Abstract
Background
In both the IMpassion 130 trial in the metastatic setting and in Keynote 522 in the neoadjuvant setting, triple negative breast cancer (TNBC) patients showed benefit from PD-1 axis immunotherapy. Here, we assess PD-L1 expression on both tumor and immune cells using quantitative immunofluorescence to assess association with benefit from neoadjuvant durvalumab concurrent with chemotherapy in TNBC.
Methods
Pre-treatment core needle biopsies (n=69) were obtained from patients who participated in a Phase I/II clinical trial (NCT02489448). The final analysis included 45 patients (pCR = 18, non-pCR = 27) due to technical issues and insufficient tissue. Slides were stained using a previously validated Ultivue DNA-based Ultimapper® kit (CD8, CD68, PD-L1, Cytokeratin/Sox10 and Hoechst counterstain). The PD-L1 expression was analyzed by molecular compartmentalization without segmentation using AQUA software (version 3.2.2.1) in three tissue compartments including tumor (cytokeratin positive cells), CD68 positive cells, and overall stroma.
Results
In patients with pCR, PD-L1 expression was significantly higher in tumor cells, in CD68 positive cells and in the stroma compared to patients non-pCR. There was no difference in the amount of CD68 positive cells in the tumor or stromal compartments between cases with pCR and non-pCR.
Conclusion
Expression of PD-L1 in tumor cells, immune cells in stroma, and co-localized with CD68 positive cells is associated with higher rates of pCR to durvalumab and chemotherapy in TNBC.
Keywords: PD1/PD-L1, immunotherapy, immune checkpoints, survival, macrophages, biomarkers, quantitative immunofluorescence, triple negative cancer
INTRODUCTION
The IMpassion 130 trial showed that patients with metastatic triple negative breast cancer (TNBC) that show >1% immune cell expression of programmed death ligand-1 (PD-L1) using the SP142 assay benefit from adding atezolizumab to nab-paclitaxel. This has led to FDA approval of atezolizumab with the SP142 assay as a companion diagnostic. In lung cancer, this assay has been shown to be least sensitive of the commonly used assays for PD-L1 expression by immunohistochemistry (IHC) and immune cell expression is the least reliable method of analysis of PD-L1 expression (1,2). Similarly, lower sensitivity has also been reported in breast cancer (Reisenbichler et al, in press). Recently, the Keynote 522 trial reported that, in the neo-adjuvant setting, addition of pembrolizumab to chemotherapy improves the pathologic complete response (pCR) rate in both PD-L1 positive and negative cancers assessed by the 22c3 IHC assay using a CPS score that includes both tumor and immune cell expression (3). But the data also showed pCR rates were higher in the PD-L1 positive patients. Since the two assays (SP142 and 22c3) use different antibodies and scoring systems, they are not comparable or interchangeable. But taken together, these results suggest that expression of PD-L1 identifies patients who are the most likely to benefit from PD-1 axis immunotherapies, but the localization of expression remains undefined. It is not yet clear if tumor cell expression contributes to the predictive value, as seen in lung cancer, of if only immune cell expression, as described in the IMpassion 130 trial, is predictive.
We hypothesize that quantitative measurement of PD-L1 expression in molecularly defined tissue compartments may bring some clarity to the role of cell type-specific PD-L1 expression in determining response to immunotherapy. In this study, we use quantitative immunofluorescence (QIF) to both quantify and localize PD-L1 protein expression in pre-treatment core needle biopsies of newly diagnosed stage I-III TNBC subsequently treated with neoadjuvant durvalumab administered concomitantly with sequential weekly nab-paclitaxel and dose dense doxorubicin/cyclophosphamide(4,5).
METHODS
Patients
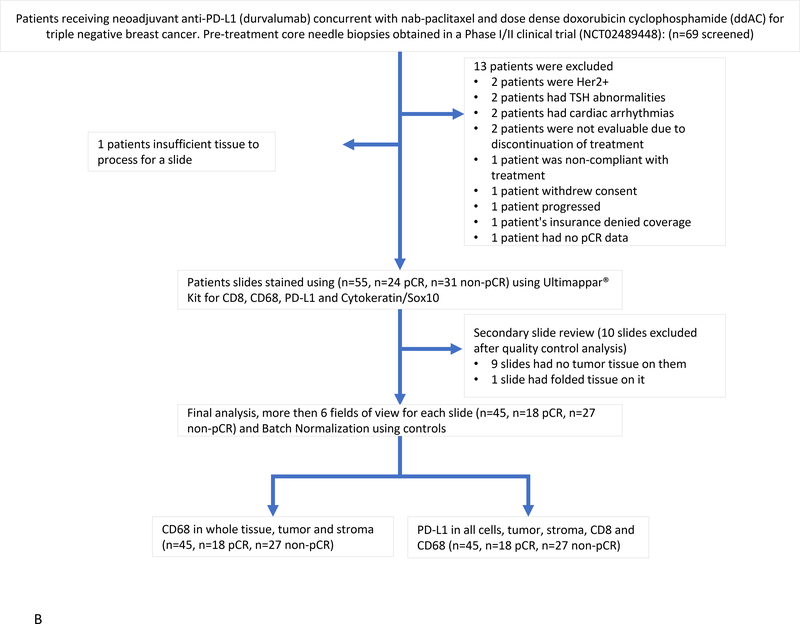
This study uses tissue from the NCT02489448, single arm Phase I/II clinical trial (figure 1A). Inclusion criteria required all patients be more than 18 years of age with newly diagnosed histologically confirmed stage I-III, ER, PR and HER2 negative invasive breast cancer as defined by the ASCO-CAP guidelines, for whom systemic chemotherapy would be indicated based on physician judgment following standard NCCN practice guidelines. Patients had to provide written informed consent for voluntary participation and undergo a baseline tumor core needle biopsy and blood draws for correlative science studies.
Figure 1.
A) Trial Schema for NCT 02489448. B) Biomarker Consort Diagram.
Major exclusion criteria included, patients who underwent partial excisional biopsy or lumpectomy, segmental mastectomy or modified radical mastectomy or sentinel node. Patients for whom anthracycline, paclitaxel or antibody therapies are contraindicated. Patients with active autoimmune disease or documented autoimmune disease within 2 years. Patients with hypothyroidism that is clinically stable and have normal TSH levels with hormone replacement, or patients with vitiligo or psoriasis not requiring treatment remain eligible for the study. Active or prior documented inflammatory bowel disease (Crohn’s disease, ulcerative colitis). Patients with known active hepatitis B or C or HIV infection or with history of tuberculosis.
Trial Design, Procedures and Outcomes
NCT02489448 was an interventional clinical trial where a pretreatment biopsy was taken, followed by nab-paclitaxel 100mg/m2 was given weekly for 12 weeks followed by doxorubicin 60mg/m2 and cyclophosphamide 600mg/m2, given every other week from 13 to 19 weeks as standard care for all patients. All patients also received a dose of MEDI4736 (durvalumab) 3mgkg or 10mg/kg every two weeks up until 19 weeks (90days). After 90 days and completion of therapy patients underwent surgery (Figure 1A).
The primary objective of the Phase I of the clinical trial was to assess the safety of MEDI4736 in combination with chemotherapy and determine if full dose of MEDI4736 can be given with a full dose weekly nab-paclitaxel followed by dose-dense AC chemotherapies, respectively.
The Phase II primary objective was to estimate the pCR rate of combination MEDI4736 and weekly nab-paclitaxel (12 doses) followed by MEDI4736 and ddAC (4 doses) for patients with clinical stages I-III TNBC. Pathologic complete response is defined as the absence of residual invasive cancer on hematoxylin and eosin evaluation of the resected breast specimen and all sampled regional lymph nodes following completion of neoadjuvant systemic therapy (i.e. ypT0/Tis ypN0).
Sixty-nine patients were screened for enrollment at Yale Cancer Center and its regional care centers and 60 patients consented for the trial between December 18, 2015 and November 21, 2018. One patient subsequently withdraw consent. Seven patients were included in the Phase I part, four at 3 mg/kg and three at 10 mg/kg dose. Fifty-two patients were enrolled in the Phase II part at 10 mg/kg dose. Two patients did not proceed to surgery; one discontinued therapy after two weekly treatments of nab-paclitaxel and one course of durvalumab due to altered mental status attributed to Guillen Barre syndrome, the other patient completed 9 weekly treatments of nab-paclitaxel (last treatment held because of peripheral neuropathy), 4 cycles of AC and 9 treatments of durvalumab and died of sudden death in her home before undergoing surgery (See figure 1B). A total of 55 patients samples underwent Ultimapper® Kit staining for CD8, CD68, PD-L1, Cytokeratin and Hoechst (nuclear staining).
QIF assessment of PD-L1 expression
Pre-treatment core needle biopsies (n=69) were obtained from patients who participated in the trial. Of these 24 patient’s samples were excluded due to a range of clinical and technical issues (Figure 2.). The final analysis had 45 patients (pCR = 18, non-pCR = 27). The slides were stained using a previously validated Ultivue DNA-based Ultimapper® kit(6) including CD8 (C8–144b), CD68 (KP-1), PD-L1(73–10), Cytokeratin (AE1/AE3) / Sox10 (BC34) and Hoechst counterstain for nuclear staining, (see supplemental figure 1 and 2). The PD-L1 positivity cut-point was validated using the PD-L1 cell line standardization TMA described previously(7). The images were collected on the PM2000 microscope (HistoRx) and analyzed by molecular compartmentalization without segmentation using AQUA software (version 3.2.2.1)(8).
Figure 2.
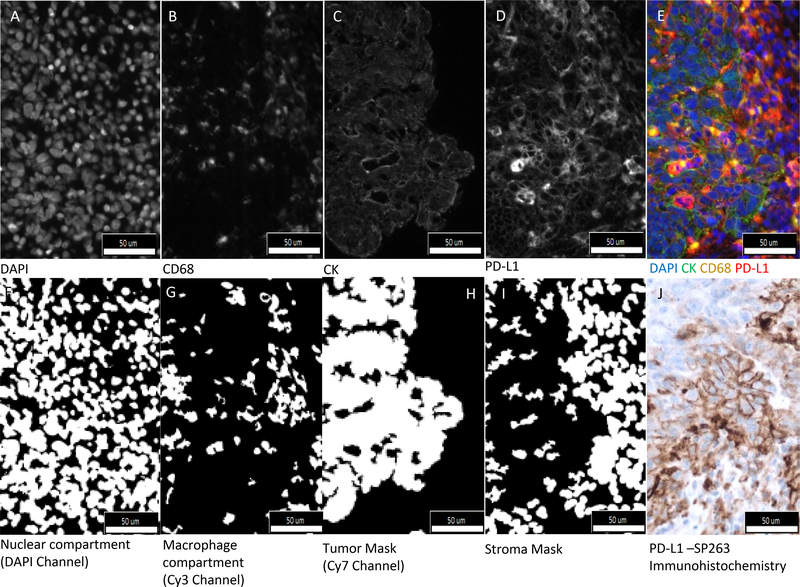
Illustration of tumor and immune cell PD-L1 expression. Fluorescent images captured in; A. DAPI channel (nuclei), B. Cy3 Channel (CD68 cells), C. Cy7 Channel (Cytokeratin), D. Cy5 Channel (PD-L1 Expression) and E. Composite image with all channels merged (Blue-Nuclear, Red-PD-L1, Yellow-Macrophages/CD68 cells, Green-Cytokeratin). Binary images that are used to define compartments are shown in F. DAPI channel (nuclear compartment), G. Cy3 channel (Macrophages compartment), H. Cy7 channel (Tumor mask), I. subtraction of Cy7 mask from a DAPI mask (Stroma mask). J. PD-L1 –SP263 Immunohistochemistry. The conventional nomenclature for AQUA defines “compartments” as just the pixels where there is signal, compared to “masks” which can be dilated and filled.
Figure 2 A–D show images taken by the PM2000 microscope, panel F. shows a pseudo-color composite of all the channels. Panel F-I show nuclear and macrophage compartments; and tumor and stroma masks. Molecular compartmentalization is achieved by AQUA using pixel-based colocalization to define compartments, rather than image-based nuclei-finding software that defines cells by segmentation of the image based on dilated distances from a central nucleus. Expression of CD 68 (figure 2B), is binarized to positive or negative pixels and then defines the molecular compartment for CD68 (figure 2G). Expression of PD-L1 is then measured in the tumor compartment (level of expression within the CK mask (figure 2H)) and in the stromal compartment (all pixels the pixels not in the CK mask (figure 2I)) and in the CD68 compartment (figure 2G) as defined above. AQUA Normalized score is dependent of the intensity of the fluorophore, time of exposure of light, wavelength channel and the area in which the target is being measured. Batch to batch reproducibility between 8 staining batches was excellent for CD68 (R2 = 0.86 to 0.94) and PD-L1 (R2 = 0.87 to 0.98) expression (supplemental figure 3).
The AQUA method of quantitative analysis was used to circumvent the reproducibility issues commonly see with cell segmentation or phenotyping. This method of molecular compartmentalization defines subcellular localization of expression of target by co-localization with biomarkers that define a region or cell type. To assess localization of PD-L1 expression, we used a fluorescence multiplex assay that allowed definition of a tumor compartment (by co-localization with cytokeratin) and a stroma compartment (tissue negative for cytokeratin). Within the stroma compartment, we could further compartmentalize PD-L1 expression into a CD68 expressing compartment (assumed to be predominantly representing macrophages). To assess lymphocytes, a CD8 compartment was attempted, but excluded from this experiment due to high tissue autofluorescence in the FITC channel technically preventing accurate classification.
IHC for SP263 comparison
The SP263 assay was performed on all slides according to the exact procedure on the label. Full tissue sections from each case were stained with the Ventana SP263 commercial assay according to manufacturer’s instructions on the package insert using the Ventana Benchmark autostainer. The slides with then read by two pathologist scoring immune cells only where positive as defined and ≥1% and negative defined as <1% as per the definitions in Schmid et al (3). Cell line arrays were used as positive control on separate slides from the study cases (9). Results were presented at the 2019 San Antonio Breast Cancer symposium(10) and are included in a parallel paper in review (Foldi et al)
Statistical analysis
For statistical analysis we used IBM-SPSS® Statistics 24.0.0.1 (64-bit, 2016), Prism 7 (GraphPad Software Inc 2016).
RESULTS
The exact localization of PD-L1 expression is controversial. Figure 2 shows PD-L1 expression on both tumor and immune cells using both the immunofluorescence assay and a chromogenic assay on a serial section. The image with the chromogen (figure 2J) illustrates the challenge of correct cellular assignment of PD-L1 expression. Figure 2A shows membranous tumor cell expression. Supplemental Figure 5B shows a stromal region where some CD68 cells express PD-L1 (red+yellow=orange) and some do not (yellow). PD-L1 expression on breast cancer cells was rarely observed in previous studies(11) using the less sensitive assay (SP142). However, the more sensitive SP263 assay with conventional IHC shows some membranous tumor cell expression pattern reminiscent of that shown in lung cancer(1,2) (Fig 2J). However, for analysis, only immune cells as defined in the FDA assay package insert, were assessed. An H&E of a corresponding serial section is shown to morphologically confirm the tumor cell assignment (supplemental Figure 5D). Analysis of SP263 showed 14 of 17 cases positive in the patients with pCR compared to 13 of 24 cases positive in patients without pCR (Fisher;’s exact test p=0.096). This work is described in detail in a separate publication (See Foldi et al, submitted)
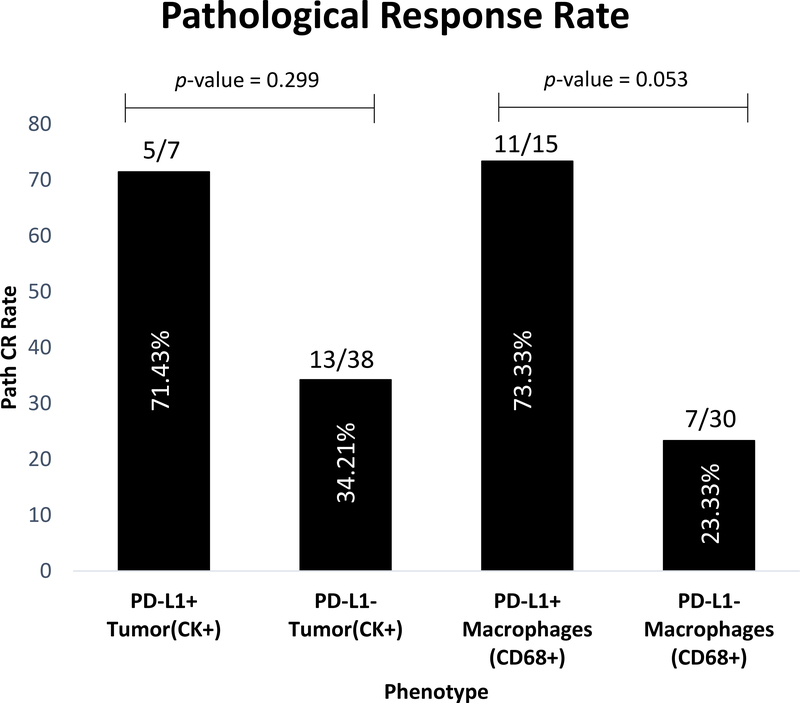
Analysis of the fluorescence images (figure 3) shows that PD-L1 expression was significantly higher in the tumor compartment (p-value <0.001), the stromal compartment (p-value <0.001) and in the CD68 compartment (p-value <0.001) in pCR patients compared to non-pCR. However, comparative analysis between the pCR and non-pCR patients showed no difference in the overall presence of CD68 cells in tumor (p-value 0.080), stroma (p-value 0.090) and both (p-value 0.140). Finally, the pCR rates in cases that had PD-L1 expression in either tumor cells or CD68 expressing immune cells were 71% and 73%, compared to 23% or 34% in tumor cells or immune cells that do not express PD-L1 (Figure 4.). As previously demonstrated(12), we also observed high positive correlation between PD-L1 expression in tumor cells and stroma. Other studies have also supported this observation (13) (14) (15). There seems to be a correlation of PD-L1 expression in the tumor, stroma and macrophages (Supplemental Figure 4.). However, the statistical analysis didn’t show any difference between the PD-L1 positivity and pathological response rate in either of tumor or macrophages. We additionally performed a univariate and multivariate (age and stage as covariates) binary logistic regression analysis, PD-L1 Expression in all cells, stromal cells and macrophages was statistically significant for both univariate and multivariate analysis (p-value <0.05; see Supplemental Figure 6.
Figure 3.
Association between PD-L1 expression within molecularly defined compartments and pathologic complete response (pCR). A. PD-L1 expression in the CK compartment (tumor). B. PD-L1 expression in the regions that do not express CK (stroma). C. PD-L1 expression in the CD68 compartment (macrophages). D. CD68 expression in the CK compartment (tumor). E. CD68 expression in the regions that do not express CK (stroma). F. CD68 expression (macrophages) in the total cellular tissue compartment (defined by DAPI). Unpaired Mann-Whitney U test was used to assess the pCR and non-pCR groups.
Figure 4.
A bar chart comparing the percentage of pCR as a function of PD-L1 expression in tumor cells as defined by the CK compartment and macrophages as defined by the CD68 compartment. A visual cut point was used to access PD-L1 Expression in tumor cells and macrophages. The visual cut-point is defined as the signal in the case were the signal can be seen “by eye”. Fisher’s Exact test was used to test for relationship between PD-L1 positive and PD-L1 negative patients in relation to pathological response.
DISCUSSION
Several studies have shown that higher PD-L1 expression is associated with higher likelihood of pCR after neoadjuvant chemotherapy with or without immune checkpoint inhibitors(16). In metastatic TNBC only patients with high expression of PD-L1 seem to greater benefit from immune checkpoint therapy in TNBC. The IMpassion 130 trial in metastatic TNBC used the Ventana SP142 assay and showed added benefit from atezolizumab only in cancer that showed PD-L1 expression in immune cells (17). Expression was predominantly localized to immune cells in the stroma, but tumor cell staining was also seen in about 9% cases. The lower sensitivity of the SP142 assay may explain the low level of tumor cell expression in most cases in the study. Notably, the patient populations were also different in that the IMpassion 130 study included only metastatic breast cancer, while our study and Keynote 522 included only patients in the neo-adjuvant setting. The I-SPY2 trial, also a neoadjuvant trial, showed both tumor and immune cell staining in TNBC using the 22c3 antibody(18).
In this study, we examined if the cellular localization of PD-L1 staining effects the predictive value of the test. We found that expression of PD-L1 on immune cells in the stroma, or on the CD68 compartment, or on tumor cells were each significantly associated with higher pCR rate to durvalumab and chemotherapy in this single arm neoadjuvant trial. This observation is concordant with two previous reports from our group that showed PD-L1 expression in CD68+ cells is associated with outcome in immunotherapy-treated melanoma(19) and lung cancer(8).
While provocative, this study has a number of limitations, including the small sample size of patients in the cohort, the reliance on response to mixed therapy in the neo-adjuvant setting rather than event free or overall survival, and the fact that all patients were from a single institution. Another limitation of this work is the fact that all interpretation was based on a single biopsy, as is the practice in neo-adjuvant studies. This may be especially problematic given the known heterogeneity of PD-L1 expression. None the less, we clarify the localization of expression of PD-L1 in TNBC using a sensitive multiplex fluorescence-based assay with molecular compartmentalization and show that both the tumor and stroma compartments express this antigen. We also demonstrate that CD68 positive cells, most likely macrophages, contribute substantially to overall PD-L1 positivity and could play a role in mediating the therapeutic effect. These results could affect the trial design of new studies designed to determine the role of PD-L1 expression in prediction of benefit from PD-1 axis therapeutics.
Supplementary Material
Translational Relevance.
PD-1 axis immunotherapy has been shown to be beneficial in triple negative breast cancer, but expression of PD-L1 as a predictive test has been equivocal. Using a quantitative approach to the definition of both cell type and amount of PD-L1 staining we find that PD-L1 expression in macrophages is associated with benefit to Durvalumab, an anti-PD-L1 therapy. This result suggests that the assay method may be important for finding predictive value for PD-1 axis drugs.
Acknowledgements
This biomarker research was supported by the Breast Cancer Research Foundation (David L. Rimm and Lajos Pusztai) and an NCI R01 grant (R01CA219647) to Lajos Pusztai. The clinical trial was supported by AstraZeneca.
Possible Conflict of Interest
Lajos Pusztai has received consulting fees and honoraria from Astra Zeneca, Merck, Novartis, Genentech, Eisai, Pieris, Immunomedics, Seattle Genetics, Almac and Syndax.
David L. Rimm has served as an advisor for Astra Zeneca, Agendia, Amgen, BMS, Cell Signaling Technology, Cepheid, Daiichi Sankyo, Genoptix/Novartis, GSK, Konica Minolta, Merck, NanoString, PAIGE.AI, Perkin Elmer, Roche, Sanofi, Ventana and Ultivue. Astra Zeneca, Cepheid, NavigateBP, NextCure, Nanostring, Lilly, and Ultivue fund research in David L. Rimm’s lab.
REFERENCES
- 1.Rimm DL, Han G, Taube JM, Yi ES, Bridge JA, Flieder DB, et al. A Prospective, Multi-institutional, Pathologist-Based Assessment of 4 Immunohistochemistry Assays for PD-L1 Expression in Non-Small Cell Lung Cancer. JAMA Oncol 2017;3:1051–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsao MS, Kerr KM, Kockx M, Beasley MB, Borczuk AC, Botling J, et al. PD-L1 Immunohistochemistry Comparability Study in Real-Life Clinical Samples: Results of Blueprint Phase 2 Project. J Thorac Oncol 2018;13:1302–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid P, Cortes J, Pusztai L, McArthur H, Kummel S, Bergh J, et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N Engl J Med 2020;382:810–21 [DOI] [PubMed] [Google Scholar]
- 4.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nature Medicine 2002;8:1323–8 [DOI] [PubMed] [Google Scholar]
- 5.Sunshine JC, Nguyen PL, Kaunitz GJ, Cottrell TR, Berry S, Esandrio J, et al. PD-L1 Expression in Melanoma: A Quantitative Immunohistochemical Antibody Comparison. Clin Cancer Res 2017;23:4938–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manesse M, Patel KK, Bobrow M, Downing SR. The InSituPlex((R)) Staining Method for Multiplexed Immunofluorescence Cell Phenotyping and Spatial Profiling of Tumor FFPE Samples. Methods Mol Biol 2020;2055:585–92 [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Morilla S, McGuire J, Gaule P, Moore L, Acs B, Cougot D, et al. Quantitative assessment of PD-L1 as an analyte in immunohistochemistry diagnostic assays using a standardized cell line tissue microarray. Laboratory investigation; a journal of technical methods and pathology 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, zugazagoitia J, Ahmed FS, Henick BS, Gettinger S, Herbst RS, et al. Immune cell PD-L1 co-localizes with macrophages and is associated with outcome in PD-1 pathway blockade therapy. Clinical Cancer Research 2019:clincanres.1040.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Morilla S, McGuire J, Gaule P, Moore L, Acs B, Cougot D, et al. Quantitative assessment of PD-L1 as an analyte in immunohistochemistry diagnostic assays using a standardized cell line tissue microarray. Lab Invest 2020;100:4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pusztai L RE, Bai Y, et al. Durvalumab (MEDI4736) concurrent with nab-paclitaxel and dose dense doxorubicin cyclophosphamide (ddAC) as neoadjuvant therapy for triple negative breast cancer (TNBC).. Presented at: 2019 San Antonio Breast Cancer Symposium; December 10–14; San Antonio, TX Abstract PD1–01.2019. [Google Scholar]
- 11.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. New England Journal of Medicine 2018;379:2108–21 [DOI] [PubMed] [Google Scholar]
- 12.Rehman JA, Han G, Carvajal-Hausdorf DE, Wasserman BE, Pelekanou V, Mani NL, et al. Quantitative and pathologist-read comparison of the heterogeneity of programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer. Mod Pathol 2017;30:340–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wimberly H, Brown JR, Schalper K, Haack H, Silver MR, Nixon C, et al. PD-L1 Expression Correlates with Tumor-Infiltrating Lymphocytes and Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancer Immunol Res 2015;3:326–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelekanou V, Carvajal-Hausdorf DE, Altan M, Wasserman B, Carvajal-Hausdorf C, Wimberly H, et al. Effect of neoadjuvant chemotherapy on tumor-infiltrating lymphocytes and PD-L1 expression in breast cancer and its clinical significance. Breast Cancer Res 2017;19:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelekanou V, Barlow WE, Nahleh ZA, Wasserman B, Lo YC, von Wahlde MK, et al. Tumor-Infiltrating Lymphocytes and PD-L1 Expression in Pre- and Posttreatment Breast Cancers in the SWOG S0800 Phase II Neoadjuvant Chemotherapy Trial. Mol Cancer Ther 2018;17:1324–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loibl S, Untch M, Burchardi N, Huober J, Sinn BV, Blohmer JU, et al. A randomised phase II study investigating durvalumab in addition to an anthracycline taxane-based neoadjuvant therapy in early triple-negative breast cancer: clinical results and biomarker analysis of GeparNuevo study. Ann Oncol 2019;30:1279–88 [DOI] [PubMed] [Google Scholar]
- 17.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med 2018;379:2108–21 [DOI] [PubMed] [Google Scholar]
- 18.Campbell MJ. Analysis of immune cell infiltrates as predictors of response to the checkpoint inhibitor pembrolizumab in the neoadjuvant I-SPY 2 TRIAL. AACR meeting 2019 2019 [Google Scholar]
- 19.Toki MI, Merritt CR, Wong PF, Smithy JW, Kluger HM, Syrigos KN, et al. High-plex predictive marker discovery for melanoma immunotherapy treated patients using Digital Spatial Profiling. Clinical Cancer Research 2019:clincanres.0104.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.