Abstract
Background
IgA nephropathy (IgAN) is the commonest glomerulonephritis worldwide. Its prevalence is difficult to estimate, as people with mild disease do not commonly receive a biopsy diagnosis. We aimed to generate an IgA nephropathy genetic risk score (IgAN-GRS) and estimate the proportion of people with hematuria who had IgAN in the UK Biobank (UKBB).
Methods
We calculated an IgAN-GRS using 14 single-nucleotide polymorphisms (SNPs) drawn from the largest European Genome-Wide Association Study (GWAS) and validated the IgAN-GRS in 464 biopsy-proven IgAN European cases from the UK Glomerulonephritis DNA Bank (UKGDB) and in 379,767 Europeans in the UKBB. We used the mean of IgAN-GRS to calculate the proportion of potential IgAN in 14,181 with hematuria and other nonspecific renal phenotypes from 379,767 Europeans in the UKBB.
Results
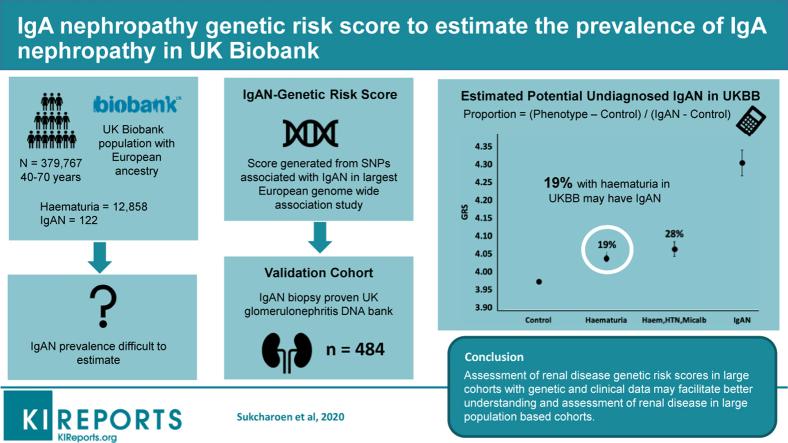
The IgAN-GRS was higher in the IgAN cohort (4.30; 95% confidence interval [95% CI: 4.23–4.38) than in controls (3.98; 3.97–3.98; P < 0.0001). The mean GRS in UKBB participants with hematuria (n = 12,858) was higher (4.04; 4.02–4.06) than UKBB controls (3.98; 3.97–3.98; P < 0.0001) and higher in those with hematuria, hypertension, and microalbuminuria (n = 1323) (4.07; 4.02–4.13) versus (3.98; 3.97–3.98; P = 0.0003). Using the difference in these means, we estimated that IgAN accounted for 19% of noncancer hematuria and 28% with hematuria, hypertension, and microalbuminuria in UKBB.
Conclusions
We used an IgAN-GRS to estimate the prevalence of IgAN contributing to common phenotypes that are not always biopsied. The noninvasive use of polygenic risk in this setting may have further utility to identify likely etiology of nonspecific renal phenotypes in large population cohorts.
Keywords: chronic kidney disease, epidemiology, Genetic Risk Scores, hematuria, IgA nephropathy
Graphical abstract
See Commentary on Page 1627
IgAN is the commonest glomerulonephritis worldwide but its true prevalence is hard to estimate and is confounded by differing biopsy practice across the world.1 A systematic review of biopsy-based studies spanning multiple countries suggests an overall incidence of at least 2.5 per 100,000.2 In the United States, Canada, and United Kingdom, renal biopsies are rarely done to investigate microscopic hematuria.3,4 In contrast, Japan, Korea, and Taiwan have urine screening programs.5 Variation in biopsy practices alone does not account for differences in geographical prevalence. Autopsy and donor registries data show Lanthanic IgA deposition in 1.3% of autopsies in trauma victims from Finland compared with 15.6% of deceased donor candidates in Japan.6,7 The variable phenotype of IgAN from mild persistent hematuria to end-stage renal disease, and variable biopsy practice, means many with mild IgAN are not diagnosed and true prevalence is very hard to estimate.
IgAN is a polygenic disease with 15 SNPs associated in the largest GWAS of a European cohort.8 GRSs sum genetic risk for disease into a single continuous variable are increasingly used for diagnosis, prediction, and mechanistic investigation of common diseases.9,10 UK Biobank (UKBB) gives a unique opportunity to study IgAN-related phenotypes in the largest population cohort. We recently used a Type 1 diabetes (T1D) GRS to identify undiagnosed T1D in the population of those older than 30 in the UKBB where misclassification of T1D is common.9,11 We aimed to generate and validate an IgAN-GRS and use this to assess the excess IgAN genetic risk in people with hematuria and IgAN-related phenotypes12,13 in UKBB, and then estimate the proportion of potential undiagnosed IgAN. Given that much of our knowledge on IgAN prevalence is derived from biopsy data, we aimed to assess the utility of an IgAN-GRS to estimate undiagnosed IgAN in those without a renal biopsy.
Methods
We generated IgAN-GRS from known SNPs associated with IgAN and validated the score with the 464 biopsy-proven IgAN cases from the UKGDB. We then generated the IgAN-GRS in 379,769 white European individuals in UKBB and assessed the score in phenotypes associated with IgAN: hematuria, hypertension, and microalbuminuria/proteinuria.
UK Glomerulonephritis DNA Bank (UKGDB)
Individuals with biopsy-proven IgAN were genotyped at 318,127 SNPs using the Illumina (San Diego, CA) Sentrix HumanHap300 BeadChip, of which 302,210 passed quality control (>90% genotyping rate, minor allele frequency >0.05, Hardy Weinberg Equilibrium P > 0.001). After quality control for ethnicity (using principal component analysis), genotyping rate, and excluding cryptic relatedness, estimated from identity-by-state information (pi-hat > 0.125),14 we genotyped 9 SNPs (rs10801555, rs6677604, rs9357155, rs2071543, rs1883414, rs4077515, rs11150612, rs3803800, rs241297) and imputed the remaining SNPs for 464 UK European patients. Genotypes for the remaining SNPs were imputed from nearby SNPs using the University of Michigan imputation server.15
UKBiobank
UKBB recruited more than 500,000 individuals aged 37 to 73 years from 2006 to 2010 across the United Kingdom.16 Recruitment was unselected.17 UKBB participants were genotyped with the UKBB Affymetrix axiom array. We studied 379,767 unrelated white Europeans, as those who self-identified as white European and confirmed as ancestrally “European” using principal components analyses of genome-wide genetic information.17
All participants completed detailed self-report health questionnaires. We used directly measured blood pressure and albumin creatinine ratio (ACR) from baseline assessment. In UKBB, a continuous measure of ACR was derived using urinary measures of albumin and creatinine. If albumin was <6.7 mg/l (the detection level of the assay in UK Biobank, http://biobank.ctsu.ox.ac.uk/crystal/docs/urine_assay.pdf), then the albumin was set at 6.7 mg/l before the calculation of the ratio. Albumin was measured in the UKBB samples using immuno-turbidimetric analysis method (Randox Bioscience, Crumlin, UK) while creatinine was measured using enzymatic analysis method (Beckman Coulter, High Wycombe, UK). ACR variable was inverse normalized before analysis. Hypertension was defined by use of blood pressure–lowering medication or a systolic blood pressure of >140 or diastolic blood pressure >90.
We used primary and secondary diagnosis codes from hospital episodes from 1996 to 2015. Unspecified hematuria was derived from diagnosis codes for any hospital episode (International Classification of Diseases, 10th Revision [ICD10], R31) of hematuria in a hospitalized participant. Those with urological malignancy were excluded. UKBB individuals without a record of IgAN, or associated phenotype, diagnosis of chronic disease, and malignancy were used as controls.
Genetic Risk Score
We generated the IgAN-GRS using 14 of 15 SNPs drawn from the largest GWAS in a European population8 (Supplementary Table S1). We excluded 1 associated variant (rs10086568) because of poor imputation (r2 0.4) (Supplementary Table S2). There was no proxy SNP to substitute the variant. It has been shown that SNP coverage at this locus is poor because of structural complexity, and the association signal is almost certainly mediated by copy-number variations (CNV) that are poorly tagged.18 Therefore, to include the full effect of DEFA variation in a GRS, one would almost certainly need to measure CNV at this locus, which has not been done in UKBB.
We generated the IgAN-GRS in all UKGDB and UKBB participants. The IgAN-GRS was calculated by summing the number of risk-increasing alleles at each SNP multiplied by the ln (odds ratio) for each allele divided by the total number of alleles. This assumes that each risk allele has a log additive effect on IgAN risk and allows for varying weight of contribution to IgAN dependent on the odds in GWAS.19,20
Statistical Analysis
Stata was used for statistical analysis (version 14; StataCorp, College Station, TX).
We used t tests to compare IgAN-GRS between cases and controls and common UKBB phenotypes versus controls.
We validated the IgAN-GRS by comparing the mean GRS between UKBB and UKGDB IgAN cases. We studied 151,103 control individuals (without hematuria, hypertension, and microalbuminuria), 12,858 with unspecified hematuria, 202,850 with hypertension, and 30,367 with microalbuminuria/proteinuria. We performed sensitivity analyses comparing the IgAN across gender and age and performed a regression of IgAN-GRS against the principle components used to define ethnicity. We calculated the reference mean IgAN-GRS as the combined mean of UKGDB and UKBB IgAN cases. We assessed the GRS in male and female individuals and different ages.
Genetic Estimates of IgAN Prevalence
The proportion of potential IgAN cases within a phenotype was estimated from the mean GRS of the phenotype relative to controls and cases (Supplementary Figure S1), using the following equation: Proportion = (phenotype GRS – control GRS) / (IgAN-GRS – control GRS). The proportion of cases with suspected IgAN was then estimated using the preceding formula in cohorts with hematuria and hematuria, hypertension, and microalbuminuria/proteinuria.
Simulated Scenarios
We first visualized estimates of suspected IgAN prevalence made within simulated mixtures of known proportions of IgAN cases and controls. The mean IgAN-GRS of artificial mixtures containing 10%, 25%, and 50% of IgAN cases was evaluated as a phenotype GRS in the preceding formula (Figure 1). For each proportion, 1000 randomly generated mixtures were made, allowing the mean and 95% CIs around estimates21 (Supplementary Figure S2). To calculate CIs using reference data, we bootstrapped around the point estimates generating 10,000 mixtures of equivalent size to the phenotype of interest.
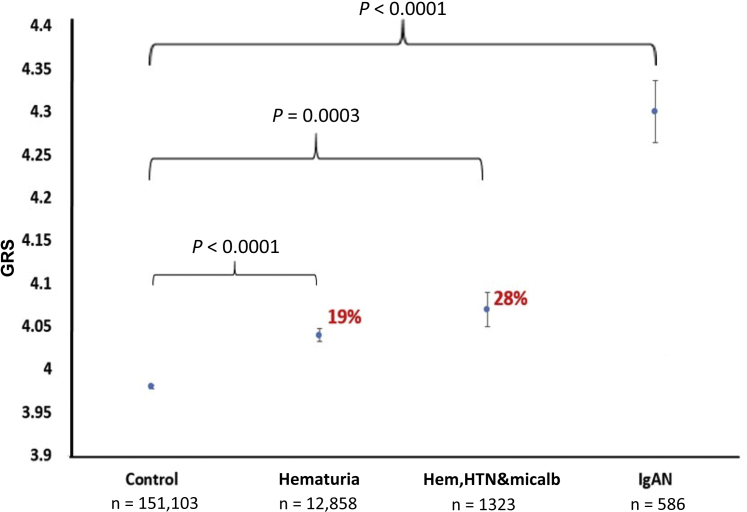
Figure 1.
Graph of estimated prevalence of IgA nephropathy (IgAN) in different mixture (hematuria, hypertension [HTN], microalbuminuria [micalb]) (with 95% confidence intervals represented by error bars). Controls: UK Biobank (UKBB) healthy individuals (without IgAN, hypertension, diabetes, albuminuria, renal disease). IgAN-related phenotypes: hematuria, hypertension, microalbuminuria. Cases: IgAN cases from UKBB and UK Glomerulonephritis DNA Bank (UKGDB) combined. Red numbers represent the genetic risk score (GRS) of phenotype that could be explained by IgAN calculated using the following formula: Proportion = (phenotype GRS – control GRS) / (IgAN-GRS – control GRS).
Sensitivity Analysis
Four HLA SNPs, shown to contribute independently to genetic risk, were in partial linkage disequilibrium (LD) (R2: 0.65–0.92). To test their contribution to the IgAN-GRS, we generated the IgAN-GRS using a subset of 11 SNPs (11 SNP score) with only the top SNP from this region included, and a 10 SNP score that excluded the 4 SNPs in high LD and a score based only on the strongest associated SNP.
Results
Cases Versus Control
The IgAN-GRS was higher in people with an ICD-10 code of IgAN in UKBB with a mean GRS of 4.18 (95% CI: 4.01–4.35) and in UKGDB cases (mean: 4.34 [4.23–4.38]) compared with controls (3.98 [3.97–3.98], both P < 0.0001). The GRS was similar between UKBB cases and UKGDB cases (4.18 vs. 4.34, P = 0.07). The mean IgAN-GRS of the IgAN cases from both cohorts combined was 4.30 (95% CI: 4.23–4.38), n = 586 (Figure 1). The IgAN-GRS was a modest discriminator of IgAN, ROC AUC of 0.6 (0.57–0.62), P < 0.0001 (Supplementary Figure S3).
Age and Gender
IgAN is known to be more common in male individuals and we observed this in UKBB IgAN cases (76% male). Male and female IgAN-GRS in IgAN cases were similar (4.16 vs. 4.23, P = 0.7). There was no association of IgAN with the principal components (UKBB comparisons, Supplementary Figures S4A and S4B) and when we adjusted for age, gender, and 5 principal components, the results were unchanged.
Simulated IgAN Cohorts
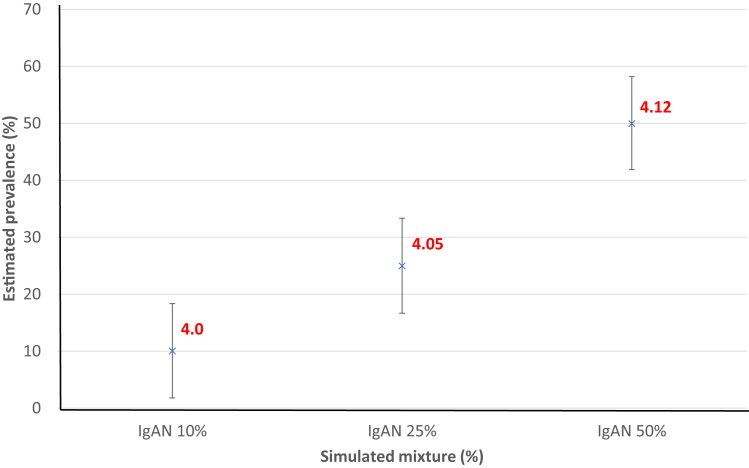
In the 3 artificial mixtures containing 10%, 25%, and 50% of IgAN cases, the mean GRS was 4.01, 4.05, and 4.12, respectively. These GRS values relative to the mean GRS of controls (3.98) and cases (4.27) allowed estimates of IgAN proportion (%) within the 3 artificial mixtures of 10% (95% CI: 2–18), 25% (17–33), and 50% (42–58), respectively (Figure 2).
Figure 2.
Proof of concept. Simulated mixture of IgA nephropathy (IgAN) and UK Biobank (UKBB) control in different percentage mixture. Data are sampled with replacement from IgAN cases (UK Glomerulonephritis DNA Bank [UKGDM] and UKBB) and UKBB control to generate these mixtures. The mean genetic risk score (GRS) is then calculated (red) and the estimated prevalence was calculated using our equation. The simulation mixture matches the calculated estimated prevalence.
Investigation of Nonspecific Phenotypes in UKBB
The IgAN-GRS was higher in individuals in UKBB with an ICD-10 code for hematuria when compared with controls, hematuria GRS was 4.04 (95% CI: 4.02–4.06) compared with control GRS of 3.98 (3.97–3.98); P < 0.0001 (Figure 1). This gave an estimate of 19% (95% CI: 13.7–24.3) of hematuria being accounted for by suspected IgAN. The GRS was slightly higher when we combined IgAN-associated phenotypes: hematuria, hypertension, and microalbuminuria/proteinuria (n = 1323) GRS 4.07 (95% CI: 4.02–4.13) versus 3.98 (3.97–3.98); P < 0.0001. This gave an estimate of 28% (95% CI: 11.6–44.6) of hematuria hypertension and microalbuminuria/proteinuria being accounted for by suspected IgAN (Figure 3).
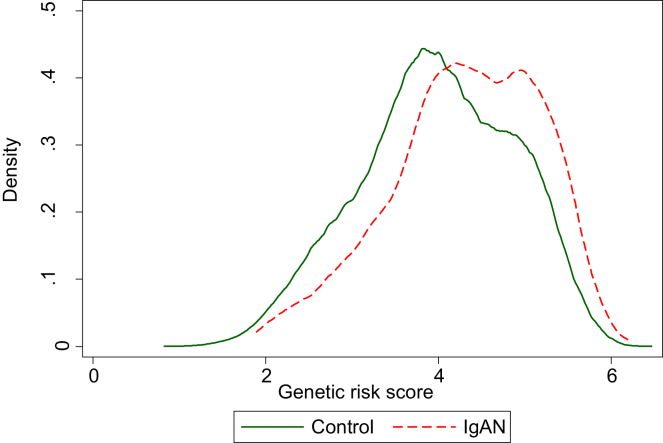
Figure 3.
Density plot demonstrating the distribution of IgA genetic risk scores of UK Biobank (UKBB) controls (green line, n = 151,103) and IgA nephropathy (IgAN) cases (red line, n = 586) from UKBB and the UK Glomerulonephritis DNA Bank cohort.
A sensitivity analysis demonstrated that most of the association of the IgAN-GRS was explained by the HLA loci. The GRS in 11 SNP (Supplementary Figure S5) and the 1 SNP scores (Supplementary Figure S6) were different between cases versus controls and in hematuria versus controls. The IgAN-GRS in the 10 SNP score was not different between cases versus controls.
Discussion
We have generated an IgAN-GRS in UKBB, shown excess IgAN genetic risk in people with hematuria, and used this to estimate potential undiagnosed IgAN. We estimated that 19% (2443) of white Europeans in UKBB with an ICD-10 code of unspecified hematuria, have suspected undiagnosed IgAN. Our simulated example highlights the validity of this approach and the CI around our estimates. Taking these findings together, our study highlights that disease-specific genetic risk scores may have utility at a population level to aid understanding of prevalence and associations of disease. With the expansion of numerous large datasets of clinical and genetic data, the utility of genetic risk scores to study undiagnosed disease is likely to increase, and the approach we have taken here could be used for other renal diseases and phenotypes.
The prevalence and incidence of IgAN vary geographically and estimates differ depending on detection from either biopsy or autopsy data. Differences between estimates from autopsy studies and biopsy studies suggest under-ascertainment of IgAN cases, which fits with variable biopsy practice worldwide and many people with mild IgAN features do not receive a biopsy diagnosis. Autopsy and donor registries additionally support at least some geographical variation of IgAN independent of biopsy practice. This is further substantiated by genome-wide reporting geospatial differences in prevalence of genetic susceptibility loci.22 Our estimate of 19% of noncancer haematuria being explained by suspected IgAN is specific to the inclusion criteria of UKBB (such as age distribution, ethnicity, demographics, and healthy volunteer bias), but is still relevant to estimates of IgAN prevalence due to the paucity of biopsy data. Our estimate, which is much higher than the number with an ICD-10 diagnosis of IgAN, adds to current knowledge due to the difficulties of assessment by other means and highlights that many with mild disease may be undiagnosed. In addition, it highlights that this genetic method could be used to assess disease prevalence in a population when the phenotype is variable, nonspecific, and does not fulfill risk benefit criteria for a renal biopsy. An important consideration in the future may be to see if estimates of IgAN prevalence, using complementary methods (GRS analyses, hospital records, biopsy registries, public health screening strategies), are congruent within other populations. The methods we use here may be directly relevant to assessment of IgAN prevalence in American and East Asian populations that are also developing large population cohort biobanks.
Ours is the first renal study to apply the use of a GRS to a question of underlying disease prevalence, and a strength of the study is the size of the population studied. This is suited to a disease such as IgAN in which ascertainment of cases is variable and there are likely undiagnosed cases due to variable biopsy practices. It also relies on robust genetic associations being described in GWASs. A recent hypertension GWAS23 of 1,000,000 people, 500,000 from UKBB, identified 535 novel blood pressure loci with several hits associated with renal disease, highlighting that undiagnosed renal disease may account for a proportion of hypertension and could potentially be explained, in part by common glomerular diseases like IgAN. The association of the 4 strongest IgAN loci with the hypertension GWAS support this finding.
GRSs are increasingly being assessed for utility in prediction, diagnosis, and common diseases.24 We recently showed that GRS can identify undiagnosed T1D in the population older than 30,10,11 and that a T1D GRS could aid diagnosis of T1D. Common disease GRSs such as cardiovascular disease10 and trait GRSs such as hypercholesterolemias can confer threefold increased risk of coronary artery disease in those with the highest polygenic burden.25 This may have utility to stratify individuals and groups of patients into risk groups with the potential to target treatment to high-risk groups26 but depends on prevalence of disease and the ability to intervene in those with high risk. We showed that although there was a clear difference in mean IgAN-GRS in cases versus controls, the individual discriminative power of the IgAN-GRS generated was modest. This is in keeping with the relatively small amount of variance explained in IgA risk by known IgA variants. Our study highlights that a GRS that is not strongly discriminative at an individual level can still have utility at a population level to aid understanding of prevalence and associations of disease.
IgAN cases and controls have improved understanding of IgAN pathogenesis, and geospatial distribution. These studies have highlighted pathways important in intestinal immunity and inflammation. Associated loci include the HLA-DR, -DQ, and -DP SNPs and alleles8,27 that are critical to antigen presentation and adaptive immunity. IgA-associated HLA-DR and HLA-DQ alleles, and non-HLA alleles, demonstrate pleiotropy with a variety of autoimmune diseases, in particular inflammatory bowel disease, but also including T1D, rheumatoid arthritis, and ankylosing spondylitis. Associated genes include ITGAM and ITGAX that encode integrins marking intestinal dendritic cells, and DEFA genes that encode alpha defensins important in mucosal defense. Associated loci also point toward roles for NF-κΒ signaling (e.g., CARD928), defense against intracellular pathogens and complement activation (e.g., CFHR3-CFHR29), and serum nonalbumin protein and IgA levels (TNFSF13 locus).28 The association of risk allele frequency with geographic location points toward multilocus adaption to environment with helminth infection a potential source of selective pressure. As larger sample of IgAN cohorts are aggregated and as more detailed SNP array and sequencing data become available, it is likely that further mechanistic insights into IgAN disease will be made using GWASs.
Limitations
UKBB is a voluntary study and participants are typically healthier and come from higher socioeconomic background compared with the general UK population.30 The identification of phenotypes and IgAN cases has come from self-report questionnaire and hospital ICD-10 codes, relying on the volunteer or the person inputting the data to be accurate. These could lead to an underestimation of the number of people with a diagnosis of IgAN. We were not able to use one known IgAN risk SNP, as it was not genotyped or imputable in the UKBB array data. We performed an analysis of IgAN-GRS using currently available published data; it is likely that as case-control GWASs increase in size and with better genome-wide coverage, newer risk loci and possibly newer approaches to GRS generation will be identified that could improve discriminative power of an IgAN-GRS and the precision around estimates of IgA disease within UKBB and other similar datasets.21
Our sensitivity analysis suggested that the IgAN-GRS could be driven by the 4 HLA SNPs in high LD, the remaining 10 SNPs are not shown to be associated with hematuria. This may be due to the modest power of our sample size of cases, and the modest effect size of the non-HLA variants associated with IgAN. The HLA-DR-DQ region had the greatest number of overlapping associations and IgAN risk alleles within this locus confer increased risk of many illnesses8; however, none that are known to cause hematuria. It is possible that the IgAN increasing HLA alleles may be enriched in those with hematuria for reasons other than IgAN, although this seems unlikely given the strong relationship with IgAN and hematuria.
It is possible that true underlying IgAN cases presenting only with hematuria have lower GRSs compared with cases with a biopsy-diagnosed IgAN, but this is not known. As our hematuria group was derived from ICD-10 codes and not urinalysis, it is possible that the number of people with hematuria in UKBB is an underestimate. However, if not all IgAN cases have hematuria in their electronic health record, this would lead to an underestimate of suspected IgAN because underascertainment of hematuria would increase the number with IgAN. If mild or undiagnosed disease is associated with a low IgAN-GRS, our estimates may further underestimate the true proportion of IgAN.
Conclusion
We have generated an IgAN-GRS that estimates that between 15% and 25% of unspecified hematuria within the UKBB population might have IgAN. This is the group that may not usually receive a renal biopsy or diagnosis. Our findings build on our assumption that the prevalence of IgAN may be an underestimate, based on biopsy registry.
This study has shown that GRS has a potential role in disease prevalence estimation in large population-based cohorts. With larger GWASs, the GRS and its power can be improved. Future work could be done in investigation of other renal diseases with genetic component. Consideration should be taken with regard to pleiotropy, more research is needed to refine the method and investigate causation of IgAN.
Disclosure
All the authors declared no competing interests.
Acknowledgments
This study was done with the UK Biobank resource (application 9072). UK Glomerulonephritis DNA Bank cohort. Piotr Słowiński, was consulted on the means method and helped with the simulation estimates and calculation. KS is funded by an Nation Institute for Health and Research (NIHR) Academic Clinical Fellowship. SAS is supported by a Diabetes UK PhD studentship (17/0005757). RAO is supported by a Diabetes UK Harry Keen Fellowship (16/0005529) MNW is supported by the Wellcome Trust Institutional Support Fund (WT097835MF). The views expressed are those of the authors and not necessarily those of the National Health Service (NHS), the NIHR, or the Department of Health.
Author Contributions
RAO, MNW, and KS designed the study. SAS, JH, JT, and MNW generated the genetic risk score. NJTconsulted on the means method. DG and JB provided the UKGDB cohort for validation. MM imputed the UKGDB data. KS, RK, JH, MNW, RAO, CB, NJT, JT, DG, and JB analyzed the data. KS wrote the first draft of the report. All authors reviewed the draft and contributed to the revision of report.
Footnotes
Table S1. SNPs used to generate IgAN genetic risk score and their basic information.
Table S2. SNPs used to generate IgAN genetic risk score and their imputation r2.
Figure S1. Density plot demonstrating the distribution of controls, hematuria, and IgAN cases. The arrows demonstrate the use of the means method to calculate the potential IgAN.
Figure S2. Comparison of Earth Mover’s Distance (EMD) between distributions; a linear combination of kernel density estimates (KDE) of distributions; a published Excess method and Means methods (mixture population ). The reference and the mixture distributions are plotted on the left (, shaded red, , shaded blue, , shaded gray, respectively). Estimated values of prevalence and 95% confidence intervals (gray dots and lines with bars at the ends) are plotted on the right. The violin plots show the distribution of the 100,000 estimates of prevalence () in the bootstrap samples. The proportion of individuals with IgAN is shown as a dashed vertical line. Calculations were based on the following participants: noncases UK Biobank, UKGDB IgAN, ICD10 IgAN in UK Biobank.
Figure S3. ROC curve assessing the IgAN-GRS to discriminate IgAN (from UKBB and UKGDB) from UKBB controls. ROC AUC was 0.6 (95% CI 0.57–0.62, P < 0.0001).
Figure S4. PCA plots (PC1 V PC2). (A) PC1VPC2 between IgAN cases and controls. (B) PC1VPC2 between UKBB Hematuria and controls. The graphs demonstrate that there were no differences by principal components.
Figure S5. 11 SNP IgAN-GRS. A regenerated 11 SNP score that included 1 of 4 HLA SNPs (with 95% confidence intervals represented by error bars).
Figure S6. 1 SNP IgAN-GRS in difference phenotypes. A regenerated 1 SNP score (with 95% confidence intervals represented by error bars).
Supplementary Material
References
- 1.Schena F., Nistor I. Epidemiology of IgA nephropathy: a global perspective. Semin Nephrol. 2018;38:435–442. doi: 10.1016/j.semnephrol.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 2.McGrogan A., Franssen C., de Vries C. The incidence of primary glomerulonephritis worldwide: a systematic review of the literature. Nephrol Dial Transplant. 2010;26:414–430. doi: 10.1093/ndt/gfq665. [DOI] [PubMed] [Google Scholar]
- 3.Donadio J., Grande J. IgA nephropathy. N Engl J Med. 2002;347:738–748. doi: 10.1056/NEJMra020109. [DOI] [PubMed] [Google Scholar]
- 4.Barratt J. IgA nephropathy. J Am Soc Nephrol. 2005;16:2088–2097. doi: 10.1681/ASN.2005020134. [DOI] [PubMed] [Google Scholar]
- 5.Imai E., Yamagata K., Iseki K. Kidney disease screening program in Japan: history, outcome, and perspectives. Clin J Am Soc Nephrol. 2007;2:1360–1366. doi: 10.2215/CJN.00980207. [DOI] [PubMed] [Google Scholar]
- 6.Varis J., Rantala I., Pasternack A. Immunoglobulin and complement deposition in glomeruli of 756 subjects who had committed suicide or met with a violent death. J Clin Pathol. 1993;46:607–610. doi: 10.1136/jcp.46.7.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki K., Honda K., Tanabe K. Incidence of latent mesangial IgA deposition in renal allograft donors in Japan. Kidney Int. 2003;63:2286–2294. doi: 10.1046/j.1523-1755.63.6s.2.x. [DOI] [PubMed] [Google Scholar]
- 8.Kiryluk K., Li Y., Scolari F. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet. 2014;46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas N., Jones S., Weedon M. Frequency and phenotype of type 1 diabetes in the first six decades of life: a cross-sectional, genetically stratified survival analysis from UK Biobank. Lancet Diabetes Endocrinol. 2018;6:122–129. doi: 10.1016/S2213-8587(17)30362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inouye M., Abraham G., Nelson C. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharp S., Rich S., Wood A. Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diabetes Care. 2019;42:200–207. doi: 10.2337/dc18-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyatt R., Julian B. IgA nephropathy. N Engl J Med. 2013;368:2402–2414. doi: 10.1056/NEJMra1206793. [DOI] [PubMed] [Google Scholar]
- 13.D'Amico G. Influence of clinical and histological features on actuarial renal survival in adult patients with idiopathic IgA nephropathy, membranous nephropathy, and membranoproliferative glomerulonephritis: survey of the recent literature. Am J Kidney Dis. 1992;20:315–323. doi: 10.1016/s0272-6386(12)70293-7. [DOI] [PubMed] [Google Scholar]
- 14.Gale D., Molyneux K., Wimbury D. Galactosylation of IgA1 is associated with common variation in C1GALT1. J Am Soc Nephrol. 2017;28:2158–2166. doi: 10.1681/ASN.2016091043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das S., Forer L., Schönherr S. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen N.E., Sudlow C., Peakman T. UK biobank data: come and get it. Science Transl Med. 2014;6:224ed4. doi: 10.1126/scitranslmed.3008601. [DOI] [PubMed] [Google Scholar]
- 17.Tyrrell J., Jones S., Beaumont R. Height, body mass index, and socioeconomic status: mendelian randomisation study in UK Biobank. BMJ. 2016;352:i582. doi: 10.1136/bmj.i582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ai Z., Li M., Liu W. Low α-defensin gene copy number increases the risk for IgA nephropathy and renal dysfunction. Sci Transl Med. 2016;8 doi: 10.1126/scitranslmed.aaf2106. 345ra88–345ra88. [DOI] [PubMed] [Google Scholar]
- 19.Hüls A., Krämer U., Carlsten C. Comparison of weighting approaches for genetic risk scores in gene-environment interaction studies. BMC Genet. 2017;18:115. doi: 10.1186/s12863-017-0586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans B.D., Hattersley A.T., Jones S.E. Estimating population level disease prevalence using genetic risk scores. medRxiv. 2020:20025528. [Google Scholar]
- 22.Kiryluk K., Li Y., Sanna-Cherchi S. Geographic differences in genetic susceptibility to IgA nephropathy: GWAS replication study and geospatial risk analysis. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evangelou E., Warren H., Mosen-Ansorena D. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torkamani A., Wineinger N., Topol E. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 25.Khera A., Chaffin M., Aragam K. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stower H. Bringing polygenic risk scores to the clinic. Nat Med. 2018;24:1303. doi: 10.1038/s41591-018-0190-8. [DOI] [PubMed] [Google Scholar]
- 27.Franke A., McGovern D., Barrett J. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osman W., Okada Y., Kamatani Y. Association of common variants in TNFRSF13B, TNFSF13, and ANXA3 with serum levels of non-albumin protein and immunoglobulin isotypes in Japanese. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W., Stambolian D., Edwards A. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107:7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith G.D., Davies N.M. Can genetic evidence help us understand why height and weight relate to social position? BMJ. 2016;352:i1224. doi: 10.1136/bmj.i1224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.