Abstract
Introduction
The present study investigated the HIV-1 subtype classification in addition to prevalence of drug resistance mutations (DRMs) in antiretroviral therapy (ART)-experienced and ART-naïve residents of Pontianak, West Kalimantan, Indonesia.
Methods
Whole blood samples collected from 30 HIV-1-infected individuals, comprising 19 ART-experienced and 11 ART-naïve individuals, were subjected to RNA and DNA extraction, followed by HIV-1 genes amplification and sequencing analysis. HIV-1 subtyping was classified on viral pol genes encoding reverse transcriptase (RT gene) and protease (PR gene) accompanied by the env and gag genes. DRMs in the RT and PR genes were also analyzed.
Results
CRF01_AE was identified as the predominant circulating recombinant form (CRF) of HIV-1 in both ART-experienced and ART-naïve individuals. In addition, CRF02_AG, subtype B, recombinant virus expressing CRF01_AE and subtype B viral genomic fragments, also recombinant virus containing CRF01_AE and CRF02_AG genomic fragments were also identified. Acquired drug resistance (ADR) was identified in 28.5% of ART-experienced individuals, while no transmitted drug resistance was identified in ART-naïve individuals.
Conclusions
This study identified CRF01_AE as the most predominant HIV-1 CRF distributing in Pontianak, Indonesia. The prevalence of ADR is considered to be high; thus, further surveillance is needed in this region.
Keywords: HIV-1, CRF01_AE, acquired drug resistance, transmitted drug resistance, Indonesia
Introduction
Indonesia is currently ranked as the fourth country with the highest population in the world, and the fourth country counted for the new human immunodeficiency virus (HIV) infection cases annually. As stated by the report of the Joint United Nations Program on HIV/AIDS, Indonesia recorded about 48,000 (in a range between 43,000-52,000) novel HIV positive cases and 38,000 (in a range between 34,000-43,000) AIDS-related deaths in 2016.1 The Ministry of Health Republic Indonesia estimated that only 11-15% of HIV-positive Indonesians were on antiretroviral therapy (ART) by the end of 2016. Furthermore, provincial estimates of HIV prevalence range between 0.1 and more than 2.0% in Indonesia.2
According to HIV epidemiological data of Indonesia in 2016, Indonesian provinces with a high HIV prevalence rate and high response by the provincial government in dealing with HIV cases were Jakarta, East Java, and Bali provinces; provinces with a high HIV prevalence rate and relatively low response were Papua and West Kalimantan provinces; and the province with a low HIV prevalence rate and relatively low response was Maluku province. Three cities, Pontianak, Singkawang, and Singkang, were acknowledged to have the most prominent HIV prevalence rate in West Kalimantan.2
Pontianak is the capital city of West Kalimantan and developed as the trading port of Borneo Island. It is located on the equator; hence, it is widely known as Kota Khatulistiwa (an equatorial city). In terms of its population, the number of residents in Pontianak has been progressively expanding each year. The population growth rate between 2013 and 2017 was approximately 6.79%.3 Due to high rates of the HIV epidemic and its crucial functions as a trading port of Borneo Island, HIV-1 epidemiology in Pontianak needs to be examined in more detail. Previous studies reported CRF01_AE as the most predominant HIV-1 circulating recombinant form (CRF) prevalent in most Indonesian regions, including the East Java, Bali, Kepulauan Riau, and Nusa Tenggara provinces, while subtype B was highly prevalent in the Papua and West Papua provinces.4-9 However, the information about HIV-1 epidemiology in Pontianak remains deficient.
Along with the identification of HIV-1 subtypes and CRFs, identification of drug resistance mutations (DRMs) is also essential. ART treatment has been proven to significantly enhance the quality of life of HIV-infected individuals and noteworthy ART has also reduced the mortality and morbidity cases associated with HIV-1 infection. The presence of acquired drug resistance (ADR) in ART-experienced patients and transmitted drug resistance (TDR) in ART-naïve patients may have undesirable effects on the clinical manifestation of HIV infection. The incidence of genotypic DRMs among ART-naïve, HIV-positive pregnant women was approximated in a range of 2.3–25%, and was currently reported to be 24% in newly infected juveniles. In Surabaya, East Java, the prevalence of TDR was found to be less than 5%, signifying that the current established first-line regimen remains efficient.4
In the present study, we accomplished genotypic analyses on HIV-1 pol genes encoding protease (PR gene) and reverse transcriptase (RT gene). Additionally, the env and gag genes originated from ART-experienced and ART-naïve patients residing in Pontianak, West Kalimantan, Indonesia were also studied. HIV-1 subtype distributions and the prevalence of both ADR and TDR in this region were examined herein.
Methods
Research participants and sample collection
Whole blood samples were obtained at the Voluntary Counseling and Testing (VCT) program in Pontianak Hospital. Thirty HIV-1-infected individuals, including 19 ART-experienced and 11 ART-naïve individuals, were enrolled in this study. Inclusion criteria of study participants were: age 18 or above, HIV-infected, as well as ART-experienced individuals who had received ART for more than one year, or ART-naïve individuals who had never received the treatment, while exclusion criteria were age <18 and pediatric patients. Thirty study participants were randomly selected. Ten milliliters of whole blood samples were stored in ethylenediaminetetraacetic acid (EDTA)-treated vacutainer tubes individually from all participants. Plasma separation was accomplished by centrifugation at 2,000 rpm for 10 min. In addition, peripheral blood mononuclear cells (PBMCs) were collected by density gradient centrifugation using Histopaque 1077 (Sigma-Aldrich, St. Louis, MO, USA). The plasma specimen from ART-naïve individuals were then subjected to RNA isolation using the QIAamp Viral RNA Mini kit (Qiagen, Hilden, Germany), while PBMCs from ART-experienced individuals were subjected to DNA isolation utilizing the QIAamp DNA blood mini kit (QIAGEN, Hilden, Germany).
Amplification of HIV-1 genomic fragments and sequencing analysis
Firstly, viral RNA was reverse transcribed into cDNA by the use of SuperScript III First-Stand Synthesis kit (Invitrogen, Carlsbad, CA, USA) and the reverse primer, K-env-R1, 5'-CCAATCAGGGAAGAAGCCTTG-3' [corresponding to nucleotides (nt) 9168 to 9148 of a HIV-1 reference strain, HXB2 (GenBank accession no. K03455), HXB2 numbering]. The HIV-1 PR, RT, env, and gag genes were amplified by using GoTaq green master mix (Promega, Madison, WI, USA) and specific primer sets corresponding to the targeted genes. All primers are described in Table 1. Detailed PCR conditions are accessible upon request. PCR products were separated by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining. The PCR products were then subjected to a sequencing analysis in order to assess the genomic fragment of the predominant viral population in a sample. A sequencing analysis of the amplified HIV-1 genomic fragment was assessed by using BigDye Terminator v3.1 Cycle Sequencing kit specifically provided for ABI PRISM 3500 xl genetic analyzer (Applied Biosystems, Foster City, CA, USA). Sequencing data were collected and aligned using Genetyx software version 10 (Genetyx, Tokyo, Japan). The collection of nucleotide sequences of targeted HIV-1 gene fragments has been submitted to the GenBank database under the following accession numbers, MN518004 - MN518016 (PR genes), MN518017 - MN518027 (RT genes), MN518039 - MN518054 (env genes), and MN518028 - MN518038 (gag genes).
Table 1. Primers used for nested PCR.
| Genes | Primer name | Primer locations (HXB2 numbering) | Sequences 5' - 3' | |
|---|---|---|---|---|
| PR | 1st round | DRPR05 | nt 2074 to 2095 | AGACAGGYTAATTTTTTAGGGA |
| DRPR02L | nt 2716 to 2691 | TATGGATTTTCAGGCCCAATTTTTGA | ||
| 2nd round | DRPR01M | nt 2148 to 2167 | AGAGCCAACAGCCCCACCAG | |
| DRPR06 | nt 2611 to 2592 | ACTTTTGGGCCATCCATTCC | ||
| RT | 1st round | RT1L | nt 2388 to 2410 | ATGATAGGGGGAATTGGAGGTTT |
| DRRT4L | nt 4402 to 4380 | TACTTCTGTTAGTGCTTTGGTTCC | ||
| 2nd round | RT7L | nt 2485 to 2509 | GACCTACACCTGTCAACATAATTGG | |
| DRRT6L | nt 4309 to 4285 | TAATCCCTGCATAAATCTGACTTGC | ||
| env | 1st round | M5 | nt 6858 to 6889 | CCAATTCCCATACATTATTGTGCCCCAGCTGG |
| M10 | nt 7661 to 7632 | CCAATTGTCCCTCATATCTCCTCCTCCAGG | ||
| 2nd round | M3 | nt 6948 to 6973 | GTCAGCACAGTACAATGIACACATGG | |
| M8 | nt 7547 to 7521 | TCCTTCCATGGGA GGGGCATACATTGC | ||
| gag | 1st round | H1G777 | nt 1231 to 1255 | TCACCTAGAACTTTGAATGCATGGG |
| H1P202 | nt 2352 to 2325 | CTAATACTGTATCATCTGCT GCTCCTGT | ||
| 2nd round | H1Gag1584 | nt 1577 to 1595 | AAAGATGGATAATCCTGGG | |
| G17 | nt 2040 to 2017 | TCCACATTTCCAACAGCCCTTTTT | ||
HIV-1 subtyping and phylogenetic analysis
HIV-1 subtyping was classified using the recombinant identification program (RIP) and jumping profile Hidden Markov Model (jpHMM)-HIV (http://jphmm.gobics.de/submission_hiv). In addition, neighbor-joining (NJ) trees with a Kimura two-parameter model were generated by MEGA 6.06 software. Subtypes A1, A2, B, C, D, and G as well as CRF01_AE and CRF02_AG are considered to be major pandemic strains of HIV-1, while 3 CRF01_AE/subtype B-recombinants, CRF15_01B, CRF33_01B, and CRF34_01B, were detected frequently in Indonesia. Therefore, reference strains representing these subtypes and CRFs were retrieved from HIV database (https://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/RefSeqs2005/RefSeqs05.html), and included in the phylogenetic analysis. HIV-1 subtyping was identified according to the successfully sequenced PR, RT, gag and/or env genes, and if the discrepancy in the subtype or CRF among those genes existed, the viral gene was accounted from a recombinant form of HIV-1.
HIV drug resistance analysis
The evaluation of drug resistance-related mutations in RT and PR genes originated from ART-experienced and ART-naïve individuals was manually assessed based on the guidelines issued by the International Antiviral Society-United States (IAS-USA).10 In addition, the data was verified by using online Genotypic Resistance Interpretation Algorithm (https://hivdb.stanford.edu/hivdb/by-mutations/).
Ethics declaration
Ethical clearance was acquired from the Institutional Ethics Committees of Universitas Airlangga (approval number: 25-995/UN3.14/PPd/2013) and Kobe University Graduate School of Medicine (approval number: 784). Inscribed informed consents were collected from all research participants prior to admission in this study.
Results
Demographic and clinical data of study participants
Among the 30 individuals enrolled in this study, 19 were already under ART for more than one year, while 11 were ART-naïve. The most commonly used ART regimen was the combination of lamivudine (3TC), zidovudine (AZT), and nevirapine (NVP) (36.7%), with an average duration of treatment of more than 3 years. The median age of study participants was 33.8 years, with the ART-experienced and ART-naïve groups being 33.9 years and 33.5 years respectively. Eighty percent of study participants were male, while 20% were female. All participants were married, and the major risk behavior for HIV-1 infection was intravenous drug use (66.7%, 20/30). Seventeen (89.5%) ART-experienced and all (100%) ART-naïve individuals were diagnosed with opportunistic infections. The demographic and clinical data of study participants are shown in Table 2.
Table 2. Demographic and clinical information on ART-experienced and ART-naïve individuals residing in Pontianak.
| Characteristics | Total | On ART | Non-ART |
|---|---|---|---|
| Sample number | 30 | 19 | 11 |
| Age, mean years (SD) | 33.8 (6.2) | 33.9 | 33.5 |
| Sex, n (%) | |||
| Male | 24 (80.0%) | 18 (94.7%) | 6 (54.5%) |
| Female | 6 (20.0%) | 1 (5.3%) | 5 (45.5%) |
| Marital status, n (%) | |||
| Married | 30 (100.0%) | 19 (100.0%) | 11 (100.0%) |
| Single | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Transmission risk category, n (%) | |||
| Heterosexual intercourse | 7 (23.3%) | 2 (10.5%) | 5 (45.5%) |
| Intravenous drug use | 20 (66.7%) | 17 (89.5%) | 3 (27.3%) |
| Homosexual intercourse | 3 (10.0%) | 0 (0.0%) | 3 (27.3%) |
| Types of ART used, n (%) | |||
| 3TC+AZT+NVP | 11 (36.7%) | 11 (57.9%) | 0 (0.0%) |
| 3TC+AZT+EFV | 2 (6.7%) | 2 (10.5%) | 0 (0.0%) |
| Others | 6 (20.0%) | 6 (31.6%) | 0 (0.0%) |
| Duration of ART, n (%) | |||
| <1 year | 0 (0%) | 0 (0.0%) | 0 (0.0%) |
| 1-3 years | 2 (6.7%) | 2 (10.5%) | 0 (0.0%) |
| >3 years | 17 (56.7%) | 17 (89.5%) | 0 (0.0%) |
| Opportunistic infection, n (%) | |||
| No | 2 (6.7%) | 2 (10.5%) | 0 (0.0%) |
| Yes | 28 (93.3%) | 17 (89.5%) | 11 (100.0%) |
HIV-1 subtyping
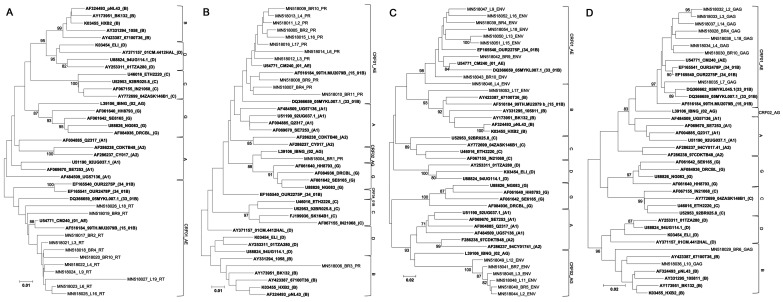
Phylogenetic trees were generated using efficiently sequenced 13 PR (296 bp), 11 RT (762 bp), 16 env (383 bp), and 9 gag gene fragments (369 bp) obtained from 26 samples (Figure 1). For ART-naïve samples, we first attempted to amplify viral gene fragments from cDNA synthesized from RNA isolated from plasma samples; however, it failed. Therefore, all viral gene fragments were amplified from DNA isolated from PBMCs derived from ART-naïve and ART-experienced individuals (data not shown). Among ART-experienced individuals, 10/16 (62.5%) were found to be infected with CRF01_AE, while 2/16 (12.5%), 1/16 (6.3%), 1/16 (6.3%), and 2/16 (12.5%) were infected with CRF02_AG, subtype B, recombinant virus containing CRF01_AE and subtype B genomic fragments, and recombinant virus containing CRF01_AE and CRF02_AG genomic fragments, respectively. Among ART-naïve individuals, the HIV-1 subtypes and CRFs detected were CRF01_AE (50%, 5/10), CRF02_AG (30%, 3/11), and subtype B (20%, 2/10). Subtyping results are summarized in Table 3.
Figure 1.
Phylogenetic trees were constructed for the HIV-1 RT (A), PR (B), env (C), and gag genes newly sequenced in the present study (D). The corresponding viral genes of reference HIV-1 strains representing subtypes A1, A2, B, C, D, and G as well as CRF01_AE (01_AE), CRF02_AG (02_AG), CRF15_01B (15_01B), CRF33_01B (33_01B), and CRF34_01B (34_01B) were included in the analyses (shown in bold letters). Sequence IDs are presented as a GenBank accession number, sample ID, or the ID of the reference HIV-1 strain, and the subtype or CRF of the reference strain (shown in parentheses) in that order. Bootstrap values were shown if they were >70.
Phylogenetic tree analysis of HIV-1 RT, PR, env, and gag gene sequences collected in Pontianak, Indonesia
Table 3. HIV-1 subtyping.
| Sample ID | Treatment | Subtype/CRF assignment | Subtyping | |||
|---|---|---|---|---|---|---|
| PR gene | RT gene | Env gene | Gag gene | |||
| BR1 | Non-ART | CRF02_AG* | CRF02_AG | NA** | NA | NA |
| BR2 | Non-ART | CRF01_AE | CRF01_AE | CRF01_AE | NA | NA |
| BR3 | Non-ART | B | B | NA | NA | NA |
| BR4 | Non-ART | CRF01_AE | CRF01_AE | CRF01_AE | CRF01_AE | CRF01_AE |
| BR5 | Non-ART | CRF02_AG | NA | NA | CRF02_AG | NA |
| BR6 | Non-ART | B | NA | NA | NA | B |
| BR8 | Non-ART | NA | NA | NANN | NA | NA |
| BR7 | Non-ART | CRF02_AG | NA | NA | CRF02_AG | NA |
| BR9 | Non-ART | CRF01_AE | CRF01_AE | CRF01_AE | CRF01_AE | NA |
| BR10 | Non-ART | CRF01_AE | CRF01_AE | CRF01_AE | CRF01_AE | CRF01_AE |
| BR11 | Non-ART | CRF01_AE | CRF01_AE | NA | NA | NA |
| L1 | On ART | NA | NA | NA | NA | NA |
| L2 | On ART | CRF01_AE | CRF01_AE | NA | CRF02_AG | CRF01_AE |
| L3 | On ART | CRF01_AE | CRF01_AE | CRF01_AE | CRF02_AG | CRF01_AE |
| L4 | On ART | CRF01_AE | CRF01_AE | CRF01_AE | CRF01_AE | CRF01_AE |
| L5 | On ART | NA | NA | NA | NA | NA |
| L6 | On ART | CRF01_AE | CRF01_AE | CRF01_AE | NA | NA |
| L7 | On ART | CRF01_AE | NA | NA | NA | CRF01_AE |
| L8 | On ART | NA | NA | NA | NA | NA |
| L9 | On ART | CRF01_AE | NA | CRF01_AE | CRF01_AE | NA |
| L10 | On ART | B | NA | NA | NA | B |
| L11 | On ART | CRF02_AG | NA | NA | CRF02_AG | NA |
| L12 | On ART | CRF02_AG | NA | NA | CRF02_AG | NA |
| L13 | On ART | CRF01_AE | NA | NA | CRF01_AE | NA |
| L14 | On ART | CRF01_AE | NA | NA | NA | CRF01_AE |
| L15 | On ART | CRF01_AE | NA | NA | CRF01_AE | NA |
| L16 | On ART | CRF01_AE | CRF01_AE | CRF01_AE | CRF01_AE | NA |
| L17 | On ART | CRF01_AE/B*** | CRF01_AE | NA | B | NA |
| L18 | On ART | CRF01_AE | NA | CRF01_AE | CRF01_AE | CRF01_AE |
| L19 | On ART | CRF01_AE | NA | CRF01_AE | NA | NA |
Recombinant form of HIV-1 containing the viral gene fragments of subtypes A and G.
Not available due to the failure of PCR.
Recombinant form of HIV-1 containing the viral gene fragments of CRF01_AE and subtype B.
Prevalence of acquired drug resistance (ADR) and transmitted drug resistance (TDR)
Seven PR and 4 RT gene sequences originated from ART-naïve individuals, while 6 PR and 7 RT gene sequences were obtained from ART-experienced individuals. DRMs were identified in the PR and RT genes as stated by the IAS-USA guidelines10 as well as detected by online Genotypic Resistance Interpretation Algorithm (https://hivdb.stanford.edu/hivdb/by-mutations/). The major DRMs remained unidentified in the PR genes, while minor DRMs were recurrently detected. All PR genes obtained from ART-naïve and ART-experienced individuals contained at least one minor DRM. Minor mutations also frequently detected in PR genes were M36I (100%, 13/13), K20I/R (69.2%, 9/13), and V77I (38.5%, 5/13) (data not shown).
No TDR was identified among ART-naïve individuals; however, ADR in the RT genes was identified in two out of seven ART-experienced individuals (28.6%) (Table 4). Three nucleoside RT inhibitor (NRTI)-associated DRMs (K65R, M184V, K219E) and 4 non-nucleoside RT inhibitor (nNRTI)-related DRMs (K130T, Y181C, G190A and P225H) were detected. These mutations confer resistance to abacavir (ABC), didanosine (ddI), lamivudine (3TC), stavudine (d4T), emtricitabine (FTC), tenofovir (TDF), zidovudine (AZT), nevirapine (NVP), etravirine (ETR), efavirenz (EFV), and rilpivirine (RPV).11
Table 4. Appearance of drug resistance mutations among ART-experienced individuals.
| Sample ID | ART regimen | Drug resistance mutations* | ||
|---|---|---|---|---|
| NRTI | NNRTI | Confer resistance to | ||
| L18 | AZT+3TC+NVP TDF+3TC+NVP |
K65R M184V K219E |
- Y181C G190A |
ABC, ddI, FTC, 3TC, d4T, TDF, AZT, EFV, ETR, NVP, RPV |
| L19 | 3TC+AZT+NVP | - | K130T P225H |
EFV |
Drug resistance mutations causing high resistance to antiretrovirals according to the guidelines published by the International Antiviral Society United States (IAS-USA) as well as detected by online Genotypic Resistance Interpretation Algorithm (https://hivdb.stanford.edu/hivdb/by-mutations/) are shown. Drug resistance-associated major mutations are shown in bold.
ABC – abacavir; ART – antiretroviral therapy; AZT – zidovudine; d4T – stavudine; ddI – didanosine; EFV – efavirenz; ETR – etravirine; FTC – emtricitabine; NNRTI – non-nucleoside reverse transcriptase inhibitor; NRTI – nucleoside reverse transcriptase inhibitor; NVP – nevirapine; TDF – tenofovir; RPV – rilpivirine; 3TC – lamivudine.
Discussion
In the present study, CRF01_AE was identified as the major CRF of HIV-1 circulating in Pontianak, West Kalimantan, Indonesia. This result was consistent with previous findings obtained in other Indonesian regions, including the East Java, Bali, Kepulauan Riau, and Nusa Tenggara provinces.4-9 CRF01_AE is also known to be predominant in Southeast-Asian countries, including Thailand, the Philippines, Singapore, and Vietnam,12,13 along with East Asian countries, consisting of Japan, China including Hong Kong and Taiwan, and South Korea.12-14 In addition, CRF01_AE was reported to be the second largest CRF circulating worldwide, with an estimated global prevalence rate of 5.3%.14 HIV-1 CRF01_AE infection correlated with a rapid CD4 decline, higher viral load, and significant risk of accelerated disease progression in China.15,16 Therefore, the consecutive and periodic surveillance of HIV-1 subtypes is fundamental to be accomplished due to the impact of different subtypes and CRFs on disease progression.
The presence of DRMs compromises the effectiveness of ART, thereby hindering treatment success.17 Several mutations in the HIV-1 RT genes have been shown to reduce viral susceptibility to ART.18 The manifestation of TDR among ART-naïve individuals resulted from the acquisition of a drug-resistant virus.19 Our previous studies in Surabaya, West Papua, and Bali revealed the occurrence of TDR in RT inhibitors counted for 4.3% (2/47), 12.9% (4/31), and 16.7% (5/30) of RT genes obtained from ART-naïve individuals, respectively.4,8-9 In other countries, various prevalence rates of TDR were identified, including 17.2% in Germany, 12% in South Carolina, USA, 20.5% in Washington DC, USA, and 3.6% in China.20-23 However, no TDR was observed among ART-naïve individuals residing in Pontianak in the present study. This result indicates that the current first-line regimen is still effective for newly infected individuals in the region.4
In contrast, ADR of the RT inhibitor was identified in 28.5% of ART-experienced individuals in Pontianak, which is considered to be high. Previous studies in Bali, Maumere, and Kepulauan Riau also identified ADR of the RT inhibitor among ART-experienced individuals.5,6 A high prevalence of ADR was also identified in other countries, such as Washington DC, USA (40.5%), and Cameroon (17.7-28.3%).22-24
Among two individuals presenting major DRMs towards an RT inhibitor, one individual (L19) was infected with HIV-1 variant(s) containing a P255H mutation, which correlated with resistance towards EFV. Another individual (L18) was infected with HIV-1 variant(s) containing several mutations that associated with resistance towards multiple drugs such as FTC, ddI, ABC, 3TC, d4T, TDF, ETR, EFV, NVP, AZT, and RPV.10 Switching to other antiretroviral drugs or regimens may be favorable for individuals with ADR in order to maintain the effectiveness of treatment.
In ART-experienced and ART-naïve individuals, no major DRMs related to PR inhibitors were identified in Pontianak. The result is consistent with previous findings reporting the absence of both ADR and TDR related to PR inhibitors in several Indonesian regions.4-8 Among ART-experienced individuals in Indonesia, only 3.3% (2,983 of 91,369) were receiving a PR inhibitor-based ART regimen in 2017.25 The limited use of a PR inhibitor-based regimen may contribute to the absence of DRMs related to PR inhibitors. Although no major mutations were found, several minor mutations were identified in the PR genes, including M36I, K20R, V77I, I93L, and G16E. These mutations have been identified as natural polymorphisms among CRF01_AE viruses.17 Finally, the present study had the following limitations. Thirty individuals were enrolled in this study, and the sample size may not be sufficiently large to provide clear research outcomes. It was due to a difficulty in recruiting more HIV-infected individuals at the site during sampling duration. In addition, we faced a low success rate of PCR amplification for viral partial gene fragments. It might be explained by sample storage and the transportation under improper conditions. Nevertheless, information on HIV epidemiology was quite limited in Pontianak, Indonesia, and we believe the data shown in the present study provide important information for revealing the situation of HIV infection in the region.
Conclusions
CRF01_AE was identified as the dominant CRF circulating in Pontianak, West Kalimantan, Indonesia. Several other subtypes identified included CRF02_AG, subtype B, recombinant virus containing CRF01_AE and subtype B genomic fragments, and recombinant virus containing CRF01_AE and CRF02_AG genomic fragments. TDR against PR and RT inhibitors was not found in ART-naïve individuals; however, ADR against an RT inhibitor was found in 28.5% of ART-experienced individuals, which is considered to be high. These results suggest the significance of the regular surveillance of HIV-1 subtypes and DRMs in both ART-experienced and ART-naïve individuals.
Acknowledgements
The authors would like to acknowledge all the study participants and the staff from Voluntary Counseling and Testing (VCT) program in Pontianak Hospital. Deepest gratitude also extends to J-GRID, AMED and ITD – Universitas Airlangga to facilitate this study. The manuscript was proofread by Medical English Service, Kyoto, Japan.
Footnotes
Authors’ contributions statement: MK and N conceived and supervised the study; SQK and TK designed experiments; DN organized study participants; SQK, NLAM and DWI conducted experiments; SQK and TK collected samples and clinical information; SQK and NLAM analyzed data; SQK wrote the manuscript; SQK and MK made manuscript revisions.
Conflicts of interest: All authors – none to declare.
Funding: Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) administered by the Ministry of Education, Culture, Sport, Science and Technology – Japan; the Japan Agency for Medical Research and Development (AMED); and the Institute of Tropical Disease as the Center of Excellence (COE) platform by the Ministry for Research and Technology (RISTEK) of Indonesia.
References
- 1.Joint United Nations Programme on HIV/AIDS (UNAIDS) Indonesia: 2019. [Accessed on: 06 March 2020]. Available at: http://www.unaids.org/en/regionscountries/countries/indonesia/ [Google Scholar]
- 2.Ministry of Health Republic Indonesia. HIV epidemiology review, Indonesia 2016. Directorate General of Disease Prevention and Control. 2017. [Accessed on: 06 March 2020]. pp. 1–66. Available at: https://id.scribd.com/document/409741880/HIV-EPIDEMIOLOGY-REVIEW-INDONESIA-2016-pdf.
- 3.Government of Pontianak. Investment profile in Pontianak, 2018. 2018. [Accessed on: 06 March 2020]. Available at: http://dpmtk.pontianakkota.go.id/layanan/docs/2019/BukuProfil2018.pdf.
- 4.Kotaki T, Khairunisa SQ, Witaningrum AM, et al. HIV-1 transmitted drug resistance mutations among antiretroviral therapy-naïuml;ve individuals in Surabaya, Indonesia. AIDS Res Ther. 2015;12:5. doi: 10.1186/s12981-015-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khairunisa SQ, Ueda S, Witaningrum AM, et al. Genotypic characterization of human immunodeficiency virus type 1 prevalent in Kepulauan Riau, Indonesia. AIDS Res Hum Retroviruses. 2018;34:555–60. doi: 10.1089/aid.2018.0040. [DOI] [PubMed] [Google Scholar]
- 6.Indriati DW, Kotaki T, Khairunisa SQ, et al. Appearance of drug resistance mutations among the dominant HIV-1 subtype, CRF01_AE in Maumere, Indonesia. Curr HIV Res. 2018;16:158–66. doi: 10.2174/1570162X16666180502114344. [DOI] [PubMed] [Google Scholar]
- 7.Yunifiar MQ, Kotaki T, Witaningrum AM, et al. Sero- and molecular epidemiology of HIV-1 in Papua Province, Indonesia. Acta Med Indones. 2017;49:205–14. [PubMed] [Google Scholar]
- 8.Witaningrum AM, Kotaki T, Khairunisa SQ, et al. Genotypic characterization of human immunodeficiency virus type 1 derived from antiretroviral therapy-naive individuals residing in Sorong, West Papua. AIDS Res Hum Retroviruses. 2016;32:812–7. doi: 10.1089/aid.2016.0054. [DOI] [PubMed] [Google Scholar]
- 9.Megasari NLA, Oktafiani D, Fitriana E, et al. The emergence of HIV-1 transmitted drug resistance mutations among antiretroviral therapy-naive individuals in Buleleng, Bali, Indonesia. Acta Med Indones. 2019;51:197–204. [PubMed] [Google Scholar]
- 10.Wensing AM, Calvez V, Günthard HF, et al. 2017 update of the drug resistance mutations in HIV-1. Top Antivir Med. 2016;24:132–3. [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Liu H, Liu L, et al. Tracing the epidemic history of HIV-1 CRF01_AE clusters using near-complete genome sequences. Sci Rep. 2017;7:4024. doi: 10.1038/s41598-017-03820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salvaña EMT, Schwem BE, Ching PR, Frost SDW, Ganchua SKC, Itable JR. The changing molecular epidemiology of HIV in the Philippines. Int J Infect Dis. 2017;61:44–50. doi: 10.1016/j.ijid.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Dey SK, Zahan N, Afrose S, et al. Molecular epidemiology of HIV in Asia. HIV AIDS Rev. 2014;13:33–9. doi: 10.1016/j.hivar.2014.02.003. [DOI] [Google Scholar]
- 14.Hemelaar J, Elangovan R, Yun J, et al. Global and regional molecular epidemiology of HIV-1, 1990-2015: a systematic review, global survey, and trend analysis. Lancet Infect Dis. 2019;19:143–55. doi: 10.1016/S1473-3099(18)30647-9. [DOI] [PubMed] [Google Scholar]
- 15.Chu M, Zhang W, Zhang X, et al. HIV-1 CRF01_AE strain is associated with faster HIV/AIDS progression in Jiangsu Province, China. Sci Rep. 2017;7:1570. doi: 10.1038/s41598-017-01858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Xue Y, Cheng H, et al. HIV-1 genetic diversity and its impact on baseline CD4+T cells and viral loads among recently infected men who have sex with men in Shanghai, China. PLoS One. 2015;10:e0129559. doi: 10.1371/journal.pone.0129559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iemwimangsa N, Pasomsub E, Sukasem C, Chantratita W. Surveillance of HIV-1 drug-resistance mutations in Thailand from 1999 to 2014. Southeast Asian J Trop Med Public Health. 2017;48:271–81. [PubMed] [Google Scholar]
- 18.Ministry of Health Republic of Indonesia (Kementerian Kesehatan Republik Indonesia) Peraturan Menteri Kesehatan Republik Indonesia Nomor 87 Tahun 2014 tentang Pedoman Pengobatan Antiretroviral No Title. 2014. [Accessed on: 06 March 2020]. Available at: http://preventcrypto.org/wp-content/uploads/2015/10/IndonesiaAdultARTguidelines20141432907982.pdf.
- 19.Trinh QM, Nguyen HL, Nguyen VN, Nguyen TV, Sintchenko V, Marais BJ. Tuberculosis and HIV co-infection-focus on the Asia-Pacific region. Int J Infect Dis. 2015;32:170–8. doi: 10.1016/j.ijid.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 20.Machnowska P, Meixenberger K, Schmidt D, et al. Prevalence and persistence of transmitted drug resistance mutations in the German HIV-1 Seroconverter Study Cohort. PLoS One. 2019;14:e0209605. doi: 10.1371/journal.pone.0209605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levintow SN, Okeke NL, Hué S, et al. Prevalence and transmission dynamics of HIV-1 transmitted drug resistance in a southeastern cohort. Open Forum Infect Dis. 2018;5:ofy178. doi: 10.1093/ofid/ofy178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aldous AM, Castel AD, Parenti DM, DC Cohort Executive Committee Prevalence and trends in transmitted and acquired antiretroviral drug resistance, Washington, DC, 1999-2014. BMC Res Notes. 2017;10:474. doi: 10.1186/s13104-017-2764-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao S, Feng Y, Hu J, et al. Prevalence of transmitted HIV drug resistance in antiretroviral treatment naïve newly diagnosed individuals in China. Sci Rep. 2018;8:12273. doi: 10.1038/s41598-018-29202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tchouwa GF, Eymard-Duvernay S, Cournil A, et al. Nationwide estimates of viral load suppression and acquired HIV drug resistance in Cameroon. EClinicalMedicine. 2018;1:21–7. doi: 10.1016/j.eclinm.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ministry of Health of Indonesia (Kementerian Kesehatan Republik Indonesia) Laporan Situasi Perkembangan HIV-AIDS & PIMS di Indonesia Januari-Desember 2017. 2018. [Accessed on: 06 March 2020]. Available at: https://id.scribd.com/document/391560045/Laporan-HIV-AIDS-TW-4-Tahun-2017-1-pdf.