Abstract
Background
On March 9, 2020, the first COVID-19 case was reported in Jodhpur, Rajasthan, in the northwestern part of India. Understanding the epidemiology of COVID-19 at a local level is becoming increasingly important to guide measures to control the pandemic.
Objective
The aim of this study was to estimate the serial interval and basic reproduction number (R0) to understand the transmission dynamics of the COVID-19 outbreak at a district level. We used standard mathematical modeling approaches to assess the utility of these factors in determining the effectiveness of COVID-19 responses and projecting the size of the epidemic.
Methods
Contact tracing of individuals infected with SARS-CoV-2 was performed to obtain the serial intervals. The median and 95th percentile values of the SARS-CoV-2 serial interval were obtained from the best fits with the weibull, log-normal, log-logistic, gamma, and generalized gamma distributions. Aggregate and instantaneous R0 values were derived with different methods using the EarlyR and EpiEstim packages in R software.
Results
The median and 95th percentile values of the serial interval were 5.23 days (95% CI 4.72-5.79) and 13.20 days (95% CI 10.90-18.18), respectively. R0 during the first 30 days of the outbreak was 1.62 (95% CI 1.07-2.17), which subsequently decreased to 1.15 (95% CI 1.09-1.21). The peak instantaneous R0 values obtained using a Poisson process developed by Jombert et al were 6.53 (95% CI 2.12-13.38) and 3.43 (95% CI 1.71-5.74) for sliding time windows of 7 and 14 days, respectively. The peak R0 values obtained using the method by Wallinga and Teunis were 2.96 (95% CI 2.52-3.36) and 2.92 (95% CI 2.65-3.22) for sliding time windows of 7 and 14 days, respectively. R0 values of 1.21 (95% CI 1.09-1.34) and 1.12 (95% CI 1.03-1.21) for the 7- and 14-day sliding time windows, respectively, were obtained on July 6, 2020, using method by Jombert et al. Using the method by Wallinga and Teunis, values of 0.32 (95% CI 0.27-0.36) and 0.61 (95% CI 0.58-0.63) were obtained for the 7- and 14-day sliding time windows, respectively. The projection of cases over the next month was 2131 (95% CI 1799-2462). Reductions of transmission by 25% and 50% corresponding to reasonable and aggressive control measures could lead to 58.7% and 84.0% reductions in epidemic size, respectively.
Conclusions
The projected transmission reductions indicate that strengthening control measures could lead to proportionate reductions of the size of the COVID-19 epidemic. Time-dependent instantaneous R0 estimation based on the process by Jombart et al was found to be better suited for guiding COVID-19 response at the district level than overall R0 or instantaneous R0 estimation by the Wallinga and Teunis method. A data-driven approach at the local level is proposed to be useful in guiding public health strategy and surge capacity planning.
Keywords: Epidemiology, SARS-CoV-2, COVID-19, serial interval, basic reproduction number, projection, outbreak response, India, mathematical modeling, infectious disease
Introduction
COVID-19 has emerged as the largest pandemic of the 21st century, with 30.7 million confirmed cases and approximately 950,000 deaths worldwide as of September 2020 [1]. India has become the second most affected country worldwide after the United States, with approximately 5.4 million confirmed COVID-19 cases [1]. COVID-19 is an emerging infectious disease, the first case being reported from Wuhan, China, in early December 2019 [2]. Various epidemiological studies are being performed to understand the transmission dynamics of the disease. Consequently, estimation of parameters such as the serial interval and basic reproduction number (R0) is being used to guide control strategies and enable disease forecasting [3-5].
In the early phase of the COVID-19 pandemic, India adopted a policy of universal health facility–based isolation of all individuals infected with SARS-CoV-2 irrespective of symptomatic status. However, in view of the increasing number of COVID-19 cases, home isolation of asymptomatic and mild cases was introduced on May 10, 2020 [6]. Due to the emerging nature of the outbreak and the evolving control measures, it is important to achieve a detailed epidemiological understanding of the COVID-19 situation at the district level to guide control measures and surge preparedness on a real-time basis.
Current mathematical modeling approaches for epidemiological understanding of COVID-19 in India are based on aggregate data reported at the national and state levels [7-13]. Very often, conclusions based on large-scale data are not appropriate for designing interventions at the local level. Therefore, we aimed to study the transmission of COVID-19 at the district level by estimating the serial interval and to determine the most suitable method for R0 estimation to support decision-making at the district level. We also aimed to demonstrate the feasibility of epidemic projection to guide COVID-19 response.
We studied the COVID-19 outbreak in the Jodhpur District of the state of Rajasthan in India. This mid-sized cultural and tourism hub is known as the gateway to the Thar Desert area in the northwestern part of India. Based on projection of 2011 census data to 2020 while assuming constant annual exponential growth, Jodhpur District has a population of 4.6 million, with an urban population of 1.6 million [14]. The first COVID-19 case in this district was reported on March 9, 2020, and at least one case has been reported daily since March 30, 2020.
Methods
Overview
We conducted a prospective observational study of the COVID-19 outbreak in Jodhpur, India. We used two data sources for the study. Firstly, serial intervals were estimated based on contact history of laboratory confirmed SARS-CoV-2 infected individuals. Secondly, the publicly available daily case count data were used together with the serial intervals to estimate R0 and project the size of the epidemic over the next 30 days. Individuals meeting the definition of a suspected case of COVID-19 were tested with the real time reverse transcriptase–polymerase chain reaction (rRT-PCR) at our institute in Jodhpur, India, as per national guidelines [15]. People who tested positive for SARS-CoV-2 were further assessed for their contact history with known COVID-19 cases in their household. The serial intervals were estimated based on the length of time between the onset of symptoms of the identified infectors and infectees. For asymptomatic individuals, the date of collection of the first positive sample was taken as a proxy of symptom onset.
The basic reproduction number (R0) is defined as the average number of susceptible individuals infected by a single primary case [16]. For R0 estimation, serial interval values along with daily case count data were taken from the official daily report released by the Jodhpur District administration. These data are also available on the internet [17].
Ethical Approval
Informed consent was obtained prior to eliciting contact history for serial interval estimation. The study was approved by the Institutional Ethics Committee (Ref: AIIMS/IEC/2020-21/3047).
Serial Interval Estimation
The mean (SD) of the serial intervals was calculated. Further, the serial interval data were fitted to weibull, log-normal, log-logistic, and generalized gamma distributions using the Flexsurv package in R software version 4.0.0 [18]. The estimates of the median serial interval were taken from the best-fitting model based on the minimum Akaike information criterion (AIC) value. The standard maximum likelihood approach was used to obtain the best model fit to the actual data.
Estimation of R0
The daily COVID-19 case data in Jodhpur District were converted to incidence objects using the Incidence package in R software [19]. The EarlyR and EpiEstim packages in R were used to estimate the overall and instantaneous values of R0, respectively, using the parameter estimates of the serial interval [20,21]. We used two main standard methods of estimation of the instantaneous R0 values to visualize their response to changes in case trends and to assess their utility for understanding real-time transmission dynamics at a local level. These methods use different mathematical modeling principles and assumptions.
Instantaneous R0 values were first calculated using the method of estimating daily incidence based on a Poisson process determined by daily infectiousness, as proposed by Jombart et al [19] and Nouvellet et al [22]. Here, λt, the force of infection observed on day t, is expressed by the following equation:
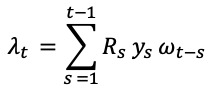
where ys is the incidence of cases on day s and Rs is the instantaneous reproduction number on day s. The value of ωt-s is the probability mass distribution of the serial interval, which represents the infectiousness of incident cases on day s to result in secondary cases on day t. In the absence of an exhaustive symptomatic history of each reported case, we approximated the day the case was reported as the day of onset, a practical approach used in earlier studies [22].
Next, we used a method described by Wallinga and Teunis [23] to estimate the time-varying R0 based on the probability of transmission between infector-infectee pairs. We adopted the parametric method of specifying the mean (SD) of the serial interval distribution for both methods. Time windows of 7 days and 14 days were used to calculate the instantaneous R0.
Forecasting of the Epidemic Size
The numbers of daily and cumulative COVID-19 cases for the next 30 days were forecasted based on the overall R0 value and the R0 value for the past 30 days as input parameters using the projections package in R [19]. The observed serial interval distribution was specified as the scale and shape parameters of the gamma distribution. Daily COVID-19 cases were predicted based on a Poisson process determined by daily infectiousness [22]. The specified serial interval distribution was taken as a prior while using the Bayesian methodology for Markov chain Monte Carlo sampling using the Metropolis algorithm. The 95% CIs of the projected daily and cumulative incidences were calculated using the bootstrap resampling method with 1000 samples.
Further, we considered two scenarios: one with a reasonable reduction of 25% SARS-CoV-2 transmission and one with an aggressive reduction of 50% transmission. A reasonable reduction would be related to compliance with strengthening of existing measures, such as contact tracing, testing, and prompt isolation of infected individuals along with physical distancing measures. Aggressive transmission reduction measures included universal mask-wearing and measures to reduce outdoor transmission through prevention of gatherings: closures of places of worship, marketplaces, restaurants, schools, and gymnasiums, along with introduction of nighttime curfews [24].
Results
Serial Interval
From the reporting of the first case of COVID-19 in Jodhpur District on March 9, 2020, to July 6, 2020, 3178 cases were reported in the district in a span of 120 days (see Figure 1). Serial interval data for 103 infector-infectee pairs were obtained through contact tracing of known infected cases (Multimedia Appendix 1).
Figure 1.
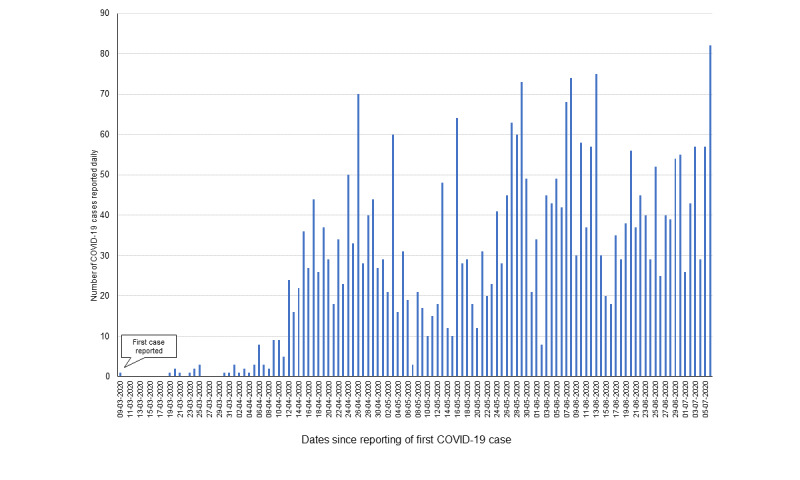
Numbers of COVID-19 cases reported daily in Jodhpur, India, from March 9 to July 6, 2020.
The mean serial interval was 6.23 days (SD 3.49). The generalized gamma distribution was found to best fit the serial interval and showed the minimum AIC value (see Table 1).
Table 1.
Fits of weibull, log-normal, log-logistic, and generalized gamma distributions with the serial interval data for SARS-CoV-2 infection in Jodhpur District, India, and the estimated median and 95 percentile values (N=103 pairs).
| Type of distribution used in the model | –2 log-likelihood |
Number of model parameters (k) |
AICa (–2 log-likelihood + 2k) |
Serial interval (days) | |
|
|
|
|
|
Median (95% CI) | 95th percentile (95% CI) |
| Weibull | 520.30 | 2 | 524.30 | 5.83 (5.18-6.54) | 12.51 (11.28-13.95) |
| Log-normal | 488.55 | 2 | 492.55 | 5.40 (5.06-6.07) | 11.96 (10.43-13.82) |
| Log-logistic | 492.46 | 2 | 496.46 | 5.44 (4.96-5.98) | 12.15 (10.44-14.39) |
| Gamma | 500.15 | 2 | 504.15 | 5.77 (5.23-6.33) | 11.79 (10.52 -13.22) |
| Generalized gamma | 482.97 | 3 | 488.97 | 5.23 (4.72-5.79) | 13.20 (10.90-18.18) |
aAIC: Akaike information criterion.
The median and 95th percentile values of the serial interval were 5.23 days (95% CI 4.72-5.79) and 13.20 days (95% CI 10.90-18.18), respectively, estimated from the fitted generalized gamma distribution (see Figure 2).
Figure 2.
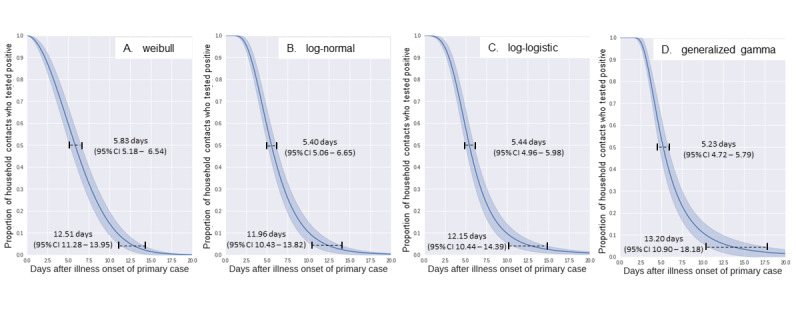
Estimates of the median and 95th percentile of the serial interval data fitted to (A) weibull, (B) log-normal, (C) log-logistic, and (D) generalized gamma distributions (N=103 pairs).
Estimation of R0
The overall R0 value in the 30 days after the first case was reported was 1.62 (95% CI 1.07-2.17), which subsequently decreased to 1.15 (95% CI 1.09-1.21). The overall R0 value for the entire outbreak duration was 1.07 (95% CI 1.04-1.11), whereas it was 1.20 (95% CI 1.14-1.27) in the last 30 days.
The time-dependent instantaneous R0 values calculated using the method by Jombart et al [19] and Nouvellet et al [22] yielded maximum values of 6.53 (95% CI 2.12-13.38) and 3.43 (95% CI 1.71-5.74) using sliding time windows of 7 days and 14 days, respectively (see Table 2 and Figure 3). Similarly, using the method described by Wallinga and Teunis [23], the maximum values of the instantaneous R0 were 2.96 (95% CI 2.52-3.36) and 2.92 (95% CI 2.65-3.22) for the 7- and 14-day time windows, respectively (see Table 2 and Figure 3). The peak R0 values corresponded with the daily rising trend in COVID-19 cases that was reported (see Figure 3).
Table 2.
Summary of the time-dependent R0 values estimated by the different methods.
| Method used and sliding time windows | Minimum value (95% CI) | Maximum value (95% CI) | Latest value as of July 6, 2020 (95% CI) | |
| Jombart et al and Nouvellet et al | ||||
|
|
7-day | 0.52 (0.42-0.62) | 6.53 (2.12-13.38) | 1.21 (1.09-1.34) |
| 14 day | 0.72 (90.64-0.80) | 3.43 (1.71-5.74) | 1.12 (1.03-1.21) | |
| Wallinga and Teunis | ||||
|
|
7-day | 0.32 (0.27-0.36) | 2.96 (2.52-3.36) | 0.32 (0.27-0.36) |
| 14-day | 0.61 (0.58-0.63) | 2.92 (2.65-3.22) | 0.61 (0.58-0.63) | |
Figure 3.
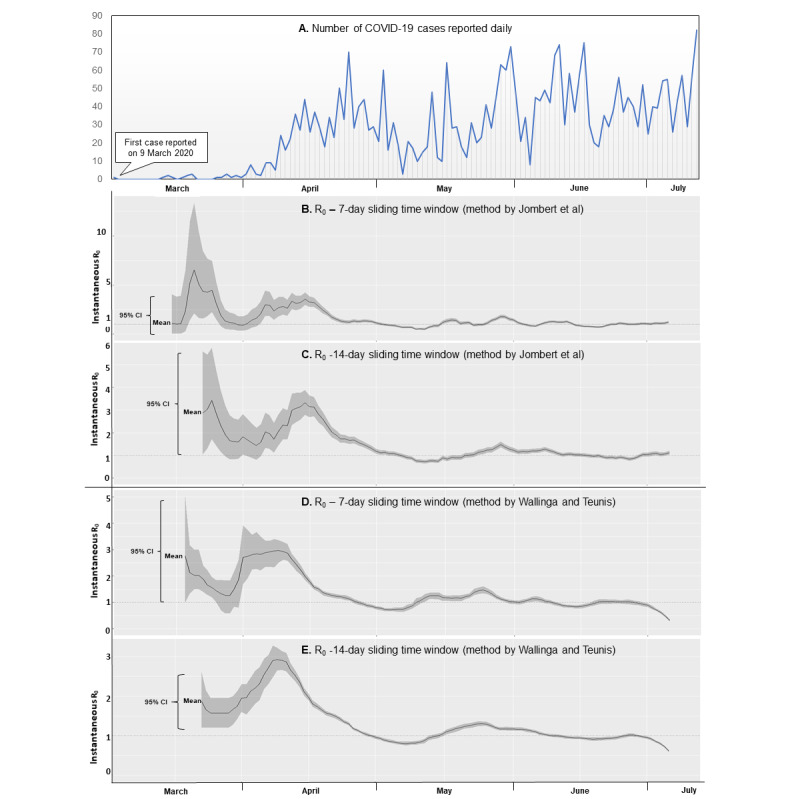
Daily COVID-19 cases in Jodhpur District, India, from March 9 to July 6, 2020 (A), instantaneous R0 values estimated using the method by Jombart et al (B-C), and instantaneous R0 values estimated using the method by Wallinga and Teunis using time windows of 7 and 14 days, respectively (D-E).
The latest instantaneous R0 values estimated on July 6, 2020, using the method by Jombart et al [19] and Nouvellet et al [22], were 1.21 (95% CI 1.09-1.34) and 1.12 (95% CI 1.03-1.21) for 7- and 14-day sliding time windows, respectively (see Table 2 and Figure 3). Similarly, the latest instantaneous R0 values estimated on July 6, 2020, using the method by Wallinga and Teunis [23] were 0.32 (95% CI 0.27-0.36) and 0.61 (95% CI 0.58-0.63), for the 7- and 14-day sliding time windows, respectively (see Table 2 and Figure 3).
Projection of Epidemic Size
The number of daily cases projected for the next month based on an overall R0 value of 1.20 (corresponding to the most recent 30 days of transmission) ranged from 55 (95% CI 38-71) on July 7, 2020 (day 1), to 143 (95% CI 110-175) on August 5, 2020, (ie, on day 30; see Figure 4). Similarly, the number of daily cases projected for the next month while taking the most recent 14-day rolling instantaneous R0 value of 1.12 ranged from 52 (95% CI 38-66) on day 1 to 91 (95% CI 66-116) on day 30 (see Figure 4). The cumulative projections of the number of COVID-19 cases over the next 30 days using the R0 values of 1.20 and 1.12 were 2817 (95% CI 2374-3259) and 2131 (95% CI 1799-1462), respectively.
Figure 4.
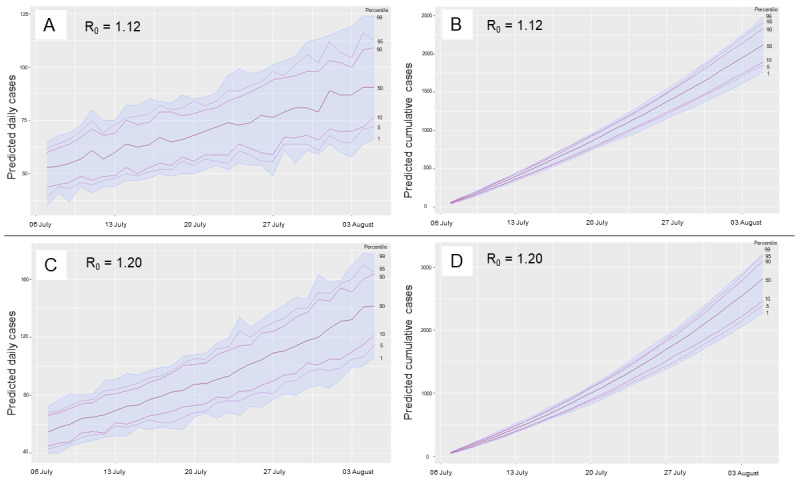
Projections of daily and cumulative COVID-19 caseloads over the next 30 days using the instantaneous R0 value of 1.12 on July 6, 2020 (A-B) and the overall R0 value of 1.20 for the most recent 30 days (C-D).
The scenarios of 25% and 50% transmission reduction of the most recent time-dependent R0 estimate (ie, reduction of R0 from 1.12 to 0.84 and 0.56, respectively) resulted in monthly projections of 880 cases (95% CI 699-1061) and 341 cases (95% CI 265-418); these projections correspond to 58.7% and 84.0% reductions in the epidemic size in Jodhpur, respectively.
Discussion
Implications of the Serial Interval and Estimated R0 Values
Our observation of the mean serial interval fell within a range of 4 to 8 days, as estimated by a meta-analysis of 7 studies conducted during the early phase of the COVID-19 pandemic [25]. Another meta-analysis including studies only from China estimated a range of serial intervals from 4.10 to 7.5 days [26]. Our experience suggests that the median and 95% CI estimates of the serial interval should be reported alongside the mean and SD, as the latter approach is more susceptible to influence by extreme values. It has also been suggested that longer serial interval intervals may be due to preventive interventions introduced during the course of the epidemic, which tend to reduce transmission [27,28]. Therefore, it is preferable to estimate the recent serial interval at a local level to better understand the transmission of SARS-CoV-2.
The distribution of R0 values was consistent with observations from other countries, indicating a similar transmission pattern [4,28]. The peak of the R0 value was reached in the first week of April 2020. The subsequent reduction toward the end of April can be attributed to aggressive testing, contact tracing, and isolation measures implemented in the urban area of Jodhpur during that month. Our R0 estimate for the first month (1.61) was slightly higher than the national estimate of 1.47 and lower than the estimate from the state of Tamil Nadu (1.88) in India during the same period of March to April 2020 [9,13]. District level R0 estimates are more likely to show pronounced fluctuations than state or national estimates, as the latter are aggregated across a wide range of epidemiological settings. Because district-level R0 estimates were not available from India, we compared our findings with those from the cities of Qom and Shahroud in Iran [29,30]. Similar to these cities, Jodhpur showed a trend of high values of R0 in the first 14-30 days, with a subsequent decrease toward 1 [29,30]. The initially high R0 values can be attributed to the suddenness of the outbreak if the surveillance system is robust. The high values may also be due to a sudden start of case reporting following an initial period of underreporting, leading to an artefactual peak in R0 [30].
Earlier detection of infection followed by isolation is known to reduce the R0 value by limiting both the duration of effective contact and the number of susceptible people an infected individual can come in contact with [16]. Our findings further support that parameters such as the serial interval, incubation period, and R0 value are likely to vary throughout the course of the epidemic and will depend on local factors influencing transmission, such as demographics, environmental conditions, modeling methodology, and stringency of control measures [16,30].
Epidemic Projections
The projected estimate of daily cases and the final outbreak size were found to depend on the value of R0 entered in the model [31-33]. The method used to estimate the R0 value and the time window over which R0 was calculated both influenced the final projection by a wide margin. The 14-day time window yielded less variable instantaneous R0 estimates compared to the 7-day time window. We found that the method by Wallinga and Teunis was more sensitive to recent fluctuations in daily case count than the method by Jombart et al in the same time window. Further, per the renewal equation stated earlier, the values of R0 are most influenced by the trend in daily cases reported within the range of the serial interval (ie, within 5 to 6 days). This model also assumes homogenous mixing, which becomes less applicable with larger populations in which cases emerge from widely separated clusters. Also, the impact of the method of R0 estimation and the time window was more pronounced when there was a fluctuating trend in cases or when the R0 value was close to 1. In research settings, R0 values should be tested through sensitivity analyses by considering variations in time windows and durations and using different methods so that reliable projections can be provided for larger populations [31]. For routine use within program settings at the district level, the method by Jombart et al may be preferable for monitoring the effectiveness of control methods and providing prior R0 values for projections compared to the method by Wallinga and Teunis and the overall R0 estimation using the EarlyR package in R.
Our study was based on contact history of infected individuals instead of on daily follow-up of contacts of infected individuals for disease onset. Therefore, we minimized underreporting of longer serial intervals, which may be due to right-truncation in the follow-up method for assessing the serial interval [30]. Further, the use of the time-varying method for daily R0 estimation and the maximum likelihood method for overall R0 estimation has the benefit of lower bias compared to the exponential growth and sequential Bayesian methods [34]. It also enables assessment of the effectiveness of control measures on a real-time basis, in contrast to other methods that only provide an aggregate R0 value [30].
Limitations
One limitation of our study is that population level estimates relying on daily official reports can underestimate the value of R0 compared to those of closed populations because many infected individuals are likely to be missed, especially if the testing capacity is limited or the proportion of asymptomatic people is high [31]. Further, modeling assumptions such as assuming a finite probability of interaction of infector-infectee pairs reported within a serial interval range may not be applicable for large population cohorts [23]. To overcome these limitations, use of both spatial and temporally structured data has been proposed [35]. The use of contact tracing applications that provide anonymized geolocated data and serial interval estimates could provide more timely and robust epidemiological understanding of emerging diseases such as COVID-19 [36,37].
Conclusions
Public health measures such as testing, contact tracing, and home isolation were found to reduce the instantaneous R0 value and could thereby reduce the final outbreak size. Instantaneous R0 estimated using the method proposed by Jombart et al is recommended for guiding COVID-19 response strategy at district level in preference to the method proposed by Wallinga and Teunis and to aggregate R0 calculation. The final epidemic size was found to be influenced by the R0 value, which in turn depended on the stringency of control measures. Even a marginal reduction in R0 as a result of strengthening control measures was found to considerably reduce the projected COVID-19 burden at the district level. Projections based on publicly released daily COVID-19 case data are feasible and could be useful in guiding a data-driven COVID-19 response strategy at a local level. This could be used for both surge capacity planning of the number of hospital beds and ventilators required and for public health responses such as the number of staff required for contact tracing and for provisioning of institutional quarantine or isolation facilities. Therefore, considering the increasing caseload and dynamic situation of COVID-19, a decentralized evidence-driven approach is currently needed.
Acknowledgments
We acknowledge the district administration of Jodhpur for providing the daily COVID-19 case data. We also thankfully acknowledge the staff involved in laboratory diagnosis of COVID-19 and the Master of Public Health scholars at All India Institute of Medical Sciences, Jodhpur, for supporting the epidemiological data collection: Dr Jay Shree Shekhawat, Mr Prasannajeet Bal, Dr Ipsa Kutlehrria, Dr Nainsi Gupta, Dr Neelam Kumari, Dr Mahima Choudhary, Dr Musarrat Siddiqui, Dr Sonali Bhattacharya, Ms Himani, Dr Uplabdhi Sahu, Dr. Shubham, Dr Oshi Chaturvedi, Ms Ashu Ranga, Dr Premlata Meghwal, Dr Chunni Lal, Dr Rupali Gupta, Dr Zeba Bano, Dr Diksha Mahajan, Ms Abhilipsa Pradhan, Dr Jinesh Saini, Dr Neha Mantri, and Dr Nishant Soni. The authors declare that no funding was received from any source for the study or the preparation of this paper. The views expressed in this article are those of the authors alone and do not necessarily represent the views of their organizations.
Abbreviations
- AIC
Akaike information criterion
- R0
basic reproduction number
- rRT-PCR
real-time reverse transcription–polymerase chain reaction
Appendix
Data used for the serial interval estimation of COVID-19 in Jodhpur, India (103 pairs).
Footnotes
Authors' Contributions: MKV, VG, and NK collected the data, and SS conducted the analysis. SS wrote the draft manuscript with further input from MKV, VG, AG, MKG, PB, and SM. PB coordinated the data collection process. SM provided overall supervision of the lab testing, clinical care, and research related to COVID-19 at the All India Institute of Medical Sciences, Jodhpur, India. All authors approved the final manuscript.
Conflicts of Interest: None declared.
References
- 1.Weekly epidemiological update for COVID-19 – 21 September 2020. World Health Organization. 2020. Sep 21, [2020-09-23]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200921-weekly-epi-update-6.pdf?sfvrsn=d9cf9496_6.
- 2.Wu Jt, Leung K, Leung Gm. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020 Feb;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alimohamadi Y, Taghdir M, Sepandi M. Estimate of the Basic Reproduction Number for COVID-19: A Systematic Review and Meta-analysis. J Prev Med Public Health. 2020 May;53(3):151–157. doi: 10.3961/jpmph.20.076. doi: 10.3961/jpmph.20.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lv M, Luo X, Estill J, Liu Y, Ren M, Wang J, Wang Q, Zhao S, Wang X, Yang S, Feng X, Li W, Liu E, Zhang X, Wang L, Zhou Q, Meng W, Qi X, Xun Y, Yu X, Chen Y, and on behalf of the COVID-19 evidence and recommendations working group Coronavirus disease (COVID-19): a scoping review. Euro Surveill. 2020 Apr;25(15):2000125. doi: 10.2807/1560-7917.ES.2020.25.15.2000125. http://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2020.25.15.2000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulut C, Kato Y. Epidemiology of COVID-19. Turk J Med Sci. 2020 Apr 21;50(SI-1):563–570. doi: 10.3906/sag-2004-172. doi: 10.3906/sag-2004-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Revised guidelines for Home Isolation of very mild/pre-symptomatic COVID-19 cases. Ministry of Health and Family Welfare, Government of India. 2020. May 10, [2020-09-23]. https://www.mohfw.gov.in/pdf/RevisedguidelinesforHomeIsolationofverymildpresymptomaticCOVID19cases10May2020.pdf.
- 7.Patrikar S, Poojary D, Basannar D, Faujdar D, Kunte R. Projections for novel coronavirus (COVID-19) and evaluation of epidemic response strategies for India. Med J Armed Forces India. 2020 Jul;76(3):268–275. doi: 10.1016/j.mjafi.2020.05.001. http://europepmc.org/abstract/MED/32773928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambikapathy B, Krishnamurthy K. Mathematical Modelling to Assess the Impact of Lockdown on COVID-19 Transmission in India: Model Development and Validation. JMIR Public Health Surveill. 2020 May 07;6(2):e19368. doi: 10.2196/19368. https://publichealth.jmir.org/2020/2/e19368/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanagarathinam K, Sekar K. Estimation of the reproduction number and early prediction of the COVID-19 outbreak in India using a statistical computing approach. Epidemiol Health. 2020 May 09;42:e2020028. doi: 10.4178/epih.e2020028. doi: 10.4178/epih.e2020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khajanchi S, Sarkar K. Forecasting the daily and cumulative number of cases for the COVID-19 pandemic in India. Chaos. 2020 Jul;30(7):071101. doi: 10.1063/5.0016240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh P, Ghosh R, Chakraborty B. COVID-19 in India: Statewise Analysis and Prediction. JMIR Public Health Surveill. 2020 Aug 12;6(3):e20341. doi: 10.2196/20341. https://publichealth.jmir.org/2020/3/e20341/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandal S, Bhatnagar T, Arinaminpathy N, Agarwal A, Chowdhury A, Murhekar M, Gangakhedkar R, Sarkar S. Prudent public health intervention strategies to control the coronavirus disease 2019 transmission in India: A mathematical model-based approach. Indian J Med Res. 2020;151(2 & 3):190–199. doi: 10.4103/ijmr.IJMR_504_20. http://www.ijmr.org.in/article.asp?issn=0971-5916;year=2020;volume=151;issue=2;spage=190;epage=199;aulast=Mandal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhaskar A, Ponnuraja C, Srinivasan R, Padmanaban S. Distribution and growth rate of COVID-19 outbreak in Tamil Nadu: A log-linear regression approach. Indian J Public Health. 2020;64(6):188. doi: 10.4103/ijph.ijph_502_20. [DOI] [PubMed] [Google Scholar]
- 14.Jodhpur District Census Handbook. Ministry of Home Affairs, Government of India. 2011. [2020-09-23]. https://censusindia.gov.in/2011census/dchb/DCHB_A/08/0815_PART_A_DCHB_JODHPUR.pdf.
- 15.Strategy for COVID-19 Testing in India – Version 5. Indian Council of Medical Research. 2020. May 18, [2020-09-03]. https://www.icmr.gov.in/pdf/covid/strategy/Testing_Strategy_v5_18052020.pdf.
- 16.Delamater PL, Street EJ, Leslie TF, Yang YT, Jacobsen KH. Complexity of the Basic Reproduction Number (R0) Emerg Infect Dis. 2019 Jan;25(1):1–4. doi: 10.3201/eid2501.171901. doi: 10.3201/eid2501.171901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.COVID-19 India. [2020-09-03]. https://www.covid19india.org/
- 18.Jackson C. flexsurv: A Platform for Parametric Survival Modeling in R. J Stat Softw. 2016 May 12;70:1–33. doi: 10.18637/jss.v070.i08. http://europepmc.org/abstract/MED/29593450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jombart T, Nouvellet P, Bhatia S, Kamvar Z, Taylor T, Ghozzi S. Projections: Project future case incidence. R Project. 2018. [2020-09-23]. https://cran.r-project.org/web/packages/projections/index.html.
- 20.Jombart T, Cori A, Nouvellet P. earlyR: Estimation of Transmissibility in the Early Stages of a Disease Outbreak. R Project. 2017. [2020-09-23]. https://cran.r-project.org/web/packages/earlyR/index.html.
- 21.Cori A, Cauchemez S, Ferguson NM, Fraser C, Dahlqwist E, Demarsh PA, Jombart T, Kamvar ZN, Lessler J, Li S, Polonsky JA, Stockwin J, Thompson R, van Gaalen R. EpiEstim: Estimate Time Varying Reproduction Numbers from Epidemic Curves. R Project. [2020-09-03]. https://cran.r-project.org/web/packages/EpiEstim/index.html.
- 22.Nouvellet P, Cori A, Garske T, Blake IM, Dorigatti I, Hinsley W, Jombart T, Mills HL, Nedjati-Gilani G, Van Kerkhove MD, Fraser C, Donnelly CA, Ferguson NM, Riley S. A simple approach to measure transmissibility and forecast incidence. Epidemics. 2018 Mar;22:29–35. doi: 10.1016/j.epidem.2017.02.012. https://linkinghub.elsevier.com/retrieve/pii/S1755-4365(17)30024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am J Epidemiol. 2004 Sep 15;160(6):509–16. doi: 10.1093/aje/kwh255. http://europepmc.org/abstract/MED/15353409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sjödin H, Wilder-Smith A, Osman S, Farooq Z, Rocklöv J. Only strict quarantine measures can curb the coronavirus disease (COVID-19) outbreak in Italy, 2020. Euro Surveill. 2020 Apr 02;25(13):2000280. doi: 10.2807/1560-7917.es.2020.25.13.2000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park M, Cook AR, Lim JT, Sun Y, Dickens BL. A Systematic Review of COVID-19 Epidemiology Based on Current Evidence. J Clin Med. 2020 Mar 31;9(4):967. doi: 10.3390/jcm9040967. https://www.mdpi.com/resolver?pii=jcm9040967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Wang T, Xie S. Meta-analysis of several epidemic characteristics of COVID-19. medRxiv. Preprint posted online on June 3. 2020 doi: 10.1101/2020.05.31.20118448. doi: 10.1101/2020.05.31.20118448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Najafi F, Izadi N, Hashemi-Nazari S, Khosravi-Shadmani F, Nikbakht R, Shakiba E. Serial interval and time-varying reproduction number estimation for COVID-19 in western Iran. New Microbes New Infect. 2020 Jul;36:100715. doi: 10.1016/j.nmni.2020.100715. https://linkinghub.elsevier.com/retrieve/pii/S2052-2975(20)30067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020 Mar 13;27(2) doi: 10.1093/jtm/taaa021. http://europepmc.org/abstract/MED/32052846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khosravi A, Chaman R, Rohani-Rasaf M, Zare F, Mehravaran S, Emamian M. The basic reproduction number and prediction of the epidemic size of the novel coronavirus (COVID-19) in Shahroud, Iran. Epidemiol Infect. 2020 Jun 10;148:e115. doi: 10.1017/S0950268820001247. http://europepmc.org/abstract/MED/32517845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aghaali M, Kolifarhood G, Nikbakht R, Saadati HM, Hashemi Nazari SS. Estimation of the serial interval and basic reproduction number of COVID-19 in Qom, Iran, and three other countries: A data-driven analysis in the early phase of the outbreak. Transbound Emerg Dis. 2020 May 30; doi: 10.1111/tbed.13656. http://europepmc.org/abstract/MED/32473049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rocklöv J, Sjödin H, Wilder-Smith A. COVID-19 outbreak on the Diamond Princess cruise ship: estimating the epidemic potential and effectiveness of public health countermeasures. J Travel Med. 2020 May 18;27(3):e. doi: 10.1093/jtm/taaa030. http://europepmc.org/abstract/MED/32109273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Diao M, Yu W, Pei L, Lin Z, Chen D. Estimation of the reproductive number of novel coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: A data-driven analysis. Int J Infect Dis. 2020 Apr;93:201–204. doi: 10.1016/j.ijid.2020.02.033. https://linkinghub.elsevier.com/retrieve/pii/S1201-9712(20)30091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tildesley MJ, Keeling MJ. Is R(0) a good predictor of final epidemic size: foot-and-mouth disease in the UK. J Theor Biol. 2009 Jun 21;258(4):623–9. doi: 10.1016/j.jtbi.2009.02.019. http://europepmc.org/abstract/MED/19269297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obadia T, Haneef R, Boëlle PY. The R0 package: a toolbox to estimate reproduction numbers for epidemic outbreaks. BMC Med Inform Decis Mak. 2012 Dec 18;12(1):147. doi: 10.1186/1472-6947-12-147. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/1472-6947-12-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng T, Wen T. Spatially Adjusted Time-varying Reproductive Numbers: Understanding the Geographical Expansion of Urban Dengue Outbreaks. Sci Rep. 2019 Dec 16;9(1):19172. doi: 10.1038/s41598-019-55574-0. doi: 10.1038/s41598-019-55574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Guo X. Global Deployment Mappings and Challenges of Contact-tracing Apps for COVID-19. SSRN. Preprint posted online on May 26. 2020 doi: 10.2139/ssrn.3609516. [DOI] [Google Scholar]
- 37.Davalbhakta S, Advani S, Kumar S, Agarwal V, Bhoyar S, Fedirko E, Misra DP, Goel A, Gupta L, Agarwal V. A Systematic Review of Smartphone Applications Available for Corona Virus Disease 2019 (COVID19) and the Assessment of their Quality Using the Mobile Application Rating Scale (MARS) J Med Syst. 2020 Aug 10;44(9):164. doi: 10.1007/s10916-020-01633-3. http://europepmc.org/abstract/MED/32779002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data used for the serial interval estimation of COVID-19 in Jodhpur, India (103 pairs).


