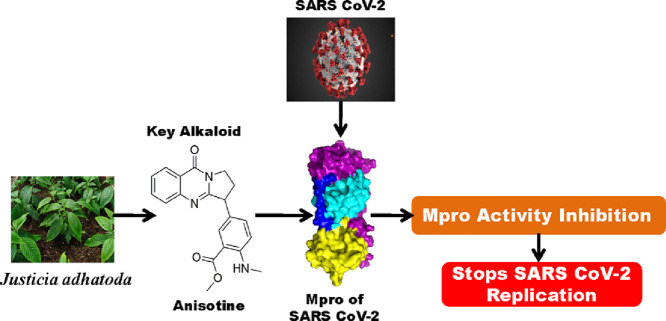
Abstract
The COVID-19 pandemic, caused by SARS CoV-2, is responsible for millions of death worldwide. No approved/proper therapeutics is currently available which can effectively combat this outbreak. Several attempts have been undertaken in the search of effective drugs to control the spread of SARS CoV-2 infection. The main protease (Mpro), key component for the cleavage of the viral polyprotein, is considered to be one of the important drug targets for treating COVID-19. Various phytochemicals, including polyphenols and alkaloids, have been proposed as potent inhibitors of Mpro. The alkaloids from leaf extracts of Justicia adhatoda have also been reported to possess anti-viral activity. But whether these alkaloids exhibit any inhibitory effect on SARS CoV-2 Mpro is far from clear. To explore this in detail, we have adopted computational approaches. Justicia adhatoda alkaloids possessing proper drug-likeness properties and two anti-HIV drugs (lopinavir and darunavir; having binding affinity -7.3 to -7.4 kcal/mol) were docked against SARS CoV-2 Mpro to study their binding properties. Only one alkaloid (anisotine) had interaction with both the catalytic residues (His41 and Cys145) of Mpro and exhibited good binding affinity (-7.9 kcal/mol). Molecular dynamic simulations (100 ns) revealed that Mpro-anisotine complex is more stable, conformationally less fluctuated; slightly less compact and marginally expanded than Mpro-darunavir/lopinavir complex. Even the number of intermolecular H-bonds and MM-GBSA analysis suggested that anisotine is a more potent Mpro inhibitor than the two previously recommended antiviral drugs (lopinavir and darunavir) and may evolve as a promising anti-COVID-19 drug if proven in animal experiments and on patients.
Keywords: COVID-19, SARS CoV-2 main protease, Docking and molecular dynamics simulation, Vasaka Alkaloids
Abbreviations: COVID-19, corona virus disease 2019; SARS CoV-2, severe acute respiratory syndrome corona virus-2; Mpro, main protease; MD, molecular dynamics; RMSD, root mean square deviation; RMSF, root mean square fluctuation; Rg, radius of gyration; SASA, solvent accessible surface area
Graphical abstract
1. Introduction
The current emergency due to the worldwide spread of the COVID-19 disease is caused by the new severe acute respiratory syndrome coronavirus-2 (SARS CoV-2). This viral disease is a great concern for global public health [1]. This disease is a lot more contagious than previous outbreaks of SARS and MERS [2,3]. This novel coronavirus was discovered in Wuhan, China in late 2019 and then spread rapidly to other countries [4,5]. On the 11th of March 2020, WHO declared COVID-19 as a pandemic disease [6]. It mostly affects the lower respiratory tract which causes pneumonia as well as the gastrointestinal system, kidney, heart and central nervous system, with common symptoms being fever, cough and diarrhea [7,8]. No approved therapeutic/effective treatment is currently available to combat this outbreak. Thus, the search of appropriate drugs and suitable vaccines are highly in demand to control COVID-19.
The SARS CoV-2 genome is composed of a long RNA strand that acts as a messenger RNA when it infects a host cell and directs the synthesis of polyproteins required for multiplication of new viruses [9], [10], [11]. The SARS CoV-2 main protease (Mpro) and papain-like protease (PLpro) are responsible for processing the viral proteins at a specific site into functional units for virus replication [9], [10], [11]. Viral replication can be blocked by inhibiting the proteolytic activity of Mpro and thus Mpro is a target for drug design. Recently, the crystal structures of the SARS CoV-2 Mpro with various inhibitors have been reported which provide useful information about the structural integrity of Mpro and also suggests that Mpro is an important drug target for developing small molecule inhibitors [12,13].
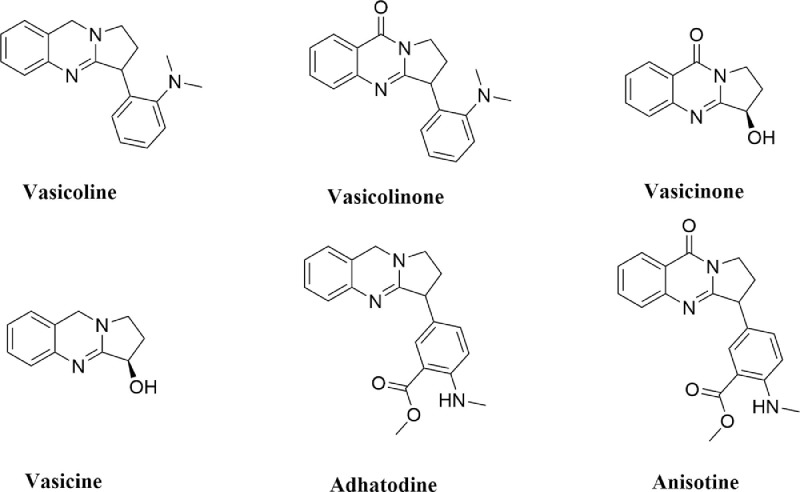
Many studies have been carried out to find suitable inhibitors of Mpro using drug repurposing strategy [14], [15], [16], [17], [18], [19], [20]. This strategy revealed that many well known antiviral drugs, vitamins, tetracyline based drugs and cholesterol-lowering drugs/statins have the potential to inhibit the activity of SARS CoV-2 Mpro [14], [15], [16], [17], [18], [19], [20]. Furthermore, various phytochemicals are also being proposed as the main protease inhibitors through screening and structure-based design approach [21], [22], [23], [24], [25], [26], [27], [28], [29]. Recently, it has been found that some plant-derived alkaloids have the potency to inhibit different components/targets of SARS CoV-2 such as RBD-S (spike glycoprotein receptor binding domain) and Mpro [21,26,27,30]. The leaves of Justicia adhatoda, commonly known as Vasaka (Vasaka plant) also contains different alkaloids (vasicoline, vasicolinone, vasicinone, vasicine, adhatodine and anisotine) (Fig. 1 ) [31]. These alkaloids are well-known for their anti-tuberculosis activity [31]. Furthermore, extracts from Justicia adhatoda leaves have also been reported to exhibit antiviral activity against influenza virus and herpes simplex virus [32,33]. But whether these alkaloids from the leaves of Justicia adhatoda exhibit any antiviral activity against SARS CoV-2 by inhibiting the enzymatic/ proteolytic activity of Mpro is far from clear. Therefore, in this study, we have examined the inhibitory potency of these six alkaloids from J. adhatoda against SARS-CoV-2 Mpro with the aid of in-silico docking studies, molecular dynamics simulations and MM-GBSA analysis. This study has revealed that only one of the alkaloids (anisotine) is more effective as a Mpro inhibitor compared to the previously recommended antiviral drugs (darunavir and lopinavir).
Fig. 1.
Chemical structure of J. adhatoda alkaloids. The two-dimensional structures of six alkaloids from J. adhatoda (vasicoline, vasicolinone, vasicinone, vasicine, adhatodine and anisotine).
2. Materials and methods
2.1. Preparation of the Mpro and ligands
The structures of Justicia adhatoda alkaloids were downloaded from PubChem database server (https://pubchem.ncbi.nlm.nih.gov) while the crystal structure of the SARS CoV-2 Mpro (PDB ID: 6LU7) [12] was downloaded from the RCSB Protein Data Bank (http://www.rcsb.org). Each of the Justicia adhatoda alkaloid structures was optimized with B3LYP/6-31G* basis set by using Gaussian09 software [34]. Standard processes were used in AutoDock Tools to obtain the pdbqt files for Mpro and Justicia adhatoda alkaloids [35,36].
2.2. Molecular docking
AutoDock Vina was used for the entire docking calculations of Mpro with two anti-HIV drugs and Justicia adhatoda alkaloids by assigning a grid box with 10.0 Å radius throughout the active site region [29,35,36]. The conformations having the lowest root mean square deviation (RMSD) values, along with the highest Vina score were selected. The output from AutoDock Vina was rendered with DS visualizer software [37].
2.3. Molecular dynamics simulation
The molecular dynamics (MD) simulations were performed in GROMACS 2019 with GROMOS9653a6 force field and SPC water model [38,39]. The ligand topologies were obtained from the PRODRG server [40]. LINCS algorithm and SETTLE algorithm were used to constrain all bond lengths of protein, anti-HIV drugs/ anisotine and water molecules, respectively [41,42]. After accommodating each system (unligated Mpro, Mpro-darunavir, Mpro-lopinavir and Mpro-anisotine complex) in a cubic box, water molecules were added to it and energy-minimization was performed using the steepest descent algorithm to achieve an equilibrated system with appropriate volume. The Particle Mesh Ewald method was used to treat the Long-range electrostatics with cut off 1.2 nm and with a Fourier grid spacing of 1.2 nm [43]. To set up a constant temperature and pressure, equilibration of each system was carried out in two main stages. First, NVT ensemble using the v-rescale algorithm for 10 ns was performed to set the temperature 300 K and then to set the pressure at 1 bar, NPT ensemble for 10 ns was carried out by positional restraining of the complexes [44]. The equilibrated systems were then subjected to unrestrained production MD simulations of 100 ns each, maintaining the same pressure (1 bar) and temperature (300 K). The root mean square deviation (RMSD), the total number of hydrogen bonds, root mean square fluctuation (RMSF), the radius of gyration (Rg), solvent accessible surface area (SASA) for each system was calculated from the MD trajectories [25,29].
2.4. MM-GBSA analysis
Several methods are used to calculate the theoretical free energies of binding of ligands to the receptor like a) the molecular mechanics generalized Born surface area (MM-GBSA) and b) molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) c) Free energy perturbation etc [29,[45], [46], [47], [48]]. Here we have used the MM-GBSA method to calculate the relative binding free energies of anti-HIV drugs and anisotine to Mpro. The free energy of binding can be calculated as ΔGbind=ΔH-TΔS.
ΔH =ΔEelec+ΔEvdW+ΔGpolar+ΔGnon-polar, where Eelec and EvdW are the electrostatic and van der Waal's contributions, and Gpolar and Gnon-polar are the polar and non-polar solvation terms, respectively. The generalized Born model with an external dielectric constant of 80 and an internal dielectric constant of 1, is for the estimation of polar contribution of the free energy, while the non-polar energy contribution is calculated from the solvent-accessible surface area (SASA). The prime module of the Schrodinger suite (Schrödinger Release 2020-1: Prime, Schrödinger, LLC, New York, NY, 2020) was used for all MM-GBSA calculations.
2.5. Pharmacokinetics study
SwissADME online server was used for the prediction of different pharmacokinetic properties of six alkaloids from J. adhatoda [49]. The drug-likeness properties of these six alkaloids such as absorption, distribution, metabolism and excretion parameters were mainly scrutinized.
3. Result and discussion
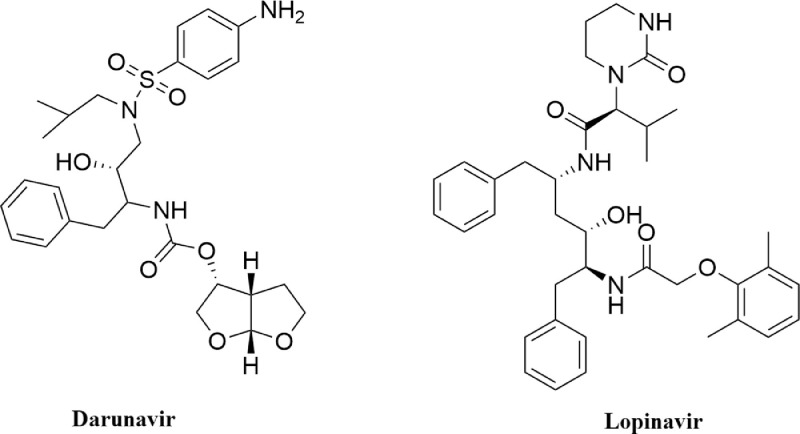
The valuable information about the structural aspects of SARS CoV-2 Mpro became available when Jin et al. and Zhang et al. solved the crystal structure of the protease [12,13]. The solved structure revealed three domains and a loop region in Mpro. Domain I consists of amino acid residues 8-101, while domain II and domain III contain residues 102-184 and 201-303, respectively. Notably, domain II is connected to domain III with a loop that consists of residues 185-200. The three-dimensional crystal structure also demonstrated an active site/ catalytic site/ substrate binding site on each of the protomer of Mpro consisting of Cys-His dyad (His41 and Cys145). This catalytic site consisting of cysteine and histidine amino acid moiety is located at the cleft of domain I and domain II. These two crystal structures also laid the basis towards the structure-based drug design against Mpro. Many small molecules are being proposed as effective SARS CoV-2 Mpro inhibitor [22], [23], [24], [25], [26], [27], [28]. Several anti-HIV drugs (darunavir, lopinavir, atazanavir, etc) display excellent binding affinity towards the active site of Mpro [50]. In the recent past, two well known anti-HIV drugs (darunavir and lopinavir) (structures mentioned in Fig. 2 ) have been selected by many investigators as standard substrates for comparing the binding affinity and/or binding modes of various small molecules [18,23,26]. Therefore, we have also decided to take these two anti-HIV drugs as standard Mpro inhibitors for this study.
Fig. 2.
Chemical structure of anti-HIV drugs. The two-dimensional structure of two anti-HIV drugs (darunavir and lopinavir).
3.1. Pharmacokinetics analysis
Prior to conducting molecular docking studies, we evaluated the drug-likeness characteristics of all six alkaloids of J. adhatoda. Pharmacokinetics analysis using SwissADME server is listed in Table 1 . This analysis revealed that the molecular weight (MW) of these six alkaloids were less than 350 (ranging from 188.23 to 349.38) which suggested that all the alkaloids may easily be transported, diffused and absorbed inside the body. The number of hydrogen bond donors (H-Do) was less than 5 and the number of hydrogen bond acceptors (H-Ac) was in the range of 0 to 1 for these alkaloids, which are in accordance with Lipinski's rules and Ghose rules. Furthermore, the topological polar surface area (TPSA) of all the alkaloids were found in the range of 18.84 to 73.72 Å2 indicating good bioavailability of these alkaloids. The high intestinal absorption (IA) signified good cell membrane permeability and oral bioavailability for all the alkaloids. Altogether, pharmacokinetic analysis clearly indicated that all J. adhatoda alkaloids possess favorable drug-likeness properties.
Table 1.
Pharmacokinetic properties of J. adhatoda alkaloids.
| Compound | MW | H-Ac | H-Do | Nrot | TPSA | GIA | BBB | LFV | GFV |
|---|---|---|---|---|---|---|---|---|---|
| Vasicoline | 291.39 | 1 | 0 | 2 | 18.84 | High | Yes | 0 | 0 |
| Vasicolinone | 305.37 | 2 | 0 | 2 | 38.13 | High | Yes | 0 | 0 |
| Vasicinone | 202.21 | 3 | 1 | 0 | 55.12 | High | No | 0 | 0 |
| Vasicine | 188.23 | 2 | 1 | 0 | 35.83 | High | No | 0 | 0 |
| Adhatodine | 335.4 | 3 | 1 | 4 | 53.93 | High | Yes | 0 | 0 |
| Anisotine | 349.38 | 4 | 1 | 4 | 73.22 | High | Yes | 0 | 0 |
MW = Molecular weight (g/mol); H-Ac = No. of hydrogen bond acceptors; H-Do = No. of hydrogen bond donors; Nrot = No. of rotatable bonds; TPSA = Topological polar surface area (Å2); LogP = Predicted octanol/water partition coefficient; GIA = Gastrointestinal absorption; BBB = Blood brain barrier permeation; LFV = Lipinski filter violation; GFV = Ghose filter violation.
3.2. Molecular docking studies
Six well-known alkaloids from J. adhatoda and two previously recommended antiviral drugs against Mpro (darunavir and lopinavir) were docked to assess if the alkaloids exhibit higher or comparable binding energy to that of “Mpro-darunavir/lopinavir interaction”. The binding energy of darunavir and lopinavir towards Mpro was -7.4 and -7.3 kcal/mol, respectively (Table 2 ). We also estimated the binding energy of six alkaloids (vasicoline, vasicolinone, vasicinone, vasicine, adhatodine and anisotine) towards Mpro using molecular docking studies. It was found that the binding energy of vasicoline, vasicolinone, vasicinone, vasicine and adhatodine was in the range of -5.5 to -6.9 kcal/mol which was much lower compared to the standards, darunavir and lopinavir (Table 2). On the contrary, anisotine exhibited higher binding affinity (-7.9 kcal/mol) towards Mpro compared to that of darunavir and lopinavir (Table 2). Interestingly, binding affinity of anisotine towards Mpro is comparable to the binding affinity of few B. papyrifera polyphenols [papyriflavonol A, broussoflavan A and kazinol J] and some cholesterol-lowering drugs/statins (rosuvastatin and fluvastatin towards Mpro [20,29]. As a whole, sinceanisotine alone had higher binding affinity than darunavir and lopinavir, we decided to proceed further with this alkaloid.
Table 2.
The binding energy of different anti-HIV drugs and J. adhatoda alkaloids with Mpro.
| Drug | Binding energy (kcal/mol) |
|---|---|
| Darunavir | -7.4 |
| Lopinavir | -7.3 |
| Vasicoline | -6.7 |
| Vasicolinone | -6.9 |
| Vasicinone | -5.8 |
| Vasicine | -5.5 |
| Adhatodine | -6.9 |
| Anisotine | -7.9 |
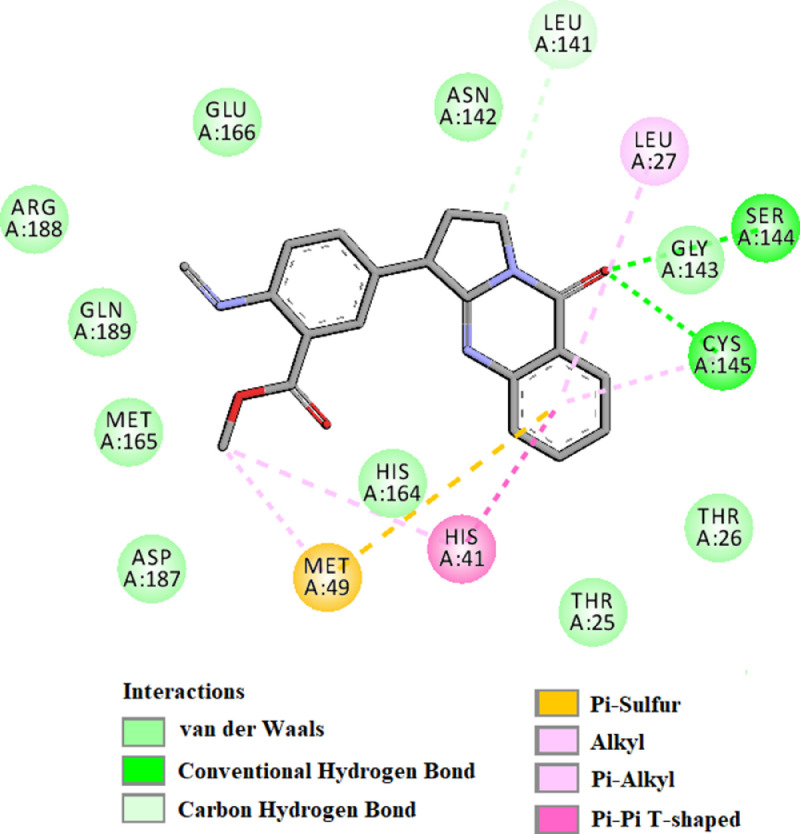
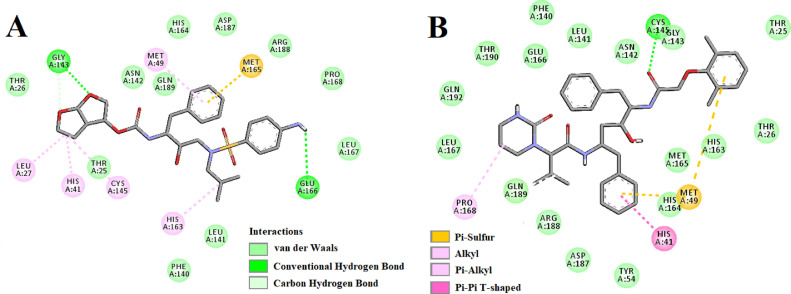
The amino acid residues within the active site of Mpro which were interacting with anisotine were carefully examined with the aid of discovery studio visualizer. It was evidenced that anisotine efficiently interacted with different amino acid residues of domain I and II of Mpro (Fig. 3 and Table 3 ). When anisotine was docked into the active site of Mpro, two hydrogen bond interactions [Ser144 (2.6 Å) and Cys145 (2.6 Å)] and ten van der Waals interactions (Thr25, Thr26, Asn142, Gly143, His164, Met165, Glu166, Asp187, Arg188 and Gln189) were evidenced (Fig. 3 and Table 3). Besides these, C-H interaction (Leu141), π-π as well as alkyl (His41), π-alkyl (Leu27) and π-sulphur (Met49) interactions were observed in the Mpro-anisotine complex (Fig. 3). Even darunavir and lopinavir also interacted with several critical residues within the active site of Mpro (Fig. 4 , Table 3). Darunavir interacted with Mpro via two hydrogen bonds [Gly143 (2.3 Å) and Glu166 (2.4 Å)], one π -sulfur bond (Met165) and multiple alkyl/ π-alkyl bonds (Leu27, His41, Met49, Cys145 and His163) (Fig. 4A and Table 3). It also formed many van der Waals interactions with different amino acid residues of Mpro (Fig. 4A). Lopinavir formed only one hydrogen bond with Cys145 and several other non-covalent bonds with various important amino acid residues (such as Thr26, His41, Met49, Phe140, Glu166, Leu167, etc) within the active site of Mpro (Fig. 4B and Table 3). Altogether, molecular docking studies revealed that anisotine interacted with both the catalytic residues (His41 and Cys145) of Mpro through non-covalent forces (Fig. 3). Most importantly, anisotine formed a single hydrogen bond with the Cys145 of Mpro. In fact, some Salvadora persica flavonoids (Isorhamnetin-3-O-β-D-glucopyranoside, Kaempferol-3-O-β-D-glucopyranoside etc.), green tea polyphenols (epigallocatechin gallate, epicatechingallate and gallocatechin-3-gallate), few B. papyrifera polyphenols [broussochalcone A, papyriflavonol A and 3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane] as well as some other phytochemicals interacted with the Cys145 of Mpro [25,29,51]. It is well known that Cys145 serves as a common nucleophile and plays a vital role in the proteolytic functioning of Mpro [52], [53], [54]. Thus, we believe that anisotine may possibly inhibit the proteolytic activity of Mpro and may potentially be used to treat patients with COVID-19. We further believe that anisotine may inhibit the proteolytic activity of Mpro in a better way than the flavonoids and other compounds which interact with Cys145 of Mpro via non-covalent interaction (other than H-bond interaction) [23,24,[55], [56], [57], [58], [59]]. Nevertheless, the Mpro-anisotine complex was further subjected to molecular dynamics simulations as well as binding free energy computations to assess the stability of this complex.
Fig. 3.
Molecular docking of anisotine with Mpro. The docked conformation of the Mpro-anisotine complex depicting the possible interactions with various amino acids of Mpro. This alkaloid interacts with many amino acid residues including His41 and Cys145 of Mpro.
Table 3.
Hydrogen bond interactions of anisotine, darunavir and lopinavir with the active site of SARS CoV-2 Mpro.
| Compounds | Number of H-bonds | Amino acids of Mpro involved in H-bonding | Hydrogen bond distance (Å) |
|---|---|---|---|
| Anisotine | 2 | Ser144 | 2.6 |
| Cys145 | 2.6 | ||
| Darunavir | 2 | Gly143 | 2.3 |
| Glu166 | 2.4 | ||
| Lopinavir | 1 | Cys145 | 2.3 |
Fig. 4.
Molecular docking of anti-HIV drugs with Mpro. Interactions of various amino acids of Mpro with darunavir (panel A) and lopinavir (panel B) are presented with the best docking pose.
3.3. Molecular dynamics (MD) simulation studies
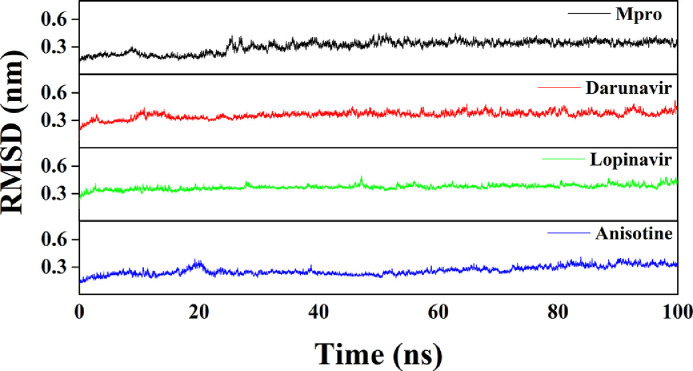
Several structural properties like overall complex stability (RMSD), conformational fluctuations (RMSF), structural compactness (Rg), and solvent accessibility (SASA) were investigated by MD simulations. We performed production run for 100 ns using GROMOS9653a6 force field of Mpro alone/Mpro (unligated) and Mpro complexed with two anti-HIV drugs, as well as with anisotine. The information about the structural stability of the protein-ligand complexes can be analyzed by RMSD. We estimated the RMSD of alpha-carbon atoms of all these systems (Fig. 5 ). The RMSD of Mpro (unligated) maintained a constant value (~0.21-0.22 nm) from 2 ns to 17 ns. Thereafter, the RMSD value gradually increased till 25 ns and reached ~0.35 nm. Then, the value was slightly decreased and persisted at ~0.31 nm from 65 ns till the end of the MD run. The RMSD values for both Mpro-darunavir and Mpro-lopinavir complexes were found to remain almost constant (~0.36-0.37 nm) from 10 ns to 100 ns with some marginal fluctuations (Fig. 5). On the other hand, the magnitude of RMSD corresponding to Mpro-anisotine complex attained an equilibrium value after 25 ns (~0.25-0.26 nm) and remained almost the same throughout the 100 ns simulation. The average RMSD values for Mpro (unligated), Mpro-darunavir complex and Mpro-lopinavir complex were found to be ~0.31 nm, ~0.36 nm and ~0.37 nm, respectively, which are in agreement with the previously reported values (Table 4 ) [23,29]. Whereas the average RMSD values of Mpro-anisotine complex was ~0.26 nm (Table 4), which was quite lower than Mpro (unligated), Mpro-darunavir and Mpro-lopinavir complexes. These results suggested that Mpro-anisotine complex was relatively more stable than that of Mpro-darunavir/ Mpro-lopinavir complexes.
Fig. 5.
RMSD plots of Mpro (unligated), Mpro-darunavir, Mpro-lopinavir and Mpro-anisotine complexes. The MD simulations for each system were performed for 100 ns. These MD trajectories were analyzed with the aid of RMSD.
Table 4.
Average values of the RMSD, RMSF, Rg, SASA and the total number of intermolecular hydrogen bonds formed during MD simulation for different systems.
| System | RMSD (nm) | RMSF (nm) | Rg(nm) | SASA (nm2) | Total number of H-bonds formed |
|---|---|---|---|---|---|
| Mpro (unligated) | 0.309 | 0.1937 | 2.195 | 151.4483 | 547 |
| Mpro-darunavir | 0.361 | 0.1952 | 2.197 | 151.1540 | 550 |
| Mpro-lopinavir | 0.371 | 0.1948 | 2.196 | 151.2825 | 551 |
| Mpro-anisotine | 0.262 | 0.1791 | 2.221 | 155.5451 | 555 |
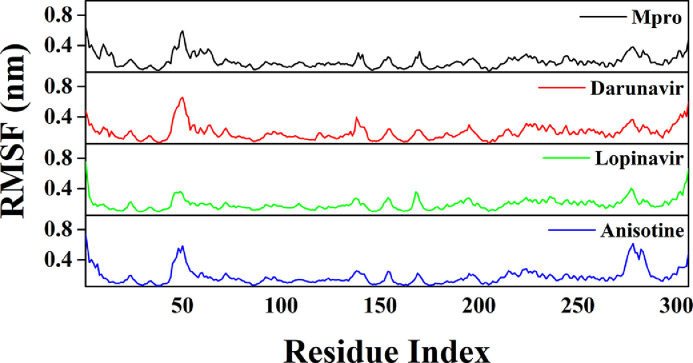
The conformational stability of the Mpro-anisotine complex was further analyzed by estimating the total number of intermolecular hydrogen bonds formed during the entire MD simulation (Table 4). The average number of intermolecular hydrogen bonds in the Mpro (unligated) system was 547, whereas, for Mpro-darunavir and Mpro-lopinavir complexes, the values were 550 and 551, respectively. A higher number of intermolecular hydrogen bonds (555) were found in the case of Mpro-anisotine complex (Table 4), which further suggests that Mpro-anisotine complex is more stable than the two selected Mpro-anti-HIV drug complexes. We have calculated the RMSF of alpha carbon atoms for all systems (Fig. 6 ). It was quite evident from the RMSF profiles that all the systems experience more conformational fluctuations in domain III. In the case of Mpro (unligated) system, we additionally observed higher fluctuations (upto ~0.6 nm) in a certain portion of domain I (residues 45-60). In fact, most of the amino acid residues within the domain I and II of this system had RMSF fluctuation below 0.3 nm. The average RMSF value for Mpro (unligated) system was ~0.194 nm (Table 4). The Mpro-darunavir and Mpro-lopinavir system experienced more or less similar conformational fluctuations to that of Mpro (unligated) system (Fig. 6). In fact, the fluctuations for the residues 45-60, were reduced upon the binding of lopinavir to Mpro (up to 0.35 nm). For both Mpro-darunavir and Mpro-lopinavir complexes, the average RMSF value was ~0.195 nm (Table 4). Upon analyzing all the RMSF profiles, it was clearly observed that Mpro-anisotine complexes showed lower fluctuations (especially in domain I and II) as compared to the Mpro (unligated) and Mpro-darunavir/lopinavir complexes (Fig. 6). The average RMSF values of Mpro-anisotine complex was ~0.179 nm (Table 4). Most importantly, the fluctuations of many key residues of the binding region of Mpro were lowered after binding to anisotine. These findings suggested that the overall conformational fluctuations of Mpro-anisotine complex are relatively less than that of Mpro-darunavir/Mpro-lopinavir complex.
Fig. 6.
RMSF profiles of Mpro (unligated), Mpro-anti-HIV drugs and Mpro-anisotine complexes. The RMSF values of Mpro (unligated) and Mpro-anti HIV drug complexes as well as Mpro-anisotine complex were plotted against the amino acid residues of Mpro.
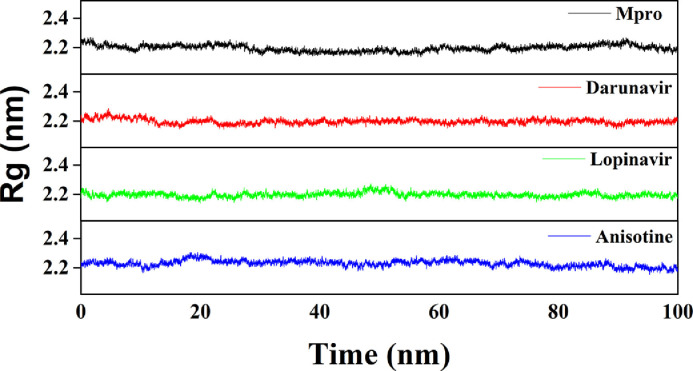
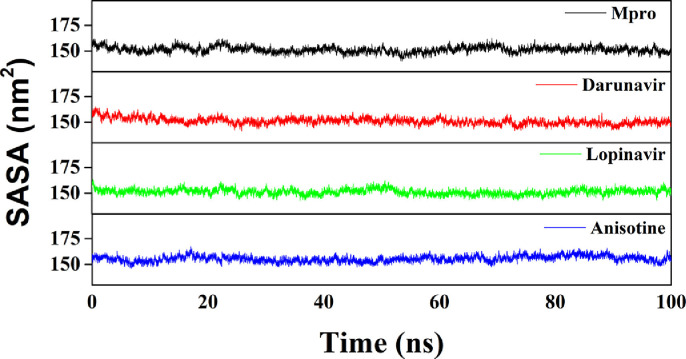
We have also estimated the Rg value to assess the compactness of all the complexes (Fig. 7 and Table 4). The average Rg value for Mpro (unligated) and the other two complexes (Mpro-darunavir and Mpro-lopinavir) was almost identical (~2.20 nm), whereas, a slightly increased average Rg value for Mpro-anisotine complex (2.221 nm) was observed (Table 4). These results suggested that the Mpro-anisotine complex was slightly less compact as compared to the two Mpro-anti-HIV drug complexes. SASA values were also calculated to assess the extent of expansion of protein volume in each system (Fig. 8 and Table 4). The average SASA values of Mpro (unligated), Mpro-darunavir complex, Mpro-lopinavir complex and Mpro-anisotine complex were 151.448 nm2, 151.154 nm2, 151.283 nm2 and 155.545 nm2, respectively. This suggests that Mpro expands a little upon its interaction with anisotine.
Fig. 7.
Estimation of Rg values of Mpro (unligated), Mpro-anti-HIV drugs and Mpro-anisotine complexes. The MD simulations for each system were performed for 100 ns. These MD trajectories were analyzed with the aid of Rg.
Fig. 8.
Determination of SASA values of Mpro (unligated), Mpro-anti-HIV drugs and Mpro-anisotine complexes. The MD simulations for each system were performed for 100 ns. These MD trajectories were analyzed with the aid of SASA.
Recently, we assessed the structural stability, conformational fluctuations, expansion of receptor (Mpro) volume and compactness of several Mpro-B. papyrifera polyphenol complexes [Mpro- broussochalcone A complex, Mpro-papyriflavonol A complex, Mpro-3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane complex, Mpro-broussoflavan A complex, Mpro-kazinol F complex and Mpro-kazinol J complex] using the similar MD simulation methodology adopted in this manuscript [29]. When we compared the above mentioned structural properties of Mpro-anisotine complex with those of Mpro-B. papyrifera polyphenol complexes, we observed that Mpro-anisotine complex is more stable than Mpro-3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane complex, Mpro-kazinol F complex and Mpro-kazinol J complex [29]. Furthermore, the conformational fluctuations of Mpro-anisotine complex is much less than Mpro-3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane complex. Interestingly, slight reduction in the complex rigidity and marginal expansion of Mpro were evidenced when anisotine and these B. papyrifera polyphenols interacted with Mpro [29].
3.4. MM-GBSA
The binding free energy of Mpro-anisotine as well as Mpro-anti-HIV drugs was calculated using MM-GBSA method. The binding free energy (ΔGbind) values of Mpro-darunavir and Mpro-lopinavir complexes were found to be -35.65 kcal/mol and -40.39 kcal/mol, respectively, whereas for Mpro-anisotine complex it was -42.23 kcal/mol (Table 5 ). The binding free energy value of “Mpro-anisotine interaction” is somewhat similar to the ΔGbind magnitude of some “Mpro-B. papyrifera polyphenol interaction” and “Mpro-pyronaridine/Mpro-pavinetant interaction” [29,60]. Altogether, it is quite evident from these MM-GBSA values that the alkaloid anisotine interacts with Mpro with a slightly higher binding free energy than that of darunavir/ lopinavir. The higher MM-GBSA value (ΔGbind) in the case of Mpro-anisotine is mostly contributed by the SASA and hydrophobic interactions.
Table 5.
MM-GBSA values of different Mpro-anti-HIV drugs and Mpro-anisotine complexes.
| System | Binding Free Energy (kcal/mol) |
|---|---|
| Mpro-darunavir | -35.65 |
| Mpro-lopinavir | -40.39 |
| Mpro-anisotine | -42.23 |
4. Conclusion
This study is aimed to test the inhibition potency of six well-known alkaloids from J. adhatoda leaves against SARS CoV-2 Mpro using a computational approach. Among the six alkaloids, only anisotine had higher AutoDock Vina energy values in compared to standard anti-HIV drugs, darunavir and lopinavir. Anisotine interacted with both the key catalytic residues (His41 and Cys145) of Mpro. The RMSD and RMSF profiles of Mpro-anisotine complex clearly suggested high stability and less conformational fluctuations. The Rg and SASA analysis revealed that Mpro-anisotine complex is slightly less compact and somewhat less expanded. Estimation of the number of intermolecular hydrogen bond formation and MM-GBSA analysis further reconfirmed that Mpro-anisotine is more stable than Mpro-darunavir/lopinavir complex. Overall, our findings revealed that anisotine has the potency to inhibit the proteolytic activity of SARS CoV-2 Mpro and may have some therapeutic effects against COVID-19 if proven in animal experiments and on patients.
CRediT authorship contribution statement
Rajesh Ghosh: Conceptualization, Data curation, Formal analysis, Investigation, Writing - original draft. Ayon Chakraborty: Conceptualization, Data curation, Formal analysis, Investigation, Writing - original draft. Ashis Biswas: Conceptualization, Formal analysis, Investigation, Resources, Writing - original draft. Snehasis Chowdhuri: Conceptualization, Formal analysis, Investigation, Resources, Software, Writing - original draft.
Declaration of Competing Interest
None.
Acknowledgments
RG acknowledges IIT Bhubaneswar for providing fellowship. The authors thank IIT Delhi HPC facility for computational resources.
References
- 1.Jamwal S., Gautam A., Elsworth J., Kumar M., Chawla R., Kumar P. An updated insight into the molecular pathogenesis, secondary complications and potential therapeutics of COVID-19 pandemic. Life Sci. 2020 doi: 10.1016/j.lfs.2020.118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanche S., Lin Y.T., Xu C., Romero-Severson E., Hengartner N., Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(7):1470–1477. doi: 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang B., Bragazzi N.L., Li Q., Tang S., Xiao Y., Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect. Dis. Modell. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382(8):727–733. doi: 10.1056/NEJMoa2001017. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta bio-medica. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. China medical treatment expert group for, clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grum-Tokars V., Ratia K., Begaye A., Baker S.C., Mesecar A.D. Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 2008;133(1):63–73. doi: 10.1016/j.virusres.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 11.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 12.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251 doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hage-Melim L., Federico L.B., de Oliveira N.K.S., Francisco V.C.C., Correia L.C., de Lima H.B., Gomes S.Q., Barcelos M.P., Francischini I.A.G., da Silva C. Virtual screening, ADME/Tox predictions and the drug repurposing concept for future use of old drugs against the COVID-19. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arun K.G., Sharanya C.S., Abhithaj J., Francis D., Sadasivan C. Drug repurposing against SARS-CoV-2 using E-pharmacophore based virtual screening, molecular docking and molecular dynamics with main protease as the target. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1779819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elmezayen A.D., Al-Obaidi A., Sahin A.T., Yelekci K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahanta S., Chowdhury P., Gogoi N., Goswami N., Borah D., Kumar R., Chetia D., Borah P., Buragohain A.K., Gogoi B. Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: an in silico based approach. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1768902. [DOI] [PubMed] [Google Scholar]
- 19.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2020:1–6. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 20.Reiner Z., Hatamipour M., Banach M., Pirro M., Al-Rasadi K., Jamialahmadi T., Radenkovic D., Montecucco F., Sahebkar A. Statins and the COVID-19 main protease: in silico evidence on direct interaction. Arch. Med. Science. 2020;16(3):490–496. doi: 10.5114/aoms.2020.94655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurung A.B., Ali M.A., Lee J., Farah M.A., Al-Anazi K.M. Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 M(pro) enzyme through in silico approach. Life Sci. 2020;255 doi: 10.1016/j.lfs.2020.117831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aanouz I., Belhassan A., El-Khatabi K., Lakhlifi T., El-Ldrissi M., Bouachrine M. Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: computational investigations. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1758790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S., Sarmah S., Lyndem S., Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors - an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyebi G.A., Ogunro O.B., Adegunloye A.P., Ogunyemi O.M., Afolabi S.O. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CL(pro)): an in silico screening of alkaloids and terpenoids from African medicinal plants. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., Chugh A., Giri A.P., Kulkarni M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umesh D.K., Selvaraj C., Singh S.K., Dubey V.K. Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1763202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1802347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorla U.S., Rao G.K., Kulandaivelu U.S., Alavala R.R., Panda S.P. Lead finding from selected flavonoids with antiviral (SARS-CoV-2) potentials against COVID-19: an in-silico evaluation. Comb. Chem. High Throughput Screen. 2020 doi: 10.2174/1386207323999200818162706. [DOI] [PubMed] [Google Scholar]
- 31.Jha D.K., Panda L., Lavanya P., Ramaiah S., Anbarasu A. Detection and confirmation of alkaloids in leaves of Justicia adhatoda and bioinformatics approach to elicit its anti-tuberculosis activity. Appl. Biochem. Biotechnol. 2012;168(5):980–990. doi: 10.1007/s12010-012-9834-1. [DOI] [PubMed] [Google Scholar]
- 32.Chavan R., Chowdhary A. In vitro inhibitory activity of justicia adhatoda extracts against influenza virus infection and hemagglutination. Int. J. Pharmaceut. Sci., Rev. Res. 2014;25:231–236. [Google Scholar]
- 33.Chavan R., Gohil D., Shah V., Kothari S., Chowdhary A. Anti-viral activity of Indian medicinal plant Justicia Adhatoda against herpes simplex virus: an in-vitro study. Int. J. Pharma Bio Sci. 2013;4:769–778. [Google Scholar]
- 34.M. Frisch, F. Clemente, Gaussian 09, Revision A. 01, MJ Frisch, GW Trucks, HB Schlegel, GE Scuseria, MA Robb, JR Cheeseman, G, Scalmani, V. Barone, B. Mennucci, GA Petersson, H. Nakatsuji, M. Caricato, X. Li, HP Hratchian, AF Izmaylov, J. Bloino, G. Zhe.
- 35.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris G.M., Huey R., Olson A.J. Using AutoDock for ligand-receptor docking. Curr. Protoc. Bioinform. Chapter. 2008;8 doi: 10.1002/0471250953.bi0814s24. Unit 8 14. [DOI] [PubMed] [Google Scholar]
- 37.Biovia D.S. Discovery studio modeling environment. Release. 2017 [Google Scholar]
- 38.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 39.Oostenbrink C., Villa A., Mark A.E., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25(13):1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 40.Schuttelkopf A.W., van Aalten D.M., PRODRG a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 41.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18(12):1463–1472. [Google Scholar]
- 42.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13(8):952–962. [Google Scholar]
- 43.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103(19):8577–8593. [Google Scholar]
- 44.Berendsen H.J.C., Postma J.P.M., Gunsteren W.F.V., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81(8):3684–3690. [Google Scholar]
- 45.Chen J. Drug resistance mechanisms of three mutations V32I, I47V and V82I in HIV-1 protease toward inhibitors probed by molecular dynamics simulations and binding free energy predictions. RSC Adv. 2016;6(63):58573–58585. [Google Scholar]
- 46.Chen J., Wang X., Zhu T., Zhang Q., Zhang J.Z. A comparative insight into amprenavir resistance of mutations V32I, G48V, I50V, I54V, and I84V in HIV-1 protease based on thermodynamic integration and MM-PBSA methods. J. Chem. Inf. Model. 2015;55(9):1903–1913. doi: 10.1021/acs.jcim.5b00173. [DOI] [PubMed] [Google Scholar]
- 47.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das B.K., Pv P., Chakraborty D. Computational insights into factor affecting the potency of diaryl sulfone analogs as Escherichia coli dihydropteroate synthase inhibitors. Comput. Biol. Chem. 2019;78:37–52. doi: 10.1016/j.compbiolchem.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Owis A.I., El-Hawary M.S., El Amir D., Aly O.M., Abdelmohsen U.R., Kamel M.S. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. 2020;10(33):19570–19575. doi: 10.1039/d0ra03582c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 53.Chou K.C., Wei D.Q., Zhong W.Z. Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochem. Biophys. Res. Commun. 2003;308(1):148–151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu M.F., Kuo C.J., Chang K.T., Chang H.C., Chou C.C., Ko T.P., Shr H.L., Chang G.G., Wang A.H., Liang P.H. Mechanism of the maturation process of SARS-CoV 3CL protease. J. Biol. Chem. 2005;280(35):31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allam A.E., Assaf H.K., Hassan H.A., Shimizu K., Elshaier Y.A.M.M. An in silico perception for newly isolated flavonoids from peach fruit as privileged avenue for a countermeasure outbreak of COVID-19. RSC Adv. 2020;10(50):29983–29998. doi: 10.1039/d0ra05265e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Havranek B., Islam S.M. An in silico approach for identification of novel inhibitors as potential therapeutics targeting COVID-19 main protease. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1776158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Islam R., Parves M.R., Paul A.S., Uddin N., Rahman M.S., Mamun A.A., Hossain M.N., Ali M.A., Halim M.A. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar A., Choudhir G., Shukla S.K., Sharma M., Tyagi P., Bhushan A., Rathore M. Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1772112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahmud S., Uddin M.A.R., Zaman M., Sujon K.M., Rahman M.E., Shehab M.N., Islam A., Alom M.W., Amin A., Akash A.S., Saleh M.A. Molecular docking and dynamics study of natural compound for potential inhibition of main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1796808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ibrahim M.A.A., Abdelrahman A.H.M., Hegazy M.F. In-silico drug repurposing and molecular dynamics puzzled out potential SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1791958. [DOI] [PMC free article] [PubMed] [Google Scholar]