Abstract
Obesity is a major epidemic in our population and has emerged as a primary health concern. Consumption of a high fat, high sugar (HFHS) diet can specifically lead to gut dysbiosis, increased inflammation, and neuroinflammation. Interestingly, sex differences in the response to a HFHS diet are emerging. In this study, we investigated the effects of a HFHS diet compared to a low fat, low sugar (LFLS) diet in 8 week old male and female C57Bl/6 mice. The diet was administered for 14 weeks; body weights and food consumption were evaluated weekly. Male and female mice fed the HFHS diet gained significantly more weight than LFLS-fed mice. However, in agreement with previous studies, males gained significantly more weight on the HFHS diet compared to females fed the same diet. Importantly, we determined significant sex and diet-induced differences to gut microbiome composition using next generation Illumina sequencing. We also observed significantly less astrocyte densitometry and no significant change to microglial morphology in the hypothalamus of Female HFHS compared to Female LFLS. On the other hand, Male HFHS revealed no change to hypothalamic astrogliosis, but increased microgliosis compared to Male LFLS. In this study, we determined sex and diet-induced differences in both the gut and the brain, however, future studies will need to be performed in order to test the direct role of the gut microbiome to weight gain and neuroinflammation in male and female mice.
Keywords: diet-induced obesity, inflammation, gliosis, gut microbiome, sex differences
Introduction
Obesity is a major epidemic in our population and has emerged as a primary health concern. The majority of the adult population in the United States is considered obese or overweight. Less than one-third of the adult population is considered of normal weight [1, 2]. Obesity is a significant risk factor for several conditions, including cardiovascular disease, hypertension, diabetes, osteoarthritis, and several forms of cancer [3-6]. Furthermore, it has previously been shown that poor diet and/or obesity are linked to the development of dementia [2, 7]. One mechanism linking obesity with these chronic conditions is inflammation. Increased adiposity can cause an immune response; macrophages surround growing adipocytes and increase release of cytokines into the bloodstream [8]. Obesity often involves a chronic, low-level of inflammation [9].
In addition to adipoctyes, the gut microbiome has emerged as another source of obesity-induced inflammation and contributor to metabolic dysfunction. The gut microbiome includes all microorganisms that are found in the gut. There are commensalisms between these microorganisms and their host which aid in many physiological processes including nutrient metabolism and drug metabolism [10, 11]. Previous studies have shown that obesity causes gut dysbiosis by changing the abundance of different microorganisms found in the gut, especially the most abundant phyla: Bacteroidetes and Firmicutes [12]. These diet-induced changes to the gut microbiome ecology subsequently change the amount of lipopolysaccharide (LPS), an inflammatory agent, in the colon which can lead to increased inflammation in the intestines [13] as well as systemic inflammation. Overall, obesity can decrease gut microbiome diversity; a lower gut microbiome diversity has been linked to diseases from depression to diabetes to celiac disease [14-18].
Sex differences have been determined in mouse models of diet-induced obesity. In previous studies, when fed a high fat diet, both male and female mice gained significant weight, but the male mice gained more weight and fat mass compared to their female counterparts on the same high fat diet [19-21]. These sex differences in obesity also play major roles in the development of health problems. In a previous study, male mice fed a high fat diet developed hyperinsulinemia, hyperglycemia, and hypercholesterolemia, while female mice showed little to no development of the same health problems [22]. Female mice fed a high fat diet have also not shown chronic low-grade inflammation while it was present in male obese mice [23]. These sex differences in obesity make male mice more vulnerable to the impacts of obesity-induced diseases and problems. However, the full impact of obesity on male versus female mice and the cause(s) for these sex differences are not yet fully understood.
In this study, we investigated the effects of a high fat, high sucrose (HFHS) diet in male and female mice; we determined sex and diet-induced differences to weight gain, food consumption, gut microbiome composition, and neurogliosis. We hypothesized that females would be more resistant to diet-induced obesity, as shown previously. Due to their resistance to diet-induced obesity, we also hypothesized that female mice would reveal a significantly different microbiome composition compared to male mice, decreased systemic inflammation, and decreased neurogliosis. Previous research has determined a correlation between obesity and gut microbiome composition, particularly increased Firmicutes and decreased Bacteroidetes [24]. However, the direct impact of the whole gut microbiome and/or specific microbes on feeding and metabolism is not yet fully understood. To start to answer this question, Turnbaugh et al. transplanted microbiota samples from obese mice (ob/ob and diet-induced obese) or lean mice to germ-free mice. The germ-free mice all started at the same weight and age but gained significantly more weight following transplantation of obese microbiota compared to lean microbiota [24]. While more research must be done, these data suggest a connection between microbiome composition and energy balance/metabolism. Therefore, we hypothesized that female mice fed the same high-fat diet as male mice, but exhibit diet-induced obesity resistance, would additionally reveal altered gut microbiome composition that may either drive the differences in weight gain or be due to the differences in weight gain. Given previous results that show increased Firmicutes and decreased Bacteroidetes due to obesity, we hypothesized these phyla to be the most significantly affected by diet and between male and female mice. Furthermore, obesity is associated with low-grade systemic inflammation. One proposed source of this increased inflammation is white adipose tissue, however, this is also not yet fully understood [25]. We additionally hypothesized that if female mice exhibited diet-induced obesity resistance, and therefore, decreased adipose tissue, they would also reveal decreased systemic inflammation, and subsequently decreased neuroinflammation compared to diet-induced obese male mice. Therefore, we evaluated weight gain, food consumption, gut microbiome composition, systemic adipokines and cytokines, and neurogliosis in this study.
Materials/Methods
Animals and diets
Forty C57Bl/6 mice (The Jackson Laboratory, Bar Harbor, ME, USA), 20 males and 20 females, were housed 1-5 to a cage and provided with ad libitum access to food and water. While all mice were initially housed in a shared environment (3-5/cage), a few animals were separated during the study due to fighting (n=5). Mice were kept on a controlled 12-hour light/12-hour dark cycle. Mice were received at 6 weeks old and were given 2 weeks to acclimate to their environment. Following the acclimation period, both the male and female mice were randomly and equally divided into 4 treatment groups: Male LFLS, Male HFHS, Female LFLS, Female HFHS. The high fat, high sugar diet (HFHS) consisted of 45% kcal from fat and the low fat, low sugar (LFLS) diet consisted of 10% kcal from fat (D12451 and D12450K, respectively; Research Diets, New Brunswick, NJ, USA). The fat sources for the diets were primarily soybean oil and lard. However, the HFHS diet contained 8.9 times more lard than the LFLS diet. The HFHS diet contained 20% (by weight) of sucrose whereas the LFLS diet contained 0% sucrose. For the LFLS diet, the main source of carbohydrate was corn starch. Both diets contained the same vitamin and mineral mixes (see Table 1 for diet compositions). Animals were kept on the diet for 14 weeks. Each week, individual animal body weights and food consumption per cage were measured. Caloric consumption was determined by multiplying the food consumption for the cage for the LFLS diet by 3.82 kcals/gram and for the HFHS diet by 4.7 kcals/gram. To account for differences in the number of animals per cage, caloric consumption was further determined by dividing food consumption per cage by the number of animals in the cage. At the end of the 14-week diet exposure, fecal samples were collected using sterile technique. Animal protocols were approved by the Furman University Institutional Care and Use Committee and carried out following the guidelines of the National Institute of Health (Approval number: 13-18-02).
Table 1.
Dietary Components. Diets were produced by Research Diets Inc.
| Components | LFLS Diet (D12450K) | HFHS Diet (D12451) |
|---|---|---|
| Casein | 200 grams | 200 grams |
| L-Cystine | 3 grams | 3 grams |
| Corn Starch | 550 grams | 72.8 grams |
| Maltodextrin 10 | 150 grams | 100 grams |
| Sucrose | 0 grams | 172.8 grams |
| Cellulose | 50 grams | 50 grams |
| Soybean Oil | 25 grams | 25 grams |
| Lard | 20 grams | 177.5 grams |
| Mineral Mix S10026 | 10 grams | 10 grams |
| DiCalcium Phosphate | 13 grams | 13 grams |
| Calcium Carbonate | 5.5 grams | 5.5 grams |
| Potassium Citrate, 1 H2O | 16.5 grams | 16.5 grams |
| Vitamin Mix V100001 | 10 grams | 10 grams |
| Choline Bitartrate | 2 grams | 2 grams |
| FD&C Red Dye #40 | 0.025 grams | 0.05 grams |
| FD&C Blue Dye#1 | 0.025 grams | 0 grams |
| Energy | 3.85 kcal/gram | 4.73 kcal/gram |
Tissue collection and preparation
Mice were anesthetized deeply using isoflurane gas and then decapitated. Trunk blood was collected for plasma analyses. Brains were dissected; one half of the brain was drop-fixed in 4% paraformaldehyde for 48 hours and then moved to 30% sucrose in 0.1 M phosphate buffered saline (PBS) for immunohistochemistry. One half of the brain was microdissected to collect the hypothalamus for a cytokine array.
Brain sectioning and immunohistochemistry
Brains were sectioned at 40 μm (Microm HM 505e cryostat). Every 6th section through the dorsal hippocampus/lateral hypothalamus was evaluated for glial fibrillary acid protein (GFAP, 1:1,000, Abcam, Cambridge, MA, USA) and ionized calcium binding adaptor protein (Iba-1, 1:1,000, FUJIFILM Wako, Osaka, Japan) immunoreactivity. Following primary antibody incubation, tissue was washed in PBST and then incubated in biotinylated goat anti-rabbit secondary antibody (1:500, Jackson ImmunoResearch, West Grove, PA, USA) for 2 hours. The tissue was washed in PBST and then incubated in ABC reagent (Vector Laboratories, Burlingame, CA, USA) for 1.5 hours. The tissue was developed with a Vector VIP kit (Vector Laboratories, Burlingame, CA, USA) and analyzed using a light microscope.
Densitometry for GFAP Analysis
The hippocampus and hypothalamus were imaged at 10x. Images were then processed using ImageJ (National Institute of Health, https://imagej.nih.gov/ij/) to semi-quantify immunoreactivity densitometry (as measured by % of photomicrograph with GFAP immunoreactivity by applying a threshold and then determining “% Area”). Analysis was performed blindly in each brain region for 3-4 sections per animal. All 40 animals (10/group) were analyzed.
FracLac for Microglia Analysis
The hippocampus and hypothalamus were imaged at 100X. Images were then processed using ImageJ (National Institute of Health, https://imagej.nih.gov/ij/). Two different plug-ins were used to analyze microglia: the Analyze Skeleton Plugin (http://imagej.net/AnalyzeSkeleton31) and the FracLac for Image J Plugin http://rsb.info.nih.gov/ij/plugins/frac-lac.html), as performed previously [26, 27]. The Analyze Skeleton Plugin was used in order to determine the number of summed microglia endpoints/soma and summed microglia branch length/soma (in the 2D field of view). Six photos from each animal were randomly taken in the dorsal hippocampus and mediobasal hypothalamus and every microglia from each photo was analyzed. A modified version of an ImageJ protocol was used [27]. FFT Bandpass Filter was applied to the image, the brightness was adjusted, and the unsharp mask and despeckle tools were used to clarify the image before thresholding. Once the image was thresholded, it was despeckled, again made binary, and outliers were removed immediately prior to all background noise being erased. Only the microglia cells remained in the image. The skeletonize and analyze skeleton plugins were then applied, which tag skeletal features relevant to microglia morphology: slab voxels (process length) and endpoints. Branches less than 1.75 mm in length were removed as described previously [28]. The FracLac Plugin was used in order to determine endpoint + branch length/soma, cell complexity (fractal dimension), and cell shape (span ratio). In each binary picture, 1-6 microglia cells were analyzed using the ImageJ FracLac plugin so that an average of 20 cells were analyzed per animal. A modified version of an ImageJ protocol was used [27]. Images were edited as indicated above so that only the microglia cells remained. Each cell was isolated and the outline feature was used. Microglia analysis was performed on 3-6 animals per group.
Plasma Adipokine and Cytokine Array
Plasma was evaluated using the RayBiotech Quantibody® Mouse Cytokine Array 8 (QAM-CYT-8, Norcross, GA, USA). The Quantibody® array is a multiplexed sandwich enzyme-linked immunosorbent assay (ELISA) on a solid (glass slide) support. Briefly, plasma was diluted 2-fold before it was added to the array for incubation; wells were washed, incubated with biotinylated antibody cocktail, washed, incubated with streptavidin, washed, and then imaged using fluorescence detection. Microarray analysis software (Quantibody® Q-Analyzer) was used to analyze the results. Each cytokine was analyzed in quadruplicate and compared to a standard curve. Data that were above or below the detection limit were excluded.
Hypothalamus Cytokine Array
Hypothalamus homogenates (0.50 mg/ml) were evaluated with the RayBiotech Mouse Neuro Discovery Array C1 (Norcross, GA, USA). This sandwich-based array provides a semi-quantitative measurement of cytokine expression. Briefly, Mouse Neuro Discovery C1 membranes were blocked, incubated with the hypothalamus homogenates, and then incubated with biotinylated detection antibody cocktail, followed by HRP-conjugated streptavidin, and followed by detection buffer. Next, the membranes were imaged and chemiluminescence was analyzed via densitometry. Data are reported as relative abundance which was determined as percent of average Male LFLS densitometry.
Gut Microbiome Analysis
Genomic DNA was isolated from fecal samples using the PowerSoil® DNA Isolation Kit (Qiagen) following the manufacturer’s instructions. As an alternative to the recommended 250mg of soil, approximately 200mg of fecal sample was added to the PowerBeads tube to undergo cell lysis. The purified DNA was eluted from the spin filter using 50uL of solution C6 and stored at −20°C until PCR amplification. The 16S universal Eubacterial primers 515F GTGCCAGCMGCCGCGGTAA and 806R GGACTACVSGGGTATCTAAT were utilized to evaluate the microbial ecology of each sample on the HiSeq 2500 with methods via the bTEFAP® DNA analysis service. Each sample underwent a single-step 30 cycle PCR using HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA, USA) and were used under the following conditions: 94°C for 3 minutes, followed by 28 cycles of 94°C for 30 seconds; 53°C for 40 seconds and 72°C for 1 minute; after which a final elongation step at 72°C for 5 minutes was performed. Following PCR, all amplicon products from different samples were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, MA, USA). Samples were sequenced utilizing the Illumina HiSeq chemistry following the manufacturer’s protocols.
The Q25 sequence data derived from the sequencing process was processed using a proprietary analysis pipeline (www.mrdnalab.com, MR DNA, Shallowater, TX, USA). Operational taxonomic units were defined after removal of singleton sequences, clustering at 3% divergence, 97% similarity; [29-32]. OTUs were then taxonomically classified using BLASTn against a curated NCBI database. Statistical analysis was performed using a variety of computer packages including XLstat, NCSS 2007, “R”, and NCSS 2010. Beta diversity and genus abundance analyses were conducted using a Kruskal-Wallis test followed by a two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli in order to control for multiple comparisons by controlling the false discovery rate (GraphPad Software, La Jolla, California, USA, www.graphpad.com). Significance reported for any analysis is defined as p<0.05.
Statistical Analysis
All of the following statistical tests were performed using GraphPad Prism (GraphPad Software, La Jolla, California, USA, www.graphpad.com). Plasma, brain morphology, and brain homogenate analyses were performed using a two-way (sex x diet) analysis of variance (ANOVA) followed by Tukey’s post-hoc test with significance of p < 0.05. Food consumption was analyzed using a three-way ANOVA (sex x diet x time). Due to a few missing values (researcher error) for body weights over the 14-week study, a mixed-effects analysis was performed for body weight (sex x diet x time). Spearman correlations were analyzed for final body weight and gut microbiome diversity indices as well as microglial FracLac results and gut microbiome diversity indices.
Results
Body Weights and Caloric Consumption
There was a significant effect of time (F(3.141, 108.7)=57.33; p<0.0001) and a significant effect of diet and sex (F(3, 35)=112.2; p<0.0001) on body weight (Figure 1A). There was also a significant interaction effect between time, diet, and sex on body weight (F(39,450)=8.209; p<0.0001) (Figure 1A). The male HFHS group gained the most weight at the end of the 14-week study. In fact, males doubled in body weight during this 14-week study (week 1: 22.09 ± 0.941 grams vs. week 14: 45.06 ± 4.02 grams).
Figure 1:
Body weights and calories consumed during the 14-week dietary study (n = 10/group). (A) HFHS-fed mice gained significantly more weight than LFLS-fed mice. Male HFHS gained significantly more weight than Female HFHS. (B) Female LFLS consumed the least amount of calories throughout the study. Female HFHS escalated caloric intake during the 14-week study and consumed similar calories to male mice by the end of the study.
There was a significant effect of time (F(13,490)=44.39; p<0.0001) and a significant effect of treatment (F(3,490) = 197.6; p<0.0001) on food consumption (Figure 1B). There was also a significant interaction effect between time and treatment (F(39,490)=5.574; p<0.0001) on caloric consumption. Interestingly, female HFHS consumed significantly more calories than female LFLS beginning at week 8 and continuing to week 14 (Fig. 1B). Female HFHS consumed a similar amount of calories as male HFHS and male LFLS during the second half of the study, despite gaining much less weight than the male mice.
Plasma Adipokine and Cytokines
There was a significant effect of diet on plasma adiponectin levels (F(1,16) = 5.254, p =0.0358; Figure 2A) and a significant interaction effect between sex and diet (F(1,16) = 8.322, p = 0.0108). Female mice fed the high fat diet had a significantly lower plasma adiponectin level compared to female mice fed the low fat diet (p = 0.0102). There was no difference in plasma adiponectin between male mice fed the low fat and high fat diet (p = 0.9744). Female HFHS revealed lower plasma adiponectin compared to Male HFHS (approaching significance; p = 0.0615).
Figure 2:
Plasma analysis following the 14-week dietary study (n = 5/group). Significant sex, diet, and/or sex x diet interaction effects were determined for a number of adipokines. However, no significant differences in plasma cytokines were determined. # approaching significance, * p<0.05, **p<0.01, ***p<0.001
There was a significant effect of diet on plasma leptin (F(1,16) = 22.38, p = 0.0002; Figure 2B) and plasma leptin receptor (F(1,16) = 16.60, p = 0.0009; Figure 2C). Additionally, there was a significant effect of sex (F(1,16) = 49.51, p<0.0001) and a significant interaction effect (F(1,16) = 4.653, p = 0.0465) for plasma leptin receptor. While both males and females revealed higher plasma leptin following consumption of the high fat diet, only males revealed a statistically significant increase. Female LFLS revealed significantly higher plasma leptin receptor levels compared to Male LFLS (p <0.0001), Male HFHS (p <0.0001), and Female HFHS (p = 0.0022).
There was a significant effect of sex on plasma C-reactive protein (CRP) levels (F(1,16) = 22.78, p = 0.0002; Figure 2D). Male LFLS revealed statistically significant higher plasma CRP levels compared to Female LFLS (p = 0.0241) and Female HFHS (p = 0.0026); Male HFHS revealed statistically significant higher plasma CRP levels compared to Female HFHS (p = 0.0137).
A statistically significant difference due to sex or diet was not determined for plasma TNFα (Figure 2E), IL-1α (Figure 2G), IL-1β (Figure 2H), IL-6 (Figure 2I), or IL-10 (Figure 2K). There was a significant interaction effect for plasma TNFα Receptor 1 (F(1,16) = 13.74, p = 0.0019; Figure 2F). Male LFLS revealed significantly lower TNFα Receptor 1 levels compared to Male HFHS (p = 0.0137) and Female LFLS (p = 0.0153).
There was a significant effect of sex on plasma IL-17 levels (F(1,16) = 9.777, p = 0.0065; Figure 2L). Male HFHS revealed higher levels of IL-17 compared to Female HFHS (approaching significance: p = 0.0549). There was also a significant effect of sex on plasma granulocyte-macrophage colony-stimulating factor (GM-CSF; F(1,16) = 8.258, p = 0.0110; Figure 2M). Male LFLS revealed higher levels of GM-CSF compared to Female LFLS (approaching significance: p = 0.0610).
Sex and Diet-Induced Differences in the Gut Microbiome
After stringent quality sequence curation, a total of 3,124,383 sequences were parsed and 2,702,795 were then clustered. 2,702,728 sequences identified within the Bacteria domain were utilized for final microbiota analyses. The average reads per sample was 84,460. For alpha and beta diversity analysis, samples were rarefied to 20,000 sequences and bootstrapped at 15,000 sequences. Data were evaluated in a multivariate manner to determine the changes between groups.
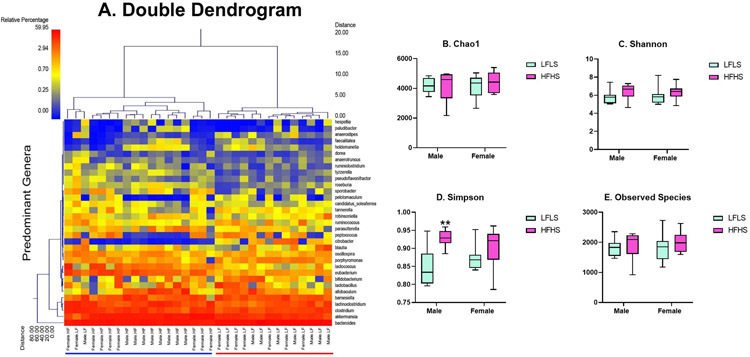
The double dendrogram (Figure 3A) reveals the greatest effect on the gut microbiome was due to diet. Additionally, four α-diversity indices were evaluated. The Chao 1 diversity index (Figure 3B), Shannon diversity index (Figure 3C), and observed species (Figure 3E) did not reveal significant differences in α diversity between the four groups (Chao 1: Kruskal-Wallis = 1.205, p = 0.7519; Shannon: Kruskal-Wallis = 3.849, p = 0.2782; Observed Species: Kruskal-Wallis: 2.102, p = 0.5515). However, the Simpson index (Figure 3D) revealed a significant difference (Kruskal-Wallis = 10.85, p = 0.0126). Male HFHS had increased species diversity compared to Male LFLS (p = 0.0170). Both the Shannon index and Simpson index estimate the species richness and evenness in a population, however the Shannon index places a greater weight on species richness and the Simpson index places a greater weight on species evenness [33]. Interestingly, the final body weights correlated with the Simpson index (r = 0.3812, p = 0.0313). However, the final body weights did not correlate with any of the other diversity indices (Chao: r = 0.1404, p = 0.4434; Shannon:r = 0.2603, p = 0.1503; Observed Species: r = 0.2210, p = 0.2241).
Figure 3:
Gut microbiome diversity (n = 8/group). To provide a visual overview combined with analysis, a dual hierarchal dendrogram is used to display the data for the predominant genera with clustering related to the different groups (A). Samples with more similar microbial populations are mathematically clustered closer together. The genera (consortium) are used for clustering. The samples with more similar consortium of genera cluster closer together with the length of connecting lines (top of heatmap) related to the similarity, shorter lines between two samples indicate closely-matched microbial consortium. The heatmap represents the relative percentages of each genus. The predominant genera are represented along the right Y-axis. The legend for the heatmap is provided in the upper left corner. Based on the clustering, there appears to be a significant difference between the mice exposed to a high fat diet and mice exposed to a low fat diet. Four α-diversity indices were evaluated. The Chao 1 diversity index (B), Shannon diversity index (C), and observed species (E) did not reveal significant differences in α diversity between the four groups. However, the Simpson index (D) revealed a significant difference.
To further evaluate sex and diet-induced differences in the gut microbiome, the relative abundance of specific genera were compared. Table 2 displays the mean relative abundance for each treatment group with significant differences indicated between Male LFLS and Male HFHS, Female LFLS and Female HFHS, Male LFLS and Female LFLS, and Male HFHS and Female HFHS. As shown in previous studies, high fat diet treatment reduced Bacteroidetes and increased Firmicutes abundance. However, baseline abundance and diet-induced changes to the Firmicutes bacteria were different between males and females for certain genera.
Table 2.
Mean relative abundance of genera from fecal analysis. Results are shown for each sex and dietary group. Statistical analyses were performed usinga Kruskal-Wallis test followed by a two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli in order to control for multiple comparisons by controlling the false discovery rate.
| Mean Relative Abundances | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genus | Phylum | Male LFLS |
Male HFHS |
Female LFLS |
Female HFHS |
Male LFLS vs. Male HFHS |
Female LFLS vs. Female HFHS |
Male LFLS vs. Female LFLS |
Male HFHS vs. Female HFHS |
| Bacteroides | Bacteroidetes | 52.13909 | 29.2137 | 48.40536 | 26.50889 | p = 0.0006 | p = 0.0018 | ||
| Akkermansia | Verrucomicrobia | 8.815587 | 22.50848 | 9.703972 | 27.8591 | p = 0.0008 | p = 0.0056 | ||
| Allobaculum | Firmicutes | 2.76258 | 7.226025 | 2.052159 | 0.442098 | p = 0.0428 | p = 0.0252 | p < 0.0001 | |
| Eubacterium | Firmicutes | 1.308409 | 3.76287 | 1.701458 | 5.619516 | p = 0.0113 | p = 0.0026 | ||
| Lactococcus | Firmicutes | 0.609975 | 2.15802 | 0.587866 | 5.326685 | p = 0.0040 | p = 0.0040 | ||
| Porphyromonas | Bacteroidetes | 1.75217 | 1.051254 | 1.645057 | 1.13796 | p = 0.0219 | |||
| Blautia | Firmicutes | 2.3117 | 0.669521 | 0.405418 | 0.389399 | p = 0.0011 | |||
| Peptococcus | Firmicutes | 0.350862 | 0.038039 | 1.640607 | 1.505609 | p = 0.0056 | p = 0.0004 | ||
| Ruminococcus | Firmicutes | 0.530486 | 0.401542 | 1.382937 | 0.584759 | p = 0.0031 | |||
| Citrobacter | Proteobacteria | 1.832578 | 0.01556 | 0.620361 | 0.020921 | p < 0.0001 | p = 0.0402 | ||
| Sporobacter | Firmicutes | 0.189 | 0.344 | 0.282 | 0.97 | p = 0.0047 | p = 0.0486 | ||
| Holdemanella | Firmicutes | 0.245 | 0.429 | 0.18 | 0.045 | p = 0.0428 | p = 0.0219 | p < 0.0001 | |
| Pelotomaculum | Firmicutes | 0.09 | 0.007 | 0.397 | 0.379 | p = 0.0113 | p = 0.0003 | ||
| Faecalitalea | Firmicutes | 0.083 | 0.265 | 0.132 | 0.028 | p = 0.0428 | p = 0.0056 | p = 0.0002 | |
| Paludibacter | Bacteroidetes | 0.12 | 0.197 | 0.013 | 0.009 | p = 0.0026 | |||
| Hespellia | Firmicutes | 0.187 | 0.136 | 0.008 | 0.008 | p = 0.0034 | p = 0.0486 | ||
| Pseudobutyrvibrio | Firmicutes | 0.044 | 0.049 | 0.016 | 0.104 | p = 0.0002 | |||
| Anaeroplasma | Tenericutes | 0.156 | 0.004 | 0.003 | 0.001 | p = 0.0082 | p=0.0021 | ||
| Sporanaerobacter | Firmicutes | 0.024 | 0.011 | 0.067 | 0.015 | p = 0.0077 | |||
| Butyrivibrio | Firmicutes | 0.029 | 0.04 | 0.01 | 0.03 | p = 0.0113 | p = 0.0252 | ||
| Acetivibrio | Firmicutes | 0.034 | 0.031 | 0.007 | 0.004 | p = 0.0014 | |||
| Enterococcus | Firmicutes | 0.002 | 0.008 | 0.022 | 0.027 | p = 0.0023 | p = 0.0197 | ||
| Moryella | Firmicutes | 0.007 | 0.021 | 0.007 | 0.022 | p = 0.0227 | p = 0.0127 | ||
| Adlercreutzia | Actinobacteria | 0.005 | 0.025 | 0.007 | 0.01 | p = 0.0001 | p = 0.0353 | ||
Astrocyte Density
There were no significant differences observed in hippocampal GFAP densitometry between the four groups (Figure 4A). However, there was a significant sex x diet interaction effect for hypothalamic GFAP densitometry (F(1,36) = 4.365, p = 0.0438; Figure 4B) and a sex difference approaching significance (F(1,36) = 3.472, p = 0.0706). Female HFHS revealed significantly less GFAP densitometry compared to Female LFLS in the hypothalamus (p = 0.0395). We did not determine a significant difference in the number of GFAP-positive astrocytes in the mediobasal hypothalamus (Male LFLS: 93.37 ± 28.1 cells; Male HFHS: 90.17 ± 29 cells; Female LFLS: 92.7 ± 13.31 cells; Female HFHS: 90.63 ± 28.7 cells; effect of sex (F(1,36) = 0.00015); p = 0.99, effect of diet (F(1,36) = 0.1055); p = 0.7472). Therefore, our data indicate diet-induced changes to astrocyte densitometry, but not astrocyte number for female mice.
Figure 4:
Hippocampal and hypothalamic GFAP densitometry (n = 10/group). (A) No significant differences in hippocampal GFAP densitometry were determined. (B) Female HFHS revealed a significant decrease in GFAP densitometry in the hypothalamus compared to Female LFLS. * p<0.05. Representative photomicrographs are shown for each group in the hippocampus and hypothalamus. Scale bar = 200 um.
Microglia Analysis
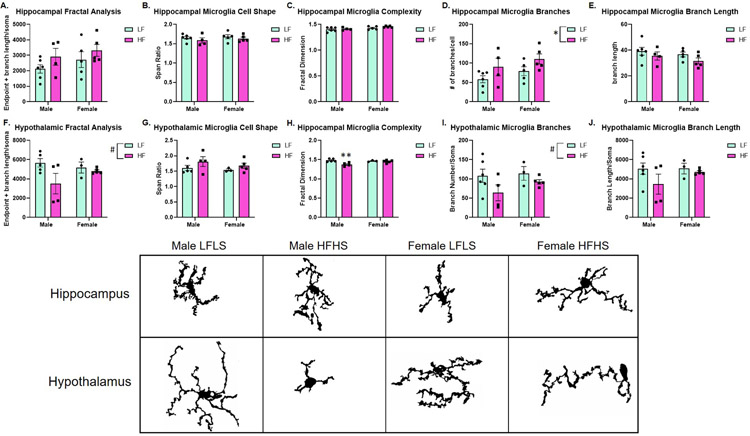
In order to evaluate microgliosis, FracLac and Skeleton analyses were performed. Microglia are very dynamic glial cells that rapidly change states between resting/ramified/surveilling, reactive, activated, and amoeboid. However, this is not necessarily a step-wise process. Instead, throughout the brain, microglia can alter their state along a spectrum between resting and amoeboid in response to the local environment [34]. Therefore, we aimed to determine features of microglia in both the hippocampus and hypothalamus such as: cell shape, cell complexity, branch length, and branch number.
In the hippocampus, a significant effect of sex on microglial branch number was determined (F(1,16) = 5.349, p = 0.0344; Figure 5D). While not statistically significant, there was an overall increase in branch length for HFHS groups as well.
Figure 5:
Hippocampal and hypothalamic microglial analysis (n=4-6/group). (A-E) Microglial analysis in the hippocampus. A significant diet effect was determined for hippocampal branch number/soma (D). (F-J) Microglial analysis in the hypothalamus. A diet effect was approaching significance for fractal analysis (F) and branch number/soma (I). Male HFHS reveal decreased microglial complexity, as measured by fractal dimension, compared to Male LFLS (H). Microglia from each group and both brain regions are shown. Microglia were imaged at 100x then converted to binary images prior to skeleton and fractal analyses.
In the hypothalamus, a sex difference in fractal analysis was approaching significance (F(1,13) = 4.277, p = 0.0591; Figure 5F) and a sex difference in branch number was approaching significance (F(1,14) = 3.930, p = 0.0674; Figure 5I). There was a significant effect of sex on cell complexity (fractal dimension; F(1,13) = 9.192, p = 0.0096; Figure 5H) and a significant sex x diet interaction (F(1,13) = 6.038, p = 0.0288). Specifically, there was a significant decrease in cell complexity for Male HFHS compared to Male LFLS (p = 0.0065). Furthermore, we determined a correlation between multiple gut microbiome diversity indices and hypothalamic Frac Lac analysis (Chao: r = 0.5780, p = 0.0333; Shannon: r = 0.6703, p = 0.0107; Observed Species: r = 0.6747, p = 0.0100). We did not determine a correlation between the Simpson index and hypothalamic Frac Lac (r = 0.4066, p = 0.1505).
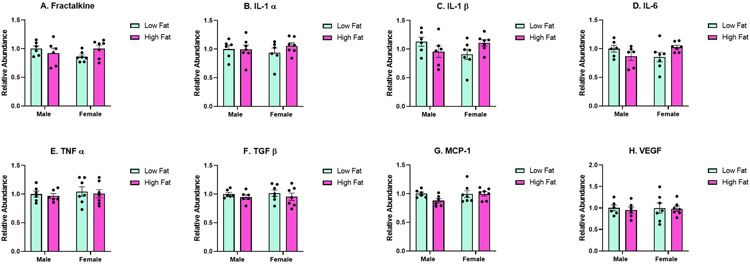
Hypothalamic Cytokine Expression
Due to the observed sex and diet-induced effects on gliosis in the hypothalamus, cytokine expression was semi-quantified using a sandwich-based membrane array in the hypothalamus. We determined an interaction effect that was approaching significance for fractalkine (Figure 6A; F(1,22) = 3.473, p = 0.0758), a significant interaction effect for IL-1β (Figure 6C: (F(1,22) = 5.032, p = 0.0353), and a significant interaction effect for IL-6 (Figure 6D; F(1,22) = 5.554, p = 0.0278). For fractalkine, IL-1β, and IL-6, the trend was for these cytokines to be decreased for Male HFHS compared to Male LFLS and the reverse for females: increased for Female HFHS compared to Female LFLS. We did not determine any significant differences due to sex, dietary treatment, or an interaction effect (sex x diet) for IL-1α (Figure 6B), TNFα (Figure 6E), TGFβ (Figure 6F), MCP-1 (Figure 6G), or VEGF (Figure 6H).
Figure 6:
Hypothalamic cytokine analysis (n = 6-7/group). A significant interaction effect was approaching significance for fractalkine (A). There was a significant interaction effect for IL-1β (C) and IL-6 (D). No other significant differences were determined.
Discussion
We determined significant effects of diet and/or sex to food consumption, weight gain, plasma adipokines, gut microbiome genera, and neurogliosis. We hypothesized that sex differences in weight gain and adiposity would lead to sex differences in the neural inflammatory response. Previous studies have shown that pro-inflammatory cytokines are elevated in specific brain regions of obese mice. Given the greater resistance to diet-induced obesity by female C57Bl/6 mice as observed in our dietary study and others [19-21], we expected decreased pro-inflammatory cytokines and decreased gliosis in female C57Bl/6 mice fed the HFHS diet compared to Male HFHS. Furthermore, we wanted to determine whether sex differences in the gut microbiome were present, providing a potential role in the sex differences in weight gain and neuroinflammation. However, future studies will need to be performed in order to test the direct role of the gut microbiome to weight gain and neuroinflammation in male and female mice fed a HFHS diet.
At the start of our experiment, no significant differences in body weights were determined, both between and within sex (p>0.05). For both sexes, HFHS dietary treatment resulted in significant weight gain compared to LFLS. We were also intrigued by the sex and diet-induced differences in calorie consumption. A challenge for mouse studies is determining individual calorie consumption while still maintaining group-housed conditions which are necessary for mouse health, including thermogenesis which contributes to weight gain and food consumption. We started our study with mice housed 3-5 per cage. However, a few mice had to be isolated (n=5) due to fighting. Food consumption was measured weekly and converted to calories consumed based on calories/gram of the specific diet. We display average calories consumed per group with “individual” values as determined by cage values divided by the number of mice per cage. We do not claim this to be an accurate measure of individual mouse calorie consumption, instead an average value to represent group calorie consumption, a limitation of our study. Previous studies have shown that housing conditions impact stress, thermogenesis, social hierarchy as well as weight gain and food consumption [35-38]. While 5 out of the 40 mice (n = 2 Male LFLS; n= 3 Female HFHS) were single-housed in the middle of our study, we did not determine any significant differences in weight gain or food consumption between these individual mice and their group-housed counterparts. As shown in Figure 1B, male LFLS and male HFHS consumed similar calories throughout the study, despite significant differences in weight gain. On the other hand, female HFHS escalated food intake over the 14 week study, revealing significant differences in calorie consumption between LFLS-fed and HFHS-fed female mice. In fact, female HFHS consumed similar calories to males despite less weight gain. Potentially, female mice do not convert consumed lipids to adiposity as males do. However, future studies will have to test this hypothesis.
We determined sex differences and diet-induced differences to specific genera of bacteria in the gut microbiome. Previous studies have shown that obesity and high fat diet consumption lead to a decrease in the Bacteroidetes phyla and an increase in the Firmicutes phyla [39, 40]. Overall, we observed the same effect in our study. For example, the HFHS fed to both male and female mice led to a significant decrease in abundance of Bacteroides (Phylum: Bacteroidetes). The HFHS diet administration led to significantly increased abundance of Eubacterium, Lactococcus, and Moryella (all Phylum: Firmicutes). However, there were interesting sex differences for other genera of bacteria. For example, Male HFHS revealed a significant increase in the abundance of Allobaculum (Phylum: Firmicutes) compared to Male LFLS whereas Female HFHS revealed a significant decrease in the same bacteria compared to males and Female LFLS. Additional effects are displayed in Table 2. It is unlikely that these sex differences in the gut microbiome are simply due to differences in adiposity given the observed sex differences in LFLS-fed mice and the fact that Female HFHS consumed similar amounts of food compared to Male HFHS. However, we did determine a correlation between final body weight and the Simpson index. The Simpson index emphasizes species evenness in the population: the balance of the taxa frequencies in a community. These data suggest that the increased final body weights were related to increased frequency of certain bacteria in the gut microbiome. We would expect direct effects of specific lipids and sugar to feed specific populations of bacteria. The adiposity itself is also revealing a contribution to this correlation. As mentioned above, previous studies have determined that obesity can lead to a lower gut microbiome diversity which is linked to various diseases. Here, we report that Male HFHS were obese, but exhibit significantly increased microbiome diversity as measured by the Simpson index. Taking into account that this diversity index heavily weighs frequency of taxa as opposed to number of different taxa, we conclude that certain microorganisms are increased for Male HFHS, likely “fed” by the fat and sugar provided by the diet. Furthermore, we did not determine a correlation between the final body weight and the Shannon index. The Shannon index emphasizes species richness in the population: a count of the number of different taxa observed in the community without regard to their frequencies [41]. These data prompt future studies to investigate differences in lipid and carbohydrate absorption/metabolism between male and female C57Bl/6 mice and the role of their gut microbiome.
Next, we investigated plasma adipokine and cytokine levels in order to better understand systemic responses to the HFHS diet. As expected, plasma leptin levels were higher for HFHS-fed mice compared to LFLS-fed mice. Furthermore, Female HFHS revealed a lower plasma leptin level compared to Male HFHS which was also expected given the significant differences in adiposity. Interestingly, we also observed significant differences in plasma leptin receptor, a soluble receptor that modulates the bioavailability of leptin [42]. Female HFHS revealed significantly higher plasma leptin receptor compared to Male HFHS. Previous studies have shown that the soluble leptin receptor affects leptin’s bioavailability and these changes are typically found with metabolic disorders [42]. In addition to sex and diet effects on leptin and leptin receptor, we observed significantly decreased adiponectin in Female HFHS compared to Female LFLS. Adiponectin influences numerous metabolic processes, including increasing insulin sensitivity and increasing beta oxidation [43]. The lower adiponectin levels, only for Female HFHS, could decrease fatty acid breakdown and favor fat accumulation. Increasing adiposity appears to be a slower process for female mice according to our data for body weight gain. These data suggest a compensatory mechanism may be necessary in female C57Bl/6 mice in order to accumulate body fat if fat absorption is altered.
We expected significant differences in plasma cytokine levels between HFHS and LFLS and we further hypothesized sex differences given differences in body weight. However, we did not determine statistically significant differences in plasma levels of TNFα, IL-1α, IL-1β, IL-6, or IL-10. We report data from five animals per group, a limitation of our study. There was a significant interaction effect (sex x diet) for plasma TNFα Receptor 1. Male LFLS revealed significantly lower TNFα Receptor 1 levels compared to Male HFHS and Female LFLS. Previous studies have revealed mixed results for cytokine measurements following high fat diet consumption. Results vary due to the type of diet, diet duration, quantification method, and whether both males and females are included in the analysis. For example, Lainez et al. determined increased serum IL-6 levels in both male and female mice fed the HFHS diet (Research Diets D12492) as used in our study, however, as compared to the Research Diets D12450J control diet. Lainez et al. also determined a statistically significant increase in serum TNFα, but only in male mice fed a high fat diet [21]. We and others have not determined significant differences in plasma cytokine levels following consumption of a high fat diet under the parameters of our current study [44-46].
Given the observed diet and sex differences in food consumption, weight gain, adipokines, and relative abundance of multiple microbiome genera, we expected a subsequent diet and sex difference in neurogliosis. The neuroglial response could be a direct response to components of the diet, metabolites following digestion, metabolites generated specifically from the gut microbiome, adipokines, or a combination of these factors. We chose to evaluate the hypothalamus given its essential role in metabolism and hormone production. We also evaluated the hippocampus given its integral role in the limbic system, but also due to reported sex differences in hippocampal functions [47] and vulnerability to diet-induced changes [2, 48]. We determined decreased astrocyte densitometry only in the hypothalamus of Female HFHS compared to Female LFLS. Astrocytes have numerous functions in the central nervous system including, but not limited to, neuroendocrine modulation, neurotransmitter uptake, injury response, blood brain barrier support, and metabolic support, particularly the astrocyte-neuron lactate shuttle [49]. It is difficult to determine the cause and/or consequence of decreased GFAP immunoreactivity in the Female HFHS hypothalamus from our current results. However, it is important to characterize sex and diet-induced effects that warrant further investigation. Previous studies have shown that high fat diet-induced obesity results in astrocyte activation, specifically in the arcuate nucleus [50, 51]. The arcuate nucleus contains anorexigenic pro-opiomelanocortin (POMC) neurons and orexigenic agouti-related peptide (AgRP)/neuropeptide Y (NPY) neurons. It has previously been determined that increased microgliosis and astrogliosis in the arcuate nucleus can lead to the loss of POMC neurons [50, 52], therefore impacting feeding behaviors. These previous studies were conducted with male mice and rats; future studies will need to be conducted in female mice as well. A previous study utilizing male and female mice determined increased astrogliosis in the arcuate nucleus for male mice fed a high fat diet compared to a control diet, but no difference in female mice fed the same high fat diet versus control diet. It was further determined that estradiol was elevated in high fat-fed female mice which decreased pro-inflammatory cytokines and increased anti-inflammatory cytokines [53]. POMC neurons were not evaluated in this study. However, these data support a sexual dimorphism in astrocyte response following high fat diet exposure, although their findings are different from our study.
We also determined sex differences in microgliosis. Previous studies have shown that consumption of a high fat diet leads to increased microglial activation in the hypothalamus of male mice as compared to female mice [20]. Using skeleton and fractal analyses, we aimed to characterize the overall landscape of microglia in the hippocampus and hypothalamus. A decrease in endpoint and branch length/soma (fractal analysis), increased span ratio, decreased cell complexity, decreased branch number/soma and decreased summed branch length/soma collectively could indicate an increase in activated or amoeboid microglial cells. On the other hand, resting/surveilling microglial cells are characterized by many, long branches and greater cell complexity [27, 34]. Male HFHS revealed a statistically significant decrease in cell complexity compared to Male LFLS in the hypothalamus. While not statistically significant, Male HFHS also revealed decreased endpoint and branch length/soma, branch number/soma, and branch length compared to all other groups in the hypothalamus. Male HFHS also revealed increased span ratio compared to all other groups. Furthermore, we determined that the microglial fractal analysis correlated with multiple gut microbiome diversity indices. As opposed to the correlation between the Simpson index (species evenness) and final body weight, here we determined that the Shannon index (species richnesss), Chao1 index (another measure of richness as well as abundance), and observed species all correlated with hypothalamic fractal analysis. We did not determine a correlation between the Simpson index and hypothalamic fractal analysis. Therefore, the measures for species richness positively correlate with increased endpoint and branch length/soma. As mentioned above, an increased endpoint and branch length is more indicative of a resting/surveilling microglia whereas the retraction of processes and therefore, decreased endpoint and branch length aligns with the activated microglia. The increased species richness in the gut microbiome may contribute to decreased hypothalamic microgliosis. For Male HFHS, significantly increased weight gain, significantly increased gut microbiome species evenness, and features of hypothalamic microglial activation were observed. Female HFHS mice did not reveal as significant weight gain, greater variability in the Simpson diversity index (species evenness), and features revealing less hypothalamic microglial activation.
Interestingly, the opposite effect to branch number and branch length was determined in the hippocampus. In fact, both males and females fed the HFHS diet revealed increased branch number/soma and increased summed branch length/soma. Previous studies have described these microglial features as hyper-ramification, an intermediate form that could precede microglial activation [26]. We did not determine any correlations between gut microbiome diversity indices and hippocampal fractal analyses.
Given the significant decrease in GFAP densitometry for Female HFHS compared to Female LFLS and increased microgliosis for Male HFHS compared to Male LFLS in the hypothalamus, we further evaluated cytokine expression in hypothalamus homogenates. We determined a significant interaction effect (sex x diet) for hypothalamic IL-6, and IL-1β. We also determined an interaction effect that was approaching significance for fractalkine. We did not determine any significant differences for the other cytokines analyzed in the hypothalamus. However, we did observe a lot of variability in these results overall. Future analyses for mRNA cytokine expression may lead to less variable results. We were surprised that IL-6, IL-1β, and fractalkine revealed decreased expression for Male HFHS compared to Male LFLS and increased expression for Female HFHS compared to Female LFLS. This is in contrast to previous findings. For example, Cavaliere et al. determined increased TNFα and IL-1β in the cortex of male C57Bl/6J mice fed a 50% high fat diet [54]. Sobesky et al. determined increased IL-1β expression in the hippocampus of male Wistar rats fed a high fat (60% calories from fat) diet [44]. However, this requires further investigation because increased astrogliosis and microgliosis does not always suggest increased pro-inflammatory cytokine expression. Instead, changes to these dynamic glia could include increased anti-inflammatory cytokine expression relative to pro-inflammatory cytokine expression, cellular apoptosis, metabolic fluctuations, among other factors.
In this study, we determined a resistance to diet-induced obesity for female C57Bl/6 mice compared to males, as shown in previous studies. We further revealed sex differences, diet-induced differences, and sex x diet interactions for gut microbiome genera which may contribute to both differences in metabolism as well as effects to the central nervous system. We hypothesized a role for increased systemic inflammation, at least in part due to gut dysbiosis, as a contributor to neuroinflammation. However, we did not determine significant differences in systemic inflammation despite gut dysbiosis (overall, increased Firmicutes and decreased Bacteroidetes bacteria following a high fat diet as compared to low fat-fed mice). Furthermore, we did determine sex and diet-induced effects to astrogliosis and microgliosis, particularly in the hypothalamus. Our current data do not support a role for peripheral inflammation as a contributor to neuroinflammation. Therefore, future studies to better understand the contribution of diet-induced gut microbiome alterations on neurogliosis must be performed. A different potential contributor to diet-induced neuroinflammation and sex differences in neuroinflammation could be gut-derived metabolites, such as short chain fatty acids.
Acknowledgments:
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM103499 (LRF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosure of interest: The authors declare that the research was conducted without any commercial or financial relationships that could be considered a potential conflict of interest.
References
- 1.Ogden CL, Carroll MD, Flegal KM. Prevalence of obesity in the United States. JAMA. 2014;312(2):189–90. [DOI] [PubMed] [Google Scholar]
- 2.Freeman LR, Haley-Zitlin V, Rosenberger DS, Granholm AC. Damaging effects of a high-fat diet to the brain and cognition: a review of proposed mechanisms. Nutr Neurosci. 2014;17(6):241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malnick SD, Knobler H. The medical complications of obesity. QJM. 2006;99(9):565–79. [DOI] [PubMed] [Google Scholar]
- 4.Bullo M, Casas-Agustench P, Amigo-Correig P, Aranceta J, Salas-Salvado J. Inflammation, obesity and comorbidities: the role of diet. Public Health Nutr. 2007;10(10A):1164–72. [DOI] [PubMed] [Google Scholar]
- 5.Myles IA. Fast food fever: reviewing the impacts of the Western diet on immunity. Nutr J. 2014;13:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haslam DW, James WP. Obesity. Lancet. 2005;366(9492):1197–209. [DOI] [PubMed] [Google Scholar]
- 7.Singh-Manoux A, Dugravot A, Shipley M, Brunner EJ, Elbaz A, Sabia S, et al. Obesity trajectories and risk of dementia: 28 years of follow-up in the Whitehall II Study. Alzheimers Dement. 2018;14(2):178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Odegaard JI, Chawla A. Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science. 2013;339(6116):172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monteiro R, Azevedo I. Chronic inflammation in obesity and the metabolic syndrome. Mediators Inflamm. 2010;2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cryan JF, O'Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, et al. The Microbiota-Gut-Brain Axis. Physiol Rev. 2019;99(4):1877–2013. [DOI] [PubMed] [Google Scholar]
- 11.Cryan JF, de Wit H. The gut microbiome in psychopharmacology and psychiatry. Psychopharmacology (Berl). 2019;236(5):1407–9. [DOI] [PubMed] [Google Scholar]
- 12.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5–11. [DOI] [PubMed] [Google Scholar]
- 13.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–72. [DOI] [PubMed] [Google Scholar]
- 14.Borre YE, Moloney RD, Clarke G, Dinan TG, Cryan JF. The impact of microbiota on brain and behavior: mechanisms & therapeutic potential. Adv Exp Med Biol. 2014;817:373–403. [DOI] [PubMed] [Google Scholar]
- 15.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Mol Psychiatry. 2014;19(2):146–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chander AM, Yadav H, Jain S, Bhadada SK, Dhawan DK. Cross-Talk Between Gluten, Intestinal Microbiota and Intestinal Mucosa in Celiac Disease: Recent Advances and Basis of Autoimmunity. Front Microbiol. 2018;9:2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aydin O, Nieuwdorp M, Gerdes V. The Gut Microbiome as a Target for the Treatment of Type 2 Diabetes. Curr Diab Rep. 2018;18(8):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Smith DL Jr., Keating KD, Allison DB, Nagy TR Variations in body weight, food intake and body composition after long-term high-fat diet feeding in C57BL/6J mice. Obesity (Silver Spring). 2014;22(10):2147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorfman MD, Krull JE, Douglass JD, Fasnacht R, Lara-Lince F, Meek TH, et al. Sex differences in microglial CX3CR1 signalling determine obesity susceptibility in mice. Nat Commun. 2017;8:14556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lainez NM, Jonak CR, Nair MG, Ethell IM, Wilson EH, Carson MJ, et al. Diet-Induced Obesity Elicits Macrophage Infiltration and Reduction in Spine Density in the Hypothalami of Male but Not Female Mice. Front Immunol. 2018;9:1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang LL, Wang CH, Li TL, Chang SD, Lin LC, Chen CP, et al. Sex differences in high-fat diet-induced obesity, metabolic alterations and learning, and synaptic plasticity deficits in mice. Obesity (Silver Spring). 2010;18(3):463–9. [DOI] [PubMed] [Google Scholar]
- 23.Pettersson US, Walden TB, Carlsson PO, Jansson L, Phillipson M. Female mice are protected against high-fat diet induced metabolic syndrome and increase the regulatory T cell population in adipose tissue. PLoS One. 2012;7(9):e46057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. [DOI] [PubMed] [Google Scholar]
- 25.Cancello R, Clement K. Is obesity an inflammatory illness? Role of low-grade inflammation and macrophage infiltration in human white adipose tissue. BJOG. 2006;113(10):1141–7. [DOI] [PubMed] [Google Scholar]
- 26.Morrison H, Young K, Qureshi M, Rowe RK, Lifshitz J. Quantitative microglia analyses reveal diverse morphologic responses in the rat cortex after diffuse brain injury. Sci Rep. 2017;7(1):13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young K, Morrison H. Quantifying Microglia Morphology from Photomicrographs of Immunohistochemistry Prepared Tissue Using ImageJ. J Vis Exp. 2018(136). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali S, Liu X, Queen NJ, Patel RS, Wilkins RK, Mo X, et al. Long-term environmental enrichment affects microglial morphology in middle age mice. Aging (Albany NY). 2019;11(8):2388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swanson KS, Dowd SE, Suchodolski JS, Middelbos IS, Vester BM, Barry KA, et al. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2011;5(4):639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS One. 2011;6(10):e26732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris EK, Caruso T, Buscot F, Fischer M, Hancock C, Maier TS, et al. Choosing and using diversity indices: insights for ecological applications from the German Biodiversity Exploratories. Ecol Evol. 2014;4(18):3514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubbelaar ML, Kracht L, Eggen BJL, Boddeke E. The Kaleidoscope of Microglial Phenotypes. Front Immunol. 2018;9:1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Heyden JA, Zethof TJ, Olivier B. Stress-induced hyperthermia in singly housed mice. Physiol Behav. 1997;62(3):463–70. [DOI] [PubMed] [Google Scholar]
- 36.Kulesskaya N, Rauvala H, Voikar V. Evaluation of social and physical enrichment in modulation of behavioural phenotype in C57BL/6J female mice. PLoS One. 2011;6(9):e24755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voikar V, Polus A, Vasar E, Rauvala H. Long-term individual housing in C57BL/6J and DBA/2 mice: assessment of behavioral consequences. Genes Brain Behav. 2005;4(4):240–52. [DOI] [PubMed] [Google Scholar]
- 38.Robertson KL, Rowland NE. Effect of two types of environmental enrichment for singly housed mice on food intake and weight gain. Lab Anim (NY). 2005;34(9):29–32. [DOI] [PubMed] [Google Scholar]
- 39.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deshpande NG, Saxena J, Pesaresi TG, Carrell CD, Ashby GB, Liao MK, et al. High fat diet alters gut microbiota but not spatial working memory in early middle-aged Sprague Dawley rats. PLoS One. 2019;14(5):e0217553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner BD, Grunwald GK, Zerbe GO, Mikulich-Gilbertson SK, Robertson CE, Zemanick ET, et al. On the Use of Diversity Measures in Longitudinal Sequencing Studies of Microbial Communities. Front Microbiol. 2018;9:1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaab M, Kratzsch J. The soluble leptin receptor. Best Pract Res Clin Endocrinol Metab. 2015;29(5):661–70. [DOI] [PubMed] [Google Scholar]
- 43.Lihn AS, Pedersen SB, Richelsen B. Adiponectin: action, regulation and association to insulin sensitivity. Obes Rev. 2005;6(1):13–21. [DOI] [PubMed] [Google Scholar]
- 44.Sobesky JL, Barrientos RM, De May HS, Thompson BM, Weber MD, Watkins LR, et al. High-fat diet consumption disrupts memory and primes elevations in hippocampal IL-1beta, an effect that can be prevented with dietary reversal or IL-1 receptor antagonism. Brain Behav Immun. 2014;42:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gzielo K, Kielbinski M, Ploszaj J, Janeczko K, Gazdzinski SP, Setkowicz Z. Long-Term Consumption of High-Fat Diet in Rats: Effects on Microglial and Astrocytic Morphology and Neuronal Nitric Oxide Synthase Expression. Cell Mol Neurobiol. 2017;37(5):783–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y, Wu T, Wu J, Zhao L, Li Q, Varghese Z, et al. Chronic inflammation exacerbates glucose metabolism disorders in C57BL/6J mice fed with high-fat diet. J Endocrinol. 2013;219(3):195–204. [DOI] [PubMed] [Google Scholar]
- 47.Koss WA, Frick KM. Sex differences in hippocampal function. J Neurosci Res. 2017;95(1-2):539–62. [DOI] [PubMed] [Google Scholar]
- 48.Underwood EL, Thompson LT. High-fat diet impairs spatial memory and hippocampal intrinsic excitability and sex-dependently alters circulating insulin and hippocampal insulin sensitivity. Biol Sex Differ. 2016;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang DD, Bordey A. The astrocyte odyssey. Prog Neurobiol. 2008;86(4):342–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thaler JP, Yi CX, Schur EA, Guyenet SJ, Hwang BH, Dietrich MO, et al. Obesity is associated with hypothalamic injury in rodents and humans. J Clin Invest. 2012;122(1):153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee D, Thaler JP, Berkseth KE, Melhorn SJ, Schwartz MW, Schur EA. Longer T(2) relaxation time is a marker of hypothalamic gliosis in mice with diet-induced obesity. Am J Physiol Endocrinol Metab. 2013;304(11):E1245–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Souza CT, Araujo EP, Bordin S, Ashimine R, Zollner RL, Boschero AC, et al. Consumption of a fat-rich diet activates a proinflammatory response and induces insulin resistance in the hypothalamus. Endocrinology. 2005;146(10):4192–9. [DOI] [PubMed] [Google Scholar]
- 53.Morselli E, Fuente-Martin E, Finan B, Kim M, Frank A, Garcia-Caceres C, et al. Hypothalamic PGC-1alpha protects against high-fat diet exposure by regulating ERalpha. Cell Rep. 2014;9(2):633–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cavaliere G, Trinchese G, Penna E, Cimmino F, Pirozzi C, Lama A, et al. High-Fat Diet Induces Neuroinflammation and Mitochondrial Impairment in Mice Cerebral Cortex and Synaptic Fraction. Front Cell Neurosci. 2019;13:509. [DOI] [PMC free article] [PubMed] [Google Scholar]