Abstract
Most neurons must last a lifetime and their microtubule cytoskeleton is an important contributor to their longevity. Neurons have some of the most stable microtubules of all cells, but the tip of every microtubule remains dynamic and, although requiring constant GTP consumption, microtubules are always being rebuilt. While some ongoing level of rebuilding always occurs, overall microtubule stability can be modulated in response to injury and stress as well as the normal developmental process of pruning. Specific microtubule severing proteins act in different contexts to increase microtubule dynamicity and promote degeneration and pruning. After axon injury, complex changes in dynamics occur and these are important for both neuroprotection induced by injury and subsequent outgrowth of a new axon. Understanding how microtubule dynamics is modulated in different scenarios, as well as the impact of the changes in stability, is an important avenue to explore for development of strategies to promote neuroprotection and regeneration.
Overview
Neurons are incredibly long-lived and long-range cells. Both aspects of their length are supported by an exquisitely regulated microtubule cytoskeleton. Microtubules in most neurons are extremely stable, and this likely supports long-distance transport. At the same time, despite the energetic cost, microtubules have dynamic ends that allow them to be rebuilt and remain responsive to stimuli including stress and damage. This balance between stability and dynamics and its relationship to injury responses is the subject we will explore.
What is meant by dynamic and stable microtubules?
Different people have different ideas in mind when they refer to microtubules as dynamic or stable. In part, this is due to differences in how microtubules are visualized and how stability and dynamicity are assayed. In most assays only one aspect of microtubule state is examined, and an inference about general dynamicity/stability is made from this. So before moving on to discuss when microtubules are more or less dynamic it is important to lay out what is being measured in different assays and how this relates to the whole microtubule. In addition to the varying assays, there have also been some differences in the inferences made from them.
General microtubule information:
Heterodimers of α and β-tubulin are the building blocks of microtubules. Both subunits bind GTP, but only β-tubulin can hydrolyze it to GDP. In vitro αβ−tubulin heterodimers and GTP are sufficient to generate microtubules that have many of the basic properties seen in cells (1). After the rate-limiting step of nucleation (2–4) is overcome, αβTubulins form a hollow cylinder of about 13 heterodimers around (Figure 1). The α subunit exposed at one end and β at the other. The end with the β exposed is termed the plus end and subunits are added relatively rapidly here (1). The minus end has the α exposed and is also a site for subunit addition, albeit more slowly (5). Both ends undergo switch-like behavior called dynamic instability; where growth phases suddenly change to shrinkage (catastrophe) and back to growth (rescue) (1, 6, 7). When tubulin heterodimers are added they are in the GTP-bound state, and after incorporation β−tubulin hydrolyzes GTP to GDP. In a growing microtubule, the plus end typically has a short region or cap of GTP-tubulin. If GDP-tubulin is exposed, the probability of depolymerization is increased (8). In vitro, proteins that bring tubulin heterodimers together (Tpx2 and XMAP215 for example) reduce the energy barrier to initiate a new microtubule and are grouped as drivers of non-templated nucleation (2). In vivo, it is thought that new microtubules are templated by the γ−Tubulin Ring Complex (γTuRC) (9), and that the normal role of the non-templated nucleators may actually be to help the γTuRC (3, 10). After nucleation, minus ends can remain capped and stabilized by the γTuRC (11, 12). Uncapped minus ends can be generated from existing microtubules by severing proteins (13, 14). In cells these newly generated minus ends are rapidly recognized by minus end-binding proteins including the CAMSAP/Patronin family members and Asp (15–19), so microtubules are thought to be associated at all times with either these or the γTuRC (20). Similarly, while plus ends in vivo undergo dynamic instability, growth parameters are heavily regulated by partner proteins (1, 21, 22).
Figure 1.
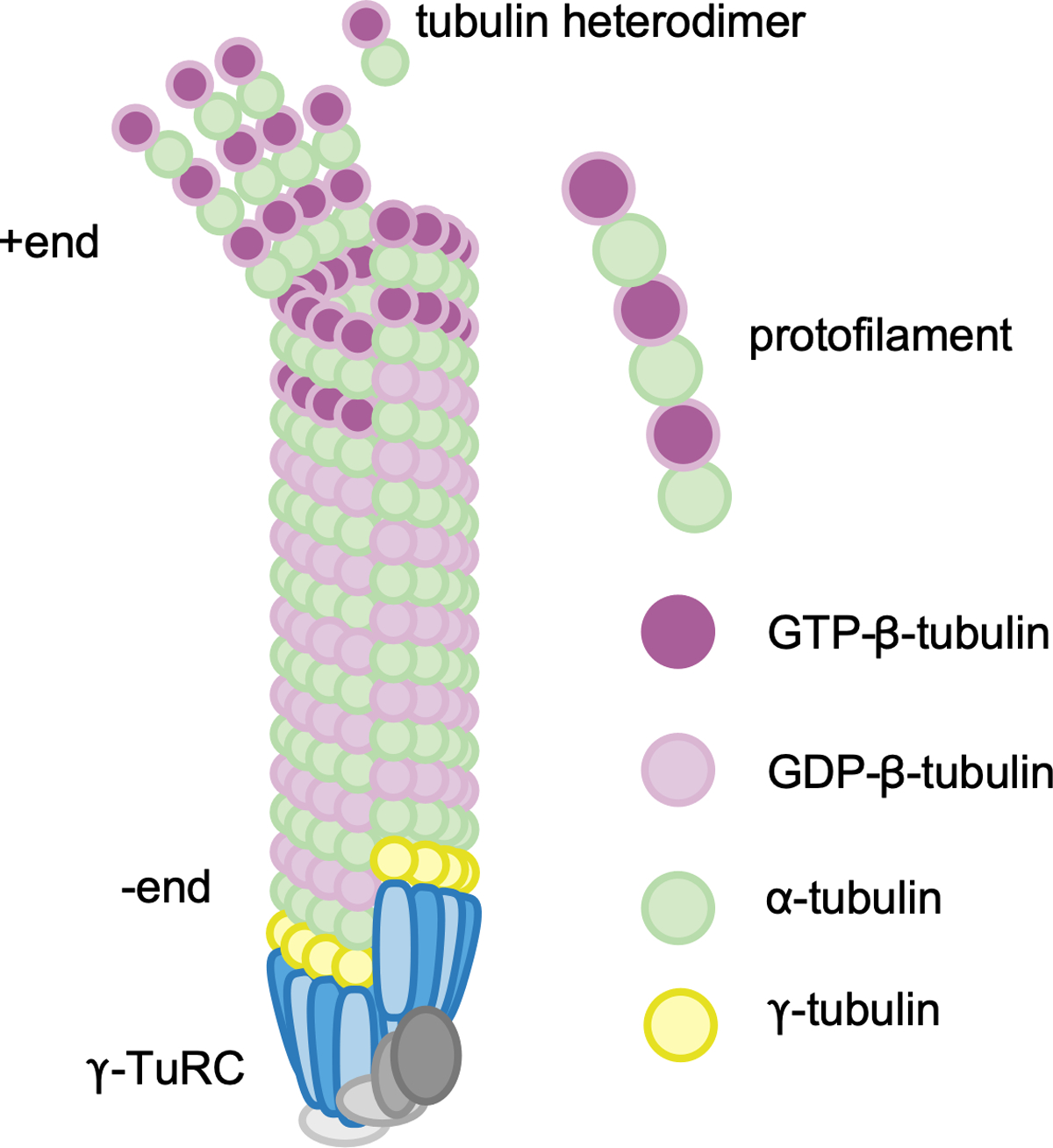
Basic building blocks of microtubules. Microtubule nucleation in vivo occurs from the γ-tubulin ring complex (γ-TuRC). Heterodimers of α-β-tubulin are added to the growing microtubule with the β end of the dimer out. The end with β-tubulin exposed is the plus end, and undergoes dynamic instability.
Types of microtubule regulators:
Several major classes of binding partners regulate microtubule behavior in cells. New microtubules are made when the γTuRC, consisting of about 13 γ−tubulin subunits scaffolded together by γ-tubulin complex proteins (GCPs) (4, 10) is activated for nucleation. In vitro, γTuRCs can stimulate nucleation, but not particularly well (2). Moreover, in vivo many γTuRCs are probably inactive (23). It has therefore been supposed that something must make γTuRCs better nucleators in cells. The best candidates for nucleation activators are proteins containing CM1 domains including CDK5RAP2/cnn (23–25). At the other end of the microtubule, plus end growth is stimulated by the microtubule polymerase XMAP215/msps (26). The growing plus end is a platform for +TIP proteins that can both tune polymerization and mediate interactions of plus ends with other cellular machinery and organelles (27–29). EB1, or end-binding protein 1, directly interacts with the GTP cap and recruits other +TIP proteins to growing microtubules. New microtubule ends can be generated by severing proteins, fidgetin, spastin and katanin, that recognize specific features of microtubules including crossovers and post-translational modification of tubulin (30). Each severing event generates a new plus end and a new minus end. Newly generated cytoplasmic minus ends are rapidly recognized by proteins of the CAMSAP/Patronin family (27, 31). While it was thought that minus ends never grow in cells (5), short stretches of minus end growth mediated by CAMSAP2 help stabilize microtubules in mammalian neurons (16) and extended minus end growth is seen in fish and fly neurons, and in fly neurons is important for polarity of dendritic microtubules (32). Plus ends generated by severing can either depolymerize or begin to grow depending on the specific cellular milieu (30). In addition to severing proteins, other microtubule associated proteins, or structural MAPs, can recognize microtubules along their length. The basic model for function of structural MAPs including tau, MAP1 and MAP2 is that their binding helps stabilize the microtubule, primarily by suppressing microtubule catastrophe and promoting rescue (1, 21). However, their function likely goes far beyond this as some MAPs promote interaction with actin, influence motor movement, or microtubule spacing (33), or in the case of tau, support long labile regions (34).
In addition to being regulated by binding partners, microtubules can be covalently modified by enzymes that add post-translational modifications (PTMs) (35, 36). The C-terminus of α-tubulin is the most heavily modified, but even lysine 40, which sits in the middle of the hollow microtubule cylinder can be acetylated. While the direct effect of most PTMs with the exception of polyamination (37) on microtubules is unclear, they can modulate affinities of microtubule regulatory proteins (36, 38) and these can in turn affect the probability of catastrophe or continued growth (39). Long-lived microtubules, including those in neurons, tend to be more heavily modified than newly polymerized ones (35), and microtubule stabilizing drugs can broadly increase modification (40). Similarly, neuronal microtubules tend to have many more MAPs along them than microtubules in other cell types (33).
The relationship between microtubule regulatory proteins and stability:
Microtubule stability is controlled by the activities of the different classes of microtubule regulatory proteins working together. The classic view of MAPs that increase stability is that they act by modulating growth, either suppressing catastrophe or promoting rescue, with a net effect of more tubulin incorporation into polymer and less is in the free pool (1, 21). By suppressing catastrophe and promoting rescue the average length of microtubules would also be expected to increase.
In vivo linking an individual regulator to specific effects on stability can be tricky, and phenotypes from loss of a regulator or PTM are often modest or surprisingly specific (41–44). However, there are some useful correlations that can help diagnose stability in specific cells. Most MAPs and PTMs are associated with stable, or long-lived microtubules (33, 35), although only polyamination has been shown to actually increase stability (37). Thus cells or regions of cells with lots of MAPs or heavily modified microtubules tend to be those with relatively stable microtubules, and neurons often stand out from other cell types based on MAP or PTM labeling. Indeed, for many years the 22C10 monoclonal antibody was used as a neuronal marker without knowing that it recognized Drosophila MAP1, futsch (45). Similarly, endogenously tagged MAPs including Drosophila tau typically label neurons much more brightly than other cells (46). Of the PTMs, acetylated tubulin is particularly prominent in Drosophila neurons compared to other cells (42, 44). In mice MAP2 is used as a specific dendritic marker and tau as an axonal one (47). Other MAPs are also particularly abundant in neurons (48), consistent with the general idea that neurons have many MAPs and overall very stable microtubules.
Measuring microtubule stability:
Relative stability of microtubules is typically measured by probing one parameter of the microtubule network in a particular cell and extrapolating from that to the overall state of the microtubule network. For example in fixed cells or tissue microtubule acetylation is often used as a readout of stability, with more acetylation correlating with higher stability. On the flip side, tyrosination can be used as an indicator of “young” or less stable microtubules (49, 50). In living cells, many more options to probe stability exist (Figure 2). For example, microtubule depolymerizing drugs like nocodazole can be added to neurons and the amount of microtubule mass resistant to depolymerization can be a rough measure of how much stable microtubule polymer is present in the cell. The use and cellular correlates of this measure of stability have been very nicely reviewed together with other aspects of neuronal microtubule stability (51). Methods relying on drug-induced depolymerization tend to work well in culture, but can be more problematic in vivo. Using fluorescently tagged proteins opens up additional ways to probe stability, that can be applied to neurons in culture, explants or in whole animal preps. Turnover of microtubules in a region of axon or dendrite can be measured using photoconvertible tubulin (40, 52). After photoconversion, free tubulin heterodimers rapidly diffuse out of the region of interest, while those that were part of a polymerized microtubule are trapped until the microtubule depolymerizes back through the area. +TIP proteins can also be used as a readout of relative dynamicity (53, 54). For a constant amount of total microtubule polymer, the number of growing microtubules in a region is higher when microtubules have short stable regions (Figure 2). In general each measure can distinguish between long, stable microtubules and more dynamic ones with a shorter stable region. This model assumes that within each microtubule there are stable regions towards the minus end and a dynamic region at the plus end (51) and that the biggest difference between a more and less dynamic population is the length of the stable region. Evidence that stable microtubules are capped with dynamic plus ends comes from staining of axonal microtubules with anti-PTM antibodies, where it was seen that within an individual microtubule regional differences were observed (55, 56). For example, staining for tyrosination (not strictly a PTM, as de-tyrosination is the modified state), which is associated with newly assembled more labile microtubule regions, was found in a stretch near the plus end, while the remainder of the microtubule was not stained (56).
Figure 2.
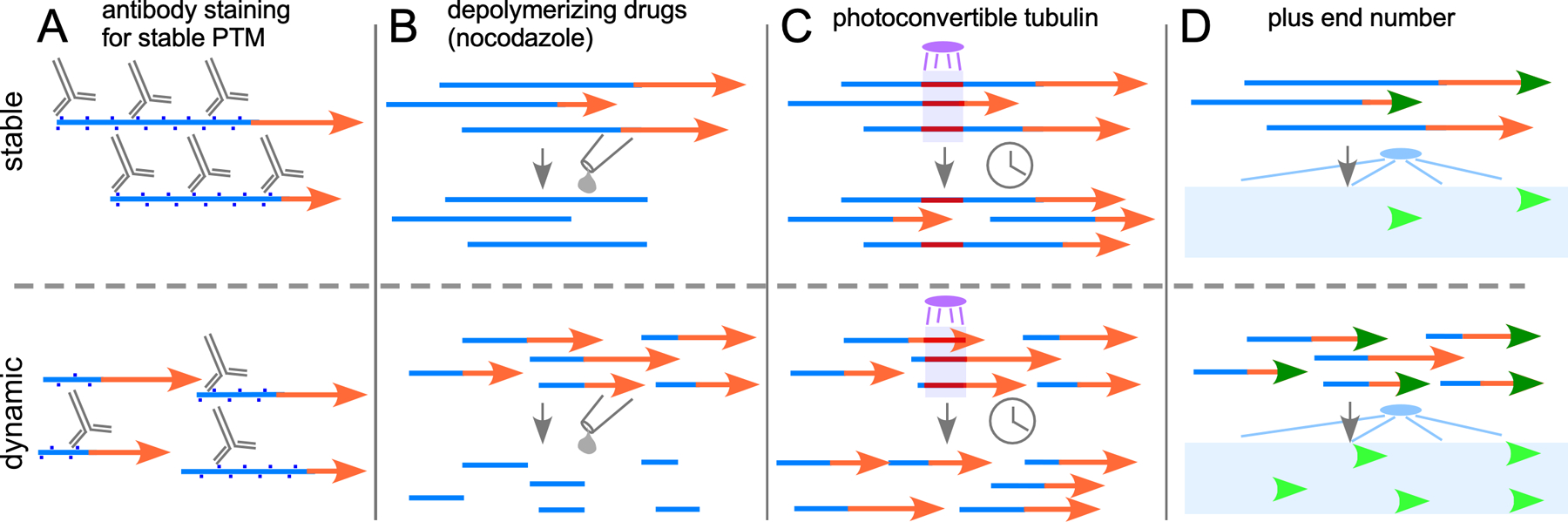
Assays to probe neuronal microtubule stability. (A) Antibodies that recognize PTMs associated with stable (blue) regions of microtubules can be used to get a relative idea of the amount of stable polymer in a region. Assuming the total amount of polymerized tubulin is similar, more antibody-binding is associated with microtubules with long stable regions (top) compared to shorter more dynamic microtubules (bottom). (B) Drugs that depolymerize microtubules initially cause loss of dynamic regions (orange) as they cycle through catastrophe and rescue. For long stable microtubules relatively little polymer is destabilized at early time points (top), while regions with short microtubules (bottom) show a greater loss. (C) Incorporation of photoconvertible, for example tdEOS, α-tubulin into microtubules allows conversion of a region of the microtubule to red over a green background using UV light. The red tubulin is trapped in the microtubule as long as it remains polymerized. Long microtubules may take hours to depolymerize back through stable regions and so the converted mark will remain (top), while complete depolymerization occurs on a much shorter timescale with more dynamic microtubules eliminating the red mark sooner (bottom). (D) Use of a labeled +TIP protein like EB1 allows visualization of each plus end that is growing, and each individual microtubule will cycle through periods of growth and shrinkage as it undergoes dynamic instability. With long stable microtubules relatively few growing plus ends will be seen at any one time (top), while shorter more dynamic microtubules will result in many more visible plus end comets (bottom).
Although it is difficult to determine whether every microtubule in a neuron has a dynamic end, the foundational model of dynamic instability of microtubules is based on this premise (1). Indeed, in vitro in the presence of GTP microtubules always grow or shrink in the absence of other factors (1). Pausing factors have been identified that modulate growth, but the periods of pausing are on the order of seconds between bouts of growth and shrinkage rather than representing a long-term non-dynamic state (57, 58). Similarly, tethered microtubules, for example at the cell cortex, have increased pausing in the context of dynamic microtubule ends (59). In neurons dynamic plus ends are distributed throughout axons and dendrites (46, 60, 61), so in the absence of strong data to support the presence of microtubules with non-dynamic plus ends, we favor a model in which neuronal microtubules have a stable region of varying length towards the minus end and a dynamic plus end.
2. How long and stable are neuronal microtubules?
All indicators of microtubule stability, from PTMs to resistance to depolymerization (Figure 2), indicate that neuronal microtubules tend to exist at the stable end of the dynamic>>stable spectrum. If the general model for microtubules is that each has a stable region towards the minus end and a labile region near the plus end (51), stability roughly translates into a long stable region. Of course, in any cell newly generated short microtubules will be present together with some longer ones, so when we think about length this is an average.
The length of neuronal microtubules has been determined using serial section electron microscopy in primary cultures of mammalian neurons and in C. elegans. In rodent sensory axons the average microtubule length was just over 100 microns (62). During initial axon outgrowth of hippocampal neurons many short microtubules are present (63), but by the time the axon extended and dendrites began to grow, most microtubules were longer than the 12 micron region reconstructed (63) consistent with the very long microtubules in sensory axons. Reconstruction of microtubules in two different C. elegans neurons indicated that microtubules in their neurites had average lengths ranging from about ten to 30 microns, and were arranged in an overlapping array (64). Neurites themselves were about 500 microns long, so the relatively short microtubule length cannot be accounted for by short neurites (64). A more recent study using living C. elegans with fluorescently labeled microtubule found that in larval worms average microtubule length was about 4 microns, and this only increased to about 10 microns in adult worms (65). It is unclear why microtubules in C. elegans axons should be so much shorter than those in mammalian axons, and there is little information on microtubule length in axons of other animals. The enrichment of PTMs associated with stability and slow turnover rate (52) of Drosophila microtubules suggests that they are stable, and therefore long, although this has not been measured directly. Turnover of microtubules in axons and dendrites of Drosophila sensory neurons is shown in Figure 3. In axons, some converted tubulin is still trapped in polymerized axonal microtubules 6h after conversion. This indicates that complete turnover of axonal microtubules takes over 6h in Drosophila, so as in mammals they are probably quite long.
Figure 3.
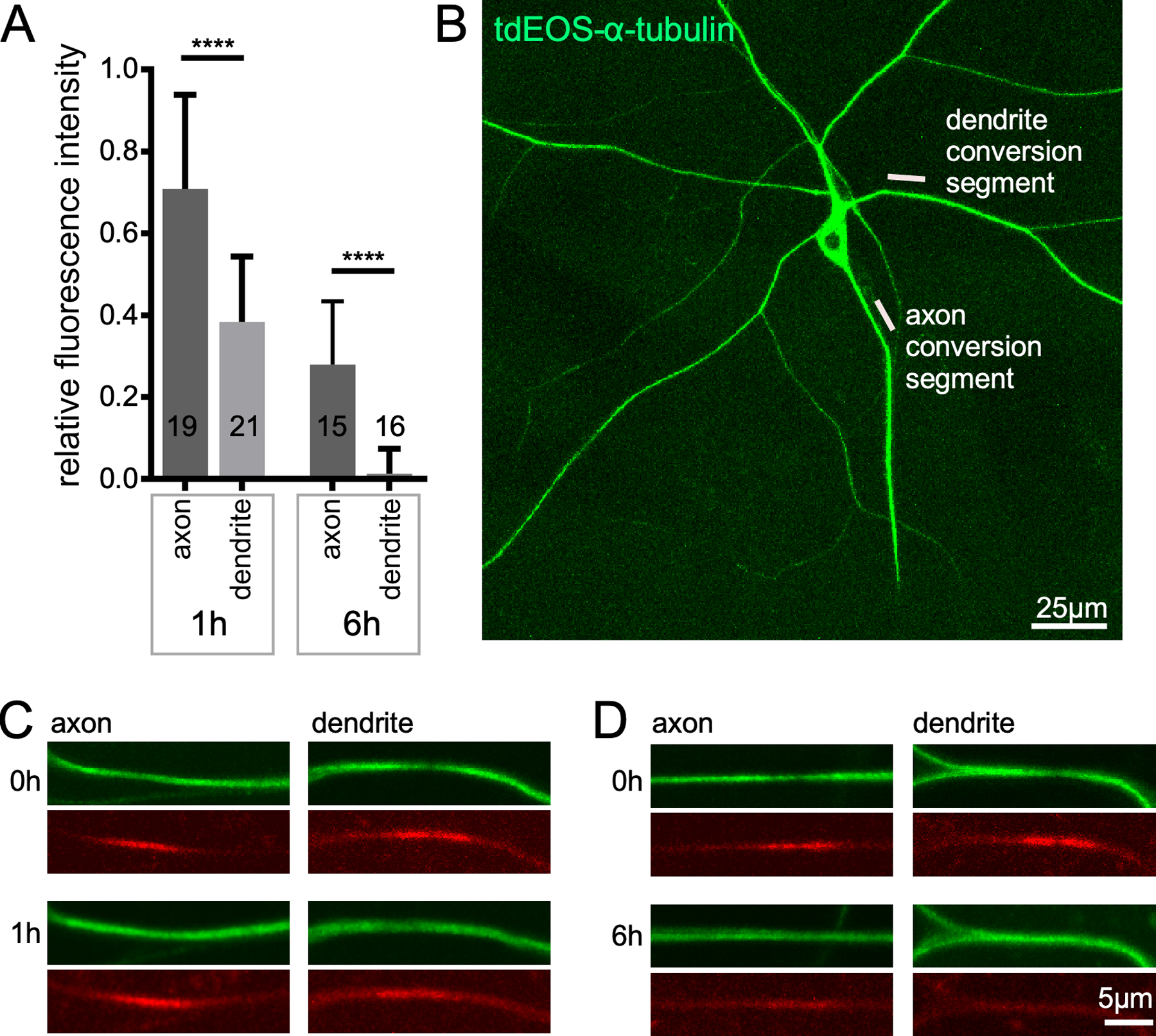
Microtubules turn over faster in dendrites than axons. Photoconvertible tdEOS-α-tubulin (111) was expressed in Drosophila class IV dendritic arborization neurons with 477-Gal4. Whole larvae were mounted for live imaging; an example image of the ddaC neurons used is shown in B. Ten micron regions in either the dendrite or axon were converted from green to red using UV light. Example images of converted segments can be seen in red channel of the 0h images in C and D. The green unconverted signal is also reduced in the conversion region. 1h after conversion the converted region can still be seen in axons and dendrites (C). However, by 6h only the axon still has a visible conversion region. The conversion signal was set at 1 in the 0h images, and the relative amount remaining at 1h and 6h is shown in A. The error bars show standard deviation and the number on the bars are the numbers of animals analyzed. Dendrite data is a re-analysis of data from (52). The statistical test used was a t-test, ****p<0.0001.
While neuronal microtubules tend to be stable in mammals and Drosophila, there are some regional differences. In general, dendritic microtubules are less stable than those in axons ((66, 67) and Figure 3). There is also a recent idea that within dendrites microtubules with opposite polarity differ in stability. Based on resistance to depolymerization after severing (61) and differing selectivity of motor binding (68) it is suggested that minus-end-out microtubules in dendrites are more stable than plus-end-out ones. Note that both the more and less stable populations still likely have dynamic plus ends, and indeed dynamic plus ends are seen at tips of microtubules in both orientations in dendrites (61).
The bottom line is that neuronal microtubules tend to be stable, and this is most likely associated with a long stable region, particularly in mature neurons. There are likely some regional differences in stability, as well as potentially different populations of microtubules within a region that may differ in stability as well as polarity. Despite the importance of microtubules to long-term neuronal function, however, there are not a lot of direct measurements of microtubule length in across neurons and animals. In part, this is because neuronal microtubules form dense bundles and so length of individual microtubules is difficult to tease out without labor-intensive methods like serial section electron microscopy reconstructions. It is also partly because the biological importance of microtubule dynamicity/stability is still emerging. It is clear, however, that neurons really “care about” microtubule stability, and respond to changes in stability with transcriptional alterations to maintain homeostasis (69, 70). It is also becoming clear that changes in stability are important in neuronal responses to injury and stress.
3. Microtubule disassembly paves the way for neurite elimination.
The most straightforward example of a link between changes in microtubule stability and neuronal stability is microtubule disassembly during elimination of specific regions of the neuron. After axons or dendrites are severed from the cell body, they undergo programmed fragmentation that is largely due to activation of the NAD cleavage enzyme Sarm (71–74). The role of Sarm in degeneration is conserved in Drosophila (71) and mammals (74). Elimination of NAD by strong Sarm activation during degeneration likely makes ATP levels fall rapidly as NAD is required for oxidative phosphorylation. However, there is some evidence that during injury-induced axon degeneration in Drosophila an mammals microtubules disappear as an early step (75, 76) that may occur before Sarm activation. However, more direct evidence for microtubule regulators or changes in dynamics that play a role in injury-induced axon degeneration remain elusive. In contrast, an early step in injury-induced dendrite degeneration is microtubule severing by the AAA ATPase fidgetin (52). During the first hour after dendrites are severed from the cell body, a dramatic increase in the number of plus ends per length is seen and this increase depends on fidgetin (52). Moreover, fidgetin reduction delays dendrite degeneration (52). It is not known how fidgetin is activated by dendrite severing, but it is likely present, and inactive, before injury.
Other types of neurite elimination or trimming have also been linked to specific microtubule destabilizers. During development, neurites often arborize exuberantly and then compete to innervate targets. The ones that lose the competition are then pruned back (77, 78). Examples of this type of developmental pruning have been linked to microtubule regulators in mice. Growth factor withdrawal from sensory neurons in culture leads to microtubule disassembly followed by degeneration (79). The depolymerizing kinesin-13 family member KIF2A is required for normal microtubule loss in cultured neurons, and consistent with a role of this protein during normal developmental pruning, the skin is hyperinnervated by sensory axons in kif2A mutant mice (79). During refinement of muscle innervation in vertebrates, multiple motor axons initially innervate individual muscle fibers. Eventually excess connections are removed and one neuron innervates one fiber (80). As the losing axon leaves the area, retraction bulbs are formed. Within these, the number of microtubule plus ends is elevated, and this depends on spastin (81), a sister AAA ATPase to fidgetin, indicating that microtubule severing may also play a role in this local pruning.
A larger scale form of developmental pruning takes place in animals like Drosophila that undergo metamorphosis during development (77, 82). As the body form is remodeled in the pupal stage, some neurons die while others survive and selectively disassemble axons and/or dendrites so that the neuron can regrow neurites that innervate new body structures (83, 84). For example, dendrites of Drosophila dendritic arborization neurons tile the larval body wall (85) and allow animals to respond to different types of mechanosensory cues (86, 87). During pupariation as the body wall is rebuilt, the sensory dendrites are completely pruned leaving a bald cell body attached to the axon; many of these cells survive and grow new dendrite arbors for use in the adult (88). Pruning has also been studied extensively in cells that make up the mushroom body, the seat of learning and memory in the Drosophila brain. Mushroom body γ neurons prune their entire dendrite arbor and distal axonal branches. In both mushroom body and dendritic arborization neurons microtubules appear to be reduced early in pruning (89, 90). Additional mechanistic studies in dendritic arborization neurons support the idea that microtubule stability is actively altered during degeneration. In a candidate screen, the AAA ATPase katanin-60 like 1 (kat-60L1) emerged as a critical regulator of the first step of dendrite pruning, detachment of dendrites from the soma associated with thinning of the microtubules in this area (91). The kinase Par-1 has also been shown to regulate this step of pruning, likely through phosphorylation of tau (92). Thus it seems that multiple microtubule regulators act together to disassemble microtubules so that dendrites can be self-severed from the soma during pruning.
One intriguing aspect of the function of microtubule regulators during various types of neurite disassembly, is the exquisite specificity of each regulator (Figure 4). For example, in mice, a kinesin-13 is required for sensory axon pruning (79) and spastin is used for refinement of motor axons (81). One could argue that these are different cell types and so perhaps express distinct suites of microtubule destabilizers that can be harnessed to take the microtubule cytoskeleton apart. However, the specificity of severing proteins in Drosophila undermines this argument. ddaC neurons are large, easily identifiable dendritic arborization neurons on the dorsal surface or Drosophila larvae. In these cells kat-60L1 is required for dendrite pruning, but does not have any role in injury-induced dendrite degeneration (91, 93). In this cell a different severing protein, fidgetin, is required to disassemble microtubules during injury-induced degeneration (52). This specificity suggests that even if multiple severing and disassembly proteins are present at the same time, they are tightly regulated so that they can be activated in distinct scenarios.
Figure 4.
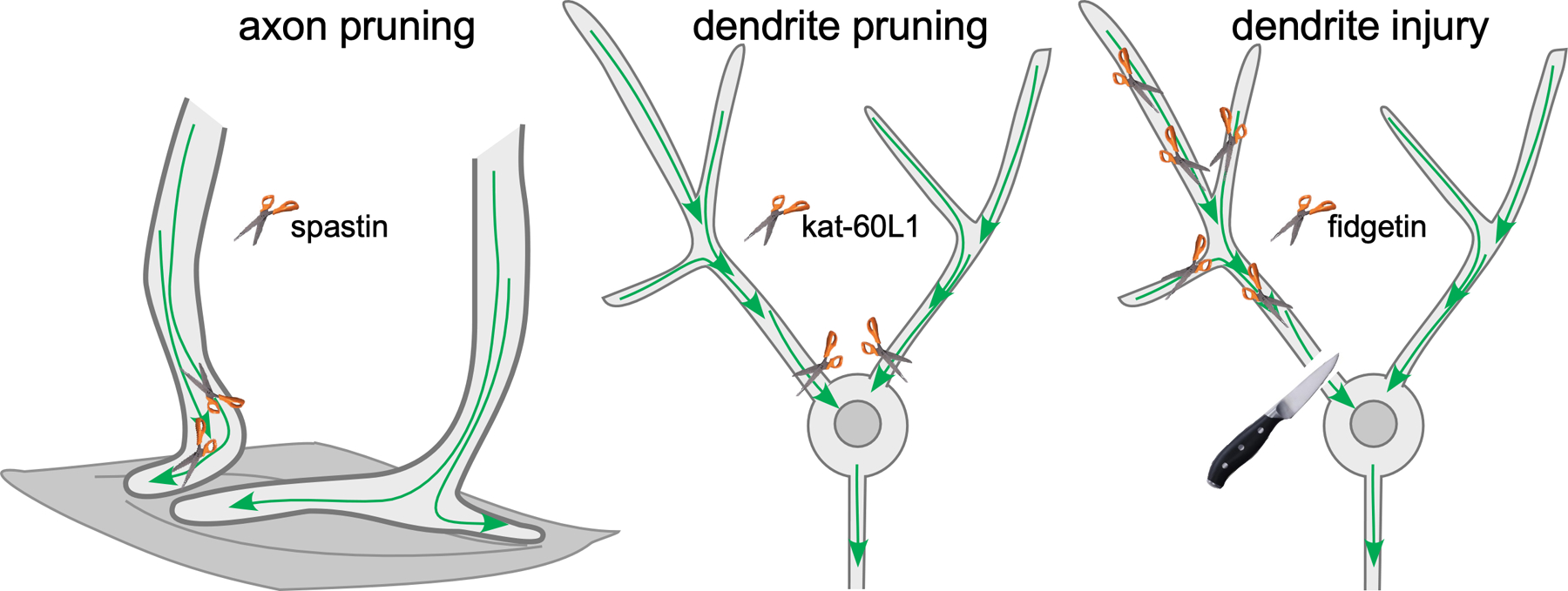
Severing proteins disassemble microtubules in neurites prior to pruning and degeneration. (Left panel) During competition to innervate muscle fibers, some motor axon terminals are eliminated from fibers. Spastin acts to increase the number of plus ends during elimination to help disassemble the “losing” terminal (81). (Middle panel) During large-scale pruning of Drosophila sensory dendrites, kat-60L1 is required to separate the dendrite from the cell body at its base (91). (Right panel) A different severing protein, fidgetin, is used to increase microtubule dynamics in a cut off dendrite after it is severed from the cell body (52).
4. Axon regeneration and microtubule stability
While the relationship between increased microtubule dynamics/reduced microtubule stability and neurite pruning and degeneration is quite intuitive, more complex changes in stability are associated with axon regeneration. First, there are local microtubule changes at the injury site that are important for setting the stage for regeneration (94, 95). In a fun study on Aplysia axons, severing was shown to cause changes in microtubule polarity just proximal to the injury site. Normally plus-end-out microtubules near the end of the remaining stump reversed polarity to form a trap where vesicles accumulated (96). Increases in dynamic microtubules occur just proximal to the injury site in C. elegans, although without any polarity reversal (97).
Changes in microtubules proximal to the injury site have also been seen in mouse sciatic nerve. After nerve ligation, microtubules are deacetylated (removal of a stability-associated PTM) by histone deacetylase 5 (HDAC5) (98), and this is associated with growth cone formation. Nuclear export of HDAC5 is also required for regeneration (99) so this protein seems to act in two places to promote axon regrowth. The microtubule severing protein fidgetin is an inhibitor of regeneration. It is proposed to reduce regeneration by targeting the dynamic ends of microtubules in the growth cone (100–102). In contrast, a different severing protein, spastin, actually promotes regeneration in Drosophila. In this case, changes in microtubule dynamics have not been associated with spastin function, and it seems to function primarily to couple concentration of the endoplasmic reticulum and microtubules near the growing axon tip (103, 104).
Together the live imaging studies on increases in microtubule dynamics near the new axon tip, and functional data on HDAC5 and severing proteins, provides substantial evidence that dynamic microtubules play an important role near the tip of an injured axon to promote regeneration. However, microtubule stabilization at the growth cone has been shown to promote regrowth on inhibitory substrates, and destabilization leads to retraction bulb formation (105). So although some increased level of dynamics at the cut site has been positively linked to outgrowth, this relationship is not straightforward and exquisite regulation of dynamics is required to facilitate growth.
In developing neurons, microtubule stability promotes axon specification, and if microtubule stabilizing drugs are added, more than one axon is made (106). At early stages of axon specification stability-associated PTMs become enriched in axons (40), and turnover of microtubules is slower in the nascent axon compared to minor neurites (40). In keeping with this idea that axons have more stable microtubules than dendrites, and their identity is intimately associated with stability one might expect that increasing stability could promote regeneration. Indeed microtubule stabilization in whole rodents leads to increased regeneration in the inhibitory environment of the central nervous system (107, 108). Surprisingly, rather than simply promoting axon growth, a major effect of the drug treatment is to reduce glial scarring and this dual neuronal and glial effect seems particularly effective in promoting growth (107, 108). How and where in the axon microtubule stability favors growth is not really understood. But it seems likely that microtubules must be dynamic near the growing tip, and then stabilized behind the tip to promote long-range transport to support growth. Regional analysis of plus end number per length (a measurement of stability; see Figure 2D) in regenerating C. elegans axons provides direct support for this model as fewer comets per length are present behind the very dynamic tip (97).
5. The riddle of dynamic microtubules and stable neurons
In addition to the local increase in microtubule dynamics seen near the axon injury site, global increases in microtubule dynamics have also been observed in systems where live imaging of microtubules at sites more distant from the injury is possible. In Drosophila neurons, severing the axon leads to more than doubling of the number of growing plus ends per length throughout the proximal axon, and even throughout the dendrite arbor of the injured neuron (53). Similar increases in microtubule dynamics have been observed in the proximal region of severed mouse intercostal nerves (54). In both cases there is a delay in this global increase, and then it is sustained for several days. The rapid genetic manipulation available in Drosophila has allowed mechanistic exploration of the increase. It requires JNK signaling and the transcription factor fos (53, 109, 110), so likely is downstream of the core axon injury transcriptional response. In this case, the increase in number of growing plus ends is due to microtubule nucleation (109). Unlike the increase in plus end number due to severing, which is associated with degeneration and pruning, the nucleation-mediated increase in dynamics after axon injury is part of a neuroprotective program that helps the remaining regions of the cell resist degeneration (109). Similar increases in dynamics are seen in neurons that express proteins that cause neurodegeneration including expanded poly-Q proteins in Drosophila (109) and disease-causing SOD1 mutants in mice (54), and in Drosophila this is also due to nucleation and is neuroprotective (109). How dynamic microtubules might promote neuronal resistance to degeneration is not understood, but it is an intriguing lead that could yield new ideas about how to help neurons win the battle against pro-degenerative genetic burden.
In keeping with the idea that microtubule stability is required somewhere in the axon for regenerative outgrowth, the global increase in microtubule dynamics is transient and largely finished by the time growth begins (109). In fact, if the high dynamics phase is prolonged by overexpression of fos or the neuroprotective protein Nmnat, then axon regeneration is dampened (110). Thus different levels of microtubule stability are beneficial in different scenarios. Paradoxically, high dynamics seems to lock down the neuron against further degeneration, but also against the plasticity required to regenerate. This paradox highlights how much more remains to be understood about the downstream impacts of changes in neuronal microtubule stability, and how important precise regulation of dynamics is for long-term neuronal function.
Acknowledgements
We are grateful to the Bloomington Drosophila Stock Center (NIH P40OD018537) and Dr. Vladimir Gelfand for the Drosophila lines used in Figure 3. Past and present members of the Rolls lab have provided valuable perspectives. Funding for microtubule work in the Rolls lab was provided by the National Institutes of Health, R01 GM085115.
References
- 1.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Roostalu J, Surrey T. Microtubule nucleation: beyond the template. Nat Rev Mol Cell Biol. 2017;18(11):702–10. doi: 10.1038/nrm.2017.75. [DOI] [PubMed] [Google Scholar]
- 3.Brouhard GJ, Rice LM. Microtubule dynamics: an interplay of biochemistry and mechanics. Nat Rev Mol Cell Biol. 2018;19(7):451–63. 10.1038/s41580-018-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol. 2011;12(11):709–21. 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dammermann A, Desai A, Oegema K. The minus end in sight. Current biology : CB. 2003;13(15):R614–24. doi: S096098220300530X [pii]. [DOI] [PubMed] [Google Scholar]
- 6.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312(5991):237–42. [DOI] [PubMed] [Google Scholar]
- 7.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009;10(8):569–74. 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 8.Manka SW, Moores CA. The role of tubulin-tubulin lattice contacts in the mechanism of microtubule dynamic instability. Nat Struct Mol Biol. 2018;25(7):607–15. doi: 10.1038/s41594-018-0087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teixido-Travesa N, Roig J, Luders J. The where, when and how of microtubule nucleation - one ring to rule them all. Journal of cell science. 2012;125(Pt 19):4445–56. doi: 10.1242/jcs.106971. [DOI] [PubMed] [Google Scholar]
- 10.Tovey CA, Conduit PT. Microtubule nucleation by gamma-tubulin complexes and beyond. Essays Biochem. 2018;62(6):765–80. doi: 10.1042/EBC20180028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nature cell biology. 2000;2(6):358–64. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 12.Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nature cell biology. 2000;2(6):365–70. 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- 13.Baas PW, Karabay A, Qiang L. Microtubules cut and run. Trends in cell biology. 2005;15(10):518–24. doi: 10.1016/j.tcb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Sharp DJ, Ross JL. Microtubule-severing enzymes at the cutting edge. Journal of cell science. 2012;125(Pt 11):2561–9. doi: 10.1242/jcs.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang K, Rezabkova L, Hua S, Liu Q, Capitani G, Altelaar AFM, Heck AJR, Kammerer RA, Steinmetz MO, Akhmanova A. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nature cell biology. 2017;19(5):480–92. doi: 10.1038/ncb3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang K, Hua S, Mohan R, Grigoriev I, Yau KW, Liu Q, Katrukha EA, Altelaar AF, Heck AJ, Hoogenraad CC, Akhmanova A. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Developmental cell. 2014;28(3):295–309. doi: 10.1016/j.devcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin SS, Vale RD. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143(2):263–74. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendershott MC, Vale RD. Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(16):5860–5. doi: 10.1073/pnas.1404133111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135(5):948–59. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 20.Akhmanova A, Hoogenraad CC. Microtubule minus-end-targeting proteins. Current biology : CB. 2015;25(4):R162–71. doi: 10.1016/j.cub.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 21.van der Vaart B, Akhmanova A, Straube A. Regulation of microtubule dynamic instability. Biochem Soc Trans. 2009;37(Pt 5):1007–13. 10.1042/BST0371007. [DOI] [PubMed] [Google Scholar]
- 22.Etienne-Manneville S. From signaling pathways to microtubule dynamics: the key players. Curr Opin Cell Biol. 2010;22(1):104–11. 10.1016/j.ceb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Farache D, Emorine L, Haren L, Merdes A. Assembly and regulation of gamma-tubulin complexes. Open Biol. 2018;8(3). doi: 10.1098/rsob.170266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. The Journal of cell biology. 2010;191(6):1089–95. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JV, Buchwalter RA, Kao LR, Megraw TL. A Splice Variant of Centrosomin Converts Mitochondria to Microtubule-Organizing Centers. Current biology : CB. 2017;27(13):1928–40 e6. doi: 10.1016/j.cub.2017.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132(1):79–88. doi: S0092–8674(07)01547–4 [pii] 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akhmanova A, Steinmetz MO. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol. 2015;16(12):711–26. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- 28.Jiang K, Akhmanova A. Microtubule tip-interacting proteins: a view from both ends. Curr Opin Cell Biol. 2011;23(1):94–101. 10.1016/j.ceb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Galjart N. Plus-end-tracking proteins and their interactions at microtubule ends. Current biology : CB. 2010;20(12):R528–37. doi: 10.1016/j.cub.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 30.McNally FJ, Roll-Mecak A. Microtubule-severing enzymes: From cellular functions to molecular mechanism. The Journal of cell biology. 2018;217(12):4057–69. doi: 10.1083/jcb.201612104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akhmanova A, Steinmetz MO. Microtubule minus-end regulation at a glance. Journal of cell science. 2019;132(11). doi: 10.1242/jcs.227850. [DOI] [PubMed] [Google Scholar]
- 32.Feng C, Thyagarajan P, Shorey M, Seebold DY, Weiner AT, Albertson RM, Rao KS, Sagasti A, Goetschius DJ, Rolls MM. Patronin-mediated minus end growth is required for dendritic microtubule polarity. The Journal of cell biology. 2019. doi: 10.1083/jcb.201810155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bodakuntla S, Jijumon AS, Villablanca C, Gonzalez-Billault C, Janke C. Microtubule-Associated Proteins: Structuring the Cytoskeleton. Trends in cell biology. 2019;29(10):804–19. doi: 10.1016/j.tcb.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Baas PW, Qiang L. Tau: It’s Not What You Think. Trends in cell biology. 2019;29(6):452–61. doi: 10.1016/j.tcb.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr Opin Cell Biol. 2008;20(1):71–6. 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, Brady ST. Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends in cell biology. 2015;25(3):125–36. doi: 10.1016/j.tcb.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song Y, Kirkpatrick LL, Schilling AB, Helseth DL, Chabot N, Keillor JW, Johnson GV, Brady ST. Transglutaminase and polyamination of tubulin: posttranslational modification for stabilizing axonal microtubules. Neuron. 2013;78(1):109–23. 10.1016/j.neuron.2013.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gadadhar S, Bodakuntla S, Natarajan K, Janke C. The tubulin code at a glance. Journal of cell science. 2017;130(8):1347–53. doi: 10.1242/jcs.199471. [DOI] [PubMed] [Google Scholar]
- 39.Peris L, Wagenbach M, Lafanechere L, Brocard J, Moore AT, Kozielski F, Job D, Wordeman L, Andrieux A. Motor-dependent microtubule disassembly driven by tubulin tyrosination. The Journal of cell biology. 2009;185(7):1159–66. doi: jcb.200902142 [pii] 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammond JW, Huang CF, Kaech S, Jacobson C, Banker G, Verhey KJ. Posttranslational modifications of tubulin and the polarized transport of kinesin-1 in neurons. Molecular biology of the cell. 2010;21(4):572–83. 10.1091/mbc.E09-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devambez I, van Dijk J, Benlefki S, Layalle S, Grau Y, Rogowski K, Parmentier ML, Soustelle L. Identification of DmTTLL5 as a Major Tubulin Glutamylase in the Drosophila Nervous System. Sci Rep. 2017;7(1):16254. doi: 10.1038/s41598-017-16586-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkins BV, Saunders HAJ, Record HL, Johnson-Schlitz DM, Wildonger J. Effects of mutating alpha-tubulin lysine 40 on sensory dendrite development. Journal of cell science. 2017;130(24):4120–31. doi: 10.1242/jcs.210203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roos J, Hummel T, Ng N, Klambt C, Davis GW. Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron. 2000;26(2):371–82. Epub 2000/06/06. [DOI] [PubMed] [Google Scholar]
- 44.Yan C, Wang F, Peng Y, Williams CR, Jenkins B, Wildonger J, Kim HJ, Perr JB, Vaughan JC, Kern ME, Falvo MR, O’Brien ET 3rd, Superfine R, Tuthill JC, Xiang Y, Rogers SL, Parrish JZ. Microtubule Acetylation Is Required for Mechanosensation in Drosophila. Cell reports. 2018;25(4):1051–65 e6. doi: 10.1016/j.celrep.2018.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hummel T, Krukkert K, Roos J, Davis G, Klambt C. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 2000;26(2):357–70. [DOI] [PubMed] [Google Scholar]
- 46.Stone MC, Roegiers F, Rolls MM. Microtubules Have Opposite Orientation in Axons and Dendrites of Drosophila Neurons. Molecular biology of the cell. 2008;19(10):4122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Craig AM, Banker G. Neuronal polarity. Annual review of neuroscience. 1994;17:267–310. [DOI] [PubMed] [Google Scholar]
- 48.Ramkumar A, Jong BY, Ori-McKenney KM. ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule-associated proteins. Developmental dynamics : an official publication of the American Association of Anatomists. 2018;247(1):138–55. doi: 10.1002/dvdy.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gundersen GG, Khawaja S, Bulinski JC. Postpolymerization detyrosination of alpha-tubulin: a mechanism for subcellular differentiation of microtubules. The Journal of cell biology. 1987;105(1):251–64. doi: 10.1083/jcb.105.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cambray-Deakin MA, Burgoyne RD. Acetylated and detyrosinated alpha-tubulins are co-localized in stable microtubules in rat meningeal fibroblasts. Cell Motil Cytoskeleton. 1987;8(3):284–91. doi: 10.1002/cm.970080309. [DOI] [PubMed] [Google Scholar]
- 51.Baas PW, Rao AN, Matamoros AJ, Leo L. Stability properties of neuronal microtubules. Cytoskeleton (Hoboken). 2016;73(9):442–60. doi: 10.1002/cm.21286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao J, Feng C, Rolls MM. The microtubule-severing protein fidgetin acts after dendrite injury to promote their degeneration. Journal of cell science. 2016;129(17):3274–81. doi: 10.1242/jcs.188540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stone MC, Nguyen MM, Tao J, Allender DL, Rolls MM. Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Molecular biology of the cell. 2010;21(5):767–77. doi: E09-11-0967 [pii] 10.1091/mbc.E09-11-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kleele T, Marinkovic P, Williams PR, Stern S, Weigand EE, Engerer P, Naumann R, Hartmann J, Karl RM, Bradke F, Bishop D, Herms J, Konnerth A, Kerschensteiner M, Godinho L, Misgeld T. An assay to image neuronal microtubule dynamics in mice. Nat Commun. 2014;5:4827. doi: 10.1038/ncomms5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baas PW, Black MM. Individual microtubules in the axon consist of domains that differ in both composition and stability. The Journal of cell biology. 1990;111(2):495–509. 10.1083/jcb.111.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmad FJ, Pienkowski TP, Baas PW. Regional differences in microtubule dynamics in the axon. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1993;13(2):856–66. Epub 1993/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakos K, Radler MR, Spiliotis ET. Septin 2/6/7 complexes tune microtubule plus-end growth and EB1 binding in a concentration- and filament-dependent manner. Molecular biology of the cell. 2019;30(23):2913–28. 10.1091/mbc.E19-07-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Riel WE, Rai A, Bianchi S, Katrukha EA, Liu Q, Heck AJ, Hoogenraad CC, Steinmetz MO, Kapitein LC, Akhmanova A. Kinesin-4 KIF21B is a potent microtubule pausing factor. eLife. 2017;6. 10.7554/eLife.24746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, Akhmanova A. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. The Journal of cell biology. 2005;168(1):141–53. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23(7):2655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yau KW, Schatzle P, Tortosa E, Pages S, Holtmaat A, Kapitein LC, Hoogenraad CC. Dendrites In Vitro and In Vivo Contain Microtubules of Opposite Polarity and Axon Formation Correlates with Uniform Plus-End-Out Microtubule Orientation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36(4):1071–85. doi: 10.1523/JNEUROSCI.2430-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bray D, Bunge MB. Serial analysis of microtubules in cultured rat sensory axons. J Neurocytol. 1981;10(4):589–605. doi: 10.1007/bf01262592. [DOI] [PubMed] [Google Scholar]
- 63.Yu W, Baas PW. Changes in microtubule number and length during axon differentiation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1994;14(5 Pt 1):2818–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chalfie M, Thomson JN. Organization of neuronal microtubules in the nematode Caenorhabditis elegans. The Journal of cell biology. 1979;82(1):278–89. doi: 10.1083/jcb.82.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yogev S, Cooper R, Fetter R, Horowitz M, Shen K. Microtubule Organization Determines Axonal Transport Dynamics. Neuron. 2016;92(2):449–60. doi: 10.1016/j.neuron.2016.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kollins KM, Bell RL, Butts M, Withers GS. Dendrites differ from axons in patterns of microtubule stability and polymerization during development. Neural Dev. 2009;4:26. 10.1186/1749-8104-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conde C, Caceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nature reviews Neuroscience. 2009;10(5):319–32. 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- 68.Tas RP, Chazeau A, Cloin BMC, Lambers MLA, Hoogenraad CC, Kapitein LC. Differentiation between Oppositely Oriented Microtubules Controls Polarized Neuronal Transport. Neuron. 2017;96(6):1264–71 e5. doi: 10.1016/j.neuron.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nechipurenko IV, Broihier HT. FoxO limits microtubule stability and is itself negatively regulated by microtubule disruption. The Journal of cell biology. 2012;196(3):345–62. 10.1083/jcb.201105154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Massaro CM, Pielage J, Davis GW. Molecular mechanisms that enhance synapse stability despite persistent disruption of the spectrin/ankyrin/microtubule cytoskeleton. The Journal of cell biology. 2009;187(1):101–17. 10.1083/jcb.200903166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, MacDonald JM, Ziegenfuss JS, Milde S, Hou YJ, Nathan C, Ding A, Brown RH Jr., Conforti L, Coleman M, Tessier-Lavigne M, Zuchner S, Freeman MR. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337(6093):481–4. 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Summers DW, Gibson DA, DiAntonio A, Milbrandt J. SARM1-specific motifs in the TIR domain enable NAD+ loss and regulate injury-induced SARM1 activation. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(41):E6271–E80. doi: 10.1073/pnas.1601506113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, Milbrandt J. The SARM1 Toll/Interleukin-1 Receptor Domain Possesses Intrinsic NAD(+) Cleavage Activity that Promotes Pathological Axonal Degeneration. Neuron. 2017;93(6):1334–43 e5. doi: 10.1016/j.neuron.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33(33):13569–80. doi: 10.1523/JNEUROSCI.1197-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50(6):869–81. [DOI] [PubMed] [Google Scholar]
- 76.Zhai Q, Wang J, Kim A, Liu Q, Watts R, Hoopfer E, Mitchison T, Luo L, He Z. Involvement of the ubiquitin-proteasome system in the early stages of wallerian degeneration. Neuron. 2003;39(2):217–25. [DOI] [PubMed] [Google Scholar]
- 77.Luo L, O’Leary DD. Axon retraction and degeneration in development and disease. Annual review of neuroscience. 2005;28:127–56. [DOI] [PubMed] [Google Scholar]
- 78.Riccomagno MM, Kolodkin AL. Sculpting neural circuits by axon and dendrite pruning. Annu Rev Cell Dev Biol. 2015;31:779–805. doi: 10.1146/annurev-cellbio-100913-013038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maor-Nof M, Homma N, Raanan C, Nof A, Hirokawa N, Yaron A. Axonal Pruning Is Actively Regulated by the Microtubule-Destabilizing Protein Kinesin Superfamily Protein 2A. Cell reports. 2013;3(4):971–7. 10.1016/j.celrep.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 80.Lichtman JW, Colman H. Synapse elimination and indelible memory. Neuron. 2000;25(2):269–78. doi: 10.1016/s0896-6273(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 81.Brill MS, Kleele T, Ruschkies L, Wang M, Marahori NA, Reuter MS, Hausrat TJ, Weigand E, Fisher M, Ahles A, Engelhardt S, Bishop DL, Kneussel M, Misgeld T. Branch-Specific Microtubule Destabilization Mediates Axon Branch Loss during Neuromuscular Synapse Elimination. Neuron. 2016;92(4):845–56. doi: 10.1016/j.neuron.2016.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu F, Schuldiner O. Axon and dendrite pruning in Drosophila. Current opinion in neurobiology. 2014;27:192–8. doi: 10.1016/j.conb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Truman JW. Developmental neuroethology of insect metamorphosis. Journal of neurobiology. 1992;23(10):1404–22. doi: 10.1002/neu.480231005. [DOI] [PubMed] [Google Scholar]
- 84.Tissot M, Stocker RF. Metamorphosis in drosophila and other insects: the fate of neurons throughout the stages. Progress in neurobiology. 2000;62(1):89–111. [DOI] [PubMed] [Google Scholar]
- 85.Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129(12):2867–78. [DOI] [PubMed] [Google Scholar]
- 86.Hughes CL, Thomas JB. A sensory feedback circuit coordinates muscle activity in Drosophila. Molecular and cellular neurosciences. 2007;35(2):383–96. 10.1016/j.mcn.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, Tracey WD. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Current biology : CB. 2007;17(24):2105–16. 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shimono K, Fujimoto A, Tsuyama T, Yamamoto-Kochi M, Sato M, Hattori Y, Sugimura K, Usui T, Kimura K, Uemura T. Multidendritic sensory neurons in the adult Drosophila abdomen: origins, dendritic morphology, and segment- and age-dependent programmed cell death. Neural Dev. 2009;4:37. doi: 1749-8104-4-37 [pii] 10.1186/1749-8104-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38(6):871–85. [DOI] [PubMed] [Google Scholar]
- 90.Williams DW, Truman JW. Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development. 2005;132(16):3631–42. 10.1242/dev.01928. [DOI] [PubMed] [Google Scholar]
- 91.Lee HH, Jan LY, Jan YN. Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(15):6363–8. doi: 0902051106 [pii] 10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Herzmann S, Krumkamp R, Rode S, Kintrup C, Rumpf S. PAR-1 promotes microtubule breakdown during dendrite pruning in Drosophila. The EMBO journal. 2017;36(13):1981–91. doi: 10.15252/embj.201695890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tao J, Rolls MM. Dendrites have a rapid program of injury-induced degeneration that is molecularly distinct from developmental pruning. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(14):5398–405. doi: 31/14/5398 [pii] 10.1523/JNEUROSCI.3826-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murillo B, Mendes Sousa M. Neuronal Intrinsic Regenerative Capacity: The Impact of Microtubule Organization and Axonal Transport. Developmental neurobiology. 2018;78(10):952–9. doi: 10.1002/dneu.22602. [DOI] [PubMed] [Google Scholar]
- 95.Blanquie O, Bradke F. Cytoskeleton dynamics in axon regeneration. Current opinion in neurobiology. 2018;51:60–9. doi: 10.1016/j.conb.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 96.Erez H, Malkinson G, Prager-Khoutorsky M, De Zeeuw CI, Hoogenraad CC, Spira ME. Formation of microtubule-based traps controls the sorting and concentration of vesicles to restricted sites of regenerating neurons after axotomy. The Journal of cell biology. 2007;176(4):497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ghosh-Roy A, Goncharov A, Jin Y, Chisholm AD. Kinesin-13 and tubulin posttranslational modifications regulate microtubule growth in axon regeneration. Developmental cell. 2012;23(4):716–28. 10.1016/j.devcel.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cho Y, Cavalli V. HDAC5 is a novel injury-regulated tubulin deacetylase controlling axon regeneration. The EMBO journal. 2012;31(14):3063–78. 10.1038/emboj.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cho Y, Sloutsky R, Naegle KM, Cavalli V. Injury-Induced HDAC5 Nuclear Export Is Essential for Axon Regeneration. Cell. 2015;161(3):691. doi: 10.1016/j.cell.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 100.Austin TO, Matamoros AJ, Friedman JM, Friedman AJ, Nacharaju P, Yu W, Sharp DJ, Baas PW. Nanoparticle Delivery of Fidgetin siRNA as a Microtubule-based Therapy to Augment Nerve Regeneration. Sci Rep. 2017;7(1):9675. doi: 10.1038/s41598-017-10250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leo L, Yu W, D’Rozario M, Waddell EA, Marenda DR, Baird MA, Davidson MW, Zhou B, Wu B, Baker L, Sharp DJ, Baas PW. Vertebrate Fidgetin Restrains Axonal Growth by Severing Labile Domains of Microtubules. Cell reports. 2015;12(11):1723–30. doi: 10.1016/j.celrep.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Matamoros AJ, Tom VJ, Wu D, Rao Y, Sharp DJ, Baas PW. Knockdown of Fidgetin Improves Regeneration of Injured Axons by a Microtubule-Based Mechanism. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2019;39(11):2011–24. doi: 10.1523/JNEUROSCI.1888-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stone MC, Rao K, Gheres KW, Kim S, Tao J, La Rochelle C, Folker CT, Sherwood NT, Rolls MM. Normal Spastin Gene Dosage Is Specifically Required for Axon Regeneration. Cell reports. 2012. 10.1016/j.celrep.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rao K, Stone MC, Weiner AT, Gheres KW, Zhou C, Deitcher DL, Levitan ES, Rolls MM. Spastin, atlastin, and ER relocalization are involved in axon but not dendrite regeneration. Molecular biology of the cell. 2016;27(21):3245–56. 10.1091/mbc.E16-05-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Erturk A, Hellal F, Enes J, Bradke F. Disorganized microtubules underlie the formation of retraction bulbs and the failure of axonal regeneration. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27(34):9169–80. doi: 27/34/9169 [pii] 10.1523/JNEUROSCI.0612-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. The Journal of cell biology. 2008;180(3):619–32. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hellal F, Hurtado A, Ruschel J, Flynn KC, Laskowski CJ, Umlauf M, Kapitein LC, Strikis D, Lemmon V, Bixby J, Hoogenraad CC, Bradke F. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science. 2011;331(6019):928–31. doi: science.1201148 [pii] 10.1126/science.1201148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ruschel J, Hellal F, Flynn KC, Dupraz S, Elliott DA, Tedeschi A, Bates M, Sliwinski C, Brook G, Dobrindt K, Peitz M, Brustle O, Norenberg MD, Blesch A, Weidner N, Bunge MB, Bixby JL, Bradke F. Axonal regeneration. Systemic administration of epothilone B promotes axon regeneration after spinal cord injury. Science. 2015;348(6232):347–52. doi: 10.1126/science.aaa2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen L, Stone MC, Tao J, Rolls MM. Axon injury and stress trigger a microtubule-based neuroprotective pathway. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(29):11842–47. 10.1073/pnas.1121180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen L, Nye DM, Stone MC, Weiner AT, Gheres KW, Xiong X, Collins CA, Rolls MM. Mitochondria and Caspases Tune Nmnat-Mediated Stabilization to Promote Axon Regeneration. PLoS Genet. 2016;12(12):e1006503. doi: 10.1371/journal.pgen.1006503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu W, Fox P, Lakonishok M, Davidson MW, Gelfand VI. Initial neurite outgrowth in Drosophila neurons is driven by Kinesin-powered microtubule sliding. Current biology : CB. 2013;23(11):1018–23. 10.1016/j.cub.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]


