Large cohort studies involving hundreds of thousands of participants have been established or launched in several regions worldwide. Cohorts provide great value for studying diverse populations and key demographic subgroups, rare genotypes and exposures, and gene-environment interactions.1 Each cohort is constrained, however, by its size, ancestral origins, and geographical boundaries, which limit the subgroups, exposures, outcomes, and interactions it can examine. Linking data across large cohorts provides a vast digital resource of diverse data to address questions that none of these cohorts can answer alone, enhancing the value of each cohort and leveraging the enormous investments made in them to date.
Leaders of large-scale cohorts, with support from the National Institutes of Health and the Wellcome Trust, and in collaboration with the Global Alliance for Genomics and Health (GA4GH) and the Global Genomic Medicine Collaborative (G2MC), have come together to form the International Hundred Thousand Plus Cohort Consortium (IHCC). As of May, 2020, IHCC comprises 103 cohorts in 43 countries involving nearly 50 million participants (figure , appendix). Collaborative efforts to date have focused on developing a queryable cohort registry and data sharing platform, identifying and piloting high-priority scientific projects, and developing a charter and governance structure to foster collaborations.
Figure.
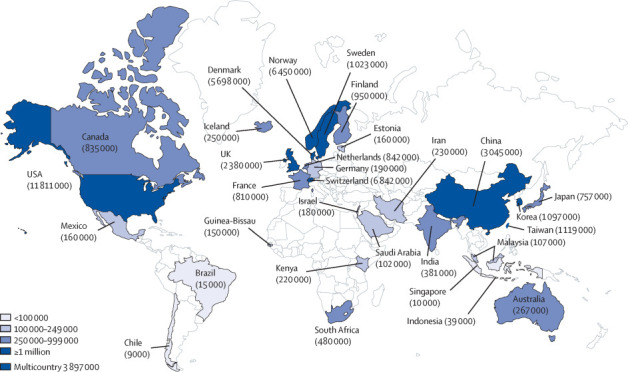
Map showing IHCC cohorts worldwide
IHCC members generally meet five criteria: greater than 100 000 enrolled participants; longitudinal follow-up in place for health outcomes; selection not based on a specific disease; biological samples collected from participants; and leaders willing and able to share data or metadata with IHCC members. Cohorts with less than 100 000 participants can apply to become full members if they include low-income and middle-income countries or disadvantaged populations in high-income countries, or if they collect data from exceptional or hard-to-accrue groups. Membership is granted by a majority vote of the Scientific Steering Committee, comprising 15 cohort leaders elected from and by the IHCC membership and representing the diversity of the IHCC. Cohorts not meeting all of the criteria for full membership, as well as people with specific expertise who do not bring a cohort into the IHCC, are eligible for affiliate membership. Industry representatives are eligible if they meet criteria for full or affiliate membership but must abide by IHCC guidelines for collaboration with industry and do not have voting rights. Policies have also been established for data sharing and collaborative publications.
An important first step in facilitating collaborations is to develop a standardised atlas or registry to share basic descriptive information about each cohort, to enhance the international visibility and engagement of the cohorts and to which cohort leaders could direct the myriad such enquiries they receive. IHCC's Data and Infrastructure Team has developed a prototype resource allowing investigators to identify IHCC member cohorts' key characteristics and standardise or harmonise key metadata elements to promote interoperability. Building on existing standards and infrastructure such as the GA4GH and Maelstrom projects,2, 3 the atlas is designed around use cases such as finding cohorts with specific measurements or particular demographic subgroups. Next steps will involve building semi-automated tools to import data dictionaries and map similar variables across datasets. Regulatory barriers to sharing of individual participant data in many countries might necessitate some analyses being done within cohorts in a federated model,4, 5 producing summary data to be shared for meta-analyses.
IHCC's Scientific Strategies Team is soliciting ideas for collaborative scientific projects from the IHCC membership, giving priority to projects involving innovative use of existing resources, broad scope across numerous cohorts, so-called quick wins within a finite timescale, and opportunities for the career development of junior researchers. A proof-of-principle scientific project involves development and testing of polygenic risk scores in four complex traits across four broad ancestry groups in seven cohorts to show the speed and robustness of this approach. With the advent of the COVID-19 pandemic, plans are underway to implement standardised collection of data and specimens to identify predictors of susceptibility to and severity of SARS-CoV2 infection as well as psychological and economic effects of the pandemic, particularly in the low-income and middle-income countries that are well-represented in the IHCC.6
Leaders of other large cohorts are invited to join the IHCC by contacting ihccinfo@ihccglobal.org. Cohort leaders are encouraged to participate in IHCC teams and annual international summits and to share their data in ways consistent with participants' consent and local regulations. IHCC views cohort independence and individuality as major strengths and is committed to ensuring that cohorts in low-income settings have sufficient resources to participate actively while maintaining their own sovereignty. Please join us!
Acknowledgments
GG reports being founder of the Global Genomic Medicine Collaborative, an independent not-for-profit 501(c)3 non-profit organisation. The authors express their appreciation for the valuable assistance of the IHCC Secretariat, and particularly Eric Plummer, Teji Rakhra-Burris, and Meredith Towery, in preparing this manuscript. We would also like to recognise the efforts of all on the IHCC Steering Committee, the teams, cohort leaders, and membership in bringing the IHCC to fruition.
Editorial note: the Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Supplementary Material
References
- 1.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 2.Global Alliance for Genomics and Health GENOMICS. A federated ecosystem for sharing genomic, clinical data. Science. 2016;352:1278–1280. doi: 10.1126/science.aaf6162. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron J, Doiron D, Marcon Y, Ferretti V, Fortier I. Fostering population-based cohort data discovery: The Maelstrom Research cataloguing toolkit. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoppers BM. Framework for responsible sharing of genomic and health-related data. HUGO J. 2014;8:3. doi: 10.1186/s11568-014-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contreras JL, Reichman JH. Sharing by design: data and decentralised commons: overcoming legal and policy obstacles. Science. 2015;350:1312–1314. doi: 10.1126/science.aaa7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbott A. Thousands of people will help scientists to track the long-term health effects of the coronavirus crisis. Nature. 2020;582:326. doi: 10.1038/d41586-020-01643-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


