Abstract
The Integrator complex is conserved across metazoans and controls the fate of many nascent RNAs transcribed by RNA polymerase II. Among its 14 subunits is an RNA endonuclease that is critical for the biogenesis of small nuclear RNAs and enhancer RNAs. Integrator is further employed to trigger premature transcription termination at many protein-coding genes, thereby attenuating gene expression. Integrator thus helps shape the transcriptome and ensures genes can be robustly induced when needed. Molecular functions for Integrator subunits beyond the RNA endonuclease remain poorly understood, but some can act independently of the multi-subunit complex. Here, we highlight recent molecular insights into Integrator and propose how mis-regulation of this complex may lead to developmental defects and disease.
Keywords: enhancer, immediate-early gene, IntS11, RNA polymerase II, snRNA, transcription termination
Discovery of the Integrator complex and its RNA endonuclease activity
The Integrator complex was serendipitously discovered in 2005 by Baillat and colleagues as part of an effort to identify interacting partners of Deleted in split hand/split foot 1 (DSS1), a protein associated with congenital limb defects in humans [1]. Immunoprecipitation of human DSS1 revealed interactions with the tumor suppressor BRCA2, the 19S proteasome, RNA polymerase II (RNAPII), and 12 previously uncharacterized, metazoan-specific proteins that the authors named Integrator subunits (IntS) 1–12 for integrating the C-terminal domain (CTD) of RNAPII (see Glossary) with 3’ end processing of small nuclear RNAs (explained more below). Two additional Integrator subunits, IntS13 and IntS14, were subsequently identified and validated [2–4], and all of these proteins together can form the >1 MDa Integrator complex in a manner independent of DSS1. Key insights into the function of this complex came from finding that (i) Integrator subunits can directly bind the RNAPII CTD, especially when Serine2 and Serine7 are phosphorylated [1, 5–8] and that (ii) IntS11 and IntS9 share sequence homology with the zinc-dependent RNA endonuclease CPSF73 (a.k.a. CPSF3) and its binding partner CSPF100 (a.k.a. CPSF2), respectively [1, 9]. CPSF73 and 100 act as part of the cleavage and polyadenylation machinery to cleave nascent mRNAs prior to addition of the poly(A) tail [10, 11] and also cleave the 3’ ends of replication-dependent histone pre-mRNAs, which are not subsequently polyadenylated [12, 13]. These observations suggested that Integrator may likewise play a role in controlling transcription and/or RNA processing events.
Using chromatin immunoprecipitation (ChIP), Baillat and colleagues found Integrator subunits bound to small nuclear RNA (snRNA) gene loci [1], including U1 and U2, which are transcribed by RNAPII to produce key components of the pre-mRNA splicing machinery [reviewed in 14] (Figure 1A). Prior work had shown that snRNA 3’ end processing is dependent on the presence of the snRNA promoter [15, 16], the CTD of RNAPII [17], and a relatively degenerate sequence, known as the 3’ box, located 9–19 nucleotides downstream of the snRNA [18]. Despite these observations, the factor(s) that processed the 3’ ends of nascent snRNA transcripts had been long unknown. Given the homology between IntS11 and the CPSF73 endonuclease, Baillat and colleagues hypothesized that Integrator may, in fact, catalyze endonucleolytic cleavage of nascent snRNAs. Indeed, increased levels of long, unprocessed snRNAs were observed when catalytically inactive IntS11 was expressed or when Integrator subunits were depleted in human cells [1, 19], Drosophila [20], planarians [21], and C. elegans [22]. Integrator is thus required across metazoans for processing of snRNA primary transcripts (Figure 1A). Integrator further enables termination of snRNA transcription in a manner independent of the Rat1/Xrn2 5’–3’ torpedo exonuclease [23, 24], which is used to terminate protein-coding gene transcription after CPSF73 cleavage [reviewed in 25]. Integrator may instead trigger termination by inducing allosteric changes in RNAPII, acting itself as an exonuclease [26], and/or by recruiting additional factors, such as the cleavage and polyadenylation machinery factors PCF11 and SSU72 [19, 27].
Figure 1. Inttegrator cleavage can terminate transcription at snRNA and protein-coding genes.
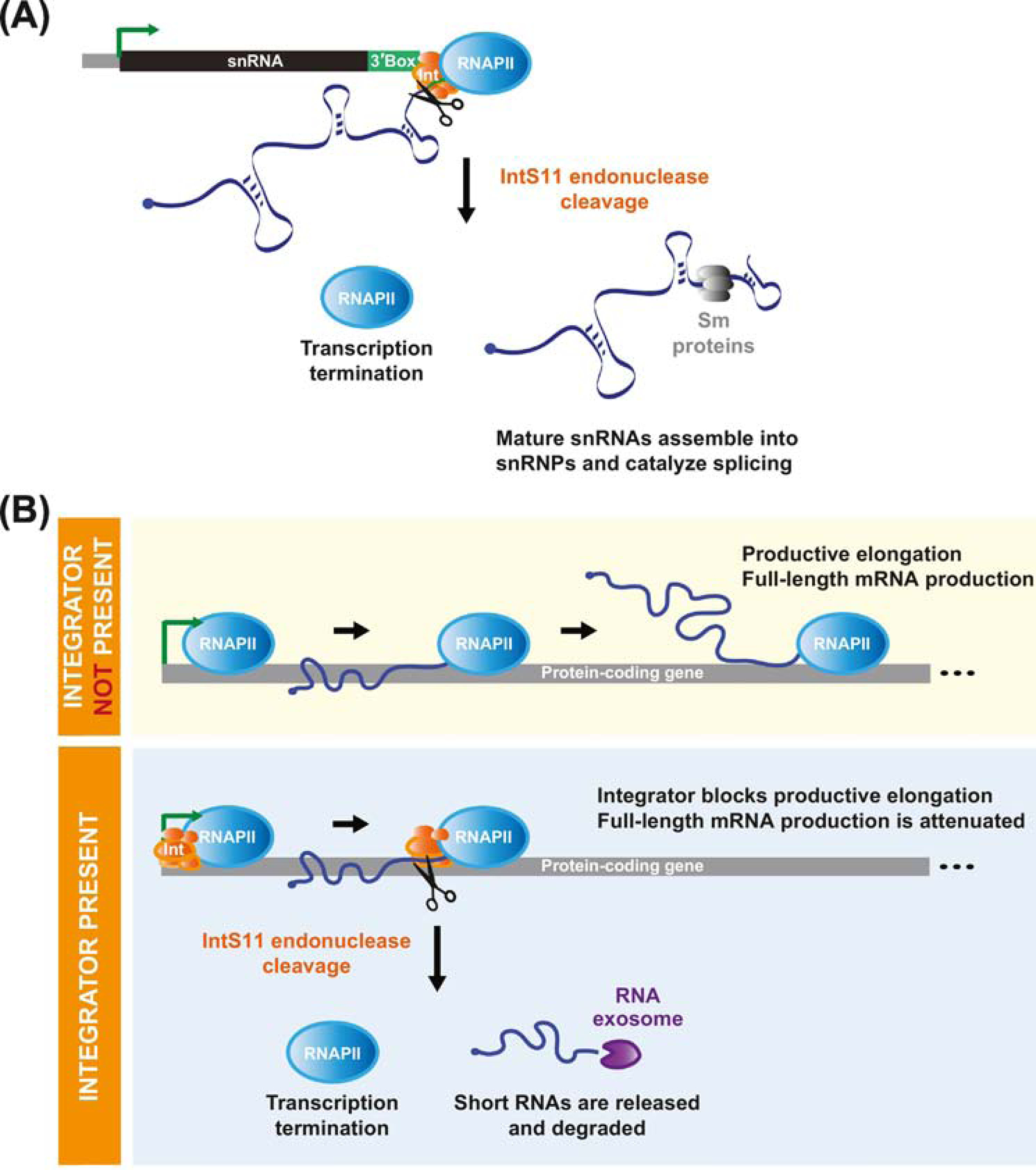
(A) Nascent snRNA transcripts are cleaved upstream of the 3’ box sequence (green) by the IntS11 RNA endonuclease component of the Integrator complex, which enables termination of RNA polymerase II (RNAPII) transcription. The cleaved snRNA transcript is released, further processed, and assembled into an RNA-protein complex, which includes Sm proteins (gray), that functions to catalyze pre-mRNA splicing reactions. (B) The Integrator complex can likewise be recruited to a subset of protein-coding loci. (Top) In the absence of Integrator, RNAPII transitions to productive elongation and full-length mRNAs are produced. (Bottom) When Integrator is recruited, IntS11 can cleave the nascent mRNA. This facilitates premature transcription termination and the resulting short RNAs are rapidly degraded from their 3’ ends by the RNA exosome (purple). Integrator thus blocks productive elongation and attenuates full-length mRNA production.
In the 15 years since the discovery of Integrator, it has become increasingly clear that this complex can control the processing and expression of additional RNAPII transcripts beyond snRNAs. This includes other non-polyadenylated RNAs, such as enhancer RNAs (eRNAs) [28, 29], telomerase RNA [30], viral microRNAs [31, 32], and replication-dependent histones [21, 33], as well as the transcription of many canonical protein-coding genes and long noncoding RNAs [29, 33–36]. It is thus perhaps not surprising that mis-regulation of Integrator expression has been associated with developmental and disease phenotypes in numerous metazoan species. Here, we highlight some of the latest findings on the molecular and physiological functions of Integrator, and propose that its role in regulating protein-coding gene expression is critical for normal metazoan development. We refer readers to earlier reviews for more details on how Integrator interacts with the CTD of RNAPII as well as how it controls snRNA processing [37–41].
The Integrator complex controls the outputs of protein-coding genes
In the initial report from Baillat and colleagues, two commonly studied human protein-coding genes (c-fos and GAPDH) were found to be unaffected by the Integrator complex [1]. However, subsequent high-throughput RNA-seq after knockdown of Integrator subunits as well as ChIP-seq experiments using higher quality antibodies revealed that Integrator subunits bind to and control the outputs of many loci besides snRNAs, including long noncoding RNAs and protein-coding genes across metazoans [22, 29, 33–36, 42, 43]. For example, in human HeLa cells [35] or Drosophila DL1 cells [29, 36], the expression of hundreds of protein-coding genes was altered after Integrator subunits were depleted by RNAi for several days. During this time frame, no significant changes in the levels of mature snRNAs were observed, consistent with these transcripts having long half-lives [44]. This suggested that the gene expression changes are not due to limiting levels of mature snRNAs causing widespread splicing defects. Instead, Integrator subunits can be detected by ChIP-seq at the 5’ ends of many of the protein-coding genes whose expression was altered by the RNAi treatments [29, 33, 34, 36]. This suggests that Integrator can have a direct effect on protein-coding gene transcription. There has nonetheless been considerable debate as to what Integrator does when bound to a protein-coding gene and, in particular, whether (and how) Integrator promotes or inhibits RNAPII elongation.
Integrator can attenuate protein-coding gene expression by catalyzing premature transcription termination
Our laboratory recently became interested in the Integrator complex when a genome-scale RNAi screen in Drosophila cells revealed that many Integrator subunits are potent inhibitors of the Metallothionein A (MtnA) promoter [36, 45]. MtnA encodes a metal chelator, and its transcription is rapidly induced when the intracellular concentration of heavy metals, such as copper or cadmium, is increased [reviewed in 46]. This induction helps ensure that copper levels are maintained in a narrow concentration range (as copper is an essential co-factor for a subset of enzymes) and that strictly toxic metals, such as cadmium, are effectively sequestered. During copper stress, we observed binding of Integrator subunits to the 5’ end of the MtnA locus, which enables the IntS11 endonuclease to directly cleave nascent MtnA mRNAs [36] (Figure 1B). Integrator does not cleave these nascent MtnA transcripts randomly, but instead appears to do so at specific locations downstream of the transcription start site (TSS), especially near nucleotides ~85 and ~110. Cleavage results in premature termination of MtnA transcription (thereby blocking the production of the full-length MtnA mRNA) as well as the release of a short RNA [36].
This role for Integrator at the Drosophila MtnA locus is mechanistically similar to how the complex functions at snRNA genes (Figure 1A), except that (i) there does not appear to be a 3’ box sequence in the vicinity of the MtnA cleavage sites and (ii) the cleaved RNAs have a very different fate. Cleaved snRNAs go on to be trimmed at their 3’ ends [47], stabilized, and assembled into snRNPs to ultimately function in pre-mRNA splicing [reviewed in 14]. In contrast, cleaved MtnA transcripts are rapidly degraded from their 3’ ends by the RNA exosome [reviewed in 48] (Figure 1B), perhaps to prevent the accumulation of R-loops at this protein-coding locus [36, 42]. Integrator thus limits or short-circuits the MtnA transcriptional program during copper stress, and recent work has revealed that Integrator also inhibits expression of a metal responsive gene in C. elegans [49]. Of note, we observed significantly less binding of Integrator subunits to the Drosophila MtnA locus during cadmium stress [36], consistent with the need to maximally produce MtnA to ensure cell survival under these conditions. How Integrator can be selectively recruited to a protein-coding gene in this system remains unknown, but the Drosophila MtnA locus highlights how the Integrator complex, particularly the IntS11 endonuclease, can be re-purposed by cells to limit transcriptional outputs from a protein-coding gene when appropriate.
Integrator-catalyzed premature termination is not unique to MtnA, and there were previous hints of this attenuation mechanism in the literature [33, 42]. It is now clear that Integrator broadly acts in Drosophila cells to control the biogenesis of eRNAs (discussed further below) and to attenuate transcription of protein-coding genes (perhaps as many as ~15% of active genes) [29, 36]. In cells depleted of IntS9, >400 mRNAs were up-regulated, some by more than 100-fold, compared to only 49 mRNAs that were down-regulated [36]. Integrator thus predominantly inhibits protein-coding gene expression in Drosophila cells, and similar observations have been made in C. elegans [22]. Direct quantification of nascent RNA synthesis using precision run-on sequencing (PRO-seq) in Drosophila confirmed that this is due to Integrator catalyzing premature transcription termination and preventing promoter-proximal RNAPII from transitioning into productive elongation [29]. Mirroring what was observed at MtnA, the IntS11 RNA endonuclease activity is critical and Integrator subunits are detected by ChIP-seq at the 5’ ends of many of these attenuated protein-coding genes [29, 36]. Nascent Drosophila mRNAs are cleaved by Integrator at well-characterized RNAPII promoter-proximal pause sites (25 to 60 nt downstream of the TSS [reviewed in 50]) [29] or slightly further downstream (e.g. at the +1 nucleosome where RNAPII can pause a second time [51]) [36], and the resulting RNAs are degraded by the RNA exosome. Integrator thereby limits full-length mRNA production and attenuates the transcriptional output of these protein-coding genes.
Integrator activity also ensures robust induction of protein-coding genes
In human cells, multiple models of Integrator function at protein-coding genes have been proposed [28, 29, 33–35, 52, 53], in part because the effect of depleting Integrator subunits appears to be more complicated than what has been observed in Drosophila or C. elegans. In HeLa cells depleted of IntS11, high-throughput sequencing of chromatin-associated RNAs revealed roughly equal numbers of protein-coding genes that were up- and down-regulated (667 and 616, respectively) [28, 29]. Current data indicate that Integrator can be critical for robust transcriptional induction of some human protein-coding genes, but there is debate as to the underlying molecular mechanism [28, 29, 33, 34, 54] (Figure 2). In particular, Integrator subunits have been detected at a subset of immediate-early genes and their associated enhancers in HeLa cells, and addition of epidermal growth factor (EGF) results in further recruitment of the Integrator complex to these loci in an ERK1/2-dependent manner [28, 34, 54]. When IntS1 or IntS11 has been depleted, a substantially diminished level of transcription induction from these genes is observed once EGF is added.
Figure 2. The Integrator complex may help ensure robust induction of genes via multiple molecular mechanisms.
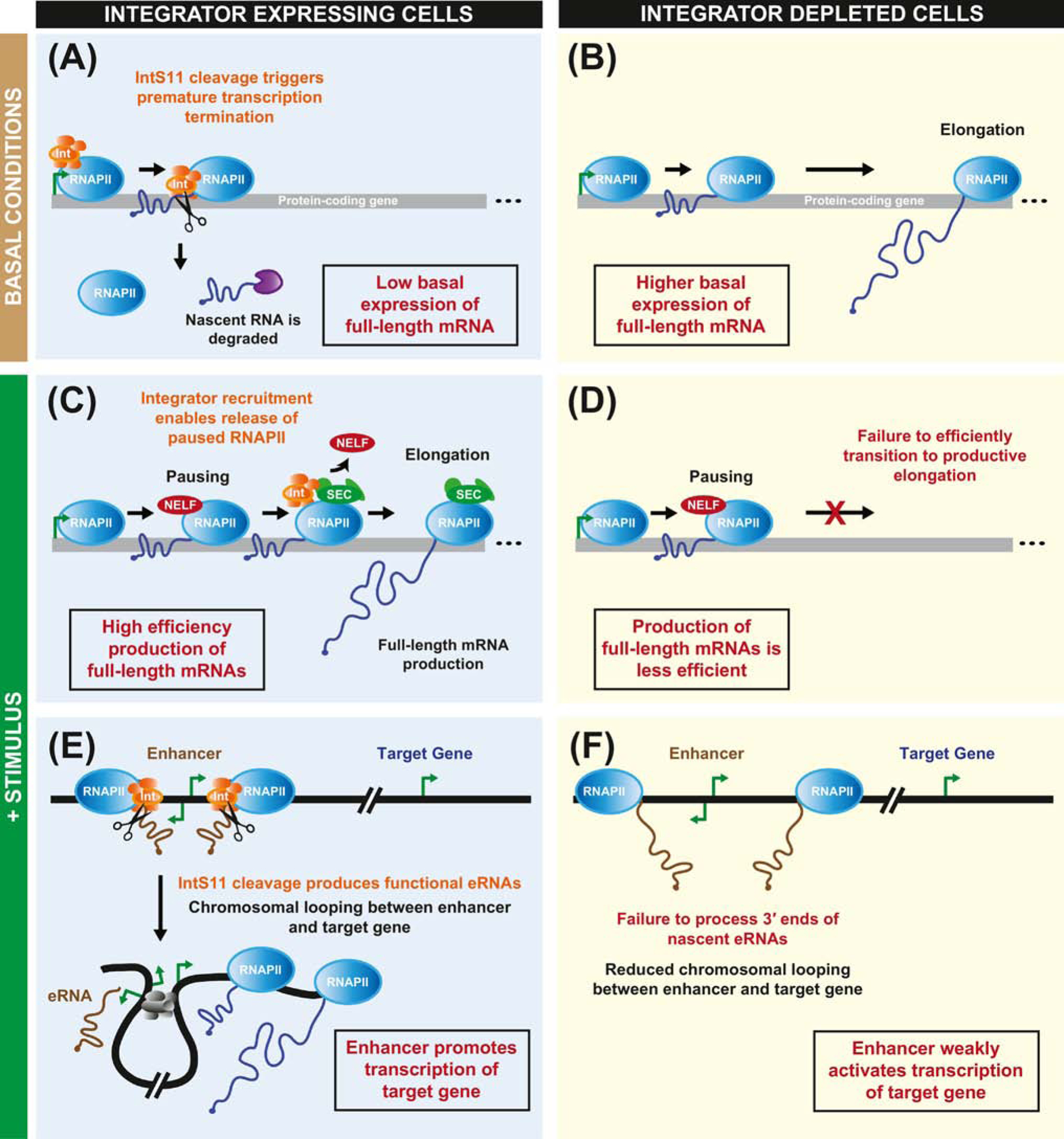
(A, B) When cells are under basal conditions, the expression of an immediate-early gene can be maintained at a low level by the Integrator complex. (A) This is because IntS11 cleaves nascent mRNAs, which are then subjected to rapid degradation. (B) In the absence of a functional Integrator complex, more RNAPII molecules transition to productive elongation, thereby resulting in a higher basal level of expression of the immediate-early gene. (C-F) Upon addition of a stimulus, the transcription of immediate-early genes is rapidly induced. (C) The Integrator complex has been proposed to facilitate induction by enabling release of paused RNAPII into productive elongation. During this transition, the super elongation complex (SEC, green) is recruited, leading to loss of NELF (red). (D) In the absence of Integrator, a greater percentage of RNAPII molecules remain paused, resulting in less efficient production of full-length mRNA. (E) The Integrator complex is also recruited to enhancer loci, which can be bi-directionally transcribed. The IntS11 endonuclease cleaves nascent enhancer RNAs (brown) to terminate their transcription. Chromosomal looping (facilitated by proteins such as Mediator, CTCF, and cohesin, depicted in gray) enables an active enhancer region to be in close proximity to a target gene and promote its transcription. (F) In the absence of Integrator, nascent enhancer RNAs are not properly processed at their 3’ ends, and this is associated with reduced chromosomal looping as well as reduced enhancer activity.
On the one hand, Integrator’s role in catalyzing premature transcription termination may underlie these observations. This is because depletion of Integrator subunits can result in the up-regulation of basal expression from inducible genes, which would cause these genes to be less induced in response to signaling [29, 33] (Figure 2A,B). Skaar and colleagues showed that the basal expression level of JUNB, an immediate-early gene, is increased ~2-fold in HeLa cells depleted of IntS3 or IntS9 [33]. Upon addition of serum, they found that JUNB transcripts reached the same level of expression regardless of whether Integrator subunits were present, but the dynamic range of induction was reduced in Integrator depleted cells (4-fold compared to 8-fold in control cells). In contrast, others have suggested that Integrator can function at these inducible genes via mechanisms that may be independent of IntS11 endonuclease activity. For example, Integrator can interact with factors that control RNAPII pausing, including NELF (negative elongation factor) and DSIF (5,6-dichloro-1β-D-ribofuranosylbenzimidazole [DRB]-sensitivity-inducing factor) [35, 52], as well as with the super elongation complex/pTEFb [55], thereby enabling release of paused RNAPII into productive elongation [34] (Figure 2C,D). These latter models thus argue that Integrator can directly promote the production of full-length mRNAs.
Another way that Integrator can alter the transcriptional output of protein-coding genes is by controlling 3’ end processing and functionality of eRNAs [28, 29]. When Integrator subunits have been depleted or when catalytically inactive IntS11 is present in HeLa cells, RNAPII termination defects are observed at enhancer loci (Figure 2E,F). This results in increased levels of primary eRNA transcripts coupled to reduced levels of mature eRNAs, which is associated with reduced chromatin looping between the enhancer region and its target promoter [28]. There is thus reduced enhancer activity when Integrator subunits have been depleted, which could further directly reduce the robustness of gene induction in response to cellular stimuli. Additional experiments are now required to distinguish between these models, but it seems likely that multiple underlying molecular mechanisms are at play (Figure 2) and their combined activities ensure that inducible genes can be robustly induced. It would, for example, be interesting to define the molecular details of how Integrator interacts with RNAPII pausing factors [52] and then examine the effect that disrupting these interactions has on cellular gene expression profiles.
snRNA and protein-coding loci have distinct dependencies on Integrator subunits
Besides IntS11 and its direct binding partners (IntS4 and 9) that make up the Integrator cleavage module [56], the exact functions of the non-catalytic Integrator subunits remain unknown. IntS12 has a PHD finger that may help recruit the complex to chromatin, but many Integrator subunits lack obvious paralogs within eukaryotic genomes and are devoid of known protein domains beyond α-helical repeats (e.g. HEAT, ARM, TPR, or VWA domains) [reviewed in 37]. Some of these domains may function as protein-protein interaction surfaces to enable assembly of the complex [56–60], and the expression of some Integrator subunits is indeed interdependent. For example, depletion of Drosophila IntS12 results in co-depletion of IntS1 (and vice versa) [56, 59, 61]. This suggests that some of the non-catalytic subunits may be only indirectly involved in Integrator’s enzymatic and biochemical functions at gene loci.
Depletion of many non-catalytic Integrator subunits, including IntS12, has only a minimal effect on 3’ end processing of snRNAs in Drosophila and human HeLa cells [20, 36, 56]. This suggests that the non-catalytic Integrator subunits may not be required for assembly or recruitment of a functional Integrator complex to snRNA loci. In stark contrast, many of the non-catalytic Integrator subunits (especially IntS1, 2, 5, 6, 7, 8, and 12) are critical for attenuating the outputs of protein-coding genes in Drosophila [36] and C. elegans [22]. This may imply that (i) distinct Integrator complexes are recruited to snRNA vs. protein-coding gene loci (discussed further below) and/or that (ii) non-catalytic Integrator subunits have specific roles at protein-coding genes, such as enabling the assembly/targeting of the complex to these loci and/or controlling RNAPII dynamics.
Individual Integrator subunits can moonlight and function outside the multi-subunit complex
Size exclusion chromatography has revealed that human Integrator subunits can be detected in a >1 MDa complex that is associated with RNAPII in the nucleus [1, 7]. How Integrator is assembled remains largely unclear, but it is unlikely that all Integrator complexes have the same components. This is because individual Integrator subunits can have distinct occupancy patterns on chromatin [7, 53], different effects when depleted [20, 22, 35, 36, 62], and even unique subcellular localization patterns in Drosophila and human cells [56, 61, 63]. The significance of these observations is incompletely understood, but they collectively suggest that Integrator subunits might have functions beyond their roles in the 14-subunit complex. Indeed, some Integrator subunits can be detected in smaller complexes in the nucleus of human cells [1, 52, 53, 60]. IntS3 and 6 have been shown to stably interact with human single-stranded DNA-binding protein 1 (SSB1, also known as nucleic acid binding protein 2 [NABP2]) to have critical roles in repair of DNA double-stranded breaks [60, 64–68] (Figure 3A). Following DNA damage, these particular Integrator subunits relocate to sites of damage and are required for the accumulation of other repair proteins, such as RAD51, TopBP1, and BRCA1, at these sites. In contrast, most other Integrator subunits, including IntS11, do not appear to significantly interact with hSSB1 or play a direct role in DNA repair [60, 64–68].
Figure 3. Integrator subunits can function independently of the multi-subunit complex.
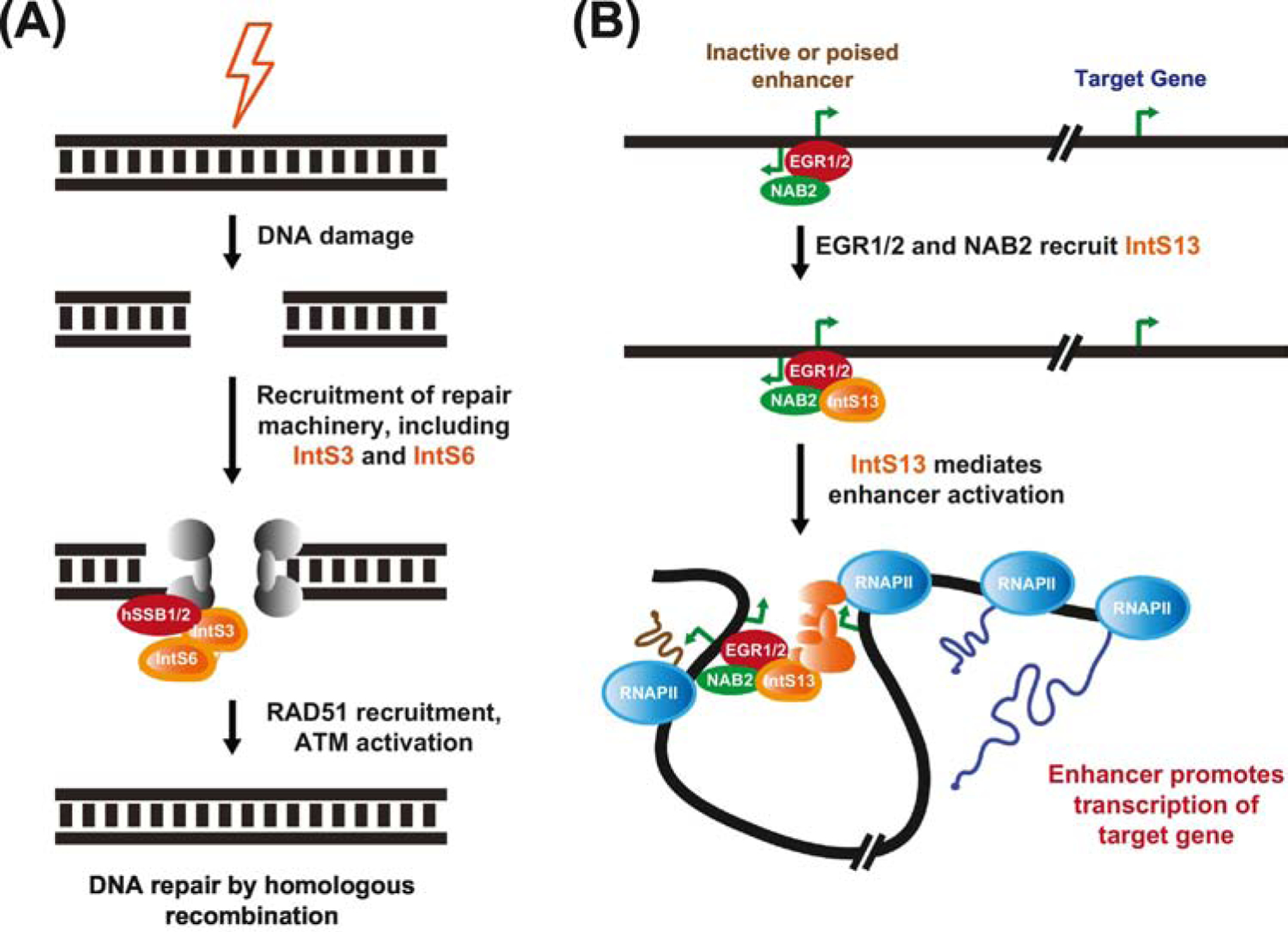
(A) Upon sensing a DNA double-stranded break, a number of proteins (including the MRE11/RAD50/NBS1 (MRN) complex, gray) are recruited to the site of damage, including IntS3 and 6. These particular Integrator subunits are found in a stable complex with hSSB1/2 and are required for downstream signaling events, including recruitment of the recombinase RAD51 and activation of the protein kinase ATM. (B) During myeloid differentiation, the IntS13 subunit is directly recruited to a subset of enhancers by the EGR1/2 transcription factors and their associated co-factor NAB2. These factors mediate enhancer activation, leading to increased transcription of the target genes (in a manner that may involve additional Integrator subunits, orange) that drive the differentiation process.
Recent work has further revealed that IntS13 can function as an independent entity to regulate human myeloid differentiation [53]. Barbieri and colleagues showed that, unlike IntS11, IntS13 is dispensable for the growth of HL-60 and primary myeloid progenitor cells. IntS13 is instead specifically required for maturation of these cells into functional monocytes and macrophages [53]. This is because IntS13 is recruited to a number of active and poised enhancers that control the expression of genes required for monocytic and macrophagic identity (Figure 3B). Other Integrator subunits (IntS1, 6, and 11) examined by ChIP were all notably absent from these enhancers. IntS13 is present in a ~200 kDa complex and appears to be directly recruited to enhancers by the DNA-binding EGR1/2 transcription factors and their associated co-factor NAB2 [53]. Once bound, IntS13 is able to mediate enhancer activation and drive the differentiation process, but further work is now required to reveal the underlying molecular details, including whether additional Integrator subunits are subsequently recruited to target gene promoters.
Mis-regulation of the Integrator complex is associated with developmental defects
Beyond the role of IntS13 in human myeloid differentiation, a growing number of studies have revealed that Integrator subunits are indispensable for normal development across metazoans [39]. Deletion or mutation of Integrator subunits in mouse (IntS1) [69], the teleost fish medaka (IntS1) [70], Drosophila (IntS4 and 7) [20, 71], C. elegans (IntS6) [22, 72], and zebrafish (IntS6) [73] have all been reported to result in abnormalities in early development and subsequent lethality. Here, it is important to point out that the subunits listed simply represent the subunits that were tested in each organism, and it is highly likely that mis-regulation of other Integrator subunits will result in similar phenotypes. Integrator is further required for normal hematopoiesis (IntS5, 9, 11) [74] and lens development (IntS1) [75] in zebrafish, adipogenesis in mouse (IntS6 and 11) [76], neoblast self-renewal and regeneration in planarians (IntS3 and 9) [21], dynein localization in cultured human cells as well as during Drosophila spermatogenesis and oogenesis (IntS13) [61, 63, 77], ciliogenesis in cultured human cells (IntS1, 3, 4, 9, 11, 12) [78], and normal brain development in multiple species, including humans (Box 1). For example, individual depletion of IntS1, 2, 5, or 8 in Drosophila results in excess immature neuroblasts due to increased de-differentiation of intermediate neural progenitors back to the stem cell fate [62]. Depletion of IntS11 did not result in a similar neuroblast phenotype [62], suggesting subunit-specific roles of Integrator components in controlling the homeostasis of neural cells. Nevertheless, depletion of IntS11 did result in cortical neuron migration defects in mice, as did depletion of IntS1 [43].
Box 1. The Integrator complex in human development and cancers.
Multiple individuals with severe neurodevelopmental delay have now been identified that carry biallelic mutations in Integrator subunits [75, 79]. Three unrelated individuals all have a homozygous nonsense mutation in IntS1 (S1784*, where * indicates a stop codon) that results in nonsense-mediated mRNA decay (NMD) [reviewed in 80] and reduced IntS1 mRNA/protein expression [79]. An additional three affected individuals are siblings that have mutations in both of their IntS8 alleles: one allele produces a transcript that stimulates NMD, while the other allele has a deletion that removes three amino acids (972–974) of IntS8. IntS8 lacking these three amino acids fails to co-purify with a number of other Integrator subunits (including IntS1 and 12) or RNAPII, suggesting that the full Integrator complex may be unable to be appropriately assembled or efficiently recruited to genes in these individuals [79]. Upon replacing the wild type IntS8 genes with the 972–974 deletion mutant in mouse P19 cells, retinoic-acid induced neuronal differentiation was disrupted, confirming that altered Integrator function is driving the phenotypes observed in patients.
IntS6 was originally identified as a tumor suppressor protein [81, 82] and it is now clear that a number of additional Integrator subunits are mis-regulated or mutated in human cancers [39, 83]. The functional significance of most of these data remains unclear. Nevertheless, in acute myeloid leukemia (AML), IntS3 was found to be mis-spliced in patients who have mutations in the metabolic enzyme IDH2 and the splicing factor SRSF2 (~5% of AML cases) [84]. The resulting IntS3 mRNA is degraded by NMD, and this down-regulation of IntS3 gene expression helps drive cancer progression as forced expression of IntS3 slows leukemia growth and induces myeloid differentiation in vivo [84]. Integrator has also been reported to be a critical transcriptional co-activator downstream of the mitogen-activated protein kinase (MAPK) signaling cascade, a pathway that is altered in >70% of human cancers [54]. Depletion of IntS11 suppressed the growth of cancer cells to a degree similar to that of direct inhibition of kinases in the MAPK pathway, and combining these treatments did not result in further growth suppression. These results are thus consistent with IntS11 participating in the MAPK cascade and suggest that Integrator may represent a therapeutic target for treating patients that are refractory to inhibitors targeting upstream kinase components of the pathway [54].
Hypothesis: Integrator’s role in controlling protein-coding gene expression is critical for development
Despite all the connections between Integrator and developmental phenotypes, the underlying molecular mechanisms remain poorly understood. Increases in unprocessed snRNA levels have been observed when Integrator subunits are depleted or mutated in multiple metazoan species [20–22, 69, 74, 79] and, at least in C. elegans, readthrough transcription downstream of snRNAs can result in the production of long chimeric snRNA-mRNA transcripts that are sometimes translated into proteins [22]. However, the levels of mature snRNAs often do not significantly change when Integrator subunits are depleted or mutated ([69] is an exception), including in the human individuals with neurodevelopmental delay [79] (Box 1). This suggests the existence of compensatory transcriptional or post-transcriptional mechanisms that ensure mature snRNAs are maintained at the appropriate levels.
Why then are developmental defects observed when Integrator subunits are depleted or mutated? It remains possible that the processing of a small subset of critical snRNA transcripts is altered (e.g. specific snRNA variants [85, 86]), but current evidence now suggests to us that Integrator’s role in fine-tuning protein-coding transcription may be critical for normal development. This is because the expression of hundreds to thousands of mRNAs can be mis-regulated when Integrator has been depleted/mutated [29, 35, 36, 79]. For example, compared to age-matched controls, the expression of more than 3,000 mRNAs was altered in human fibroblasts from patients with IntS8 mutations (215 genes also showed alternative splicing changes) [79]. It remains largely unknown which of these gene expression changes represent direct vs. secondary effects, and further work is needed to define the frequency at which each of the molecular mechanisms depicted in Figures 1–3 are used to regulate expression patterns across development. Nevertheless, modulating the expression of a single mRNA can, in some cases, be sufficient to rescue the developmental defects. Injection of Lefty1 or Bmp2b mRNA was sufficient to rescue the embryonic patterning defects observed in IntS6 mutant zebrafish [73]. Likewise, introduction of Smad1 or Smad5 mRNA was sufficient to rescue the hematopoietic defects observed in zebrafish embryos depleted of IntS5 [74]. These examples suggest that the developmental phenotypes observed are not due to Integrator’s role in snRNA processing, but instead because Integrator directly controls the outputs of key protein-coding genes that control differentiation processes. Identifying more of these key genes (and their exact mode of regulation, Figures 1–3) will likely not only explain how Integrator mis-regulation can contribute to such a wide variety of physiological phenotypes, including cancer (Box 1), but also may suggest new therapeutic opportunities.
Concluding Remarks
In summary, recent work has revealed that the Integrator complex controls the expression of many gene loci that are transcribed by RNA polymerase II, and we propose that mis-regulation of these processes can result in developmental defects or disease. Nevertheless, many of the molecular details of how Integrator is assembled, recruited, and functions are still poorly understood, thereby leaving a number of key questions unanswered (see Outstanding Questions). It is now clear that Integrator can function as a transcriptional tuner or almost the equivalent of an on/off switch at some protein-coding genes, and yet very little is known about how this complex is regulated so that the appropriate amounts of full-length mRNAs can be made depending on the cellular state. This may be achieved through direct modulation of IntS11 RNA endonuclease activity, but it is important to keep in mind that most of the other subunits within the complex are also critical for Integrator function and represent potential points of regulation [87]. Few of the other Integrator subunits have known protein domains [reviewed in 37] so it is currently difficult to hypothesize their molecular roles, let alone explain exactly why point mutations in these subunits can cause developmental defects. It nevertheless seems likely that many of the characterized mutations affect protein-protein interactions and/or Integrator complex stability, as has already been shown for Ints8 mutations observed in human patients (Box 1).
OUTSTANDING QUESTIONS.
What is the molecular function of each of the non-catalytic Integrator subunits?
How is the Integrator complex assembled and what are all the molecular interactions between the subunits?
How is the Integrator complex recruited to every snRNA locus but only to a subset of protein-coding genes and enhancers?
Are all 14 Integrator subunits recruited to snRNAs, enhancers, and protein-coding genes or are distinct complexes present at each of these loci?
How is Integrator recruitment/activity at specific protein-coding loci tuned depending on the state of the cell and its transcriptional needs?
Is Integrator activity at protein-coding loci and enhancers always dependent on the IntS11 RNA endonuclease?
Why and how does the Integrator complex fine-tune the expression of some genes (~2 fold inhibition) while also robustly (>100 fold inhibition) controlling the expression of other genes?
How are IntS11 endonucleolytic cleavage sites in nascent pre-mRNAs and eRNAs defined?
Integrator cleavage products from protein-coding genes are generally rapidly degraded, but are some stabilized and functionally important?
Mutations in Integrator subunits are associated with developmental abnormalities, but what are the underlying molecular mechanisms that have been altered?
Focused efforts are now needed to reveal the structures and molecular binding partners of Integrator subunits, as this will provide the field with a much deeper perspective on why the Integrator complex has so many subunits. In addition, it likely will be informative to look more closely for parallels between Integrator and the cleavage/polyadenylation machinery [reviewed in 11] as well as with complexes in lower eukaryotes that catalyze premature termination events, including the Nrd1-Nab3-Sen1 (NNS) complex in S. cerevisiae (which, like Integrator, also catalyzes termination at snRNA loci) [reviewed in 88] and the MTREC (Mtl1-Red1 core) complex in S. pombe [89, 90]. Finally, Integrator is likely not a single homogenous complex, and experiments are now needed to clarify what exact subunits are/are not recruited to protein-coding genes, snRNAs, enhancers, and other cellular locations, as well as how these patterns change over developmental time courses, across tissues, or in disease. Beyond identifying the genes bound by Integrator, it may be possible to determine the exact binding sites at high resolution using techniques like CUT&RUN [91], ChIP-exo [92], or ChIP-nexus [93]. Much has been recently learned about Integrator’s role in transcriptional control and development, and it is certain that many more insights into this complex will be revealed in the coming years.
HIGHLIGHTS.
The multi-subunit Integrator complex is conserved across metazoans and harbors the IntS11 RNA endonuclease that can cleave many nascent RNAs transcribed by RNA polymerase II.
Cleavage by Integrator is required for the biogenesis of functional small nuclear RNAs and enhancer RNAs, but attenuates the expression of protein-coding mRNAs by triggering premature transcription termination.
Integrator activity helps ensure that immediate-early genes are robustly induced upon sensing a stimulus.
The molecular function of most of the non-catalytic Integrator subunits remains unknown, but some can act independently of the multi-subunit complex, including at a subset of enhancers and upon activation of the DNA damage response.
Depletion or mutation of Integrator subunits often results in developmental defects, including in humans, and can contribute to cancer progression.
ACKNOWLEDGMENTS
We thank all members of the Wilusz laboratory for helpful discussions and suggestions. Research on the Integrator complex in our laboratory is supported by NIH grants R35-GM119735 (to J.E.W.) and K99-GM131028 (to D.C.T.). J.E.W. is a Rita Allen Foundation Scholar.
GLOSSARY
- 3’ box
A conserved, but relatively degenerate A/T-rich sequence that is located 9–19 nt downstream of the sequence specifying the mature snRNA. The 3’ box is necessary for processing of nascent snRNA transcripts [18]
- C-terminal domain (CTD) of RNA polymerase II
A sequence of heptad repeats (consensus Tyr-Ser-Pro-Thr-Ser-Pro-Ser), varying in number from 26 in yeast to 52 in vertebrates that is present on the end of the largest subunit of RNA polymerase II. The CTD is subjected to extensive post-translational modifications, especially phosphorylation, during the transcription cycle and serves as a binding scaffold for factors that control RNA synthesis and maturation [reviewed in 94]
- Enhancer
Cis-acting DNA sequences that can increase the transcription of genes by binding to transcription factors and co-factors as well as modifying the nearby chromatin. Active enhancers are transcribed to yield enhancer RNAs (eRNAs) and often make looping contacts with their target promoters [reviewed in 95]
- Immediate-early gene
A gene whose transcription is rapidly induced (within minutes) in response to a variety of cellular stimuli, without the need for de novo protein synthesis. Many immediate-early genes encode transcription factors or other DNA-binding proteins that enable changes in cellular physiology, such as proliferation, differentiation, or storage of a memory
- Integrator cleavage module
A heterotrimeric complex comprised of IntS4, 9, and 11 that is likely responsible for Integrator endonuclease activity [56]. IntS11 contains the active site responsible for catalysis, but its associations with IntS4 and 9 are also critical for activity. The Integrator cleavage module is analogous to the core cleavage complex (CPSF73, CPSF100, and Symplekin) that is required for 3’ end processing of all pre-mRNAs [12, 13]
- Precision Run-On Sequencing (PRO-seq)
A high-throughput technique that provides base-pair-resolution mapping of transcriptionally engaged RNA polymerase II molecules across the genome [96]
- R-loop
a three-stranded nucleic acid structure that naturally forms during transcription and is comprised of a DNA:RNA hybrid, with the nontemplate DNA strand being single-stranded [97]
- RNA exosome
A multi-subunit protein complex with 3’−5’ ribonuclease activity that controls the maturation, quality, and turnover of nearly all types of eukaryotic RNAs [reviewed in 98]
- Small nuclear RNA (snRNA)
A class of abundant RNAs in eukaryotic cells that are typically 60–200 nucleotides in length and transcribed by RNAPII. The lone exception is U6, which is transcribed by RNA polymerase III. snRNAs are critical components of the major (U1, U2, U4, U5, and U6) and minor (U11, U12, U4atac, U6, and U6atac) spliceosomes, whereas U7 snRNA is involved in histone 3’ end processing [reviewed in 14]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Baillat D et al. (2005) Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123 (2), 265–76. [DOI] [PubMed] [Google Scholar]
- 2.Chen J et al. (2012) An RNAi screen identifies additional members of the Drosophila Integrator complex and a requirement for cyclin C/Cdk8 in snRNA 3’-end formation. RNA 18 (12), 2148–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malovannaya A et al. (2010) Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A 107 (6), 2431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malovannaya A et al. (2011) Analysis of the human endogenous coregulator complexome. Cell 145 (5), 787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egloff S et al. (2007) Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318 (5857), 1777–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egloff S et al. (2010) The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem 285 (27), 20564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egloff S et al. (2012) Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell 45 (1), 111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah N et al. (2018) Tyrosine-1 of RNA Polymerase II CTD Controls Global Termination of Gene Transcription in Mammals. Mol Cell 69 (1), 48–61 e6. [DOI] [PubMed] [Google Scholar]
- 9.Dominski Z et al. (2005) A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol 25 (4), 1489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandel CR et al. (2006) Polyadenylation factor CPSF-73 is the pre-mRNA 3’-end-processing endonuclease. Nature 444 (7121), 953–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y and Manley JL (2015) The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev 29 (9), 889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan KD et al. (2009) A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34 (3), 322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y et al. (2020) Structure of an active human histone pre-mRNA 3’-end processing machinery. Science 367 (6478), 700–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matera AG and Wang Z (2014) A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15 (2), 108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez N and Weiner AM (1986) Formation of the 3’ end of U1 snRNA requires compatible snRNA promoter elements. Cell 47 (2), 249–58. [DOI] [PubMed] [Google Scholar]
- 16.de Vegvar HE et al. (1986) 3’ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell 47 (2), 259–66. [DOI] [PubMed] [Google Scholar]
- 17.Medlin JE et al. (2003) The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3’ processing of U2 snRNA. EMBO J 22 (4), 925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez N (1985) Formation of the 3’ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J 4 (7), 1827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Reilly D et al. (2014) Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Res 42 (1), 264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ezzeddine N et al. (2011) A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3’-end formation. Mol Cell Biol 31 (2), 328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt D et al. (2018) The Integrator complex regulates differential snRNA processing and fate of adult stem cells in the highly regenerative planarian Schmidtea mediterranea. PLoS Genet 14 (12), e1007828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez-Orte E et al. (2019) Disruption of the Caenorhabditis elegans Integrator complex triggers a non-conventional transcriptional mechanism beyond snRNA genes. PLoS Genet 15 (2), e1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fong N et al. (2015) Effects of Transcription Elongation Rate and Xrn2 Exonuclease Activity on RNA Polymerase II Termination Suggest Widespread Kinetic Competition. Mol Cell 60 (2), 256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eaton JD et al. (2018) Xrn2 accelerates termination by RNA polymerase II, which is underpinned by CPSF73 activity. Genes Dev 32 (2), 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eaton JD and West S (2020) Termination of Transcription by RNA Polymerase II: BOOM! Trends Genet. doi: 10.1016/j.tig.2020.05.008 [DOI] [PubMed] [Google Scholar]
- 26.Yang XC et al. (2009) Studies of the 5’ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol Cell Biol 29 (1), 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hallais M et al. (2013) CBC-ARS2 stimulates 3’-end maturation of multiple RNA families and favors cap-proximal processing. Nat Struct Mol Biol 20 (12), 1358–66. [DOI] [PubMed] [Google Scholar]
- 28.Lai F et al. (2015) Integrator mediates the biogenesis of enhancer RNAs. Nature 525 (7569), 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elrod ND et al. (2019) The Integrator Complex Attenuates Promoter-Proximal Transcription at Protein-Coding Genes. Mol Cell 76 (5), 738–752 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubtsova MP et al. (2019) Integrator is a key component of human telomerase RNA biogenesis. Sci Rep 9 (1), 1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cazalla D et al. (2011) A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol Cell 43 (6), 982–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie M et al. (2015) The host Integrator complex acts in transcription-independent maturation of herpesvirus microRNA 3’ ends. Genes Dev 29 (14), 1552–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skaar JR et al. (2015) The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res 25 (3), 288–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardini A et al. (2014) Integrator regulates transcriptional initiation and pause release following activation. Mol Cell 56 (1), 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadelmayer B et al. (2014) Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun 5, 5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tatomer DC et al. (2019) The Integrator complex cleaves nascent mRNAs to attenuate transcription. Genes Dev 33 (21–22), 1525–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baillat D and Wagner EJ (2015) Integrator: surprisingly diverse functions in gene expression. Trends Biochem Sci 40 (5), 257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peart N et al. (2013) Non-mRNA 3’ end formation: how the other half lives. Wiley Interdiscip Rev RNA 4 (5), 491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rienzo M and Casamassimi A (2016) Integrator complex and transcription regulation: Recent findings and pathophysiology. Biochim Biophys Acta 1859 (10), 1269–80. [DOI] [PubMed] [Google Scholar]
- 40.Guiro J and Murphy S (2017) Regulation of expression of human RNA polymerase II-transcribed snRNA genes. Open Biol 7 (6), 170073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi H et al. (2020) The role of Mediator and Little Elongation Complex in transcription termination. Nat Commun 11 (1), 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nojima T et al. (2018) Deregulated Expression of Mammalian lncRNA through Loss of SPT6 Induces R-Loop Formation, Replication Stress, and Cellular Senescence. Mol Cell 72 (6), 970–984 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Berg DLC et al. (2017) Nipbl Interacts with Zfp609 and the Integrator Complex to Regulate Cortical Neuron Migration. Neuron 93 (2), 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fury MG and Zieve GW (1996) U6 snRNA maturation and stability. Exp Cell Res 228 (1), 160–3. [DOI] [PubMed] [Google Scholar]
- 45.Tatomer DC and Wilusz JE (2020) Attenuation of Eukaryotic Protein-Coding Gene Expression via Premature Transcription Termination. Cold Spring Harb Symp Quant Biol. doi: 10.1101/sqb.2019.84.039644 [DOI] [PubMed] [Google Scholar]
- 46.Gunther V et al. (2012) The taste of heavy metals: gene regulation by MTF-1. Biochim Biophys Acta 1823 (9), 1416–25. [DOI] [PubMed] [Google Scholar]
- 47.Lardelli RM et al. (2017) Biallelic mutations in the 3’ exonuclease TOE1 cause pontocerebellar hypoplasia and uncover a role in snRNA processing. Nat Genet 49 (3), 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmid M and Jensen TH (2019) The Nuclear RNA Exosome and Its Cofactors. Adv Exp Med Biol 1203, 113–132. [DOI] [PubMed] [Google Scholar]
- 49.Wu CW et al. (2019) RNA processing errors triggered by cadmium and integrator complex disruption are signals for environmental stress. BMC Biol 17 (1), 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Core L and Adelman K (2019) Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33 (15–16), 960–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aoi Y et al. (2020) NELF Regulates a Promoter-Proximal Step Distinct from RNA Pol II Pause-Release. Mol Cell 78 (2), 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto J et al. (2014) DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat Commun 5, 4263. [DOI] [PubMed] [Google Scholar]
- 53.Barbieri E et al. (2018) Targeted Enhancer Activation by a Subunit of the Integrator Complex. Mol Cell 71 (1), 103–116 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yue J et al. (2017) Integrator orchestrates RAS/ERK1/2 signaling transcriptional programs. Genes Dev 31 (17), 1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo Z et al. (2012) The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13 (9), 543–7. [DOI] [PubMed] [Google Scholar]
- 56.Albrecht TR et al. (2018) Integrator subunit 4 is a ‘Symplekin-like’ scaffold that associates with INTS9/11 to form the Integrator cleavage module. Nucleic Acids Res 46 (8), 4241–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albrecht TR and Wagner EJ (2012) snRNA 3’ end formation requires heterodimeric association of integrator subunits. Mol Cell Biol 32 (6), 1112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Y et al. (2017) Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance. Proc Natl Acad Sci U S A 114 (17), 4394–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen J et al. (2013) Functional analysis of the integrator subunit 12 identifies a microdomain that mediates activation of the Drosophila integrator complex. J Biol Chem 288 (7), 4867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang F et al. (2013) A core hSSB1-INTS complex participates in the DNA damage response. J Cell Sci 126 (Pt 21), 4850–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jodoin JN et al. (2013) Nuclear-localized Asunder regulates cytoplasmic dynein localization via its role in the integrator complex. Mol Biol Cell 24 (18), 2954–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y et al. (2019) The Integrator Complex Prevents Dedifferentiation of Intermediate Neural Progenitors back into Neural Stem Cells. Cell Rep 27 (4), 987–996 e3. [DOI] [PubMed] [Google Scholar]
- 63.Anderson MA et al. (2009) Asunder is a critical regulator of dynein-dynactin localization during Drosophila spermatogenesis. Mol Biol Cell 20 (11), 2709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang J et al. (2009) SOSS complexes participate in the maintenance of genomic stability. Mol Cell 35 (3), 384–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skaar JR et al. (2009) INTS3 controls the hSSB1-mediated DNA damage response. J Cell Biol 187 (1), 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y et al. (2009) HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J Biol Chem 284 (35), 23525–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang F et al. (2009) Integrator3, a partner of single-stranded DNA-binding protein 1, participates in the DNA damage response. J Biol Chem 284 (44), 30408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu P et al. (2013) Single strand DNA binding proteins 1 and 2 protect newly replicated telomeres. Cell Res 23 (5), 705–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hata T and Nakayama M (2007) Targeted disruption of the murine large nuclear KIAA1440/Ints1 protein causes growth arrest in early blastocyst stage embryos and eventual apoptotic cell death. Biochim Biophys Acta 1773 (7), 1039–51. [DOI] [PubMed] [Google Scholar]
- 70.Iwanami N et al. (2009) Ethylnitrosourea-induced thymus-defective mutants identify roles of KIAA1440, TRRAP, and SKIV2L2 in teleost organ development. Eur J Immunol 39 (9), 2606–16. [DOI] [PubMed] [Google Scholar]
- 71.Rutkowski RJ and Warren WD (2009) Phenotypic analysis of deflated/Ints7 function in Drosophila development. Dev Dyn 238 (5), 1131–9. [DOI] [PubMed] [Google Scholar]
- 72.Han SM et al. (2006) Deleted in cancer 1 (DICE1) is an essential protein controlling the topology of the inner mitochondrial membrane in C. elegans. Development 133 (18), 3597–606. [DOI] [PubMed] [Google Scholar]
- 73.Kapp LD et al. (2013) The integrator complex subunit 6 (Ints6) confines the dorsal organizer in vertebrate embryogenesis. PLoS Genet 9 (10), e1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tao S et al. (2009) The Integrator subunits function in hematopoiesis by modulating Smad/BMP signaling. Development 136 (16), 2757–65. [DOI] [PubMed] [Google Scholar]
- 75.Krall M et al. (2019) Biallelic sequence variants in INTS1 in patients with developmental delays, cataracts, and craniofacial anomalies. Eur J Hum Genet 27 (4), 582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Otani Y et al. (2013) Integrator complex plays an essential role in adipose differentiation. Biochem Biophys Res Commun 434 (2), 197–202. [DOI] [PubMed] [Google Scholar]
- 77.Sitaram P et al. (2014) asunder is required for dynein localization and dorsal fate determination during Drosophila oogenesis. Dev Biol 386 (1), 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jodoin JN et al. (2013) The snRNA-processing complex, Integrator, is required for ciliogenesis and dynein recruitment to the nuclear envelope via distinct mechanisms. Biol Open 2 (12), 1390–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oegema R et al. (2017) Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS Genet 13 (5), e1006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lykke-Andersen S and Jensen TH (2015) Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 16 (11), 665–77. [DOI] [PubMed] [Google Scholar]
- 81.Wieland I et al. (1999) Isolation of DICE1: a gene frequently affected by LOH and downregulated in lung carcinomas. Oncogene 18 (32), 4530–7. [DOI] [PubMed] [Google Scholar]
- 82.Filleur S et al. (2009) INTS6/DICE1 inhibits growth of human androgen-independent prostate cancer cells by altering the cell cycle profile and Wnt signaling. Cancer Cell Int 9, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Federico A et al. (2017) Pan-Cancer Mutational and Transcriptional Analysis of the Integrator Complex. Int J Mol Sci 18 (5), 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoshimi A et al. (2019) Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574 (7777), 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu Z and Matera AG (2014) Developmental analysis of spliceosomal snRNA isoform expression. G3 (Bethesda) 5 (1), 103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marz M et al. (2008) Evolution of spliceosomal snRNA genes in metazoan animals. J Mol Evol 67 (6), 594–607. [DOI] [PubMed] [Google Scholar]
- 87.Tan CSH et al. (2018) Thermal proximity coaggregation for system-wide profiling of protein complex dynamics in cells. Science 359 (6380), 1170–1177. [DOI] [PubMed] [Google Scholar]
- 88.Arndt KM and Reines D (2015) Termination of Transcription of Short Noncoding RNAs by RNA Polymerase II. Annu Rev Biochem 84, 381–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee NN et al. (2013) Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 155 (5), 1061–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chalamcharla VR et al. (2015) Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc Natl Acad Sci U S A 112 (51), 15548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Skene PJ and Henikoff S (2017) An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rhee HS and Pugh BF (2011) Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147 (6), 1408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He Q et al. (2015) ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol 33 (4), 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harlen KM and Churchman LS (2017) The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat Rev Mol Cell Biol 18 (4), 263–273. [DOI] [PubMed] [Google Scholar]
- 95.Li W et al. (2016) Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17 (4), 207–23. [DOI] [PubMed] [Google Scholar]
- 96.Kwak H et al. (2013) Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339 (6122), 950–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Skourti-Stathaki K and Proudfoot NJ (2014) A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 28 (13), 1384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Puno MR et al. (2019) SnapShot: The RNA Exosome. Cell 179 (1), 282–282 e1. [DOI] [PubMed] [Google Scholar]


