, England) 2007;134:635–46. [DOI] [PubMed] [Google Scholar]
- [7].Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–76. [DOI] [PubMed] [Google Scholar]
- [8].Radzisheuskaya A, Silva JC. Do all roads lead to Oct4? The emerging concepts of induced pluripotency. Trends Cell Biol 2014;24:275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang YJ, Herlyn M. The emerging roles of Oct4 in tumor-initiating cells. Am J Physiol Cell Physiol 2015;309:C709–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Radzisheuskaya A, Silva JCR. Do all roads lead to Oct4? The emerging concepts of induced pluripotency. Trends Cell Biol 2014;24:275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kim R-J, Nam J-S. OCT4 expression enhances features of cancer stem cells in a mouse model of breast cancer. Lab Anim Res 2011;27:147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].PeirisPagès M, Martinez Outschoorn UE, Pestell RG, et al. Cancer stem cell metabolism. Breast Cancer Res 2016;18:55–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim BW, Cho H, Choi CH, et al. Clinical significance of OCT4 and SOX2 protein expression in cervical cancer. BMC Cancer 2015;15:1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Matsuoka J, Yashiro M, Sakurai K, et al. Role of the stemness factors sox2, oct3/4, and nanog in gastric carcinoma. J Surg Res 2012;174:130–5. [DOI] [PubMed] [Google Scholar]
- [15].Zhou J, Dong D, Cheng R, et al. Aberrant expression of KPNA2 is associated with a poor prognosis and contributes to OCT4 nuclear transportation in bladder cancer. Oncotarget 2016;7:72767–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mohiuddin IS, Wei SJ, Kang MH. Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim Biophys Acta Mol Basis Dis 2020;1866:165432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dong Z, Zeng Q, Luo H, et al. Increased expression of OCT4 is associated with low differentiation and tumor recurrence in human hepatocellular carcinoma. Pathol Res Pract 2012;208:527–33. [DOI] [PubMed] [Google Scholar]
- [18].He W, Li K, Wang F, et al. Expression of OCT4 in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. World J Gastroenterol 2012;18:712–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu CG, Lu Y, Wang BB, et al. Clinical implications of stem cell gene Oct-4 expression in breast cancer. Ann Surg 2011;253:1165–71. [DOI] [PubMed] [Google Scholar]
- [20].Miyoshi N, Fujino S, Ohue M, et al. The POU5F1 gene expression in colorectal cancer: a novel prognostic marker. Surg Today 2018;48:709–15. [DOI] [PubMed] [Google Scholar]
- [21].Wang QH, Zhang M, Shi CT, et al. High Oct4 predicted worse prognosis of right-sided colon cancer patients. Future Oncol 2018;14:2279–91. [DOI] [PubMed] [Google Scholar]
- [22].Sodja E, Rijavec M, Koren A, et al. The prognostic value of whole blood SOX2, NANOG and OCT4 mRNA expression in advanced small-cell lung cancer. Radiol Oncol 2016;50:188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huang CF, Xu XR, Wu TF, et al. Correlation of ALDH1, CD44, OCT4 and SOX2 in tongue squamous cell carcinoma and their association with disease progression and prognosis. J Oral Pathol Med 2014;43:492–8. [DOI] [PubMed] [Google Scholar]
- [24].Ge N, Lin HX, Xiao XS, et al. Prognostic significance of Oct4 and Sox2 expression in hypopharyngeal squamous cell carcinoma. J Transl Med 2010;8:94–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fu TY, Hsieh IC, Cheng JT, et al. Association of OCT4, SOX2, and NANOG expression with oral squamous cell carcinoma progression. J Oral Pathol Med 2016;45:89–95. [DOI] [PubMed] [Google Scholar]
- [26].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [28].Greenland S. Quantitative methods in the review of epidemiologic literature. Epidemiol Rev 1987;9:1–30. [DOI] [PubMed] [Google Scholar]
- [29].Gwak JM, Kim M, Kim HJ, et al. Expression of embryonal stem cell transcription factors in breast cancer: Oct4 as an indicator for poor clinical outcome and tamoxifen resistance. Oncotarget 2017;8:36305–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huang P, Qiu J, Li B, et al. Role of Sox2 and Oct4 in predicting survival of hepatocellular carcinoma patients after hepatectomy. Clin Biochem 2011;44:582–9. [DOI] [PubMed] [Google Scholar]
- [31].Kim BW, Cho H, Choi CH, et al. Clinical significance of OCT4 and SOX2 protein expression in cervical cancer. BMC Cancer 2015;15:1015–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kong D, Su G, Zha L, et al. Coexpression of HMGA2 and Oct4 predicts an unfavorable prognosis in human gastric cancer. Med Oncol 2014;31:130–130. [DOI] [PubMed] [Google Scholar]
- [33].Krogh Petersen J, Jensen P, Dahl Sørensen M, et al. Expression and prognostic value of Oct-4 in astrocytic brain tumors. PLoS One 2016;11:e0169129–1169129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li C, Yan Y, Ji W, et al. OCT4 positively regulates Survivin expression to promote cancer cell proliferation and leads to poor prognosis in esophageal squamous cell carcinoma. PLoS One 2012;7:e49693–149693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li N, Deng W, Ma J, et al. Prognostic evaluation of Nanog, Oct4, Sox2, PCNA, Ki67 and E-cadherin expression in gastric cancer. Med Oncol 2015;32:433–1433. [DOI] [PubMed] [Google Scholar]
- [36].LiuF Cg, Lu Y, Wang Bb, et al. Clinical implications of stem cell gene Oct-4 expression in breast cancer. Ann Surg 2011;253:1165–71. [DOI] [PubMed] [Google Scholar]
- [37].Wang Q-H, Zhang M, Shi CT, et al. High Oct4 predicted worse prognosis of right-sided colon cancer patients. Future Oncol 2018;14:2279–91. [DOI] [PubMed] [Google Scholar]
- [38].Yang Y, Wang Y, Yin C, et al. Clinical significance of the stem cell gene Oct-4 in cervical cancer. Tumour Biol 2014;35:5339–45. [DOI] [PubMed] [Google Scholar]
- [39].You L, Guo X, Huang Y. Correlation of Cancer Stem-Cell Markers OCT4, SOX2, and NANOG with Clinicopathological Features and Prognosis in Operative Patients with Rectal Cancer. Yonsei Med J 2018;59:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yu B, Cai H, Xu Z, et al. Expressions of stem cell transcription factors Nanog and Oct4 in renal cell carcinoma tissues and clinical significance. Artif Cells Nanomed Biotechnol 2016;44:1818–23. [DOI] [PubMed] [Google Scholar]
- [41].Zhang JM, Wei K, Jiang M. OCT4 but not SOX2 expression correlates with worse prognosis in surgical patients with triple-negative breast cancer. Breast Cancer 2018;25:447–55. [DOI] [PubMed] [Google Scholar]
- [42].Zhang X, Wang H, Jin B, et al. Correlations between OCT4 expression and clinicopathological factors and prognosis of patients with lung adenocarcinoma. Zhongguo Fei Ai Za Zhi 2013;16:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang XY, Dong QG, Huang JS, et al. The expression of stem cell-related indicators as a prognostic factor in human lung adenocarcinoma. J Surg Oncol 2010;102:856–62. [DOI] [PubMed] [Google Scholar]
- [44].Zhao RC, Zhou J, Chen KF, et al. The prognostic value of combination of CD90 and OCT4 for hepatocellular carcinoma after curative resection. Neoplasma 2016;63:288–98. [DOI] [PubMed] [Google Scholar]
- [45].Zou Q, Yang L, Yang Z, et al. PSCA and Oct-4 expression in the benign and malignant lesions of gallbladder: implication for carcinogenesis, progression, and prognosis of gallbladder adenocarcinoma. Biomed Res Int 2013;2013:648420–1648420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jiang XD, Luo G, Wang XH, et al. Expression of Oct4 and Sox2 and their clinical significance in tongue squamous cell carcinoma. Zhonghua Kou Qiang Yi Xue Za Zhi 2017;52:27–33. [DOI] [PubMed] [Google Scholar]
- [47].Yin X, Li YW, Jin JJ, et al. The clinical and prognostic implications of pluripotent stem cell gene expression in hepatocellular carcinoma. Oncol Lett 2013;5:1155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kosaka T, Mikami S, Yoshimine S, et al. The prognostic significance of OCT4 expression in patients with prostate cancer. Hum Pathol 2016;51:1–8. [DOI] [PubMed] [Google Scholar]
- [49].Shen L, Huang X, Xie X, et al. High expression of SOX2 and OCT4 indicates radiation resistance and an independent negative prognosis in cervical squamous cell carcinoma. J Histochem Cytochem 2014;62:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Huang P, Chen J, Wang L, et al. Implications of transcriptional factor, OCT-4, in human bladder malignancy and tumor recurrence. Med Oncol 2012;29:829–34. [DOI] [PubMed] [Google Scholar]
- [51].Lai SC, Su YT, Chi CC, et al. DNMT3b/OCT4 expression confers sorafenib resistance and poor prognosis of hepatocellular carcinoma through IL-6/STAT3 regulation. J Exp Clin Cancer Res 2019;38:474–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Roy S, Kar M, Roy S, et al. KLF4 expression in the surgical cut margin is associated with disease relapse of oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol 2019;128:154–65. [DOI] [PubMed] [Google Scholar]
- [53].Zhang MX, Gan W, Jing CY, et al. High expression of Oct4 and Nanog predict poor prognosis in intrahepatic cholangiocarcinoma patients after curative resection. J Cancer 2019;10:1313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Basati G, Mohammadpour H, Emami Razavi A. Association of high expression levels of SOX2, NANOG, and OCT4 in gastric cancer tumor tissues with progression and poor prognosis. J Gastrointest Cancer 2020;51:41–7. [DOI] [PubMed] [Google Scholar]
- [55].Yang F, Zhang J, Yang H. OCT4, SOX2, and NANOG positive expression correlates with poor differentiation, advanced disease stages, and worse overall survival in HER2(+) breast cancer patients. Onco Targets Ther 2018;11:7873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ahmad G, Amiji MM. Cancer stem cell-targeted therapeutics and delivery strategies. Expert Opin Drug Deliv 2017;14:997–1008. [DOI] [PubMed] [Google Scholar]
- [57].Kumar SM, Liu S, Lu H, et al. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene 2012;31:4898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kim WT, Ryu CJ. Cancer stem cell surface markers on normal stem cells. BMB Rep 2017;50:285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cai S, Geng S, Jin F, et al. POU5F1/Oct-4 expression in breast cancer tissue is significantly associated with non-sentinel lymph node metastasis. BMC Cancer 2016;16:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zou Q, Yang L, Yang Z, et al. PSCA and Oct-4 expression in the benign and malignant lesions of gallbladder: implication for carcinogenesis, progression, and prognosis of gallbladder adenocarcinoma. Biomed Res Int 2013;2013:648420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Curtarelli RB, Goncalves JM, Dos Santos LGP, et al. Expression of cancer stem cell biomarkers in human head and neck carcinomas: a systematic review. Stem Cell Rev 2018;14:769–84. [DOI] [PubMed] [Google Scholar]
- [62].Dai X, Ge J, Wang X, et al. OCT4 regulates epithelial-mesenchymal transition and its knockdown inhibits colorectal cancer cell migration and invasion. Oncol Rep 2013;29:155–60. [DOI] [PubMed] [Google Scholar]
- [63].Wang G, Zhou H, Gu Z, et al. Oct4 promotes cancer cell proliferation and migration and leads to poor prognosis associated with the survivin/STAT3 pathway in hepatocellular carcinoma. Oncol Rep 2018;40:979–87. [DOI] [PubMed] [Google Scholar]
- [64].Wang XQ, Ongkeko WM, Chen L, et al. Octamer 4 (Oct4) mediates chemotherapeutic drug resistance in liver cancer cells through a potential Oct4-AKT-ATP-binding cassette G2 pathway. Hepatology 2010;52:528–39. [DOI] [PubMed] [Google Scholar]
- [65].Shao M, Bi T, Ding W, et al. OCT4 potentiates radio-resistance and migration activity of rectal cancer cells by improving epithelial-mesenchymal transition in a ZEB1 dependent manner. Biomed Res Int 2018;2018:3424956–13424956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wen K, Fu Z, Wu X, et al. Oct-4 is required for an antiapoptotic behavior of chemoresistant colorectal cancer cells enriched for cancer stem cells: effects associated with STAT3/Survivin. Cancer Lett 2013;333:56–65. [DOI] [PubMed] [Google Scholar]
- [67].Li SW, Wu XL, Dong CL, et al. The differential expression of OCT4 isoforms in cervical carcinoma. PLoS One 2015;10:e0118033–1118033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Li C, Zhu M, Lou X, et al. Transcriptional factor OCT4 promotes esophageal cancer metastasis by inducing epithelial-mesenchymal transition through VEGF-C/VEGFR-3 signaling pathway. Oncotarget 2017;8:71933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dong P, Xiong Y, Yue J, et al. Tumor-intrinsic PD-L1 signaling in cancer initiation, development and treatment: beyond immune evasion. Front Oncol 2018;8:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Comisso E, Scarola M, Rosso M, et al. OCT4 controls mitotic stability and inactivates the RB tumor suppressor pathway to enhance ovarian cancer aggressiveness. Oncogene 2017;36:4253–66. [DOI] [PubMed] [Google Scholar]
- [71].Lu Y, Zhu H, Shan H, et al. Knockdown of Oct4 and Nanog expression inhibits the stemness of pancreatic cancer cells. Cancer Lett 2013;340:113–23. [DOI] [PubMed] [Google Scholar]
- [72].Hu T, Liu S, Breiter DR, et al. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res 2008;68:6533–40. [DOI] [PubMed] [Google Scholar]


