Abstract
TRIM (Tripartite motif) and TRIM-like proteins have emerged as an important class of E3 ligases in innate immunity. Their functions range from activation or regulation of innate immune signaling pathway to direct detection and restriction of pathogens. Despite the importance, molecular mechanisms for many TRIM/TRIM-like proteins remain poorly characterized, in part due to challenges of identifying their substrates. In this review, we discuss several TRIM/TRIM-like proteins in RNA sensing pathways and viral restriction functions. We focus on those containing PRY-SPRY, the domain most frequently used for substrate recognition, and discuss emerging mechanisms that are commonly utilized by several TRIM/TRIM-like proteins to tightly control their interaction with the substrates.
Keywords: Innate immunity, Tripartite motif protein, PRY-SPRY domain, Substrate recognition, RIG-I like receptors, Oligomer
1. Introduction
The innate immune system is the first line of defense against a broad range of microbial pathogens. Innate immune receptors, so called pattern recognition receptors (PRRs), recognize conserved pathogen-associated molecular patterns (PAMPs) and activate a variety of innate immune responses that mediate immune cell recruitment and pathogen restriction. Over the last three decades or so, several families of germ-line encoded PRRs have been identified and extensively characterized. These include membrane-bound Toll Like Receptors (TLRs) that survey extracellular or endosomal space for the presence of infection, and soluble receptors, such as RIG-I like receptors (RLRs), cGAS and NOD-like receptors (NLRs), that monitor the cytosolic compartment [1]. Upon recognition of cognate PAMPs, these receptors engage with their downstream adaptor proteins, such as MAVS (also known as VISA, IPS-1, and Cardif [2], [3], [4], [5]), STING (also known as MITA, MPYS, ERIS and TMEM173 [6], [7], [8], [9]) and ASC, to initiate signaling cascades that culminate in transcriptional or post-translational activation of antiviral and pro-inflammatory cytokines [10]. While their functions are critical for proper defense against pathogens, accumulating evidence suggests that inappropriate activation of these receptors or inefficient suppression of their signaling pathways can lead to a spectrum of autoimmune and inflammatory disorders. As such, multiple regulatory mechanisms are in place to tightly regulate the activities of these innate immune pathways, while ensuring rapid amplification of the immune signaling cascades upon pathogen detection.
One primary mechanism of immune regulation is through post-translational modifications, which allows rapid and often reversible modulation of the activities of immune signaling molecules. Among those, ubiquitin (Ub) and Ub-like (Ubl) protein modifications play central roles in regulating nearly all innate immune signaling pathways [11], [12]. Ub/Ubl modification is mediated by sequential actions of an E1 activating enzyme, E2 conjugating enzyme and E3 ligase. The E3 ligase is the one that carries out the final step of Ub transfer from E2 to the substrate, and thus dictates the substrate specificity. There are over 600 E3 ligases in primates, as opposed to 2 E1s and ~40 E2s [13]. Accordingly, much effort has been made to identify E3 ligases that modulate any given immune pathway.
Tripartite motif proteins (TRIMs) are a family of E3 ligases that are emerging as key players in innate immunity. One of the first clues for their innate immune functions came from the fact that many TRIMs are induced by interferons or pro-inflammatory cytokines [14]. While E3 ligases are often thought to negatively regulate the stability of the target molecule by Ub-mediated proteasomal targeting, many TRIMs have been shown to enhance innate immune signaling pathways [15], through both proteasome-dependent and -independent mechanisms. Furthermore, some TRIMs were shown to play more active immune functions as pathogen sensors or restriction factors [16], [17], instead of simply up- or down- regulating the activities of immune signaling molecules.
Despite their importance in a broad range of innate immune functions, precise modes of action of many TRIM proteins remain elusive. This limitation largely reflects the challenge of identifying substrate proteins, which is the key to understanding their functions and mechanisms. We here review several TRIM and related E3 ligases (broadly defined as TRIM-like proteins) in innate immunity, with a particular focus on their substrate recognition mechanisms. We first begin by introducing the general properties of TRIM and TRIM-like proteins. We here apologize in advance to those whose studies were not included in the review due to the space limitation.
2. TRIM and TRIM-like proteins: shared domain architectures
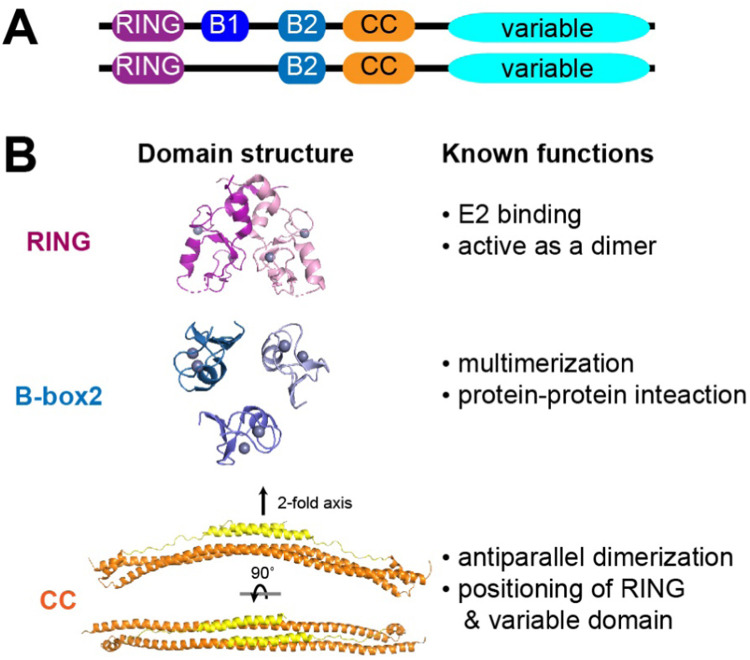
TRIMs are characterized by three distinct domains at the N-terminus: a RING domain that binds E2 conjugating enzymes, followed by one or two B-box domains and a coiled-coil (CC) domain ( Fig. 1A). They are also known as RBCC (RING-B-box-Coiled-Coil) proteins because the order at which these domains appear is conserved [18]. The TRIM family is one of the largest subclasses of RING-E3 ligases, consisting of more than 70 members in human [19]. It is also an ancient family of E3 ligases as members of the family can be found in almost all metazoans [19]. Not all TRIM proteins, however, have all three domains; some TRIMs have linkers replacing one of the three domains, while preserving the order of the rest of the domains [18], [20]. There are also many ligases (to be called TRIM-like proteins) not formally categorized into the TRIM family, but harboring two out of three domains with the same conserved order. The strict conservation of the domain orders implicates that functions of these domains are intimately coordinated with one another and their spatial arrangement in three dimensions is also likely conserved.
Fig. 1.
TRIM proteins display a common domain architecture. A. Common domain architectures of TRIMs, which consist of RING, B-boxes 1 and 2 (B1 and B2) and coiled-coil (CC). C-terminal domains are often involved in substrate recognition and are variable. B. Structures and functions of RING (PDB:5FEY, [23]), B-box (PDB: 5IEA, [25]) and coiled-coil domains (PDB: 6FLN, [29]). Yellow regions in CC (orange) indicate the linker C-terminal to TRIM CC that often folds back onto CC.
As RING-E3 ligases, TRIM/TRIM-like proteins function as an adaptor that bridges E2 and substrate. The RING domain carries out the E2 binding activity, while the C-terminal variable domain is often responsible for substrate binding (see Section 3). While most TRIM/TRIM-like proteins have been shown to function as Ub E3 ligases, some can conjugate Ubl proteins, such as SUMO and ISG15 [21], [22]. Many RING domains in TRIM/TRIM-like proteins require homo-dimerization to induce the “closed” E2~Ub conformation that stimulates Ub transfer from E2 to the substrate [23], [24] (Fig. 1B). While some RINGs form a constitutive dimer, others may form the dimer transiently when bound with the E2~Ub complex.
B-box is a zinc-finger domain, and is further categorized into B-box1 and B-box2 depending on the residues coordinating the Zn ions [20] (Fig. 1B). While many TRIMs have both B-boxes, with B-box1 preceding B-box2, some harbor only a single B-box, in which case it is always B-box2 [18] (Fig. 1A). While this suggests an essential function of B-box2 that cannot be replaced by B-box1, the precise function of B-box2 remains unclear. In TRIM5α, B-box2 was found to form a trimer (Fig. 1B), which leads to higher-order multimerization of TRIM5α [25]. It remains to be examined whether this is a generalizable property of B-box2 and what role B-box1 plays for those TRIM/TRIM-like proteins with both B-boxes.
Previous studies have shown that CCs in many TRIM proteins form a homodimer in vitro [26] (Fig. 1B), but its function appears to be more complex than simple dimerization. Structures of several TRIM CC’s showed an antiparallel dimeric architecture, which would place the two RING domains near the opposite ends of the CC [26], [27]. Since RING dimerization is required for many TRIMs, this geometric restraint raises the question of whether a RING:RING contact occurs within a TRIM dimer or through an inter-dimeric interaction. For certain TRIMs, the two RINGs may be too far apart to allow an intra-dimeric RING:RING contact, in which case higher-order oligomerization is likely required for their E3 ligase activities. Additionally, TRIM CC also appears to play roles in positioning the C-terminal variable domain. Many TRIMs harbor a characteristic linker following CC, which folds back onto the CC [23], [26], [27], [28], [29], [30] (Fig. 1B) and tethers the C-terminal domain near the center of CC [23], [26], [27], [28], [29], [30]. Such tethering appears important at least in some cases (e.g. TRIM5α [30] and TRIM25 [29]), as further evidenced by viral antagonists that target this function of CC [29]. Finally, CC was also proposed to promote higher-order oligomerization [23], [31], [32] or to form hetero-oligomeric complex with other related TRIMs [18], [33]. Thus, TRIM CC plays diverse functions in regulating the overall architecture and functions of TRIM proteins.
3. PRY-SPRY domain: structure and function
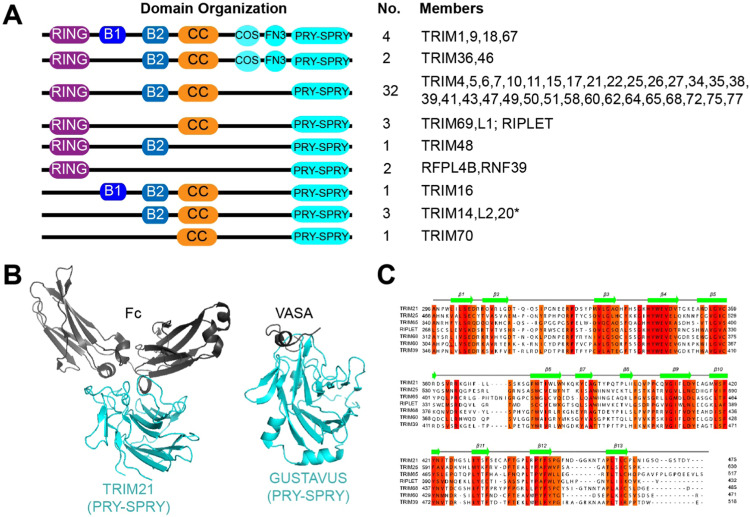
The C-terminal variable domain of TRIM/TRIM-like proteins often play important roles in substrate recognition. The PRY-SPRY domain is the most common C-terminal domain, accounting for that of about half of all known TRIMs [34], [35] ( Fig. 2A). TRIMs with PRY-SPRY are vertebrate-specific, emerged recently, evolves more rapidly, and are more intimately involved in host-pathogen interactions than other TRIMs [19], [27], [36].
Fig. 2.
The PRY-SPRY domain is a common domain for substrate recognition among TRIM/TRIM-like proteins. A. Domain architectures of TRIM/TRIM-like proteins with a PRY-SPRY domain. COS and FN3 indicate cos box and fibronectin type III repeat domains, respectively. *TRIM20 has a PYRIN domain in the N-terminus besides the domains illustrated. B. Structures of PRY-SPRY recognizing the cognate substrate using the cluster of variable loops. Structures show that PRY-SPRY can detect a three-dimensional structure of a substrate (left) or a linear peptide sequence (right). Left: TRIM21 PRY-SPRY in complex with Fc (PDB: 2IWG, [39]), Right: GUSTAVUS PRY-SPRY in complex with the VASA peptide (PDB: 2IHS, [43]). C. Sequence alignment of PRY-SPRY among several TRIM/TRIM-like proteins, showing the high sequence conservation in β-strands and variable loops between the β-strands.
The PRY-SPRY (a.k.a B30.2) domain is a ~200 amino acids-long, single globular domain (Fig. 2B). It is formed by appendage of the N-terminal PRY (~60 amino acids) to the C-terminal SPRY (~140 amino acids). The SPRY domain is widely conserved from yeast to human. By contrast, the PRY-SPRY domain is only found in vertebrates [37]. Previous structural analyses showed that PRY-SPRY domains display a highly conserved beta sandwich structure where two beta sheets are packed against each other [38] (Fig. 2B). While the sequences in the core β-strands are well-conserved, those of loops between β-strands are highly variable [39] (Fig. 2C). It is the cluster of these variable loops that bind substrates [39], [40], [41], [42] (Fig. 2B). The high sequence variability in these loops allows recognition of a diverse set of substrates, ranging from a linear peptide to three dimensional protein structure [39], [41], [42], [43] and to even an RNA molecule [44]. The utilization of variable loops for substrate recognition is akin to that of immunoglobulins, which utilize the complementary determine regions (CDRs) for substrate specificity.
Below, we focus on PRY-SPRY-containing TRIM/TRIM-like proteins and their substrate recognition mechanisms. We begin with TRIM/TRIM-like proteins involved in innate immune signaling pathways and expand our discussion to include those involved in antiviral effector immunity.
4. TRIMs as modulators of innate immune signaling pathways
4.1. TRIMs and innate immune receptors
4.1.1. RIPLET and RIG-I
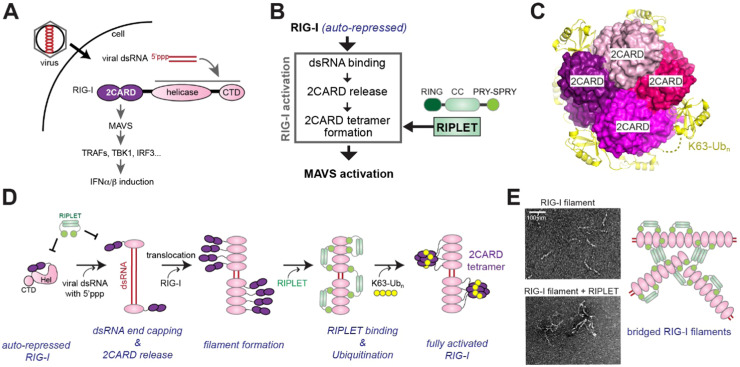
RIG-I is the founding member of the RLR family that recognizes viral RNAs and activates the signaling pathways to upregulate type I/III interferons and pro-inflammatory cytokines [45]. RIG-I consists of two tandem caspase activation recruitment domains (2CARD) at the N-terminus, a DEXD/H helicase domain in the middle, and a characteristic C-terminal domain (CTD) ( Fig. 3A). 2CARD is responsible for downstream signal activation, while the helicase domain and CTD cooperate to recognize viral RNAs. RIG-I relies on the presence of 5’-triphosphate (5’ppp) or 5’-diphosphate and dsRNA structure for distinguishing viral RNAs from cellular RNAs [46], [47]. For cellular RNAs, 5’ppp is typically removed from nascent transcripts during 5’-processing before they are exported to the cytoplasm, but this does not occur for many viral RNAs that are synthesized in the cytoplasm. In addition to 5’ppp, dsRNA length also plays an important role in self vs. non-self-discrimination by RIG-I. dsRNA duplex of > 20 bp is required and ~40–150 bp are ideal for activating RIG-I [31] (to be discussed more later). Although a short hairpin RNA with a ~10 bp stem was found to activate RIG-I [48], < ~20 bp dsRNA without a hairpin loop do not [48]. Given our current understanding that RIG-I does not bind the loop, the precise reason for the distinct RIG-I-stimulatory activities of hairpin vs. standard dsRNA remains unclear. It is possible that concatemerization of a hairpin RNA extends the duplex region, allowing it to mimic longer dsRNAs, as was shown before [49]. In addition to 5’ppp and 5’pp dsRNA, there are several other features of RNA that have been reported to stimulate RIG-I, such as features of 3’ end or sequence composition [50], [51], [52]. In many cases, however, the mechanisms remain unclear and require further investigations.
Fig. 3.
RIPLET is an essential E3 ligase for RIG-I signaling pathway. A. Schematic of RIG-I signaling pathway and domain architecture of RIG-I. During viral infection, viral RNAs accumulate in the cytoplasm. RIG-I detects viral dsRNA with a 5’-triphosphate group (5’ppp), and activates the downstream adaptor molecule MAVS. Activated MAVS in turn induces type I interferon production by triggering TRAF2/3/5/6, TBK1 and IRF3. B. Three steps involved in RIG-I activation. RIG-I binding to dsRNA releases 2CARD auto-repression, but this is not sufficient for signal activation. 2CARD must be tetramerized in order to activate MAVS. RIPLET promotes 2CARD tetramerization by conjugating K63-Ubn to RIG-I, which binds 2CARD and stabilizes its tetramer structure (see C). C. The crystal structure of RIG-I 2CARD (purple-shade colors) in complex with K63-Ubn (yellow) (PDB:4NQK, [54]). K63-Ubn wraps around the 2CARD tetramer, stabilizing its assembly architecture. D. In the absence of viral RNA, RIG-I is in the auto-repressed state. In the presence of viral dsRNA, RIG-I binds dsRNA end recognizing 5’ppp. dsRNA binding triggers ATP hydrolysis, which in turn stimulates translocation along dsRNA, and subsequent recruitment of additional RIG-I molecules. Iterations of the end recruitment and translocation of RIG-I molecules then results in RIG-I filament formation near dsRNA ends. RIPLET binds RIG-I only in the filamentous state, leading to K63-Ubn conjugation, 2CARD tetramerization and MAVS activation. E. RIPLET binding also leads to RIG-I filament bridging and clustering, which results in the amplification of RIG-I signaling. Negative stain electron micrographs are taken from [31].
Previous biochemical and structural studies provided detailed pictures of how RIG-I selectively recognizes viral RNAs and how it activates the downstream antiviral immune response. Upon viral RNA binding, RIG-I undergoes at least two distinct conformational changes (Fig. 3B). Before RNA engagement, RIG-I is in the auto-repressed state, in which 2CARD is bound by the helicase domain [53]. The auto-repression is released by viral dsRNA binding as RNA competes with 2CARD for the helicase domain [53]. Release of 2CARD, however, is insufficient to activate RIG-I signaling. The second necessary conformational change is homo-tetramerization of 2CARD [54], [55] (Fig. 3B). Only tetramerized 2CARD can stably interact with the downstream adaptor MAVS [56]. The 2CARD tetramer then acts as a nucleus to induce filament formation of MAVS, which serves as the signaling scaffold for activating the further downstream signaling pathway [56], [57].
Studies showed that the tetramerization of RIG-I 2CARD is greatly stimulated by K63-linked Ub chains (K63-Ubn), which wraps around the 2CARDs tetramer to stabilize the core 2CARD tetramer [54] (Fig. 3C). Recent genetic and biochemical studies showed that the E3 ligase RIPLET (a.k.a. RNF135) is responsible for RIG-I ubiquitination and activation [31], [58], [59]. While it has long been thought that another E3 ligase, TRIM25, was responsible for RIG-I activation, this notion was established largely based on the effect of TRIM25 on isolated 2CARDs, an artificial construct that also activates MAVS upon overexpression [60]. In fact, few studies have demonstrated the positive impact of TRIM25 on the signaling activity of full-length RIG-I. Furthermore, TRIM25 does not directly bind or ubiquitinate full-length RIG-I in a manner dependent on RNA ligand, the condition that must be satisfied in order to be functionally relevant [31]. This is because any E3 ligase would have a non-specific ubiquitination activity at high protein concentrations. Thus, specificity control, such as RIG-I in the absence of stimulatory RNA, is essential to validate the assay condition. Consistent with the notion that RIPLET is the bona fide E3 ligase for RIG-I, RIPLET binds and ubiquitinates RIG-I only in the presence of RIG-I-stimulatory RNA, recapitulating the cellular activation condition [31]. Interestingly, multiple studies have shown that TRIM25 can inhibit a broad range of viruses in a manner independent of RIG-I; TRIM25 binds influenza A nucleocapsid and blocks its replication [61], and cooperates with the antiviral protein ZAP to inhibit viral gene expression [62], [63]. TRIM25 was also shown to directly interact with the linear ubiquitin chain assembly complex (LUBAC), which often plays a negative regulatory role in innate immune signaling [64]. Altogether, the antiviral functions of TRIM25 appears complex, but is not directly involved in the RIG-I signaling pathway. Instead, RIPLET mediates ubiquitination and activation of RIG-I.
Another area of controversy has been on the importance of covalent Ub conjugation vs. non-covalent binding in RIG-I signaling. Previous biochemical and structural studies [54], [55] have showed that unanchored K63-Ubn alone can induce 2CARD tetramerization and activate RIG-I signaling in vitro. However, whether non-covalent interaction alone is indeed sufficient in cells has been controversial. In order to address this issue, multiple studies have examined the effect of mutations of potential conjugation sites of RIG-I, but such efforts frequently involved mutations of multiple lysines, which alone can significantly affect protein folding and other activities besides ubiquitination. Multiple lines of evidence support the importance of covalent Ub conjugation as well as non-covalent interaction. First, non-covalent interaction between K63-Ubn and RIG-I 2CARD is weak, while covalent conjugation greatly stimulates the stability of their complex and the resultant 2CARD tetramer [54]. Second, a recent biochemical analysis suggests that RIPLET can constitutively generate unanchored K63-Ubn without RIG-I, but the presence of RIG-I filaments converts its activity to the RIG-I conjugation mode, suggesting that the conjugated K63-Ubn is more likely to be relevant to RIG-I functions [31]. Third, in a reconstituted signaling system, RIPLET-dependent RIG-I signaling was maintained even after treatment with isoT, which selectively degrades unanchored Ub chains [31]. While another previous study reported an abrogation of RIG-I signaling upon isoT treatment [65], one important difference is that the latter study utilized TRIM25, which unlike RIPLET, only generates unanchored K63-Ubn with or without RIG-I. TRIM25 in this in vitro system likely generates a sufficient level of unanchored K63-Ubn to activate RIG-I. Since TRIM25 does not bind RIG-I [31], its activity to constitutively synthesize unanchored K63-Ubn is unlikely to be relevant for RIG-I signaling. In summary, these observations collectively suggest the importance of both anchored and unanchored K63-Ubn for RIG-I signaling, and may serve as a model to study the importance of covalent Ub conjugation in other systems.
RIPLET is a TRIM-like E3 ligase, harboring RING, CC and PRY-SPRY, but not B-boxes (Fig. 3B). As mentioned above, RIPLET binds and ubiquitinates RIG-I only in the presence of dsRNA [31], which parallels dsRNA-dependent RIG-I signaling in cells. A more detailed analysis showed that RIPLET:RIG-I binding requires dsRNA to be at least ~20 bp in length, again in agreement with the cellular requirement for RIG-I activation [31]. This is because individual PRY-SPRY has low affinity for monomeric RIG-I, and a high affinity interaction requires both PRY-SPRY domains of dimeric RIPLET to simultaneously bind multimeric RIG-I assembled on > 20 bp dsRNA [31]. This avidity-driven binding explains why RIG-I signaling is more efficient with longer dsRNA (~40–150 bp), on which RIG-I forms filaments [31], [66]. RIG-I forms filaments through two steps (Fig. 3D): initial recruitment of RIG-I to a dsRNA end via 5’ppp moiety, and subsequent translocation of RIG-I from the dsRNA end to the interior through an ATP hydrolysis-driven motor activity. Iterations of this process with multiple RIG-I molecules result in accumulation of filamentous oligomers of RIG-I near the dsRNA end [31], [66]. The reason RIG-I and RIPLET’s activity declines on > ~0.5 kb dsRNA is that RIG-I filament formation is dependent on 5’ppp, which is diluted with the increasing length of dsRNA [31].
Intriguingly, RIPLET binding to RIG-I filaments can also lead to filament cross-bridging, creating aggregate-like assemblies with a heightened signaling potential [31] (Fig. 3E). The CC domain of RIPLET plays an important role in filament bridging, presumably by forming higher-order oligomers [31]. In this clustered assembly structure, RIG-I signaling is further amplified, likely because there are more RIPLET RING domains to form an active RING:RING dimer, and/or there is higher concentration of RIG-I molecules to form the active 2CARD tetramer. The study of RIG-I and RIPLET interaction provides a detailed example of how TRIM-like proteins utilize bivalency and CC for regulating substrate selectivity, higher-order oligomerization and innate immune function.
4.1.2. TRIM65 and MDA5
MDA5 is another member of the RLR helicase family that functions as a cytosolic receptor for viral dsRNAs. MDA5 shares with RIG-I the same domain architecture and the downstream signaling pathway, including the adaptor molecule MAVS. However, MDA5 and RIG-I play non-redundant roles as they recognize largely distinct groups of viruses and viral RNAs [67]. While RIG-I detects a broad range of negative strand RNA viruses by recognizing 5’ppp-containing dsRNA with a preference for mid-long lengths (~40–150 bp), MDA5 detects several positive strand RNA viruses, such as picornaviruses and coronaviruses [68], [69] through recognition of much longer (>~1 kb) dsRNAs independent of 5’ppp [70], [71]. While MDA5 also forms filaments along the length of dsRNA, it utilizes different mechanisms for assembling filaments and regulating its stability; while RIG-I filament propagates from a dsRNA end to an interior in a 5’ppp- and ATP-dependent manner, MDA5 filament is nucleated directly from a dsRNA interior, and disassembles from its termini during ATP hydrolysis [66], [72]. These differences ensure MDA5 to form filaments more efficiently on much longer dsRNAs than those preferred by RIG-I, and its signaling activity to progressively increase with dsRNA length.
Like RIG-I, MDA5 signaling also requires homo-tetramerization of its 2CARD, and the efficiency of 2CARD tetramerization is enhanced by K63-Ubn [54], [73]. MDA5 signaling and ubiquitination is independent of RIPLET [31], but instead relies on TRIM65 [74], [75]. Knocking out TRIM65 impairs MDA5 signaling activity without affecting other antiviral signaling pathways, such as that of RIG-I. Analogous to how RIPLET binds RIG-I, TRIM65 utilizes the PRY-SPRY domain to bind the helicase domain of MDA5 [75]. Furthermore, TRIM65 also binds MDA5 only in its filamentous form on long dsRNA, and this filament-specific recognition stems from the requirement for bivalent PRY-SPRY engagement, as was the case for RIPLET [76] .
Thus, for both RIPLET and TRIM65, avidity-dependent substrate recognition allows more precise control of immune activation and ensures immune signaling to occur only in the filamentous form, i.e. in the presence of the viral RNA ligand. Given that an increasing number of receptors and signaling molecules in the innate immune system are shown to multimerize upon activation [77], it is tempting to speculate that TRIM/TRIM-like proteins may utilize multimer-specific substrate recognition as a common mechanism for regulating their immune functions.
4.1.3. TRIM/TRIM-like proteins for RLRs and other helicases
Besides RIPLET and TRIM65, several other TRIM/TRIM-like proteins have been proposed to act on RLRs and modulate their stabilities and/or antiviral signaling activities. One such E3 ligase is TRIM38, which was reported to promote antiviral signaling of both RIG-I and MDA5 through SUMO conjugation [22]. TRIM38 uses its PRY-SPRY domain to interact with both 2CARDs and the helicase domains of RIG-I and MDA5, and these interactions were found to be dependent on viral infection. This study further reports that SUMO modification is dynamically regulated over the course of infection, and delays K48-linked ubiquitination of RIG-I and MDA5, thereby temporarily stabilizing these receptors and potentiating their signaling activities [22]. It is yet unclear how SUMOylation competes with K48-linked ubiquitination and how the SUMOylation of RLRs changes during the course of infection. Intriguingly, TRIM38-mediated SUMOylation was later found to also regulate cGAS, a foreign DNA sensor that functions parallel to RLRs [78]. Considering that cGAS also forms filament-like structure during viral DNA sensing [79], it is possible that a multimer-specific substrate recognition mechanism, akin to those of RIPLET and TRIM65, is involved in virus-dependent SUMOylation of cGAS and RLRs by TRIM38.
Besides RLRs, several other DExD/H-box helicases have been reported to play roles in sensing viral nucleic acids. One such helicase is DDX41, which is an RNA helicase previously known to be involved in splicing, translation and many other cellular RNA processes [80], [81]. Recent studies found that DDX41 is also involved in innate immune response to foreign dsDNA [82], [83], [84]. A systematic siRNA screen of DEXD/H helicases showed that knock-down of DDX41 greatly reduces the level of IFN-β induction in response to poly(dA:dT) or HSV-1 [85]. The DEAD domain of DDX41 can directly bind both dsDNA and STING, the signaling adaptor molecule for cGAS [85]. The interaction between DDX41 and STING was proposed to result in the activation of TBK1 and the IKK complex for the induction of type I IFNs and pro-inflammatory cytokines. Interestingly, the stability of DDX41 was reported to be regulated by TRIM21 [85]. TRIM21 utilizes PRY-SPRY to bind the DEAD domain and conjugates DDX41 with K48-linked Ub chains (K48-Ubn) for proteasomal degradation [85]. Knocking down TRIM21 enhances the IFN expression upon dsDNA stimulation or DNA virus infection [85]. Much remains to be investigated as to whether TRIM21 also regulates the stability of DDX41 during its constitutive function in RNA processes, and whether their interaction is regulated differently during RNA processing vs. DNA sensing. More detailed analyses of how TRIM21 acts on DDX41 and whether this interaction depends on the conformational or oligomeric state of DDX41 upon DNA or RNA binding may bring new mechanistic insights.
4.2. TRIMs for innate immune signaling molecules
4.2.1. TRIMs and MAVS
MAVS is the signaling adaptor molecule for RIG-I and MDA5. Upon interaction with activated RIG-I and MDA5, MAVS forms filaments on the surface of the mitochondria, serving as a signaling platform to recruit and activate further downstream molecules, such as TRAFs and TBK1 [56], [86]. At least six TRIM proteins have been reported to regulate MAVS. Among them, TRIM21, TRIM25, and TRIM14 utilize PRY-SPRY for interaction with MAVS [87], [88], [89], [90]. TRIM21 was reported to conjugate MAVS with K27-linked Ub chains to facilitate recruitment of TBK1 [88], while TRIM25 was found to modify MAVS with K48-linked Ub chain to promote its degradation [87], [91]. Unlike TRIM21 and TRIM25, TRIM14 lacks the RING domain and does not ubiquitinate MAVS. Instead, TRIM14 was proposed to function as an adaptor molecule to recruit NF-kB essential modulator (NEMO), WHIP and PPP6C to MAVS to potentiate its signaling activity [89], [90]. Viral infection was shown to promote TRIM14’s interaction with MAVS [90], raising the possibility that their interaction may depend upon MAVS filament formation. Additionally, TRIM40, TRIM44 and TRIM31, which do not contain PRY-SPRY, were also reported to interact with MAVS and alter MAVS’s stability and/or signaling activity in both Ub-dependent and –independent manners [92], [93], [94]. However, given that MAVS forms filaments, which could attract a large number of molecules through specific or non-specific binding, more detailed investigation is necessary to understand precisely how each TRIM protein acts on MAVS, and how these TRIM proteins collectively work together for proper functioning of MAVS.
4.2.2. TRIM38 and TRAF6
TRAF6 is an E3 ligase involved in multiple immune signaling pathways, including those of RLRs, TLRs, IL-1 receptor, TGFβ receptors and B-cell receptors. Upon activation of these upstream receptors, TRAF6 is recruited to the receptors or downstream signaling complexes [95], [96], which triggers its E3 ligase activity to synthesize K63-Ubn. While the precise activation mechanism and ubiquitination target of TRAF6 has been unclear, the current model is that K63-Ubn conjugation within the signaling complex (either on TRAF6 or other targets) enables recruitment of various signaling molecules, leading to the activation of NF-kB, MAPK and/or IRF pathways [96], [97].
Recent studies suggest that TRIM38 is an important negative regulator of TRAF6 [98]. TRIM38 utilizes its PRY-SPRY domain to recognize TRAF6 and conjugates TRAF6 with K48-Ubn for proteasomal degradation [98]. How the activity of TRIM38 on TRAF6 is regulated remains to be further investigated. One potential mechanism is a temporal control of the level of TRIM38. TRIM38 is known to be up-regulated by a variety of immune signals, including RLR and TLR pathway activation, which may ensure TRIM38 to act on TRAF6 only upon immune signal activation [98], [99], [100]. Alternatively, TRAF6 was proposed to form a higher-order assembly upon its activation [101], [102], which may function as a signal for TRIM38 to engage and ubiquitinate TRAF6 only upon its activation. It also remains to be addressed how, during RLR signaling, TRIM38 functions as an Ub E3 ligase for TRAF6, while acting as a SUMO E3 ligase for RIG-I/MDA5, and how its positive effect on RIG-I/MDA5 interplays with its negative effect on TRAF6. Mechanistic details of TRIM38 functions in the context of the RLR pathway require future studies.
4.2.3. TRIM26 and IRF3
IRF3 is an essential transcription factor for expressing type I IFNs and is broadly involved in many PRR signaling pathways in innate immunity, including RLRs, TLRs and cGAS pathways. Upon activation of these receptors, their respective downstream adaptor molecules (MAVS, TRIF, and STING, respectively) recruit TBK1, which then phosphorylates the C-terminal tail of IRF3 [103], [104], [105]. Phosphorylated IRF3 in turn forms a homodimer and translocates to the nucleus, which results in the transcriptional activation of type I IFNs [106].
IRF3 has long been known to undergo rapid turnover upon its activation [106], [107], [108]. One mechanism by which the stability of IRF3 is regulated is through TRIM26. A study showed that, upon TLR activation, TRIM26 interacts with IRF3, and conjugates IRF3 with K48-Ubn for proteasomal degradation [105]. Intriguingly, TRIM26 is translocated to the nucleus upon infection or IFN-β treatment, which allows TRIM26 to specifically target only the activated IRF3, leaving inactive cytosolic IRF3 intact. Similarly, constitutively active IRF3 variants are targeted by TRIM26, while phosphorylation-defective IRF3 or that lacking the nuclear localization signal is not [105]. By targeting only the activated IRF3, cells may be able to rapidly shut down the immune signal, while still maintaining the ability to elicit a subsequent immune response in the case of uncontrolled infection or re-infection. The spatial control of TRIM26 thus shows yet another mechanism by which TRIM/TRIM-like proteins are regulated.
5. TRIMs as virus sensors and effectors
5.1. TRIM5α and retroviral capsid
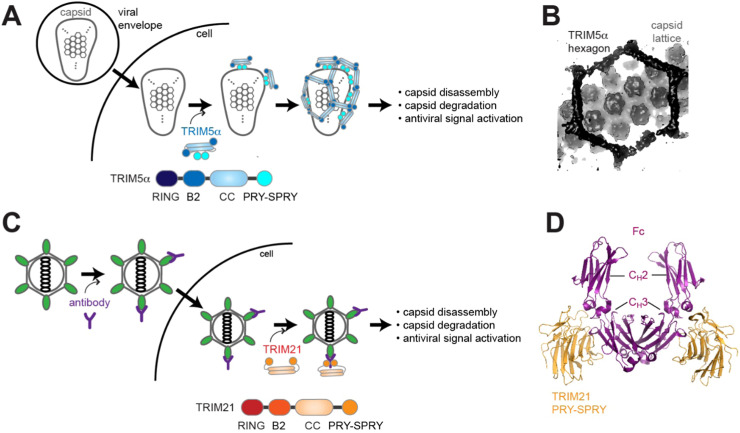
Some TRIM proteins can function as a direct sensor of viruses and a restriction factor. One well-known example is TRIM5α, an isoform of TRIM5 that contains the PRY-SPRY domain [32]. Previous studies showed that TRIM5α from Rhesus monkey restricts HIV-1 replication by inducing premature capsid disassembly and inhibiting reverse transcription [109], [110]. It was later found that the restriction activity of TRIM5α is highly dependent on the species and viral strains/types; human TRIM5α cannot restrict HIV-1, but it can restrict another retrovirus, N-tropic murine leukemia virus (N-MLV) [111], [112]. The viral specificity of TRIM5α is determined by the viral capsid and the PRY-SPRY domain of TRIM5α [109], [113]. A single amino acid change in the HIV-2 capsid rendered it more susceptible to human TRIM5α [114]. Similarly, three amino acid substitution in PRY-SPRY to mimic those of Rhesus TRIM5α allowed human TRIM5α to restrict HIV-1 [113], [114].
More detailed study of TRIM5α revealed avidity-dependent binding of HIV-1 capsid, analogous to that of RIPLET and TRIM65. HIV-1 capsid protein forms a fullerene-shaped cone, through hexagonal assembly of the capsid hexamers and pentamers [32], [115]. TRIM5α does not bind free capsid molecules, and instead, recognizes only the pre-assembled capsid lattice [110], [116] ( Fig. 4A). Viral capsid binding further induces higher-order oligomerization of TRIM5α into a hexagonal lattice with the symmetry that matches that of the viral capsid (Fig. 4A & Fig. 4B), further strengthening their avidity-driven interaction [117], [118]. The lattice formation of TRIM5α appears to be driven by B-box2, which forms a trimer at the three-fold symmetric vertex of the hexagonal lattice [25], [117] (Fig. 4A). Mutations in the B-box2 or CC of TRIM5α impair its lattice formation and its viral restriction function [119], [120], [121].
Fig. 4.
TRIM5α and TRIM21 have dual functions as innate immune receptors and effectors. A. Schematic of TRIM5α function. TRIM5α binds incoming retroviral capsid in a sequence-specific and avidity-dependent manner. Capsid then serves as a template to further assemble hexagonal lattice of TRIM5α. This results in both the effector function of capsid disassembly and degradation as well as the receptor function of antiviral signal activation. Ubiquitination activity of TRIM5α is thought to mediate both of these functions, although some studies proposed a direct role of TRIM5α as an autophagy receptor [125], [126]. B. Cryo-electron microscope reconstruction map of TRIM5α hexagonal lattice bound to HIV-1 capsid lattice (EMD-20565, [32]). C. Schematic of TRIM21 function. Microbial pathogen partially coated with antibody may evade neutralization, and enter the cytosolic compartment. TRIM21 recognizes the Fc portion of the antibody, which triggers the auto-ubiquitination activity and subsequently its antiviral functions similar to those of TRIM5α.(D.) Crystal structure of TRIM21 PRY-SPRY in complex with a dimeric Fc portion of IgG (PDB:2IWG, [39]).
Precise mechanism by which TRIM5α’s capsid binding leads to viral restriction is yet unclear. Upon capsid binding and lattice formation, the E3 ligase activity of TRIM5α was proposed to be stimulated as the lattice formation would cluster the RING domain and promote its dimerization, the requirement for efficient Ub transfer from E2 to the substrate [122]. Although whether TRIM5α can ubiquitinate the capsid is unclear, TRIM5α binding and lattice formation on the viral capsid is important for proteasomal or autophagic degradation of the capsid proteins and capsid disassembly [123], [124], [125], [126]. TRIM5α has been shown to conjugate K63-Ubn to itself or generate unanchored Ub chains [123], [127], which was proposed to assemble signaling complexes to induce antiviral signaling cascades. While much of these proposed mechanisms remain to be further investigated, TRIM5α serves as an intriguing example of TRIMs that can function as both a PRR and a restriction factor.
5.2. TRIM21 as an intracellular Fc receptor
TRIM21 (a.k.a. Ro52 or SS-A) was first identified as an autoantigen in autoimmune diseases, such as Sjogren’s syndrome and systemic lupus erythematosus [128], but more recent studies showed various other immune functions of TRIM21 in the cytoplasm. When non-enveloped viruses or bacteria are opsonized by antibody, they can be neutralized and cleared by phagocytes. However, opsonized microbial pathogens, especially those partially coated with antibody, can evade extracellular neutralization and can still invade cells. TRIM21 appears to act on such microbes that enter the cytosolic compartment with antibody attached to their surface [129] (Fig. 4C). TRIM21 tightly binds Fc of IgG antibodies, although it also binds Fc’s of IgM and IgA with lower affinities [130]. This engagement leads to the auto-ubiquitination of TRIM21, first with K63-Ubn, but gradually involving K48-Ubn in the form of mixed or branched chains [131]. Auto-ubiquitinated TRIM21 then recruits the proteasome for degradation of pathogen-associated molecules, such as viral capsid, and neutralization of the pathogen infectivity [129], [131] (Fig. 4C). In parallel, the proteasome-associated deubiquitinating enzyme Poh1 liberates K63-Ubn from TRIM21, allowing unanchored K63-Ubn to serve as a signal to activate antiviral signaling pathways [131] (Fig. 4C). Thus, as with TRIM5α, TRIM21 also displays a dual function as a PRR and an effector. They, however, differs in that TRIM5α directly recognizes PAMPs, while TRIM21 indirectly recognizes PAMPs through antibodies.
As with other PRY-SPRY containing TRIM/TRIM-like proteins, TRIM21 utilizes PRY-SPRY to bind Fc. While individual PRY-SPRY has a low affinity for Fc, its bivalency within the dimeric TRIM21 dramatically increases the affinity for dimeric Fc (K d decreases from ~200 to 0.6 nM) [129]. Thus, as a dimer, TRIM21 is the tightest Fc receptor known to date. The crystal structure of PRY-SPRY in complex with Fc showed that PRY-SPRY binds the hinge interface between the CH2 and CH3 domains, recognizing the three-dimensional structure of Fc [39], [130] (Fig. 4D). This is unlike other PRY-SPRY domains that recognize a linear peptide sequence [41]. More detailed study suggests that TRIM21 PRY-SPRY recognizes both the sequence and structure of Fc that are conserved across a broad range of vertebrate species [40]. It would be interesting to examine the evolutionary origin of the immune defense mechanism of TRIM21 that utilizes both innate and adaptive immune components. Another intriguing question is whether the Fc binding activity of TRIM21 is also involved in the pathogenesis of the autoimmune diseases, that are associated with anti-TRIM21 autoantibodies. Note that anti-TRIM21 autoantibodies target the RING and B-box domains of TRIM21, not the PRY-SPRY domain [132]. The multiple possible modes of interaction between TRIM21 and anti-TRIM21 antibodies would undoubtedly endow their immune complex with the unique property to form large insoluble aggregates, which may contribute to the pathogenesis.
6. Conclusions and perspectives
We here summarized several TRIM/TRIM-like proteins that are involved in innate immunity, in particular in RNA sensing and viral restriction pathways. From the survey of TRIM/TRIM-like proteins with PRY-SPRY, the domain most widely used by TRIM/TRIM-like proteins for substrate recognition, a common mechanism for substrate recognition emerges. That is, TRIM/TRIM-like proteins often utilize bivalency or multivalency to tightly control their substrate specificity and activity during immune response. Many targets of TRIM/TRIM-like proteins are the host immune signaling molecules that form oligomers or large assemblies in their activated state [77], [133]. TRIM/TRIM-like proteins also target viral molecules that form large assemblies (e.g. capsid) in their pathogenic state. The avidity-driven substrate recognition mechanism of TRIM/TRIM-like proteins would thus ensure more precise control of innate immune signaling and restriction functions. Furthermore, we also reviewed TRIM/TRIM-like proteins that utilize spatial and temporal control of their protein levels as an additional measure to control their activities. These observations thus highlight the complex layers of mechanisms that regulate “the regulators” of innate immunity.
Despite the increasing appreciation of the importance of TRIM/TRIM-like proteins in innate immunity, molecular mechanisms for many TRIM/TRIM-like proteins remain enigmatic. In particular, substrate identity, which is the key to understanding their functions, has been one of the most controversial topic for many TRIMs. While co-immunoprecipitation and in vitro or cellular ubiquitination assay have often been used for validating E3 ligase-substrate relationships, it is important to recognize caveats and limitations in each of these methods. In our own experience, pulling-down full-length E3 ligase to identify or validate a substrate is less ideal than pulling-down ubiquitination-deficient variant (e.g. RING-deletion TRIM). This is because once ubiquitinated, substrates often recruit many additional partners that would confound efforts to identify direct substrates of the TRIM/TRIM-like protein. For cellular ubiquitination assay, one often pulls down a target protein of interest under non-denaturing or mildly denaturing condition (e.g. RIPA) followed by anti-Ub blot to examine its ubiquitination state. However, proteins purified under such conditions are often not pure enough to allow confident assignment of anti-Ub signal to the target protein of interest. Thus, more stringent, fully-denaturing condition is necessary to disrupt large and stable assemblies that are expected for TRIM’s substrates. For in vitro ubiquitination assays, proper specificity control is crucial as the assays often require high protein concentrations to achieve robust ubiquitination signal. While one may choose any arbitrary protein for the control, the best control, in our opinion, is the same target protein in a different conformational or activity state that allows direct comparison of substrate specificity in cells vs. in vitro. Such close controls allow more confident assessment of whether the in vitro ubiquitination indeed recapitulates cellular events.
Besides substrate identity, many exciting questions remain to be addressed. For example, some TRIM/TRIM-like proteins have been reported to bind one another [111], [134], [135] or with other E3 ligases (e.g. TRIM25:LUBAC, and TRIM38:TRAF6), raising a question of how their interactions modify functions of individual E3 ligases. Additionally, the role of TRIM/TRIM-like proteins has so far been attributed predominantly to Ub/Ubl modification. However, recent studies suggest that Ub/Ubl-independent activities also exist [31], [123]. We anticipate many new discoveries on functions, mechanisms and regulations of TRIM/TRIM-like proteins as the field continues to move forward.
Declaration of competing interest
Authors declare no conflict of interests.
Acknowledgements
The authors acknowledge NIH R01s (AI154653 and AI111784 to SH) and Roche post-doctoral fellowship (to HW) for supporting this work.
References
- 1.Kumar H., Kawai T., Akira S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011;30(1):16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6(10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 3.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437(7062):1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 4.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., Shu H.B. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19(6):727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., Shu H.B. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Jin L., Hill K.K., Filak H., Mogan J., Knowles H., Zhang B., Perraud A.L., Cambier J.C., Lenz L.L. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J. Immunol. 2011;187(5):2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 2009;106(21):8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X., Xu H., Chen Z.J. Prion-Like polymerization in immunity and inflammation. Cold Spring Harb. Perspect. Biol. 2017;9(4) doi: 10.1101/cshperspect.a023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X., Wang Q., Chen W., Wang C. Dynamic regulation of innate immunity by ubiquitin and ubiquitin-like proteins. Cytokine Growth Factor Rev. 2013;24(6):559–570. doi: 10.1016/j.cytogfr.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Hu H., Sun S.C. Ubiquitin signaling in immune responses. Cell Res. 2016;26(4):457–483. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clague M.J., Heride C., Urbe S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25(7):417–426. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Carthagena L., Bergamaschi A., Luna J.M., David A., Uchil P.D., Margottin-Goguet F., Mothes W., Hazan U., Transy C., Pancino G., Nisole S. Human TRIM gene expression in response to interferons. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Versteeg G.A., Rajsbaum R., Sánchez-Aparicio M.T., Maestre A.M., Valdiviezo J., Shi M., Inn K.S., Fernandez-Sesma A., Jung J., García-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38(2):384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G., Ke D., Vu T., Ahn J., Shah V.B., Yang R., Aiken C., Charlton L.M., Gronenborn A.M., Zhang P. Rhesus TRIM5alpha disrupts the HIV-1 capsid at the inter-hexamer interfaces. PLoS Pathog. 2011;7(3) doi: 10.1371/journal.ppat.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McEwan W.A. Surveillance for intracellular antibody by cytosolic Fc receptor TRIM21. Antibodies. 2016;5(4) doi: 10.3390/antib5040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reymond A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20(9):2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sardiello M., Cairo S., Fontanella B., Ballabio A., Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 2008;8:225. doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Micale L., Chaignat E., Fusco C., Reymond A., Merla G. The tripartite motif: structure and function. Adv. Exp. Med Biol. 2012;770:11–25. [PubMed] [Google Scholar]
- 21.Zou W., Zhang D.E. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J. Biol. Chem. 2006;281(7):3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 22.Hu M.M., Liao C.Y., Yang Q., Xie X.Q., Shu H.B. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J. Exp. Med. 2017;214(4):973–989. doi: 10.1084/jem.20161015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koliopoulos M.G., Esposito D., Christodoulou E., Taylor I.A., Rittinger K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 2016;35(11):1204–1218. doi: 10.15252/embj.201593741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez J.G., Chiang J.J., Sparrer K.M.J., Alam S.L., Chi M., Roganowicz M.D., Sankaran B., Gack M.U., Pornillos O. Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway. Cell Rep. 2016;16(5):1315–1325. doi: 10.1016/j.celrep.2016.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner J.M. Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5alpha. Elife. 2016:5. doi: 10.7554/eLife.16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esposito D., Koliopoulos M.G., Rittinger K. Structural determinants of TRIM protein function. Biochem. Soc. Trans. 2017;45(1):183–191. doi: 10.1042/BST20160325. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Wu H., Wu W., Zhuo W., Liu W., Zhang Y., Cheng M., Chen Y.G., Gao N., Yu H., Wang L., Li W., Yang M. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res. 2014;24(6):762–765. doi: 10.1038/cr.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez J.G., Okreglicka K., Chandrasekaran V., Welker J.M., Sundquist W.I., Pornillos O. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc. Natl. Acad. Sci. USA. 2014;111(7):2494–2499. doi: 10.1073/pnas.1318962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koliopoulos M.G., Lethier M., van der Veen A.G., Haubrich K., Hennig J., Kowalinski E., Stevens R.V., Martin S.R., Reis e Sousa C., Cusack S., Rittinger K. Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition. Nat. Commun. 2018;9(1):1820. doi: 10.1038/s41467-018-04214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roganowicz M.D., Komurlu S., Mukherjee S., Plewka J., Alam S.L., Skorupka K.A., Wan Y., Dawidowski D., Cafiso D.S., Ganser-Pornillos B.K., Campbell E.M., Pornillos O. TRIM5alpha SPRY/coiled-coil interactions optimize avid retroviral capsid recognition. PLoS Pathog. 2017;13(10) doi: 10.1371/journal.ppat.1006686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cadena C., Ahmad S., Xavier A., Willemsen J., Park S., Park J.W., Oh S.W., Fujita T., Hou F., Binder M., Hur S. Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell. 2019;177(5):1187–1200. doi: 10.1016/j.cell.2019.03.017. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skorupka K.A., Roganowicz M.D., Christensen D.E., Wan Y., Pornillos O., Ganser-Pornillos B.K. Hierarchical assembly governs TRIM5alpha recognition of HIV-1 and retroviral capsids. Sci. Adv. 2019;5(11):eaaw3631. doi: 10.1126/sciadv.aaw3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Napolitano L.M., Meroni G. TRIM family: pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64(1):64–71. doi: 10.1002/iub.580. [DOI] [PubMed] [Google Scholar]
- 34.Ozato K., Shin D.M., Chang T.H., Morse H.C. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 2008;8(11):849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci. 2017;42(4):297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Meroni G. Genomics and evolution of the TRIM gene family. Adv. Exp. Med. Biol. 2012;770:1–9. doi: 10.1007/978-1-4614-5398-7_1. [DOI] [PubMed] [Google Scholar]
- 37.Rhodes D.A., de Bono B., Trowsdale J. Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence? Immunology. 2005;116(4):411–417. doi: 10.1111/j.1365-2567.2005.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Cruz A.A., Babon J.J., Norton R.S., Nicola N.A., Nicholson S.E. Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci. 2013;22(1):1–10. doi: 10.1002/pro.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.James L.C., Keeble A.H., Khan Z., Rhodes D.A., Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. USA. 2007;104(15):6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keeble A.H., Khan Z., Forster A., James L.C. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc. Natl. Acad. Sci. USA. 2008;105(16):6045–6050. doi: 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Y., Liang L., Jin Y., Yin Y. The TRIM14 PRYSPRY domain mediates protein interaction via its basic interface. FEBS Lett. 2019;593(10):1122–1129. doi: 10.1002/1873-3468.13386. [DOI] [PubMed] [Google Scholar]
- 42.Biris N., Yang Y., Taylor A.B., Tomashevski A., Guo M., Hart P.J., Diaz-Griffero F., Ivanov D.N. Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module. Proc. Natl. Acad. Sci. USA. 2012;109(33):13278–13283. doi: 10.1073/pnas.1203536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo J.S., Suh H.Y., Park S.Y., Oh B.H. Structural basis for protein recognition by B30.2/SPRY domains. Mol. Cell. 2006;24(6):967–976. doi: 10.1016/j.molcel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 44.Choudhury N.R., Heikel G., Trubitsyna M., Kubik P., Nowak J.S., Webb S., Granneman S., Spanos C., Rappsilber J., Castello A., Michlewski G. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol. 2017;15(1):105. doi: 10.1186/s12915-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 46.Schlee M., Hartmann G. The chase for the RIG-I ligand--recent advances. Mol. Ther. 2010;18(7):1254–1262. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J., Iskarpatyoti J.A., Barchet W., Ludwig J., Dermody T.S., Hartmann G., Reis e Sousa C. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5’-diphosphates. Nature. 2014;514(7522):372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linehan M.M., Dickey T.H., Molinari E.S., Fitzgerald M.E., Potapova O., Iwasaki A., Pyle A.M. A minimal RNA ligand for potent RIG-I activation in living mice. Sci. Adv. 2018;4(2) doi: 10.1126/sciadv.1701854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinicke L.A., Wong C.J., Lary J., Nallagatla S.R., Diegelman-Parente A., Zheng X., Cole J.L., Bevilacqua P.C. RNA dimerization promotes PKR dimerization and activation. J. Mol. Biol. 2009;390(2):319–338. doi: 10.1016/j.jmb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito T., Owen D.M., Jiang F., Marcotrigiano J., Gale M. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454(7203):523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uzri D., Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J. Virol. 2009;83(9):4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manivannan P., Siddiqui M.A., Malathi K. RNase L amplifies interferon signaling by inducing protein kinase R-mediated antiviral stress granules. J. Virol. 2020;94:13. doi: 10.1128/JVI.00205-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147(2):423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 54.Peisley A., Wu B., Xu H., Chen Z.J., Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509(7498):110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang X., Kinch L.N., Brautigam C.A., Chen X., Du F., Grishin N.V., Chen Z.J. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36(6):959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu B., Peisley A., Tetrault D., Li Z., Egelman E.H., Magor K.E., Walz T., Penczek P.A., Hur S. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol. Cell. 2014;55(4):511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu S., Chen J., Cai X., Wu J., Chen X., Wu Y.T., Sun L., Chen Z.J. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2 doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayman T.J., Hsu A.C., Kolesnik T.B., Dagley L.F., Willemsen J., Tate M.D., Baker P.J., Kershaw N.J., Kedzierski L., Webb A.I., Wark P.A., Kedzierska K., Masters S.L., Belz G.T., Binder M., Hansbro P.M., Nicola N.A., Nicholson S.E. RIPLET, and not TRIM25, is required for endogenous RIG-I-dependent antiviral responses. Immunol. Cell Biol. 2019;97(9):840–852. doi: 10.1111/imcb.12284. [DOI] [PubMed] [Google Scholar]
- 59.Oshiumi H., Miyashita M., Inoue N., Okabe M., Matsumoto M., Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8(6):496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 60.Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J.U. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 61.Meyerson N.R., Zhou L., Guo Y.R., Zhao C., Tao Y.J., Krug R.M., Sawyer S.L. Nuclear TRIM25 specifically targets influenza virus ribonucleoproteins to block the onset of RNA chain elongation. Cell Host Microbe. 2017;22(5):627–638. doi: 10.1016/j.chom.2017.10.003. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng X., Wang X., Tu F., Wang Q., Fan Z., Gao G. TRIM25 is required for the antiviral activity of zinc finger antiviral protein. J. Virol. 2017;91:9. doi: 10.1128/JVI.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li M.M.H., Lau Z., Cheung P., Aguilar E.G., Schneider W.M., Bozzacco L., Molina H., Buehler E., Takaoka A., Rice C.M., Felsenfeld D.P., MacDonald M.R. TRIM25 enhances the antiviral action of zinc-finger antiviral protein (ZAP) PLoS Pathog. 2017;13(1) doi: 10.1371/journal.ppat.1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inn K.S., Gack M.U., Tokunaga F., Shi M., Wong L.Y., Iwai K., Jung J.U. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell. 2011;41(3):354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z.J. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141(2):315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peisley A., Wu B., Yao H., Walz T., Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell. 2013;51(5):573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 67.Loo Y.M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., García-Sastre A., Katze M.G., Gale M. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zalinger Z.B., Elliott R., Rose K.M., Weiss S.R. MDA5 is critical to host defense during infection with murine coronavirus. J. Virol. 2015;89(24):12330–12340. doi: 10.1128/JVI.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. USA. 2017;114(21):E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., Yamaguchi O., Otsu K., Tsujimura T., Koh C.S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 72.Peisley A., Jo M.H., Lin C., Wu B., Orme-Johnson M., Walz T., Hohng S., Hur S. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc. Natl. Acad. Sci. USA. 2012;109(49):E3340–E3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152(1–2):276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 74.Kamanova J., Sun H., Lara-Tejero M., Galán J.E. The Salmonella Effector Protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PLoS Pathog. 2016;12(4) doi: 10.1371/journal.ppat.1005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lang X., Tang T., Jin T., Ding C., Zhou R., Jiang W. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J. Exp. Med. 2017;214(2):459–473. doi: 10.1084/jem.20160592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kato K., Ahmad S., Zhu Z., Young J.M., Mu X., Park S., Malik H.S., Hur S. Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. bioRxiv. 2020 doi: 10.1016/j.molcel.2020.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kagan J.C., Magupalli V.G., Wu H. SMOCs: supramolecular organizing centres that control innate immunity. Nat. Rev. Immunol. 2014;14(12):821–826. doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu M.M., Yang Q., Xie X.Q., Liao C.Y., Lin H., Liu T.T., Yin L., Shu H.B. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity. 2016;45(3):555–569. doi: 10.1016/j.immuni.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 79.Andreeva L., Hiller B., Kostrewa D., Lässig C., de Oliveira Mann C.C., Jan Drexler D., Maiser A., Gaidt M., Leonhardt H., Hornung V., Hopfner K.P. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature. 2017;549(7672):394–398. doi: 10.1038/nature23890. [DOI] [PubMed] [Google Scholar]
- 80.Polprasert C., Schulze I., Sekeres M.A., Makishima H., Przychodzen B., Hosono N., Singh J., Padgett R.A., Gu X., Phillips J.G., Clemente M., Parker Y., Lindner D., Dienes B., Jankowsky E., Saunthararajah Y., Du Y., Oakley K., Nguyen N., Mukherjee S., Pabst C., Godley L.A., Churpek J.E., Pollyea D.A., Krug U., Berdel W.E., Klein H.U., Dugas M., Shiraishi Y., Chiba K., Tanaka H., Miyano S., Yoshida K., Ogawa S., Müller-Tidow C., Maciejewski J.P. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27(5):658–670. doi: 10.1016/j.ccell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peters D., Radine C., Reese A., Budach W., Sohn D., Jänicke R.U. The DEAD-box RNA helicase DDX41 is a novel repressor of p21(WAF1/CIP1) mRNA translation. J. Biol. Chem. 2017;292(20):8331–8341. doi: 10.1074/jbc.M116.772327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011;12(10):959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parvatiyar K., Zhang Z., Teles R.M., Ouyang S., Jiang Y., Iyer S.S., Zaver S.A., Schenk M., Zeng S., Zhong W., Liu Z.J., Modlin R.L., Liu Y., Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 2012;13(12):1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quynh N.T., Hikima J., Kim Y., Fagutao F.F., Kim M.S., Aoki T., Jung T.S. The cytosolic sensor, DDX41, activates antiviral and inflammatory immunity in response to stimulation with double-stranded DNA adherent cells of the olive flounder, Paralichthys olivaceus. Fish. Shellfish Immunol. 2015;44(2):576–583. doi: 10.1016/j.fsi.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 85.Zhang Z., Bao M., Lu N., Weng L., Yuan B., Liu Y.J. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 2013;14(2):172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu B., Hur S. How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 2015;12:91–98. doi: 10.1016/j.coviro.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu W., Li J., Zheng W., Shang Y., Zhao Z., Wang S., Bi Y., Zhang S., Xu C., Duan Z., Zhang L., Wang Y.L., Jiang Z., Liu W., Sun L. Cyclophilin A-regulated ubiquitination is critical for RIG-I-mediated antiviral immune responses. Elife. 2017;6:6. doi: 10.7554/eLife.24425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xue B., Li H., Guo M., Wang J., Xu Y., Zou X., Deng R., Li G., Zhu H. TRIM21 promotes innate immune response to RNA viral infection through Lys27-linked polyubiquitination of MAVS. J. Virol. 2018;92(14) doi: 10.1128/JVI.00321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tan P., He L., Cui J., Qian C., Cao X., Lin M., Zhu Q., Li Y., Xing C., Yu X., Wang H.Y., Wang R.F. Assembly of the WHIP-TRIM14-PPP6C mitochondrial complex promotes RIG-I-mediated antiviral signaling. Mol. Cell. 2017;68(2):293–307. doi: 10.1016/j.molcel.2017.09.035. e5. [DOI] [PubMed] [Google Scholar]
- 90.Zhou Z., Jia X., Xue Q., Dou Z., Ma Y., Zhao Z., Jiang Z., He B., Jin Q., Wang J. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc. Natl. Acad. Sci. USA. 2014;111(2):E245–E254. doi: 10.1073/pnas.1316941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Castanier C., Zemirli N., Portier A., Garcin D., Bidère N., Vazquez A., Arnoult D. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 2012;10:44. doi: 10.1186/1741-7007-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang B., Wang J., Wang Y., Zhou H., Wu X., Tian Z., Sun B. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J. Immunol. 2013;190(7):3613–3619. doi: 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- 93.Liu B., Zhang M., Chu H., Zhang H., Wu H., Song G., Wang P., Zhao K., Hou J., Wang X., Zhang L., Gao C. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat. Immunol. 2017;18(2):214–224. doi: 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- 94.Zhao C., Jia M., Song H., Yu Z., Wang W., Li Q., Zhang L., Zhao W., Cao X. The E3 ubiquitin ligase TRIM40 attenuates antiviral immune responses by targeting MDA5 and RIG-I. Cell Rep. 2017;21(6):1613–1623. doi: 10.1016/j.celrep.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 95.Xie P. TRAF molecules in cell signaling and in human diseases. J. Mol. Signal. 2013;8(1):7. doi: 10.1186/1750-2187-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dainichi T., Matsumoto R., Mostafa A., Kabashima K. Immune control by TRAF6-mediated pathways of epithelial cells in the eime (epithelial immune microenvironment) Front. Immunol. 2019;10:1107. doi: 10.3389/fimmu.2019.01107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z.J. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 98.Zhao W., Wang L., Zhang M., Yuan C., Gao C. E3 ubiquitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated factor 6 in macrophages. J. Immunol. 2012;188(6):2567–2574. doi: 10.4049/jimmunol.1103255. [DOI] [PubMed] [Google Scholar]
- 99.Zhao W., Wang L., Zhang M., Wang P., Yuan C., Qi J., Meng H., Gao C. Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-beta production and antiviral response by targeting NAP1. J. Immunol. 2012;188(11):5311–5318. doi: 10.4049/jimmunol.1103506. [DOI] [PubMed] [Google Scholar]
- 100.Hu M.M., Xie X.Q., Yang Q., Liao C.Y., Ye W., Lin H., Shu H.B. TRIM38 negatively regulates TLR3/4-mediated innate immune and inflammatory responses by two sequential and distinct mechanisms. J. Immunol. 2015;195(9):4415–4425. doi: 10.4049/jimmunol.1500859. [DOI] [PubMed] [Google Scholar]
- 101.Yin Q., Lin S.C., Lamothe B., Lu M., Lo Y.C., Hura G., Zheng L., Rich R.L., Campos A.D., Myszka D.G., Lenardo M.J., Darnay B.G., Wu H. E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 2009;16(6):658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu L., Xu J., Xie X., Zhou Y., Tao P., Li H., Han X., Wang C., Liu J., Xu P., Neculai D., Xia Z. Oligomerization-primed coiled-coil domain interaction with Ubc13 confers processivity to TRAF6 ubiquitin ligase activity. Nat. Commun. 2017;8(1):814. doi: 10.1038/s41467-017-01290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hiscott J., Pitha P., Genin P., Nguyen H., Heylbroeck C., Mamane Y., Algarte M., Lin R. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 1999;19(1):1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- 104.Sharma S. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300(5622):1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 105.Wang P., Zhao W., Zhao K., Zhang L., Gao C. TRIM26 negatively regulates interferon-beta production and antiviral response through polyubiquitination and degradation of nuclear IRF3. PLoS Pathog. 2015;11(3) doi: 10.1371/journal.ppat.1004726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin R., Heylbroeck C., Pitha P.M., Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell Biol. 1998;18(5):2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bibeau-Poirier A., Gravel S.P., Clément J.F., Rolland S., Rodier G., Coulombe P., Hiscott J., Grandvaux N., Meloche S., Servant M.J. Involvement of the IkappaB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J. Immunol. 2006;177(8):5059–5067. doi: 10.4049/jimmunol.177.8.5059. [DOI] [PubMed] [Google Scholar]
- 108.Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K.P., Yamaoka S. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 2006;7(6):598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 109.Stremlau M., Owens C.M., Perron M.J., Kiessling M., Autissier P., Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 110.Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H., Diaz-Griffero F., Anderson D.J., Sundquist W.I., Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. USA. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li X., Gold B., O’hUigin C., Diaz-Griffero F., Song B., Si Z., Li Y., Yuan W., Stremlau M., Mische C., Javanbakht H., Scally M., Winkler C., Dean M., Sodroski J. Unique features of TRIM5alpha among closely related human TRIM family members. Virology. 2007;360(2):419–433. doi: 10.1016/j.virol.2006.10.035. [DOI] [PubMed] [Google Scholar]
- 112.Nakayama E.E., Shioda T. Role of Human TRIM5alpha in Intrinsic Immunity. Front. Microbiol. 2012;3:97. doi: 10.3389/fmicb.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stremlau M., Perron M., Welikala S., Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J. Virol. 2005;79(5):3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yap M.W., Nisole S., Stoye J.P. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 2005;15(1):73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 115.Ganser-Pornillos B.K., Yeager M., Sundquist W.I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008;18(2):203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sebastian S., Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li X., Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol. 2008;82(23):11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ganser-Pornillos B.K., Chandrasekaran V., Pornillos O., Sodroski J.G., Sundquist W.I., Yeager M. Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. USA. 2011;108(2):534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Diaz-Griffero F., Kar A., Perron M., Xiang S.H., Javanbakht H., Li X., Sodroski J. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J. Virol. 2007;81(19):10362–10378. doi: 10.1128/JVI.00703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Diaz-Griffero F., Qin X., Hayashi F., Kigawa T., Finzi A., Sarnak Z., Lienlaf M., Yokoyama S., Sodroski J. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 2009;83(20):10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sakuma R., Ohmine S., Ikeda Y. Determinants for the rhesus monkey TRIM5alpha-mediated block of the late phase of HIV-1 replication. J. Biol. Chem. 2010;285(6):3784–3793. doi: 10.1074/jbc.M109.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yudina Z., Roa A., Johnson R., Biris N., de Souza Aranha Vieira D.A., Tsiperson V., Reszka N., Taylor A.B., Hart P.J., Demeler B., Diaz-Griffero F., Ivanov D.N. RING dimerization links higher-order assembly of TRIM5alpha to synthesis of K63-linked polyubiquitin. Cell Rep. 2015;12(5):788–797. doi: 10.1016/j.celrep.2015.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fletcher A.J., Vaysburd M., Maslen S., Zeng J., Skehel J.M., Towers G.J., James L.C. Trivalent RING assembly on retroviral capsids activates TRIM5 ubiquitination and innate immune signaling. Cell Host Microbe. 2018;24(6):761–775. doi: 10.1016/j.chom.2018.10.007. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Campbell E.M., Weingart J., Sette P., Opp S., Sastri J., O’Connor S.K., Talley S., Diaz-Griffero F., Hirsch V., Bouamr F. TRIM5alpha-mediated ubiquitin chain conjugation is required for inhibition of HIV-1 reverse transcription and capsid destabilization. J. Virol. 2016;90(4):1849–1857. doi: 10.1128/JVI.01948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mandell M.A., Jain A., Arko-Mensah J., Chauhan S., Kimura T., Dinkins C., Silvestri G., Münch J., Kirchhoff F., Simonsen A., Wei Y., Levine B., Johansen T., Deretic V. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell. 2014;30(4):394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mandell M.A., Kimura T., Jain A., Johansen T., Deretic V. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy. 2014;10(12):2387–2388. doi: 10.4161/15548627.2014.984278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pertel T., Hausmann S., Morger D., Züger S., Guerra J., Lascano J., Reinhard C., Santoni F.A., Uchil P.D., Chatel L., Bisiaux A., Albert M.L., Strambio-De-Castillia C., Mothes W., Pizzato M., Grütter M.G., Luban J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472(7343):361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oke V., Wahren-Herlenius M. The immunobiology of Ro52 (TRIM21) in autoimmunity: a critical review. J. Autoimmun. 2012;39(1–2):77–82. doi: 10.1016/j.jaut.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 129.Mallery D.L., McEwan W.A., Bidgood S.R., Towers G.J., Johnson C.M., James L.C. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc. Natl. Acad. Sci. USA. 2010;107(46):19985–19990. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Foss S., Watkinson R., Sandlie I., James L.C., Andersen J.T. TRIM21: a cytosolic Fc receptor with broad antibody isotype specificity. Immunol. Rev. 2015;268(1):328–339. doi: 10.1111/imr.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fletcher A.J., Mallery D.L., Watkinson R.E., Dickson C.F., James L.C. Sequential ubiquitination and deubiquitination enzymes synchronize the dual sensor and effector functions of TRIM21. Proc. Natl. Acad. Sci. USA. 2015;112(32):10014–10019. doi: 10.1073/pnas.1507534112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ottosson L., Hennig J., Espinosa A., Brauner S., Wahren-Herlenius M., Sunnerhagen M. Structural, functional and immunologic characterization of folded subdomains in the Ro52 protein targeted in Sjogren’s syndrome. Mol. Immunol. 2006;43(6):588–598. doi: 10.1016/j.molimm.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 133.Cadena C., Hur S. Filament-like assemblies of intracellular nucleic acid sensors: commonalities and differences. Mol. Cell. 2019;76(2):243–254. doi: 10.1016/j.molcel.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yamauchi K., Wada K., Tanji K., Tanaka M., Kamitani T. Ubiquitination of E3 ubiquitin ligase TRIM5 alpha and its potential role. FEBS J. 2008;275(7):1540–1555. doi: 10.1111/j.1742-4658.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- 135.Herquel B., Ouararhni K., Khetchoumian K., Ignat M., Teletin M., Mark M., Bechade G., Van Dorsselaer A., Sanglier-Cianferani S., Hamiche A., Cammas F., Davidson I., Losson R. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA. 2011;108(20):8212–8217. doi: 10.1073/pnas.1101544108. [DOI] [PMC free article] [PubMed] [Google Scholar]