Abstract
Background:
Lead (Pb) is an environmentally ubiquitous heavy metal associated with a wide range of adverse health effects in children. Both lead exposure and the early life microbiome—which plays a critical role in human development—have been linked to similar health outcomes, but it is unclear if the adverse effects of lead are partially driven by early life gut microbiota dysbiosis. The objective of this study was to examine the association between in utero and postnatal lead levels (measured in deciduous baby teeth) and early life bacterial and fungal gut microbiota in the first year of life.
Methods:
Data from the Wayne County Health, Environment, Allergy and Asthma Longitudinal Study (WHEALS) birth cohort were analyzed. Tooth lead levels during the 2nd and 3rd trimesters and postnatally (<1 year of age) were quantified using high-resolution microspatial mapping of dentin growth rings. Early life microbiota were measured in stool samples collected at approximately 1 and 6 months of age, using both 16S rRNA (bacterial) and ITS2 (fungal) sequencing. Of the 1,258 maternal-child pairs in WHEALS, 146 had data on both tooth metals and early life microbiome.
Results:
In utero tooth lead levels were significantly associated with gut fungal community composition at 1-month of age, where higher levels of 2nd trimester tooth lead was associated with lower abundances of Candida and Aspergillus and higher abundances of Malassezia and Saccharomyces; 3rd trimester lead was also associated with lower abundances of Candida. Though lead did not significantly associate with the overall structure of the infant gut bacterial community, it associated with the abundance of some specific bacterial taxa, including the increased abundance of Collinsella and Bilophila and a decreased abundance of Bacteroides taxa.
Conclusions:
The observed associations between lead exposure and infant gut microbiota could play a role in the impact of lead on childhood development. Given the paucity of research examining these associations in humans—particularly for fungal microbiota—further investigation is needed.
Keywords: lead, microbiome, epidemiology, birth cohort
1. Introduction
The human gut microbiome can be considered an ancillary organ capable of directly influencing host health status (Evans et al. 2013). It produces a vast range of bioactive metabolites that affect metabolic function and is involved in both energy harvest and storage, as well as immune system development (Clemente et al. 2012). Early life gut microbiota in particular are thought to affect health later in life, with dysbiosis associated with an increased risk of allergy and asthma (Johnson and Ownby 2017), obesity (Koleva et al. 2015), and neurodevelopmental outcomes (Sordillo et al. 2019). The human microbiome poses a unique research challenge because, unlike many environmental factors that are analyzed as external exposures, the microbiome exists within us, and thus the human physiology serves as the environment for the microbiota. However, with the advent of high-throughput approaches, we have only recently begun to appreciate the role that microbiota play and have perhaps only scratched the surface. Human microbiomes are dynamic and complex, and studies examining how common epidemiological factors relate to the infant gut microbiota only explain a small percentage of its variability, even when considered jointly (Levin et al. 2016). Therefore, taking into account environmental exposures may help to explain additional variability and improve prediction of infant gut microbiota assembly. In particular, the gut microbiome is influenced by environmental toxicants (Adamovsky et al. 2018). Environmental xenobiotic agents may act directly or indirectly on the microbiome to influence its composition; the gut microbiome may also modulate the influence of a toxic exposure by altering the pharmacokinetics of chemicals (National Academies of Sciences Engineering and Medicine 2018). In short, there is growing recognition of the need to study the role of environmental toxicants on the microbiome (National Academies of Sciences Engineering and Medicine 2018).
Lead (Pb) is considered the most important toxic heavy element and is ubiquitous in the environment, with potential sources of exposure including food, water, air, dust, soil, and lead-based paint (Wani et al. 2015). Upon exposure, Pb bioaccumulates throughout the human body and therefore has been implicated in a wide range of adverse health effects, including neurological, renal, immunological, and developmental (Abadin et al. 2019). There is no safe blood Pb level according to the Centers for Disease Control and Prevention (Betts 2012), particularly for children, in which the toxicity of lead has the largest impact due to high soft tissue absorption (Wani et al. 2015). Both Pb exposure and microbiome composition have been linked to similar health outcomes in humans, including neurological (Abadin et al. 2019; Chu et al. 2019) and immune function (Abadin et al. 2019; Gao et al. 2018), and the microbiome may play a role in mediating the association between lead exposure and health outcomes. Therefore, it is important to understand how lead exposure impacts microbiota development in the infant gut.
In addition to lead exposure in children themselves, it is not yet clear how prenatal exposure to lead may influence the development and trajectory of the human gut microbiome. In a study using a mouse model (Wu et al. 2016), pregnant mice were dosed with lead through drinking water, and their offspring showed altered gut microbiomes and body weight in adulthood, compared to controls. Potential mechanisms of these effects could be through preprogramming of the immune system or by altering offspring microbiome through microbial vertical transmission (either before, during, or after birth). However, it is unknown how prenatal exposures to lead or other environmental toxicants may influence the gut microbiome in humans. This analysis is the first to examine this relationship, focusing on associations between prenatal (2nd and 3rd trimester) and early postnatal (birth to approximately 1 year of age) lead exposure and the human gut microbiome in infancy (bacterial and fungal microbiota at 1 and 6 months of age). Lead levels were measured in naturally shed deciduous teeth, which uniquely allows for retrospective quantification of fetal and early life lead levels with precise windows of exposure.
2. Methods
2.1. Study Information
Analyses were based on data from the Wayne County Health, Environment, Allergy and Asthma Longitudinal Study (WHEALS), a racially-diverse unselected birth cohort of 1,258 maternal-child pairs based in Detroit, Michigan (Wegienka et al. 2015; Wegienka et al. 2016). WHEALS recruited pregnant women with due dates from September 2003 through December 2007, and who were seeing a Henry Ford Health System (HFHS) obstetrics practitioner at one of five clinics to establish an unselected birth cohort (Cassidy-Bushrow et al. 2012; Havstad et al. 2011; Wegienka et al. 2011). All women were in their second trimester or later, were aged 21-49 years, and were living in a predefined geographic area in Wayne and Oakland counties of Michigan that included the city of Detroit as well as the suburban areas immediately surrounding the city. All participants provided written, informed consent. Study protocols were approved by the HFHS Institutional Review Board.
Details of the Tooth Fairy Study, a sub-study of the WHEALS cohort, have previously been reported (Cassidy-Bushrow et al. 2017; Cassidy-Bushrow et al. 2019). Briefly, advertisements for the WHEALS Tooth Fairy Study were placed in study newsletters, a save-the-tooth refrigerator magnet was sent in a holiday mailer to all participants, and families were asked if they wanted to donate a tooth for the study during planned recruitment phone calls for other WHEALS activities. A total of 512 teeth were received from 203 participants between December 2011 and September 2019. Teeth were selected for metal measurement if (1) the child had at least some outcome data available (birth outcomes and/or a 2-year clinic visit) or early life microbiome data; and (2) the tooth sample met laboratory quality control/quality assurance guidelines. A total of 180 participants had tooth metals quantified.
2.2. Stool Specimens
Home visits with participants were conducted targeting infant ages 1 and 6 months. Families were asked to retain the most recent soiled diaper prior to the home visit and stool samples were banked at −80 °C. Detailed information on DNA extraction methods are presented elsewhere (Fujimura et al. 2016).
2.3. Polymerase Chain Reaction Conditions and Library Preparation for Sequencing
The V4 region of the 16S rRNA gene was amplified, as described in detail elsewhere (Caporaso et al. 2012). Briefly, 16S rRNA amplification was performed in 25-μL reactions using 0.025 U Takara Hot Start ExTaq (Takara Mirus Bio Inc., Madison, WI), 1X Takara buffer with MgCl2, 0.4 pmol/μL of F515 and R806 primers, 0.56 mg/mL of bovine serum albumin (Roche Applied Science, Indianapolis, IN), 200 μM of dNTPs and 10 ng of genomic DNA. Reactions were performed in triplicate with the following: initial denaturation (98°C, 2 min), 30 cycles of 98°C (20 sec), annealing at 50°C (30 sec), extension at 72°C (45 sec), and final extension at 72°C (10 min). Amplicons were verified using a 2% Tris/Borate/EDTA agarose e-gel (Life Technologies, Grand Island, NY), cleaned and normalized using SequalPrep Normalization Plates (Applied Biosystems, Foster City, CA), and further quantified using the Qubit 2.0 Fluorometer and the double-stranded DNA HS Assay Kit (Life Technologies). Samples were pooled in equal moles at concentrations of 5 ng, purified using AMPure SPRI beads (Beckman Coulter, Brea, C A), denatured and diluted to 2 nM, and 5 pM was loaded onto the Illumina Nextseq cartridge with 40% (v/v) of denatured 12.5 pM PhiX spike-in control.
The internal transcribed spacer region (ITS) 2 of the rRNA gene was amplified using the primer pair fITS7 (5’- GTGARTCATCGAATCTTTG-3’) and ITS4 (5’-TCCTCCGCTTATT GATATGC-3’). PCR reactions were performed in triplicate in 25 μl reaction with 1X Takara buffer (Takara Mirus Bio), 200 nM of each primer, 200 μM dNTPs, 2.75 mM of MgCl2, 0.56 mg ml-1 of BSA (Roche Applied Science), 0.025 U Takara Hot Start ExTaq and 50 ng of gDNA. Reactions were conducted under the following conditions: initial denaturation (94 °C for 5 min) followed by 30 cycles of 94 °C (30 sec), annealing at 54 °C (30 sec), extension at 72 °C (30 sec) and a final extension at 72 °C (7 min). PCR verification and purification were performed as described above for bacterial library preparation. Samples were quantified using KAPA SYBR (KAPA Biosystems, Wilmington, MA) qPCR as recommended by the manufacturers. Samples were then pooled in equal moles at 5 ng, purified, denatured, and diluted similar to the 16S amplicon library described above. 10pM of the ITS2 amplicon library was loaded onto the Illumina MiSeq cartridge with 25% (v/v) of denatured 10pM PhiX spike-in control.
2.4. Sequence Data Processing and Quality Control
Bacterial paired-end sequences were assembled using FLASH v1.2.7(Magoč and Salzberg 2011) requiring a minimum base pair overlap of 25bp and demultiplexed by barcode using QIIME v1.8 (Caporaso et al. 2010b). Quality filtering was performed using USEARCH v8.0.1623 (Edgar 2010) to remove reads with >2 expected errors. Quality reads were dereplicated at 100% sequence identity, clustered at 97% sequence identity into operational taxonomic units (OTUs), filtered of chimeric sequences by UCHIME (Edgar et al. 2011), and mapped back to resulting OTUs using UPARSE (Edgar 2013); sequence reads that failed to cluster with a reference sequence were clustered de novo. Taxonomy was assigned to the OTUs using the Greengenes database v13_5 (McDonald et al. 2012). Sequences were aligned using PyNAST (Caporaso et al. 2010a), and FastTree 2.1.3 (Price et al. 2009) was used to build a phylogenetic tree. A rarefaction depth of 60,000 reads per sample was selected by using rarefaction curves, based on the plateau of reads and maximizing sample inclusion. Note that those included versus excluded from the current analysis did not have significantly different total read depths (p=0.42), making it unlikely that a different rarefaction depth would be more appropriate if only done within the analysis subset. Additionally, rather than selecting a single rarefaction table, we multiply rarefied the data in order to ensure a representative community was selected for each sample (Fujimura et al. 2016). Briefly, multiple rarefied OTU tables are calculated, and for each sample, the distance between the sample-specific rarefied vectors calculated. The rarefied vector that is the minimum average distance from itself to all other rarefied vectors is selected, as it is considered the most representative for that sample.
Fungal sequences were quality trimmed (Q score, <25) and removed of adaptor sequences using cutadapt (Martin 2011). Paired-end sequences were assembled, demultiplexed by barcode, clustered into OTUs at 97% identity and filtered of chimeras using similar methods as described for 16S amplicons. Taxonomy was assigned using UNITE v7.0 (Koljalg et al. 2013). Resulting sequence reads were normalized by again multiply rarefying to 1,000 reads per sample as described previously to assure representative reduced data. Sequencing data was uploaded to the NCBI Sequence Read Archive (SRA) BioProject PRJNA648818 under the accession number SUB7839619.
A total of 580 children had at least 1 stool sample in the final rarefied OTU table; of these, 146 (25%) had tooth metal levels quantified, providing the final analysis subset sample size (Supplemental Figure 1). Of these 146 children, 43 had 1-month bacterial microbiota only, 35 had 6-month bacterial microbiota only, and 68 had bacterial microbiota at both time points. Sample sizes were smaller for fungal microbiota: as is common in early life cohorts, many samples failed to produce an ITS2 amplicon (53%), with 33 children having 1-month fungal microbiota only, 29 having 6-month fungal microbiota only, and 19 having it at both time points. In the analytical dataset, stool specimens from the 1-month visit were collected at a mean ± SD of 38 ± 16 days (minimum = 16, maximum = 107) and stool specimens from the 6-month visit were collected at a mean ± SD of 205 ± 27 days (minimum = 172, maximum = 290). Throughout, “1 month” and “6 month” are used as labels of the targeted time period of sample collection.
2.5. Covariate Measurement
Maternal race, household income, marital status, exposure to environmental tobacco smoke (ETS), smoking during pregnancy, year home was built, and exposure to indoor pets prenatally were self-reported. Year residence was built was dichotomized as 1980 or after or before 1980, to indicate risk of lead exposure due to lead-based paint (Dixon et al. 2009). Address during pregnancy was recorded and used to define whether the fetus was mainly exposed to an urban residence (defined as within the confines of the city of Detroit) or a suburban residence. Prenatal and delivery records for WHEALS women and children were abstracted to obtain body mass index (BMI) at the first prenatal care visit, delivery type, gestational age at delivery, and birthweight. Breastfeeding status was determined via questionnaires at the 1-month and 6-month study visits.
2.6. Analysis of lead in tooth samples
We directly measured lead in baby teeth using laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) and assigned developmental times as detailed elsewhere (Arora et al. 2014; Arora et al. 2012). Teeth were sectioned and the neonatal line (a histological feature formed in enamel and dentine at the time of birth) and incremental markings were used to assign temporal information to sampling points. Second trimester, third trimester, postnatal (birth through 1-year), and childhood (age 1 to tooth shedding) lead levels were distinguished using previously validated methods that rely on incremental markings in teeth, akin to growth rings in trees (Arora et al. 2014; Arora et al. 2012). The childhood time point was excluded from this analysis because exposure time was after microbiota quantification. We used an ArF excimer laser ablation system (ESI, USA) attached to an Agilent Technologies 8800 triple-quad ICP-MS. Data were analysed as metal-to-calcium ratios (e.g. 208Pb:43Ca) to control for variations in mineral content within a tooth and between samples. Samples were analysed in three batches. Tooth attrition, which is the amount of tooth lost due to grinding or wear, was also measured; teeth with excessive attrition that would impede the chemical analysis are excluded. National Institute of Standards and Technology SRM 612 was used for calibration and quality control. The detection limit was 0.05 μg/g for lead. Across all time points, approximately 2-7% of lead measurements were below the detection limit; values below the detection limit are excluded from statistical analysis.
A small number of children (N=17) had two teeth analyzed for quality control procedures; in these cases, metals levels were averaged over teeth within each child. We have previously shown moderate to excellent agreement for lead concentrations between teeth from the same child (second trimester intraclass correlation coefficients [ICC]=0.55; third trimester ICC=0.74; postnatal ICC=0.87) (Cassidy-Bushrow et al. 2017).
2.7. Statistical Analysis
Tooth lead levels were tested for differences by maternal and child characteristics using the Kruskal-Wallis test for categorical covariates and Spearman correlations for continuous covariates. For these tests, tooth lead levels were shown as metal-to-calcium ratios; for further modelling, lead levels were divided by the standard deviation to represent a one standard deviation increase. Additionally, all tests of the relationship between lead levels and infant gut microbiota were adjusted for tooth type, tooth attrition, tooth batch, exact age at stool sample collection, and child race, for which we have previously shown associations with tooth lead levels (Cassidy-Bushrow et al. 2017) and infant gut microbiota (Levin et al. 2016) in this cohort. Note that sex-specific effects were not explored. In addition to the relatively small sample size, while there is clear evidence of a sex-specific effect of lead on health (Polanska et al. 2018; Wu et al. 2016), there is a lack of evidence of a sex-specific effect of metals on the gut microbiota (Wu et al. 2016).
Infant gut alpha diversity metrics (bacterial: richness, Pielou’s evenness, Faith’s phylogenetic diversity, Shannon’s diversity; fungal: richness, Pielou’s evenness, Shannon’s diversity) were tested for differences in lead levels using covariate-adjusted linear regression models. The association between fungal amplification failure and lead levels was modelled using covariate-adjusted logistic regression models. Adjusted permutational multivariate analysis of variance (PERMANOVA) was used to test for infant gut compositional differences by lead levels (Anderson 2001) using the R package vegan (Oksanen et al. 2019). In order to capture different features of community composition, both phylogenetic [bacterial only: unweighted and weighted UniFrac (Lozupone et al. 2011)] and non-phylogenetic [bacterial and fungal: Bray- Curtis, Canberra] distances were used in PERMANOVA tests. Principal coordinates analysis (PCoA) was performed on these distances using the R package labdsv (Roberts 2019); plots of these distances are provided in Supplemental Figure 2.
Tests of differential genera and OTU abundance were performed using covariate-adjusted zero-inflated negative binomial regression using the R package pscl (Zeileis et al. 2008); in cases where zero-inflated models failed to converge, the standard negative binomial was implemented as a secondary modelling strategy using the R package MASS (Venables and Ripley 2002). Only genera and OTUs found in 10% or more of samples were tested. Statistical significance was assessed after accounting for multiple testing using the Benjamini & Hochberg false discovery rate (FDR) adjustment (Benjamini and Hochberg 1995), with pFDR<0.05 considered significant. Analyses were performed using SAS 9.4 and R 3.6.1. Covariate and lead data are provided in Supplemental Data 1.
3. Results
3.1. Factors associated with lead exposure
In order to describe the data and assess potential confounders for the main association of interest (i.e., between lead levels and early life microbiota), lead levels were first compared across a variety of maternal and child characteristics (Table 1). Location of residence, prenatal indoor pets, birthweight z-score, and child race were all significantly associated with lead levels. Specifically, black children (all p<0.001) and children living in urban residences had significantly higher lead levels at all three time points (p=0.024, 0.005, 0.003, respectively). Children whose mothers had a pet during pregnancy had significantly lower lead levels, but only during the 3rd trimester (p=0.019). Higher lead levels at all three time points were also associated with lower birthweight z-scores (p=0.021, 0.019, 0.036, respectively). After adjusting for race, however, associations of tooth lead level with location of residence and prenatal pets was diminished and no longer significant (all p≥0.067). Of note, no associations were observed between lead levels and mode of delivery or breastfeeding status, which we have previously shown have a large impact on infant gut microbiota in these children (Levin et al. 2016).
Table 1:
Maternal and child characteristics by in utero and postnatal lead levels (N=146)
| Variable | Level | 2nd Trimester Leada | 3rd Trimester Leada | Postnatal Leada | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Median (IQR) or p |
p-valueb | N | Median (IQR) or p |
p-valueb | N | Median (IQR) or p |
p-valueb | ||
| Maternal education | dHS diploma | 13 | 0.024 (0.018) | 0.26 | 13 | 0.026 (0.013) | 0.087 | 13 | 0.023 (0.026) | 0.077 |
| Some college | 61 | 0.027 (0.037) | 64 | 0.033 (0.047) | 65 | 0.030 (0.029) | ||||
| ≥Bachelor’s Degree | 62 | 0.022 (0.026) | 65 | 0.031 (0.034) | 65 | 0.024 (0.030) | ||||
| Household income | <$20,000 | 9 | 0.024 (0.011) | 0.98 | 9 | 0.033 (0.030) | 0.89 | 9 | 0.027 (0.021) | 0.49 |
| $20,000-<$40,000 | 24 | 0.026 (0.032) | 25 | 0.032 (0.033) | 26 | 0.028 (0.031) | ||||
| $40,000-<$80,000 | 40 | 0.029 (0.024) | 44 | 0.035 (0.040) | 45 | 0.032 (0.036) | ||||
| $80,000-<$ 100,000 | 25 | 0.021 (0.030) | 25 | 0.027 (0.041) | 24 | 0.022 (0.024) | ||||
| ≥$100,000 | 23 | 0.021 (0.028) | 24 | 0.032 (0.034) | 24 | 0.025 (0.028) | ||||
| Refused to Answer | 15 | 0.020 (0.053) | 15 | 0.028 (0.045) | 15 | 0.022 (0.046) | ||||
| Married | No | 36 | 0.028 (0.051) | 0.089 | 38 | 0.034 (0.066) | 0.14 | 39 | 0.034 (0.040) | 0.063 |
| Yes | 100 | 0.023 (0.026) | 104 | 0.030 (0.033) | 104 | 0.024 (0.026) | ||||
| Location of residence | Suburban | 85 | 0.021 (0.026) | 0.024 | 88 | 0.027 (0.030) | 0.005 | 89 | 0.022 (0.028) | 0.003 |
| Urban | 51 | 0.028 (0.039) | 54 | 0.041 (0.056) | 54 | 0.033 (0.027) | ||||
| Maternal age at birth | --- | 136 | 0.056 | 0.52 | 142 | 0.094 | 0.27 | 143 | 0.054 | 0.52 |
| Maternal BMI at first prenatal visit | --- | 135 | −0.014 | 0.87 | 141 | 0.031 | 0.72 | 142 | 0.064 | 0.45 |
| Mother Atopic | No | 62 | 0.025 (0.037) | 0.79 | 64 | 0.032 (0.047) | 0.69 | 66 | 0.024 (0.035) | 0.55 |
| Yes | 71 | 0.024 (0.026) | 75 | 0.030 (0.033) | 74 | 0.029 (0.023) | ||||
| Prenatal antibiotic use | No | 54 | 0.023 (0.027) | 0.37 | 57 | 0.033 (0.040) | 0.74 | 57 | 0.024 (0.027) | 0.16 |
| Yes | 77 | 0.025 (0.032) | 78 | 0.031 (0.044) | 79 | 0.029 (0.032) | ||||
| Prenatal antifungal use | No | 108 | 0.026 (0.029) | 0.20 | 112 | 0.033 (0.045) | 0.18 | 113 | 0.029 (0.031) | 0.097 |
| Yes | 23 | 0.024 (0.030) | 23 | 0.026 (0.025) | 23 | 0.021 (0.022) | ||||
| Mother smoked during pregnancy | No | 127 | 0.024 (0.028) | 0.33 | 133 | 0.031 (0.038) | 0.41 | 134 | 0.027 (0.028) | 0.84 |
| Yes | 9 | 0.035 (0.045) | 9 | 0.042 (0.075) | 9 | 0.024 (0.033) | ||||
| ETS during pregnancy | No | 107 | 0.024 (0.026) | 0.13 | 112 | 0.030 (0.037) | 0.18 | 112 | 0.027 (0.028) | 0.34 |
| Yes | 29 | 0.035 (0.044) | 30 | 0.039 (0.059) | 31 | 0.027 (0.033) | ||||
| Prenatal indoor pets | No | 76 | 0.029 (0.042) | 0.058 | 80 | 0.038 (0.048) | 0.019 | 82 | 0.028 (0.030) | 0.19 |
| Yes | 60 | 0.021 (0.020) | 62 | 0.026 (0.024) | 61 | 0.024 (0.027) | ||||
| Year House was Built | 1980 or after | 26 | 0.023 (0.030) | 0.40 | 28 | 0.026 (0.036) | 0.16 | 28 | 0.022 (0.024) | 0.099 |
| Before 1980 | 104 | 0.024 (0.028) | 108 | 0.032 (0.038) | 109 | 0.027 (0.031) | ||||
| First born child | No | 79 | 0.025 (0.019) | 0.90 | 83 | 0.032 (0.032) | 0.64 | 85 | 0.027 (0.027) | 0.81 |
| Yes | 57 | 0.024 (0.033) | 59 | 0.030 (0.051) | 58 | 0.027 (0.031) | ||||
| Season of birth | Winter | 29 | 0.022 (0.022) | 0.52 | 30 | 0.029 (0.035) | 0.65 | 31 | 0.029 (0.033) | 0.88 |
| Spring | 34 | 0.029 (0.048) | 36 | 0.034 (0.058) | 37 | 0.025 (0.039) | ||||
| Summer | 35 | 0.021 (0.028) | 37 | 0.030 (0.032) | 37 | 0.024 (0.026) | ||||
| Fall | 38 | 0.026 (0.032) | 39 | 0.032 (0.045) | 38 | 0.029 (0.021) | ||||
| Mode of delivery | Vaginal | 83 | 0.023 (0.026) | 0.31 | 87 | 0.029 (0.034) | 0.29 | 88 | 0.026 (0.026) | 0.49 |
| C-Section | 52 | 0.028 (0.033) | 54 | 0.034 (0.045) | 54 | 0.028 (0.030) | ||||
| Gestational age at birth | --- | 133 | 0.013 | 0.88 | 142 | −0.023 | 0.79 | 139 | 0.01 | 0.91 |
| Birthweight z-score | --- | 124 | −0.208 | 129 | −0.207 | 0.019 | 131 | −0.184 | 0.036 | |
| Child race | White | 50 | 0.018 (0.018) | <0.001 | 52 | 0.022 (0.025) | <0.001 | 52 | 0.020 (0.019) | <0.001 |
| Black | 56 | 0.031 (0.050) | 58 | 0.044 (0.061) | 59 | 0.032 (0.033) | ||||
| Other/Mixed | 30 | 0.024 (0.022) | 32 | 0.031 (0.041) | 32 | 0.031 (0.032) | ||||
| Child sex | Male | 66 | 0.029 (0.026) | 0.072 | 69 | 0.036 (0.039) | 0.065 | 70 | 0.029 (0.020) | 0.33 |
| Female | 70 | 0.021 (0.026) | 73 | 0.027 (0.035) | 73 | 0.024 (0.036) | ||||
| Breastfeeding status at 1-month | Formula Feeding | 25 | 0.032 (0.030) | 0.32 | 26 | 0.042 (0.048) | 0.58 | 26 | 0.028 (0.033) | 0.60 |
| Mixed Feeding | 81 | 0.023 (0.028) | 85 | 0.031 (0.049) | 87 | 0.027 (0.029) | ||||
| Exclusive Breastfeed | 30 | 0.029 (0.025) | 31 | 0.029 (0.023) | 30 | 0.026 (0.031) | ||||
| Breastfeeding status at 6-months | Formula Feeding | 25 | 0.032 (0.030) | 0.38 | 26 | 0.042 (0.048) | 0.58 | 26 | 0.028 (0.033) | 0.93 |
| Mixed Feeding | 95 | 0.024 (0.027) | 99 | 0.031 (0.042) | 100 | 0.027 (0.026) | ||||
| Exclusive Breastfeed | 16 | 0.024 (0.023) | 17 | 0.026 (0.029) | 17 | 0.025 (0.033) | ||||
tooth lead levels shown as metal-to-calcium ratios.
Kruskal-Wallis or Spearman correlation p-value.
3.2. Association between lead exposure and bacterial gut microbiota
We next evaluated the association between lead levels and infant gut bacterial alpha diversity metrics (richness, evenness, phylogenetic/Shannon's diversity), a high-level summary statistic of microbiota structure. After adjusting for tooth type, attrition, batch, exact age at stool sample collection, and child race, in utero and postnatal lead levels did not significantly associate with bacterial alpha diversity, at both 1 and 6 months of age (Table 2). When microbiota beta diversity (microbiota composition) was considered, lead did not significantly explain the overall variability in bacterial microbiota composition, after adjusting for covariates (Table 3). Though lead exposure did not appear to profoundly influence the structure or composition of the early life bacterial gut microbiome, it may nevertheless relate to the presence and abundance of specific bacterial taxa. Therefore, we also performed genus (Figure 1) and OTU (Supplemental Table 1) tests of differences by tooth lead levels. Of note, at the genus level, 2nd and 3rd trimester lead levels were positively associated with Collinsella abundance at 1-month of age (pFDR<0.001, 0.022, respectively), as well as Bilophila abundance at 6-months of age (pFDR=0.023, 0.008, respectively) (Figure 1). At the OTU level, the positive association between lead levels and Collinsella abundance appeared to be driven by four Collinsella aerofaciens OTUs; lead levels also positively associated with two Bilophila OTUs, but these did not have classification available beyond genus (Supplemental Table 1). Additionally, in utero and postnatal lead levels negatively associated with several Bacteroides OTUs, at both 1 and 6 months of age (Supplemental Table 1).
Table 2:
Association between tooth lead levels and alpha diversity metrics.
| Time of Metal Measurement |
Outcome | Time of Stool Measurement |
Bacterial | Fungal | ||||
|---|---|---|---|---|---|---|---|---|
| N | β (SE)b | p-value | N | β (SE)b | p-value | |||
| 2nd trimester | Richness | 1-Month | 104 | 5.53 (6.98) | 0.43 | 48 | 0.29 (1.65) | 0.86 |
| 6-Months | 96 | −7.77 (7.31) | 0.29 | 45 | 1.7 (1.51) | 0.27 | ||
| Evenness | 1-Month | 104 | 0 (0.01) | 0.97 | 37 | 0.03 (0.05) | 0.56 | |
| 6-Months | 96 | −0.02 (0.01) | 0.092 | 40 | −0.02 (0.05) | 0.73 | ||
| Faith’s Diversitya | 1-Month | 104 | 0.16 (0.39) | 0.67 | ||||
| 6-Months | 96 | −0.19 (0.37) | 0.61 | |||||
| Shannon’s Diversity | 1-Month | 104 | 0.01 (0.08) | 0.89 | 48 | 0.06 (0.15) | 0.67 | |
| 6-Months | 96 | −0.11 (0.07) | 0.11 | 45 | 0 (0.14) | 0.99 | ||
| 3rd trimester | Richness | 1-Month | 107 | 2.52 (6.37) | 0.69 | 49 | 0.69 (1.82) | 0.71 |
| 6-Months | 101 | −13.11 (8.36) | 0.12 | 47 | 2.54 (1.56) | 0.11 | ||
| Evenness | 1-Month | 107 | −0.01 (0.01) | 0.43 | 38 | 0.03 (0.05) | 0.52 | |
| 6-Months | 101 | −0.02 (0.01) | 0.19 | 42 | 0.03 (0.05) | 0.58 | ||
| Faith’s Diversitya | 1-Month | 107 | 0.03 (0.35) | 0.93 | ||||
| 6-Months | 101 | −0.52 (0.42) | 0.22 | |||||
| Shannon’s Diversity | 1-Month | 107 | -0.05 (0.07) | 0.50 | 49 | 0.09 (0.16) | 0.57 | |
| 6-Months | 101 | −0.12 (0.08) | 0.15 | 47 | 0.15 (0.15) | 0.34 | ||
| Postnatal | Richness | 1-Month | 109 | 2.18 (7.16) | 0.76 | 50 | −1.85 (2.54) | 0.47 |
| 6-Months | 100 | −2.55 (6.42) | 0.69 | 46 | −0.35 (1.05) | 0.74 | ||
| Evenness | 1-Month | 109 | −0.02 (0.01) | 0.21 | 39 | 0.07 (0.1) | 0.47 | |
| 6-Months | 100 | −0.01 (0.01) | 0.40 | 41 | 0.06 (0.06) | 0.36 | ||
| Faith’s Diversitya | 1-Month | 109 | −0.08 (0.39) | 0.84 | ||||
| 6-Months | 100 | 0.11 (0.32) | 0.74 | |||||
| Shannon’s Diversity | 1-Month | 109 | −0.1 (0.08) | 0.22 | 50 | −0.07 (0.23) | 0.78 | |
| 6-Months | 100 | −0.05 (0.06) | 0.46 | 46 | −0.05 (0.1) | 0.59 | ||
Faith’s phylogenetic diversity not able to be calculated for fungal data as no phylogenetic tree can be derived.
linear regression coefficient, which represents the change in alpha diversity metric, for a 1-SD increase in tooth lead level, after adjusting for tooth type, attrition, batch, exact age at stool sample collection, and child race.
Table 3:
Association between tooth lead levels and beta diversity metrics.
| Time of Metal | Outcome | Time of Stool Measurement | PERMANOVAa | ||
|---|---|---|---|---|---|
| N | R2 | p-value | |||
| Bacterial | |||||
| 2nd trimester | Unweighted UniFrac | 1-Month | 104 | 0.007 | 0.87 |
| 6-Months | 96 | 0.011 | 0.32 | ||
| Weighted UniFrac | 1-Month | 104 | 0.012 | 0.28 | |
| 6-Months | 96 | 0.006 | 0.70 | ||
| Canberra | 1-Month | 104 | 0.009 | 0.70 | |
| 6-Months | 96 | 0.011 | 0.46 | ||
| Bray-Curtis | 1-Month | 104 | 0.010 | 0.40 | |
| 6-Months | 96 | 0.013 | 0.21 | ||
| 3rd trimester | Unweighted UniFrac | 1-Month | 107 | 0.008 | 0.75 |
| 6-Months | 101 | 0.011 | 0.25 | ||
| Weighted UniFrac | 1-Month | 107 | 0.008 | 0.47 | |
| 6-Months | 101 | 0.006 | 0.66 | ||
| Canberra | 1-Month | 107 | 0.009 | 0.66 | |
| 6-Months | 101 | 0.010 | 0.33 | ||
| Bray-Curtis | 1-Month | 107 | 0.006 | 0.80 | |
| 6-Months | 101 | 0.009 | 0.55 | ||
| Postnatal | Unweighted UniFrac | 1-Month | 109 | 0.008 | 0.64 |
| 6-Months | 100 | 0.008 | 0.84 | ||
| Weighted UniFrac | 1-Month | 109 | 0.008 | 0.49 | |
| 6-Months | 100 | 0.006 | 0.75 | ||
| Canberra | 1-Month | 109 | 0.008 | 0.84 | |
| 6-Months | 100 | 0.010 | 0.59 | ||
| Bray-Curtis | 1-Month | 109 | 0.007 | 0.75 | |
| 6-Months | 100 | 0.009 | 0.58 | ||
| Fungalb | |||||
| 2nd trimester | Canberra | 1-Month | 48 | 0.018 | 0.59 |
| 6-Months | 45 | 0.016 | 0.94 | ||
| Bray-Curtis | 1-Month | 48 | 0.040 | 0.048 | |
| 6-Months | 45 | 0.014 | 0.65 | ||
| 3rd trimester | Canberra | 1-Month | 49 | 0.019 | 0.51 |
| 6-Months | 47 | 0.014 | 0.96 | ||
| Bray-Curtis | 1-Month | 49 | 0.039 | 0.049 | |
| 6-Months | 47 | 0.006 | 0.94 | ||
| Postnatal | Canberra | 1-Month | 50 | 0.016 | 0.78 |
| 6-Months | 46 | 0.021 | 0.52 | ||
| Bray-Curtis | 1-Month | 50 | 0.021 | 0.39 | |
| 6-Months | 46 | 0.028 | 0.22 | ||
adjusted for tooth type, attrition, batch, exact age at stool sample collection, and child race.
UniFrac metrics not able to be calculated for fungal data as no phylogenetic tree can be derived.
Figure 1:
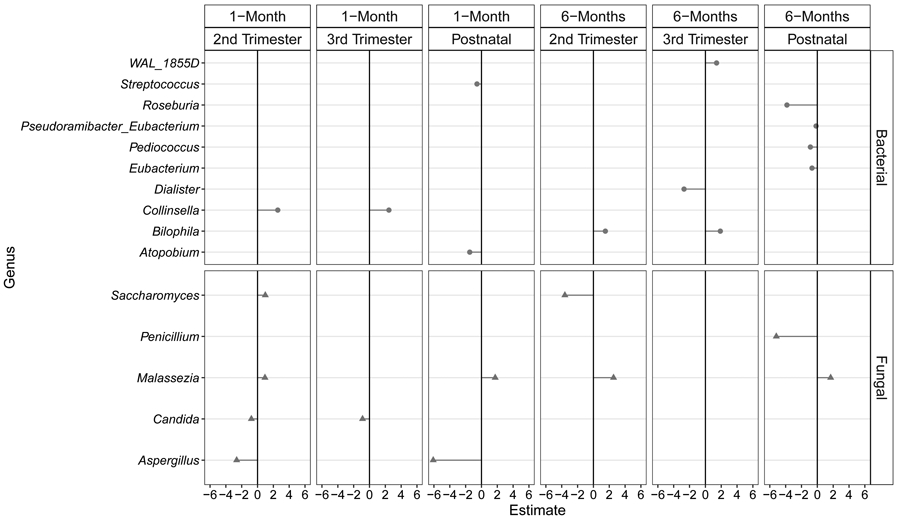
Bacterial and fungal genera significantly associated with in utero and postnatal lead levels (pFDR<0.05). Estimates shown are coefficients from zero-inflated negative binomial models or standard negative binomial models, after adjusting for tooth type, attrition, batch, exact age at stool sample collection, and child race. These can be interpreted as the change in log genera abundance, for a 1-standard deviation increase in lead levels.
3.3. Association between lead exposure and fungal gut microbiota
After covariate adjustment, lead levels did not significantly associate with fungal alpha diversity, at both 1 and 6 months of age (Table 2). In tests of the association between tooth lead levels and fungal amplification failure, no associations were detected after covariate adjustment (all p≥0.16). However, 2nd trimester and 3rd trimester lead levels significantly associated with fungal community composition at 1-month of age (Table 3; Bray-Curtis p-value=0.048, 0.049, respectively), with the association explaining approximately 4% of the variability in community composition.
In tests of differential fungal genera abundance by tooth lead levels (after covariate adjustment and FDR correction), in utero and postnatal lead levels significantly associated with a total of five genera (Figure 1): Candida, Malassezia, Penicillinm, Saccharomyces, and Aspergillus. Specifically, higher tooth lead levels in the 2nd and 3rd trimesters were associated with significantly lower Candida abundances at 1-month of age (PFDR=0.006, 0.003, respectively); 2nd trimester and postnatal lead levels also negatively correlated with Aspergillus abundance at 1-month of age (Figure 1; PFDR=0.002, <0.001, respectively). Additionally, postnatal lead levels negatively correlated with Penicillium abundance at 6-months of age (Figure 1; PFDR=0.002). In contrast, higher tooth lead levels in the 2nd trimester and postnatally were associated with significantly higher Malassezia abundances at both 1-month (Figure 1; PFDR=0.002, 0.013, respectively) and 6-months of age (Figure 1; both PFDR<0.001). Additionally, higher tooth lead levels in the 2nd trimester was associated with significantly higher Saccharomyces abundance at 1-month of age (PFDR=0.006), but significantly lower Saccharomyces abundance at 6-months of age (Figure 1; PFDR=0.006). Tests of specific fungal OTUs (Supplemental Table 1) suggested that the positive association between lead and Malassezia is primarily driven by Malassezia restricta and Malassezia globosa OTUs, whereas the negative association with Candida appears to be primarily driven by a Candida parapsilosis OTU.
4. Discussion
Our study identified an association between tooth lead levels and overall gut fungal community composition at 1-month of age, where lead levels were negatively correlated with the abundance of Candida parapsilosis, which has been shown to be sensitive to lead (Bansal et al. 2019), and positively correlated with the abundance of Malassezia restricta and M. globosa as well as Saccharomyces species, which has been shown to be lead-resistant (Jaroslawiecka and Piotrowska-Seget 2014). Interestingly, compositional differences were only found for in utero but not postnatal lead levels, highlighting the advantage of capturing multiple exposure time points both before and after birth using novel tooth-matrix biomarkers. In contrast, we did not detect an association between in utero or early life tooth lead levels and overall infant gut bacterial community composition, though specific bacterial taxa including Collinsella, Bilophila, and Bacteroides were identified after covariate adjustment and FDR correction.
Several animal studies have shown that lead is associated with the bacterial gut microbiome. In female C57BL/6 mice (~7 weeks old), those exposed to lead in their drinking water (10 ppm) for 13 weeks had significantly different alpha and beta diversity trajectories of the gut microbiome compared to control mice (Gao et al. 2017). Mice (~6 week old female Balb/C) treated for 8 weeks with cadmium or lead had statistically significant lower levels of Lachnospiraceae and higher numbers of Lactobacillaceae and Erysipelotrichaceacae in their gut than control animals. Finally, Wu et al. examined the impact of maternal lead exposure in drinking water (exposed pre-conception through lactation) on the gut microbiome of the adult offspring of non-agouti wild-type mice (Wu et al. 2016). While there were no differences in the richness or diversity of the exposed offspring compared to control mice, Bacteroidetes were significantly reduced while Firmicutes and Desulfovibrionales were significantly increased in the mice exposed to lead through their mothers (Wu et al. 2016). Consistent with these findings, we also observed a significant reduction of specific Bacteroides OTUs as well as a significant increase in Bilophila (which is of the order Desulfovibrionales) with higher levels of lead exposure; of note, Desulfovibrio desulfuricans has been shown to be resistant to Pb (Sani et al. 2003).
In addition to animal models, a small number of studies have examined the association between lead levels and bacterial community composition in humans. One such study was a randomized trial aimed to evaluate the effectiveness of a probiotic yogurt in lowering heavy metal levels among at-risk pregnant women and school-aged children in Tanzania (Bisanz et al. 2014). In the children, they found that elevated blood lead was associated with the increased abundance of Succinivibrionaceae and Gammaproteobacteria (Bisanz et al. 2014). While no OTUs were classified as Succinivibrionaceae in the current study, a total of 10 Gammaproteobacteria OTUs were significantly associated with lead levels, but the direction of association depended on more refined taxonomic classification (Supplemental Table 1). Specifically, Acinetobacter and Xanthomonadaceae OTUs negatively associated with lead levels, while Enterobacteriaceae OTUs were primarily positively associated with lead levels. A study by Eggers et al. examined urinary lead concentration in relation to gut microbiota composition in US adults (Eggers et al. 2019), finding that lead levels were significantly associated with increased bacterial richness and an increased abundance of Proteobacteria. In addition to observing an increased abundance of Bilophila with lead exposure (which is of the phylum Proteobacteria), we also identified 9 Proteobacteria OTUs significantly and positively associated with lead levels (Supplemental Table 1), providing support for this finding.
A comparison of our finding that lead associates with fungal gut microbiota to that of others is limited due to the paucity of published epidemiological studies. In fact, to our knowledge, no other studies have examined the association between lead levels and fungal community composition in humans. More work has been done in environmental studies, however. For example, in an observational study of how lead levels impact native communities of fungi in soil surrounding an abandoned lead smelting factory, lead levels were associated with decreased fungal richness and distinct community composition (Faggioli et al. 2019); bacterial community composition was not examined. A similar study examining both the bacteria and fungi in the soil of a shooting range heavily polluted by lead found that lead did not alter bacterial microbiota, but was associated with clear changes in the fungal community structure relative to controls (Hui et al. 2012). They also noted significantly higher fungal diversity in the polluted samples, in contrast to the previous study. The authors hypothesized that bacteria may be more capable of avoiding or adapting to lead exposure by acquiring new traits, whereas replacement may be the typical strategy for fungi (Hui et al. 2012). In a study of experimental rats, yeast levels were significantly decreased following lead exposure; however, yeast levels were determined using a culture-based approach rather than using high-throughput methods (Reddy et al. 2018).
The observed associations between lead exposure and infant gut microbiota could play a role in the impact of lead on childhood development. For example, lead exposure has been implicated as a risk factor for neurocognitive disorders, including Autism Spectrum Disorder (ASD) (Kim et al. 2016; Lakshmi Priya and Geetha 2011). More recently, studies are beginning to investigate how gut microbiota can affect neurodevelopment disorders through the “gut-brain axis”, in which signals from intestinal microbiota traffic to the central nervous system through neural, endocrine, and immune pathways (Sampson and Mazmanian 2015). Indeed, a study comparing the gut microbiota of 40 individuals with ASD to that of 40 neurotypical controls found that individuals with ASD had a reduced abundance of Bacteroidetes and an increased abundance of Collinsella (Strati et al. 2017). Given that we found that lead exposure associates with reduced Bacteroidetes abundance and increased Collinsella abundance, it is plausible that these microbiota lie in the causal pathway between lead exposure and ASD. A mechanistic study using a mouse model of ASD provides further support for this hypothesis, as it demonstrated that oral treatment of Bacteroides fragilis corrected autism-related behavioral impairments and gut dysbiosis (Hsiao et al. 2013). The gut microbiome may play a similar role in other disorders affected by lead exposure.
Our study is not without limitations. Given the observational nature of this study, results are associative and not necessarily causative. However, two of the three tooth lead measurements were quantified prior to birth and therefore preceded the measurement of infant microbiota composition. Though birth is a central driver of infant gut microbiota colonization (Milani et al. 2017), there is evidence suggesting that highly limited, but viable bacteria are present in the fetal intestine at mid-gestation (Rackaityte et al. 2020). Thus, fetal lead exposure may alter early life microbiota either directly or through the fetal microbiota; however, reverse causation cannot be ruled out, as it is unknown if fetal microbiota can impact the uptake of lead by the fetus. Indeed, in addition to previous studies demonstrating that lead can directly alter microbiota composition (Gao et al. 2017), it is also known that microbes can act as bioremediators, capable of directly binding to, detoxifying, and preventing the absorption of lead (Monachese et al. 2012; Tian et al. 2012). Additionally, lead has been detected in breast milk samples (Vollset et al. 2019) and therefore may also impact microbial communities; however, this could not be assessed as breast milk lead levels were not measured in the WHEALS cohort (though, the postnatal tooth measurement may capture this exposure).
The small sample size used in this analysis—particularly for fungal microbiota—limited the ability to extensively adjust for covariates and may result in spurious findings. Though we adjusted for race—which is highly associated with both lead levels and microbiota composition and explained away the association between other hypothesized confounders and lead levels—residual confounding is still possible. In fact, the only covariate examined that remained associated with lead exposure after adjusting for race was birthweight z-score; however, we posit that this is a potential mediator rather than a confounder, as birth outcomes may be directly affected by lead exposure (Taylor et al. 2015) and lie within the causal pathway. Lastly, metagenomic sequencing was not performed on the infant stool samples, so the functional capacity of organisms was not quantified.
5. Conclusions
Using a U.S. birth cohort of racially diverse children, we identified an association between in utero tooth lead levels and gut fungal community composition at 1-month of age, where higher levels of 2nd trimester tooth lead was associated with lower abundances of Candida and Aspergillus and higher abundances of Malassezia and Saccharomyces. Third trimester lead was also associated with lower abundances of Candida. Though lead did not significantly impact the overall structure of the infant gut bacterial community, it associated with the abundance of some specific bacterial taxa, including an increase in Collinsella and Bilophila, as well as a decrease in Bacteroides taxa. Additional studies are needed to confirm these findings, ideally in both epidemiological studies of humans as well as animal models.
Supplementary Material
Highlights.
We examined associations of Pb (measured in baby teeth) and infant gut microbiota.
Pb was measured before and after birth; stool was collected at 1- and 6-months.
Fetal Pb associated with 1-month Candida, Aspergillus, Malassezia, and Saccharomyces.
Pb did not associate with overall structure of infant gut bacterial communities.
Yet, some bacteria (e.g., Collinsella, Bilophila, Bacteroides) associated with Pb.
Acknowledgements:
The authors would like to acknowledge the continued dedicated participation of the WHEALS families in this longstanding birth cohort study.
Funding: This work was supported by the National Institutes of Health [grant numbers R21 ES022321, R01 HD082147, P01 AI089473, R01 AI050681, R01 HL113010]; and the Fund for Henry Ford Hospital. Study sponsors had no role in the study design, data collection or interpretation, or in the writing of this manuscript.
Abbreviations:
- Pb
Lead
- WHEALS
Wayne County Health, Environment, Allergy and Asthma Longitudinal Study
- HFHS
Henry Ford Health System
- ITS
Internal Transcribed Spacer Region
- OTU
Operational Taxonomic Unit
- BMI
Body Mass Index
- ETS
Environmental Tobacco Smoke
- ICC
Intraclass Correlation Coefficient
- PERMANOVA
Permutational Multivariate Analysis of Variance
- FDR
False Discovery Rate
- ASD
Autism Spectrum Disorder
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abadin H; Taylor J; Buser MC; Scinicariello F; Przybyla I; Klotzbach JM; Diamond GL; Chappell LL; Mcllroy LA Toxicological profile for lead: draft for public comment. 2019; [Google Scholar]
- Adamovsky O; Buerger AN; Wormington AM; Ector N; Griffith RJ; Bisesi JH Jr.; Martyniuk CJ The gut microbiome and aquatic toxicology: An emerging concept for environmental health. Environmental toxicology and chemistry 2018;37:2758–2775 [DOI] [PubMed] [Google Scholar]
- Anderson MJ A new method for non-parametric multivariate analysis of variance. Austral ecology 2001;26:32–46 [Google Scholar]
- Arora M; Austin C; Sarrafpour B; Hernandez-Avila M; Hu H; Wright RO; Tellez-Rojo MM Determining prenatal, early childhood and cumulative long-term lead exposure using micro-spatial deciduous dentine levels. PLoS One 2014;9:e97805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora M; Bradman A; Austin C; Vedar M; Holland N; Eskenazi B; Smith DR Determining fetal manganese exposure from mantle dentine of deciduous teeth. Environmental science & technology 2012;46:5118–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S; Garg M; Chatteijee M Evaluation of heavy metal resistance profile of Candida parapsilosis. Indian J Biotechnol 2019; [Google Scholar]
- Benjamini Y; Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological) 1995:289–300 [Google Scholar]
- Betts KS CDC updates guidelines for children's lead exposure. Environ Health Perspect 2012;120:a268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisanz JE; Enos MK; Mwanga JR; Changalucha J; Burton JP; Gloor GB; Reid G Randomized open-label pilot study of the influence of probiotics and the gut microbiome on toxic metal levels in Tanzanian pregnant women and school children. MBio 2014;5:e01580–01514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG; Bittinger K; Bushman FD; DeSantis TZ; Andersen GL; Knight R PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010a;26:266–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG; Kuczynski J; Stombaugh J; Bittinger K; Bushman FD; Costello EK; Fierer N; Pena AG; Goodrich JK; Gordon JI; Huttley GA; Kelley ST; Knights D; Koenig JE; Ley RE; Lozupone CA; McDonald D; Muegge BD; Pirrung M; Reeder J; Sevinsky JR; Turnbaugh PJ; Walters WA; Widmann J; Yatsunenko T; Zaneveld J; Knight R QIIME allows analysis of high-throughput community sequencing data. Nature methods 2010b;7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG; Lauber CL; Walters WA; Berg-Lyons D; Huntley J; Fierer N; Owens SM; Betley J; Fraser L; Bauer M Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal 2012;6:1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Bushrow A; Wegienka G; Barone C; Valentini R; Yee J; Havstad S; Johnson C Race-specific relationship of birth weight and renal function among healthy young children. Pediatric nephrology (Berlin, Germany) 2012;27:1317–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Bushrow AE; Sitarik AR; Havstad S; Park SK; Bielak LF; Austin C; Johnson CC; Arora M Burden of higher lead exposure in African-Americans starts in utero and persists into childhood. Environ Int 2017;108:221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Bushrow AE; Wu KH; Sitarik AR; Park SK; Bielak LF; Austin C; Gennings C; Curtin P; Johnson CC; Arora M In utero metal exposures measured in deciduous teeth and birth outcomes in a racially-diverse urban cohort. Environ Res 2019;171:444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C; Murdock MH; Jing D; Won TH; Chung H; Kressel AM; Tsaava T; Addorisio ME; Putzel GG; Zhou L; Bessman NJ; Yang R; Moriyama S; Parkhurst CN; Li A; Meyer HC; Teng F; Chavan SS; Tracey KJ; Regev A; Schroeder FC; Lee FS; Liston C; Artis D The microbiota regulate neuronal function and fear extinction learning. Nature 2019;574:543–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC; Ursell LK; Parfrey LW; Knight R The impact of the gut microbiota on human health: an integrative view. Cell 2012;148:1258–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SL; Gaitens JM; Jacobs DE; Strauss W; Nagaraja J; Pivetz T; Wilson JW; Ashley PJ Exposure of U.S. children to residential dust lead, 1999-2004: II. The contribution of lead-contaminated dust to children's blood lead levels. Environ Health Perspect 2009;117:468–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–2461 [DOI] [PubMed] [Google Scholar]
- Edgar RC UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature methods 2013;10:996–998 [DOI] [PubMed] [Google Scholar]
- Edgar RC; Haas BJ; Clemente JC; Quince C; Knight R UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S; Safdar N; Sethi AK; Suen G; Peppard PE; Kates AE; Skarlupka JH; Kanarek M; Malecki KMC Urinary lead concentration and composition of the adult gut microbiota in a cross-sectional population-based sample. Environment international 2019;133:105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JM; Morris LS; Marchesi JR The gut microbiome: the role of a virtual organ in the endocrinology of the host. The Journal of endocrinology 2013;218:R37–47 [DOI] [PubMed] [Google Scholar]
- Faggioli V; Menoyo E; Geml J; Kemppainen M; Pardo A; Salazar MJ; Becerra AG Soil lead pollution modifies the structure of arbuscular mycorrhizal fungal communities. Mycorrhiza 2019;29:363–373 [DOI] [PubMed] [Google Scholar]
- Fujimura KE; Sitarik AR; Havstad S; Lin DL; Levan S; Fadrosh D; Panzer AR; LaMere B; Rackaityte E; Lukacs NW; Wegienka G; Boushey HA; Ownby DR; Zoratti EM; Levin AM; Johnson CC; Lynch SV Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016;22:1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B; Chi L; Mahbub R; Bian X; Tu P; Ru H; Lu K Multi-Omics Reveals that Lead Exposure Disturbs Gut Microbiome Development, Key Metabolites, and Metabolic Pathways. Chemical research in toxicology 2017;30:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J; Xu K; Liu H; Liu G; Bai M; Peng C; Li T; Yin Y Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Frontiers in cellular and infection microbiology 2018;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havstad S; Wegienka G; Zoratti EM; Lynch SV; Boushey HA; Nicholas C; Ownby DR; Johnson CC Effect of prenatal indoor pet exposure on the trajectory of total IgE levels in early childhood. J Allergy Clin Immunol 2011;128:880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY; McBride SW; Hsien S; Sharon G; Hyde ER; McCue T; Codelli JA; Chow J; Reisman SE; Petrosino JF; Patterson PH; Mazmanian SK Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013;155:1451–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui N; Liu X-X; Kurola J; Mikola J; Romantschuk M Lead (Pb) contamination alters richness and diversity of the fungal, but not the bacterial community in pine forest soil. Boreal environment research 2012;17 [Google Scholar]
- Jaroslawiecka A; Piotrowska-Seget Z Lead resistance in micro-organisms. Microbiology 2014;160:12–25 [DOI] [PubMed] [Google Scholar]
- Johnson CC; Ownby DR The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Translational Research 2017;179:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN; Kwon HJ; Hong YC Low-level lead exposure and autistic behaviors in school-age children. Neurotoxicology 2016;53:193–200 [DOI] [PubMed] [Google Scholar]
- Koleva PT; Bridgman SL; Kozyrskyj AL The infant gut microbiome: evidence for obesity risk and dietary intervention. Nutrients 2015;7:2237–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koljalg U; Nilsson RH; Abarenkov K; Tedersoo L; Taylor AF; Bahram M; Bates ST; Bruns TD; Bengtsson-Palme J; Callaghan TM; Douglas B; Drenkhan T; Eberhardt U; Duenas M; Grebenc T; Griffith GW; Hartmann M; Kirk PM; Kohout P; Larsson E; Lindahl BD; Lucking R; Martin MP; Matheny PB; Nguyen NH; Niskanen T; Oja J; Peay KG; Peintner U; Peterson M; Poldmaa K; Saag L; Saar I; Schussler A; Scott JA; Senes C; Smith ME; Suija A; Taylor DL; Telleria MT; Weiss M; Larsson KH Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 2013;22:5271–5277 [DOI] [PubMed] [Google Scholar]
- Lakshmi Priya MD; Geetha A Level of trace elements (copper, zinc, magnesium and selenium) and toxic elements (lead and mercury) in the hair and nail of children with autism. Biol Trace Elem Res 2011; 142:148–158 [DOI] [PubMed] [Google Scholar]
- Levin AM; Sitarik AR; Havstad SL; Fujimura KE; Wegienka G; Cassidy-Bushrow AE; Kim EL; Zoratti EM; Lukacs NW; Boushey HA; Ownby DR; Lynch SV; Johnson CC Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Scientific Reports 2016;6:31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C; Lladser ME; Knights D; Stombaugh J; Knight R UniFrac: an effective distance metric for microbial community comparison. The ISME journal 2011;5:169–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T; Salzberg SL FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal 2011;17:10–12 [Google Scholar]
- McDonald D; Price MN; Goodrich J; Nawrocki EP; DeSantis TZ; Probst A; Andersen GL; Knight R; Hugenholtz P An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C; Duranti S; Bottacini F; Casey E; Turroni F; Mahony J; Belzer C; Delgado Palacio S; Arboleya Montes S; Mancabelli L; Lugli GA; Rodriguez JM; Bode L; de Vos W; Gueimonde M; Margolles A; van Sinderen D; Ventura M The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev 2017;81:e00036–00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monachese M; Burton JP; Reid G Bioremediation and tolerance of humans to heavy metals through microbial processes: a potential role for probiotics? Applied and environmental microbiology 2012;78:6397–6404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences Engineering and Medicine. Environmental Chemicals, the Human Microbiome, and Health Risk: A Research Strategy. Washington, DC: The National Academies Press; 2018 [PubMed] [Google Scholar]
- Oksanen J; Blanchet FG; Friendly M; Kindt R; Legendre P; McGlinn D; Minchin PR; O'Hara RB; Simpson GL; Solymos P; Henry M; Stevens H; Szoecs E; Wagner H vegan: Community Ecology Package. 2019 [Google Scholar]
- Polanska K; Hanke W; Pawlas N; Wesolowska E; Jankowska A; Jagodic M; Mazej D; Dominowska J; Grzesiak M; Mirabella F; Chiarotti F; Calamandrei G Sex-Dependent Impact of Low-Level Lead Exposure during Prenatal Period on Child Psychomotor Functions. Int J Environ Res Public Health 2018;15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN; Dehal PS; Arkin AP FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009;26:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackaityte E; Halkias J; Fukui EM; Mendoza VF; Hayzelden C; Crawford ED; Fujimura KE; Burt TD; Lynch SV Viable bacterial colonization is highly limited in the human intestine in utero. Nature Medicine 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy YS; Srivalliputturu SB; Bharatraj DK The effect of lead (Pb) exposure and iron (Fe) deficiency on intestinal lactobacilli, E. coli and yeast: A study in experimental rats. Journal of occupational health 2018;60:475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DW LabDSV: ordination and multivariate analysis for ecology. R package version 2.0-1. https://CRAN.R-project.org/package=labdsv. 2019 [Google Scholar]
- Sampson TR; Mazmanian SK Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 2015;17:565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani RK; Peyton BM; Jandhyala M Toxicity of lead in aqueous medium to desulfovibrio desulfuricans G20. Environ Toxicol Chem 2003;22:252–260 [PubMed] [Google Scholar]
- Sordillo JE; Korrick S; Laranjo N; Carey V; Weinstock GM; Gold DR; O'Connor G; Sandel M; Bacharier LB; Beigelman A; Zeiger R; Litonjua AA; Weiss ST Association of the Infant Gut Microbiome With Early Childhood Neurodevelopmental Outcomes: An Ancillary Study to the VDAART Randomized Clinical Trial. JAMA network open 2019;2:e190905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati F; Cavalieri D; Albanese D; De Felice C; Donati C; Hayek J; Jousson O; Leoncini S; Renzi D; Calabrò A; De Filippo C New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CM; Golding J; Emond AM Adverse effects of maternal lead levels on birth outcomes in the ALSPAC study: a prospective birth cohort study. Bjog 2015;122:322–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F; Zhai Q; Zhao J; Liu X; Wang G; Zhang H; Zhang H; Chen W Lactobacillus plantarum CCFM8661 alleviates lead toxicity in mice. Biol Trace Elem Res 2012;150:264–271 [DOI] [PubMed] [Google Scholar]
- Venables WN; Ripley BD Modern applied statistics with S.Fourth Edition. New York: Springer; 2002 [Google Scholar]
- Vollset M; Iszatt N; Enger Ø; Gjengedal ELF; Eggesbo M Concentration of mercury, cadmium, and lead in breast milk from Norwegian mothers: Association with dietary habits, amalgam and other factors. Sci Total Environ 2019;677:466–473 [DOI] [PubMed] [Google Scholar]
- Wani AL; Ara A; Usmani JA Lead toxicity: a review. Interdisciplinary toxicology 2015;8:55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegienka G; Havstad S; Joseph CL; Zoratti E; Ownby D; Woodcroft K; Johnson CC Racial Disparities in Allergic Outcomes in African Americans Emerge as Early as Age 2 Years. Clinical & Experimental Allergy 2011;42:909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegienka G; Havstad S; Zoratti EM; Kim H; Ownby DR; Johnson CC Association between vitamin D levels and allergy-related outcomes vary by race and other factors. J Allergy Clin Immunol 2015;136:1309–1314 e1301-1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegienka G; Kaur H; Sangha R; Cassidy-Bushrow AE Maternal-Cord Blood Vitamin D Correlations Vary by Maternal Levels. Journal of pregnancy 2016;2016:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J; Wen XW; Faulk C; Boehnke K; Zhang H; Dolinoy DC; Xi C Perinatal Lead Exposure Alters Gut Microbiota Composition and Results in Sex-specific Bodyweight Increases in Adult Mice. Toxicol Sci 2016;151:324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeileis A; Kleiber C; Jackman S Regression models for count data in R. Journal of Statistical Software 2008;27 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


