Abstract
Background:
Pulmonary atelectasis is frequent in clinical settings. Yet, there is limited mechanistic understanding and substantial clinical and biological controversy on its consequences. We hypothesize that atelectasis produces local transcriptomic changes related to immunity and alveolar-capillary barrier function conducive to lung injury and further exacerbated by systemic inflammation.
Methods:
Female sheep underwent unilateral lung atelectasis using a left bronchial blocker and thoracotomy while the right lung was ventilated, with (n=6) or without (n=6) systemic lipopolysaccharide infusion. Computed tomography guided samples were harvested for NextGen RNA sequencing from atelectatic and aerated lung regions. The Wald test was used to detect differential gene expression as an absolute fold change >1.5 and adjusted P-value (Padj, Benjamini-Hochberg) <0.05. Functional analysis was performed by gene set enrichment analysis.
Results:
Lipopolysaccharide-unexposed atelectatic vs aerated regions presented 2,363 differentially expressed genes. Lipopolysaccharide exposure induced 3,767 differentially expressed genes in atelectatic lungs but only 1,197 genes in aerated lung relative to the corresponding lipopolysaccharide-unexposed tissues. Gene set enrichment for immune response in atelectasis vs aerated tissues yielded negative normalized enrichment scores without lipopolysaccharide (<−1.23, Padj<0.05) but positive scores with lipopolysaccharide (>1.33, Padj<0.05). Leukocyte-related processes (e.g., leukocyte migration, activation and mediated immunity) were enhanced in lipopolysaccharide-exposed atelectasis partly through interferon-stimulated genes. Furthermore, atelectasis was associated with negatively enriched gene sets involving alveolar-capillary barrier function irrespective of lipopolysaccharide (normalized enrichment scores<−1.35, Padj<0.05). Yes-associated protein signaling was dysregulated with lower nuclear distribution in atelectatic vs aerated lung (lipopolysaccharide-unexposed: 10.0±4.2 vs 13.4±4.2 arbitrary units, lipopolysaccharide-exposed: 8.1±2.0 vs 11.3±2.4 arbitrary units, effect of lung aeration, P=0.003).
Conclusions:
Atelectasis dysregulates the local pulmonary transcriptome with negatively enriched immune response and alveolar-capillary barrier function. Systemic lipopolysaccharide converts the transcriptomic immune response into positive enrichment but does not affect local barrier function transcriptomics. Interferon-stimulated genes and Yes-associated protein might be novel candidate targets for atelectasis-associated injury.
Introduction
Atelectasis, a condition characterized by complete or partial collapse of lung regions with loss of aeration, is widely pervasive in clinical settings. Approximately 90% of 230 million patients undergoing major surgery with general anesthesia and mechanical ventilation develop atelectasis1-3. It is also found in bedridden patients and in virtually all patients with the acute respiratory distress syndrome (ARDS), which affects 10 to 86 patients per 100,000 each year in the United States, accounting for 10% of intensive care unit admissions4. Considered an injurious process5,6, atelectasis is usually reduced by open lung strategies which are thought to be beneficial for patient outcomes7. However, recent clinical trials have suggested “permissive atelectasis” as a favorable management strategy in conditions such as abdominal surgery8,9 and extra-corporeal membrane oxygenation10.
Biological and molecular information on mechanisms underlying atelectasis that might be conducive to lung injury are limited and contradictory. A few reports have shown increased inflammatory activity with neutrophil infiltration and potentiation of the neutrophil oxidative response in atelectatic lung11,12. In contrast, others found no increase in neutrophil recruitment and inflammatory cytokine production13,14 or even an inflammatory dysfunction for alveolar macrophages15. In vitro studies suggest that pulmonary endothelial cell immobility, as present in atelectatic tissue, impaired pulmonary barrier recovery ability and increased lung permeability in response to thrombin16. However, such findings, derived mostly from a small number of rodent studies, are limited in describing the structural and functional heterogeneity of the human lung, and could have been affected by the stress response, hypoxia and mechanical injury beyond atelectasis. In vitro pulmonary cell studies cannot provide the in vivo understanding of atelectatic lung tissue exposed not only to mechanical stretch but also to systemic and local chemical stimuli. Furthermore, no study identified the molecular basis for the observed changes in cellular function accompanying atelectasis. Therefore, there is a remarkable lack of knowledge on the early molecular processes underlying atelectatic lung tissue. This gap markedly limits not only the mechanistical understanding of this common potential morbid entity, but also the advance of therapeutic interventions.
We hypothesized that atelectasis would produce local transcriptomic changes in lung tissue conducive to lung injury and that these changes would be exacerbated by systemic inflammation. To investigate this hypothesis, we constructed a clinically relevant “pure” atelectasis large animal (sheep) model of one-lung collapse and mechanical ventilation with global and regional physiological properties comparable to that of humans. In vivo regional lung endophenotypes were assessed by tracer kinetics modeling of positron emission tomography showing metabolic and inflammatory activity, and barrier dysfunction, and by computed tomography images showing lung expansion. We used these imaging techniques to guide tissue sampling for deep next-generation sequencing and transcriptome analysis.
Materials and Methods
Experimental Protocol
Protocols were approved by the Subcommittee on Research Animal Care and the Institutional Animal Care and Use Committee of the Massachusetts General Hospital (Boston, Massachusetts), and followed National Institutes of Health and ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines.
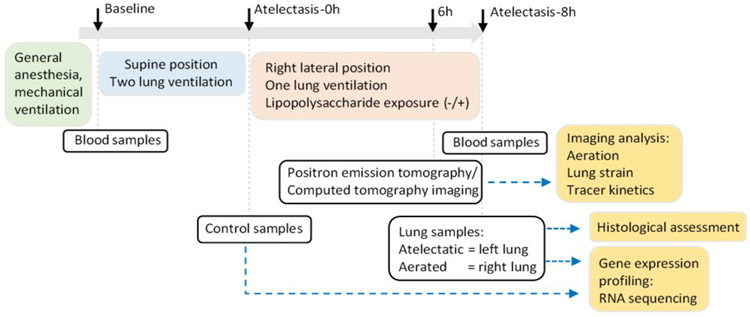
The experimental approach to imaging and tissue analysis is summarized in Figure 1 and detailed in the Supplemental Digital Content 1. Twelve female sheep (18.2±2.3 kg) were anesthetized, paralyzed, and intubated. One-lung atelectasis (left lung) was achieved using a bronchial blocker (Arndt, Cook Medical, Bloomington, IN, USA) and lateral thoracotomy, while the right lung was ventilated (tidal volume (VT)=10ml/kg, positive end-expiratory pressure (PEEP=2 cmH2O) for eight hours. Animals were divided into lipopolysaccharide-unexposed (LPS(−), n=6, 18.1±2.4 kg) or LPS-exposed (LPS(+), n=6, 18.3±2.4 kg, 5-10 ng·kg−1·min−1; Escherichia coli O55: B5, List Biologic Laboratories Inc.) group. Physiological data and samples were acquired at baseline, the beginning of lung collapse (Atelectasis-0h) and the end of the study (Atelectasis-8h).
Figure 1. Schematic diagram of experiment.
Black arrows indicate time points for sample collection.
Image acquisition and analysis
Computed tomography images were acquired for aeration and strain analysis (end-inspiration and end-expiration) and at mean lung volume for positron emission tomography attenuation correction and delineation of regions-of-interest17. Voxel gas fraction was quantified as = voxel Hounsfield units/−1000 with air = −1000 Hounsfield units and tissue = 0 Hounsfield units. Tidal strain at the voxel-level was computed by image registration of the end-inspiratory to the end-expiratory computed tomography images (voxel volume change/end-expiratory volume including lung tissue and air)17. Dynamic positron emission tomography images of 2-deoxy-2-[18F] fluoro-D-glucose (18F-FDG) were acquired to measure parameters of tissue metabolism representative of local inflammation: phosphorylation rate (k3), tissue-normalized net uptake rate (Kis) and volumes of distribution (Fes and Fees)18,19. Mean aeration, median strain and 18F-FDG kinetics were quantified in two regions of interest: the atelectatic left lung and a homogeneously aerated region of the ventilated right lung.
Tissue samples
Lung tissue samples were collected from atelectatic and aerated lung regions based on computed tomography images within 15 min after euthanasia, and from the atelectatic lung at the time of thoracotomy (control). We assessed lung water content by wet/dry ratios, Yes-associated protein (YAP) distribution by immunofluorescent staining, histological lung injury by hematoxylin and eosin stain, myeloperoxidase by immunohistochemistry, protein levels by immunoblotting, and other cytokines by enzyme-linked immunosorbent assay.
RNA-Sequencing Analysis and Validation
Total RNA from lung tissues was extracted using PAXgene tissue RNA kit (Qiagen, Germany). Gene expression was quantified by RNA-sequencing (Illumina HiSeq2500). Differentially expressed genes were defined by an absolute fold change>1.5 and an adjusted P-value (Benjamini-Hochberg)<0.05, assessed with DESeq2 Bioconductor package20. Functional analysis of enriched biological processes was performed with gene set enrichment analysis (Padj<0.05) in R, based on templates from the bcbioRNASeq package21. Transcription factor prediction analysis was performed in iRegulon (version 1.3)22. Plots were visualized in Cytoscape (http://www.cytoscape.org), GraphPad Prism software v.7.0 (GraphPad Software, USA) or Rstudio (R version 3.4.4). Gene expression was validated using real-time polymerase chain reaction (PCR).
Experimental Outcomes
Our primary outcomes were the differential regional (atelectatic vs aerated) lung transcriptomic signatures (differential gene expression, functional analysis) as assessed by RNA-sequencing. Secondary outcomes were the imaging measures of regional aeration, strain, and 18F-FDG kinetics. Exploratory outcomes and associated confirmatory variables consisted of the immunofluorescence assessment of tissue YAP, lung injury score and its components, tissue myeloperoxidase, and protein levels.
Statistical Analysis
No statistical power calculation was conducted prior to the study and sample size was determined based on a previous study indicating six biological replicates as a general guideline for RNA-seq experiments23. Data are presented as mean±SD if normally distributed and median and interquartile interval [25%, 75%] otherwise. Two-way ANOVA for repeated measurements was used for comparisons within and between groups. The interaction between conditions was included when significant. Post hoc Tukey test was used for normally distributed data and Kruskal–Wallis test otherwise. The statistical significance of mRNA expression was calculated by a paired, two-tailed Student’s t-test. Tests were two tailed and performed in R (R version 3.4.4). Significance was considered at P<0.05.
Results
Cardiopulmonary function is compromised by atelectasis and exacerbated by systemic inflammation.
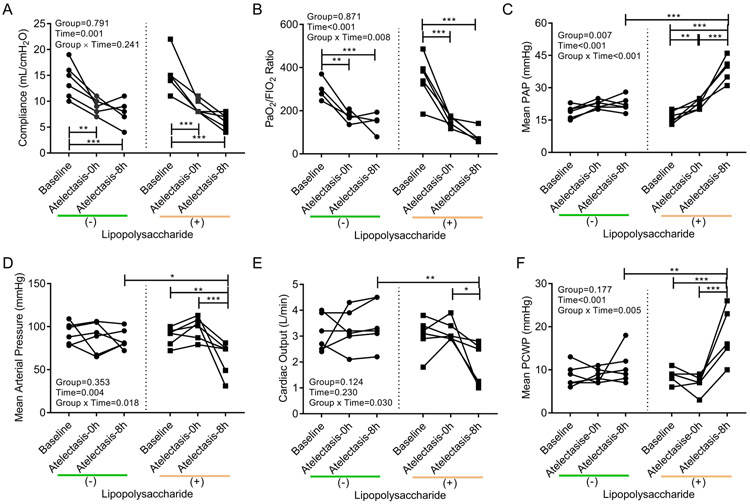
Baseline cardiopulmonary function was normal (Figure 2 and Supplemental Digital Content 1, Table S1). Eight hours of one-lung atelectasis resulted in decreased respiratory system compliance (Figure 2A) and PaO2/FIO2 ratio (Figure 2B). This hypoxemia resulted per protocol in increased FIO2 (P<0.001) and PEEP (P=0.029) at 8h with LPS exposure (Supplemental Digital Content 1, Table S1). Addition of systemic LPS to one-lung atelectasis impaired cardiovascular function with high pulmonary artery pressure (Figure 2C), low mean arterial pressure (Figure 2D), low cardiac output (Figure 2E) and high pulmonary capillary wedge pressure (Figure 2F).
Figure 2. Compromised cardiopulmonary function along 8 hours of one-lung atelectasis and systemic lipopolysaccharide (LPS) exposure.
Atelectasis and systemic LPS significantly affected respiratory system compliance (A), ratio of arterial oxygen tension (PaO2) and inspired oxygen fraction (FIO2) (B), mean pulmonary artery pressure (PAP) (C), mean arterial pressure (D), cardiac output (E), and mean pulmonary capillary wedge pressure (PCWP) (F). Lines connect data from each animal in the group exposed (+) or not (−) to LPS. P-values corresponding to analyzed effects (LPS exposure (Group), time point (Time), and their interaction (Group × Time)) are indicated. * P<0.05; ** P<0.01; *** P<0.001.
Metabolic activity is affected by atelectasis and systemic inflammation.
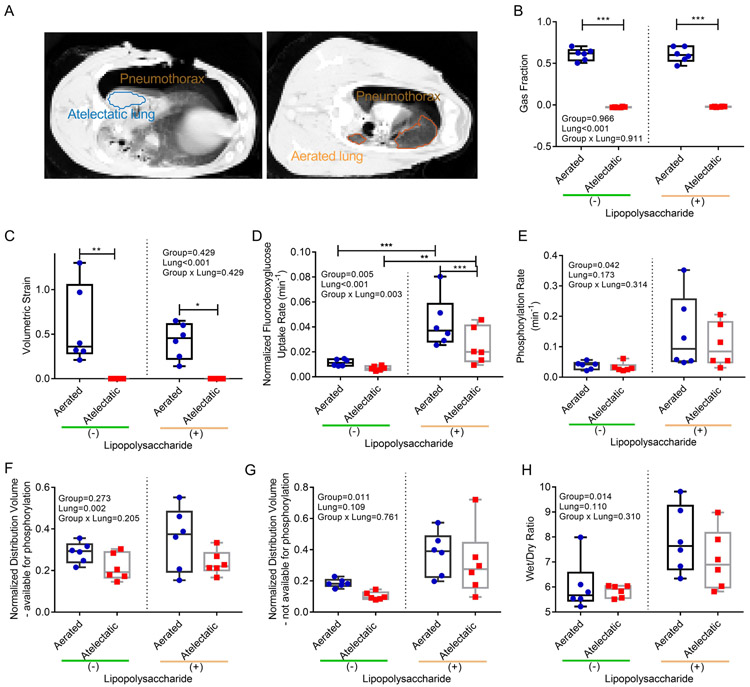
To explore processes specific to atelectasis, we compared atelectatic to normally aerated regions of the same lung (Figure 3A). As expected, the atelectatic lung had a gas fraction less than 0.1 and no volumetric strain (Figure 3B and C). The gas fraction of the aerated regions was consistent with normal-aeration (LPS(−): 0.61±0.07, LPS(+): 0.61±0.09) (Figure 3B). Volumetric strain in those regions (LPS(−): 0.58±0.44, LPS(+): 0.42±0.20) (Figure 3C) was below levels propose as injurious24.
Figure 3. Regional lung function at 6 hours of one-lung atelectasis and systemic lipopolysaccharide (LPS) exposure.
(A) Computed tomography images revealed homogeneous atelectasis throughout the left lung (blue region) and normally-aerated regions in the right lung (orange region). (B) Gas fractions were virtually zero in the atelectatic lung and normal in aerated regions both in LPS(−) and LPS(+) sheep. (C) Tidal volumetric strain showed non-injurious levels in aerated regions and no strain in the atelectatic lung. Cellular metabolic activity measured by fluorodeoxyglucose (18F-FDG) kinetics parameters showed significant effects of aeration, LPS and their interaction on tissue-normalized fluorodeoxyglucose uptake rate (Kis) (D), LPS on 18F-FDG phosphorylation rate (k3) (E), aeration on tissue-normalized distribution volume of 18F-FDG immediately available for phosphorylation (Fes) (F) , and LPS on tissue-normalized distribution volume of 18F-FDG not immediately available for phosphorylation (Fees) (G). The latter consistent with LPS effect on wet/dry ratios (H). P-values corresponding to analyzed effects (LPS exposure (Group), lung region (Lung), and their interaction (Group × Lung)) are indicated. * P<0.05; ** P<0.01; *** P<0.001.
The tissue-normalized 18F-FDG uptake rate (Kis), an in vivo measure of cellular metabolic activity, was affected by aeration (Figure 3D) and systemic inflammation (Figure 3D). LPS exposure increased Kis by 358% in atelectatic regions compared to LPS-unexposed atelectasis. It also increased cellular phosphorylation rate (k3) in lung tissue (Figure 3E). The tissue-normalized volume of distribution of 18F-FDG (Fes) was lower in atelectatic than aerated regions (Figure 3F), and contributed most to the lower Kis in atelectasis with LPS.
Tissue-normalized Fee (Fees), a measure of edema, was larger in LPS-exposed than LPS-unexposed conditions (Figure 3G), without significant difference in atelectatic vs aerated regions (Figure 3G). LPS exposure increased Fees by 206% in atelectasis (LPS(+) vs (−): 0.316±0.218 vs 0.103±0.025) and 101% in aerated regions (LPS(+) vs (−): 0.374±0.140 vs 0.186±0.027). These results were corroborated by increased wet/dry ratios with LPS exposure (Figure 3H) and significant correlation of Fees with wet/dry ratios (R=0.75, Supplemental Digital Content 1, Figure S1). Such increased wet/dry ratios and Fees with LPS occurred in the absence of significant different fluid volume administered to each group (LPS(−)=1000±316ml, LPS(+)=1250±524ml, P=0.341).
LPS exposure increased histological lung injury scores with significant capillary congestion and infiltration of neutrophils (Supplemental Digital Content 1, Figure S2 and Table S2) as well as the densities of myeloperoxidase (Supplemental Digital Content 1, Figure S3) in atelectatic and aerated regions. Circulating peripheral white blood cells decreased after LPS exposure (Supplemental Digital Content 1, Figure S4).
Regional transcriptomics are altered by aeration state and systemic LPS exposure.
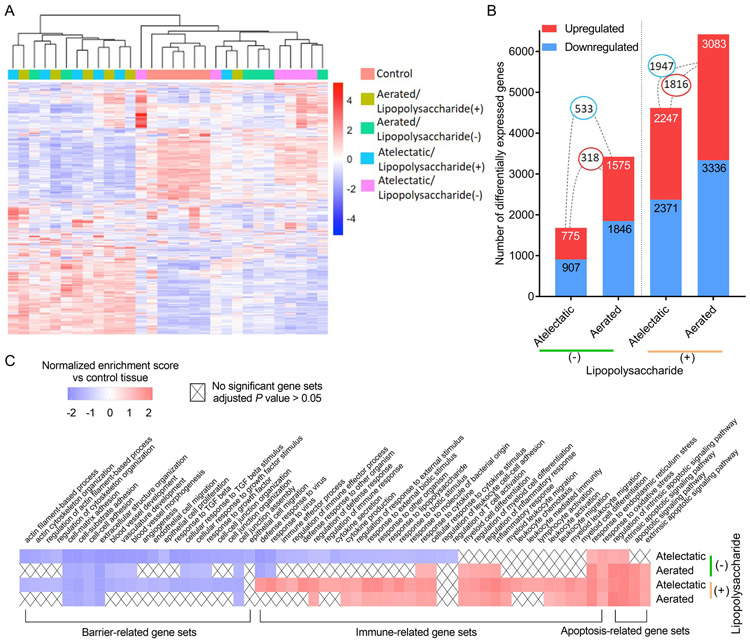
Differential expression was tested in 26,867 transcripts. Relative to control, gene expression differed according to regional aeration (atelectasis vs normal-aeration) and to LPS exposure (Fig. 4A). The number of differentially expressed genes in lung tissues increased with the presumed magnitude of injurious stimuli, i.e., level of local aeration and LPS exposure (Fig. 4B). Accordingly, LPS-unexposed atelectatic lung presented 1,682 differentially expressed genes, while 4,618 were found in LPS-exposed atelectasis. The corresponding numbers for the aerated lung were 3,421 and 6,419. Without LPS, only 17% differentially expressed genes were common to both atelectatic and aerated regions (up: 318 and down: 533), while the overlap increased to 34% with LPS (up: 1,816 and down: 1,947).
Figure 4. Regional transcriptomics differ according to regional aeration and systemic lipopolysaccharide (LPS) exposure.
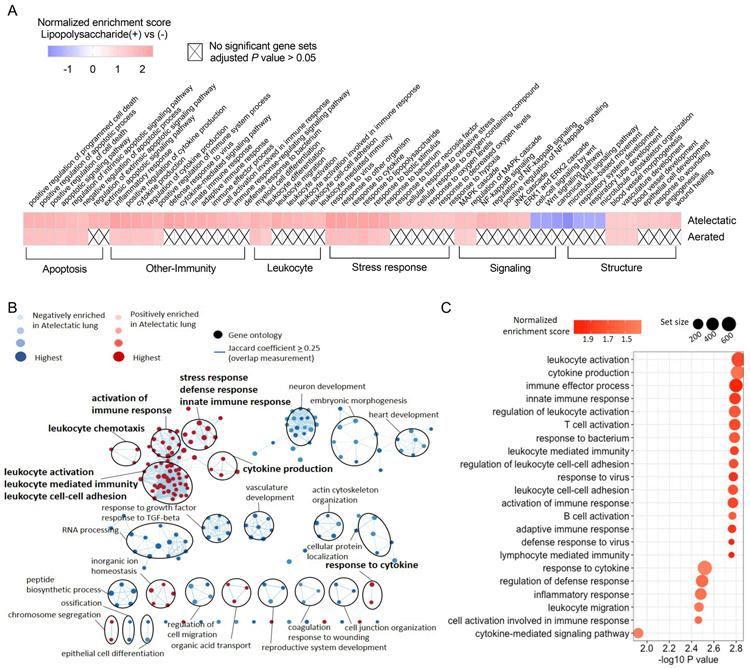
(A) Hierarchical clustering shows distinct gene expression profiles according to lung aeration and LPS exposure. Gene expression values (log2-transformed intensities) are scaled and depicted in color code format: Red: upregulated; Blue: downregulated. (B) Number of differentially expressed genes (absolute fold change>1.5 and Padj<0.05) in atelectatic and aerated tissues in LPS(−) or LPS(+) conditions vs control. Red: upregulated; Blue: downregulated. Connections between bars show the number of common genes between lung regions. (C) Gene sets differentially enriched in combination of aeration state and LPS exposure vs control. Cells are colored according to normalized enrichment scores, and "X" denotes no significance (Padj>0.05).
Functional analysis in each condition vs control (Fig. 4C and Supplemental Digital Content 2, Table S3-S6) revealed transcriptomic changes involved in apoptosis, immunity and barrier integrity. As expected, systemic LPS induced transcriptomic changes associated with immune response in both atelectatic and aerated lung regions, characterized by positive enrichment for gene sets such as cytokine production, response to lipopolysaccharide and cytokine, inflammatory response and leukocyte-associated processes. Interestingly, immune-related gene sets were negatively enriched in LPS-unexposed atelectasis. Furthermore, transcriptomic patterns associated with alveolar-capillary barrier function were dysregulated in all lung regions, independent of aeration and LPS exposure. Such dysregulation was characterized by negatively enriched gene sets including cell-matrix adhesion, extracellular structure organization and cell junction assembly. Of note, transcriptomic dysregulation in organization of actin cytoskeleton and cell junction occurred only in atelectasis, regardless of LPS exposure.
Atelectasis is associated with transcriptomic changes in immune regulation.
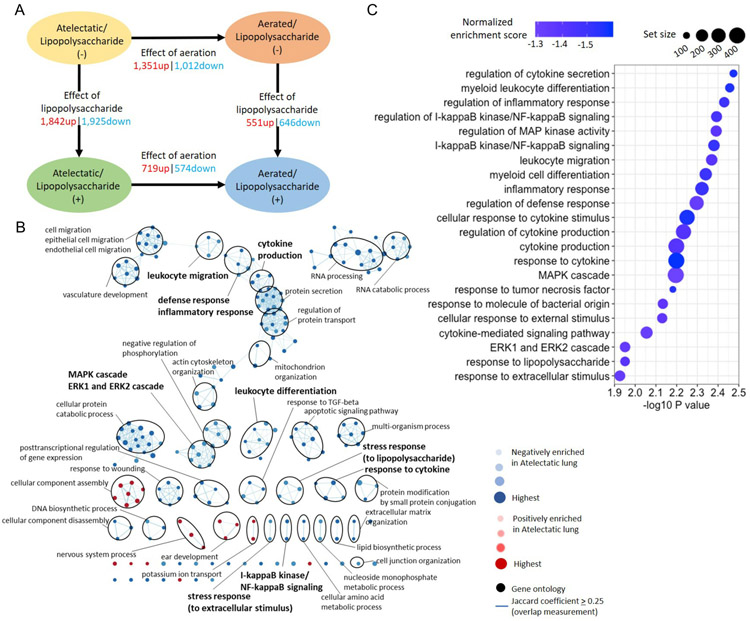
Without LPS, atelectatic vs aerated regions of the same lung presented 2,363 differentially expressed genes (8.8% of total transcripts) (Figure 5A and Supplemental Digital Content 1, Figure S5). Functional analysis revealed 206 of 226 significantly altered biological processes negatively enriched in atelectatic tissue (Figure 5B and Supplemental Digital Content 2, Table S7). For immune processes, normalized enrichment scores were negative (<−1.23, Padj<0.05, Figure 5C and Supplemental Digital Content 1, Figure S6A, S7), consistent with negative enrichment in atelectatic vs control tissues (Figure 4C). Such processes included inflammatory response, cytokine production and leukocyte migration (Figure 5C). Also negatively enriched in atelectasis were gene sets for cellular stress responses (e.g., response to cytokine, TNF, lipopolysaccharide and extracellular stimulus) and immune-associated signaling (e.g., MAPK cascade, ERK1 and ERK2 cascade, and NF-κB signaling) (Figure 5C).
Figure 5. Dysregulated transcriptomic responses of local immunity in atelectasis without systemic lipopolysaccharide (LPS) exposure.
(A) Number of differentially expressed genes (absolute fold change>1.5 and Padj<0.05) in atelectatic and aerated lung in LPS(−) or LPS(+) conditions. Up: upregulated; down: downregulated. (B) Network visualization of gene sets in atelectasis vs aerated regions. Node color and darkness indicate the signal and absolute value of normalized enrichment scores in atelectasis: red: positive; blue: negative. Node size represents the false discovery rate adjusted enrichment P-value. Overlap of genes between nodes is indicated by an edge. Immune-related gene sets are highlighted in bold. (C) Gene sets in LPS(−) relevant to immune response negatively enriched in atelectatic vs aerated lung. Node colors represent normalized enrichment score and node sizes represent the number of genes in the set.
Atelectasis increases sensitivity of lung tissue to systemic inflammation.
Transcriptomic changes due to LPS exposure were more pronounced in atelectatic than aerated tissue. LPS exposure induced 3,767 differentially expressed genes (14% of total transcripts) in atelectasis (Figure 5A), which was more than three-fold the 1,197 (4.5% of total transcripts) genes induced in aerated tissue. As expected, systemic LPS induced positive enrichment of gene sets associated with apoptosis, immune and stress response in both lung regions relative to the corresponding LPS-unexposed regions (Figure 6A and Supplemental Digital Content 2, Table S8 and S9). Interestingly, LPS produced positive enrichment of gene sets related to leukocyte function, NF-κB signaling and hypoxia (i.e., response to oxygen levels and hypoxia) only in the atelectatic lung (Figure 6A).
Figure 6. Increased immune response of the atelectatic lung to systemic lipopolysaccharide (LPS) exposure.
(A) Gene sets differentially enriched in LPS(+) vs LPS(−) conditions in the atelectatic and aerated lung. Cells are colored according to normalized enrichment scores, and "X" denotes no significance (Padj>0.05). (B) Network visualization of gene sets in atelectasis vs aerated regions during LPS(+) condition. Node color and darkness indicate the signal and absolute value of normalized enrichment scores in atelectasis: red: positive; blue: negative. Node size represents the false discovery rate adjusted enrichment P-value. Overlap of genes between nodes is indicated by an edge. Immune-related gene sets are highlighted in bold. (C) Gene sets in LPS(+) relevant to immune response positively enriched in atelectatic vs aerated regions. Node colors represent normalized enrichment score and node sizes represent the number of genes in the set.
Atelectatic vs aerated tissues with LPS yielded 1,293 differentially expressed genes (4.8% of total transcripts) (Figure 5A). Functional analysis revealed 113 of 200 significantly altered biological processes negatively enriched in atelectasis (Figure 6B and Supplemental Digital Content 2, Table S10). Of note, LPS exposure led to a shift toward enhanced immune transcriptomic response in atelectatic tissue (normalized enrichment scores>1.33, Padj<0.05, Figure 6C and Supplemental Digital Content 1, Figure S6B, S8), in contrast to the negatively enriched response without LPS (Figure 5C). Cellular stress responses and immune processes were positively enriched in LPS-exposed atelectasis (Figure 6C). These included leukocyte-related processes such as leukocyte migration, activation, mediated immunity and leukocyte cell-cell adhesion (Figure 6C).
Atelectasis dysregulates the transcriptome related to pulmonary barrier function.
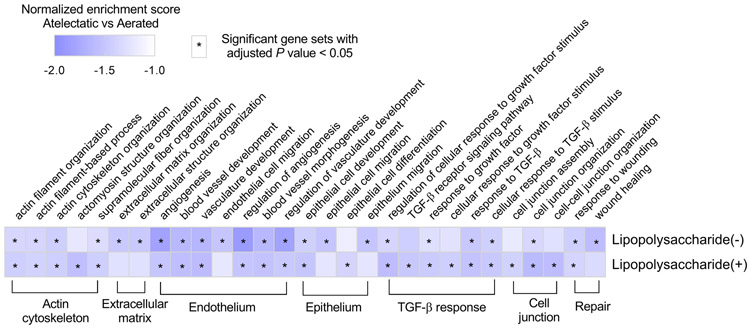
Notably, atelectasis by itself resulted in remarkable transcriptomic changes involved in pulmonary barrier function (normalized enrichment scores<−1.35, Padj<0.05, Figure 7 and Supplemental Digital Content 1, Figure S9A). Processes including angiogenesis, vasculature development, and epithelial cell development, related to major cellular structures of the alveolar-capillary barrier, were negatively enriched. Also, gene sets for actin cytoskeleton and cell junction, structural elements for the architectural integrity of the pulmonary barrier, and for tissue repair function, such as wound healing and response to wounding, were negatively enriched in atelectatic vs aerated tissue. Additionally, TGF-β signaling, relevant in wound healing, cell proliferation and differentiation, was dysregulated in atelectasis. LPS-exposed atelectatic regions showed negative enrichment for barrier integrity-related processes similar to LPS-unexposed atelectasis (normalized enrichment scores<−1.51, Padj<0.05, Figure 7 and Supplemental Digital Content 1, Figure S9B).
Figure 7. Dysregulated transcriptome of pulmonary barrier function in atelectasis independent of systemic lipopolysaccharide (LPS) exposure.
Heat map showing the similar degree of dysregulated barrier-related biological processes in atelectatic vs aerated lung in LPS(−) or LPS(+) conditions. Biological processes are grouped into functional categories. Normalized enrichment scores are scaled and depicted in color code format. Cells with asterisk denote gene sets with adjusted P value < 0.05.
ISGs are related to the response of atelectatic tissue to systemic inflammation.
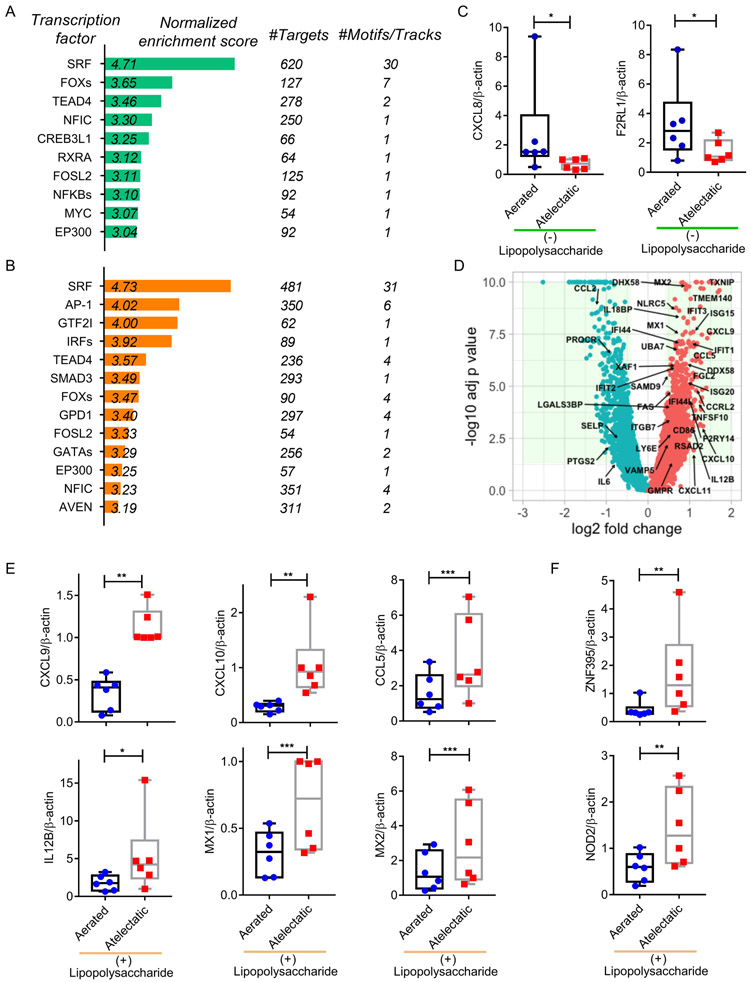
To explore the regulatory mechanisms triggering transcriptomic response to atelectasis and systemic inflammation, we performed transcription factor prediction analysis (Figure 8A, B and Supplemental Digital Content 1, Figure S10A, B). This revealed enrichment for NF-κB with low gene expression in LPS-unexposed atelectasis (Figure 8A and Supplemental Digital Content 1, Figure S10A). While total protein levels for the NF-κB component p65 were not affected by lung aeration (P=0.443, Supplemental Digital Content 1, Figure S11), NF-κB-related factors, such as CXCL8 (P=0.018) and F2RL1 (P=0.025), validated by real-time PCR, were less expressed in LPS-unexposed atelectatic tissue than in aerated regions (Figure 8C). Protein levels of CXCL8 tended to be lower in atelectatic than aerated regions in the absence of LPS (P=0.086, Supplemental Digital Content 1, Figure S12A).
Figure 8. Association of ISGs with the response of atelectatic tissue to systemic lipopolysaccharide (LPS) exposure.
Enriched transcription factors in the atelectatic vs aerated lung in LPS(−) (A) or LPS(+) (B) conditions. (C) Real-time polymerase chain reaction (PCR) validation showing lower mRNA expression for NF-κB-associated factors CXCL8 and F2RL1 in LPS-unexposed atelectasis (n=6 animals). (D) Volcano plot presenting a large subset of interferon-stimulated genes (ISGs) upregulated in LPS-exposed atelectatic. (E, F) Real-time PCR validation showing mRNA expression for upregulated ISGs (E) and ISGs regulators (F) in the atelectatic vs aerated lung during LPS exposure (n=6 animals). * P<0.05; ** P<0.01; *** P<0.001.
Predicted transcription factors differed according to systemic inflammation. LPS exposure produced significant enrichment for IRFs and STAT1/2 with relative higher gene expression in atelectasis (Figure 8B, Supplemental Digital Content 1, Figure S10B). Due to their essential function in regulation of interferon-stimulated genes (ISGs)25, IRFs/STAT1/2 were targeted by a subset of ISGs, which were more strongly expressed in LPS-exposed atelectasis (Figure 8D and Supplemental Digital Content 1, Figure S10D). We also found changes in ISGs related to bacterium response (e.g., CXCL10, ISG15, IL12B and CCL5) and leukocyte-associated processes, such as leukocyte migration (e.g., CXCL10, CXCL11 and CCL5) and activation (e.g., IL12B, CCL5 and RSAD2) (Supplemental Digital Content 1, Figure S6B), consistent with their function in immune responses and host defenses26.
Real-time PCR validation confirmed the increase of mRNA expression for ISGs, including CXCL9, CXCL10, CCL5, IL12B, MX1 and 2, in LPS-exposed atelectasis (Figure 8E). With LPS exposure, protein levels were larger for CXCL10 (P=0.022) and numerically larger for CCL5 (P=0.087) in atelectasis than in aerated regions (Supplemental Digital Content 1, Figure S12B, C). Furthermore, mRNA expression of ISGs activators was increased in LPS-exposed atelectasis (Figure 8F): ZNF395 (P=0.005), a hypoxia induced transcription factor and ISGs activator27, and NOD2 (P=0.002), a pro-inflammatory receptor with roles in interferon response and subsequent induction of ISGs28, while STAT1 protein levels were comparable in atelectatic vs aerated regions (P=0.117, Supplemental Digital Content 1, Figure S11).
YAP signaling is associated with barrier dysfunction in atelectasis.
Interestingly, transcription factor prediction analysis revealed that SRF, TEAD4 and AP-1 binding sites exhibited enrichment in the altered genes independent of systemic LPS (Figure 8A, B and Supplemental Digital Content 1, Figure S10A, B). These transcription factors were associated with a subset of differentially expressed genes, including CYR61/CCN1, CTGF/CCN2, ANKRD1, THBS1, SERPINE1, ESM1 and F3 (Supplemental Digital Content 1, Figure S10C, D). Moreover, the activity of these transcription factors is regulated by the transcriptional effector YAP, which directly controls the expression of the identified genes.
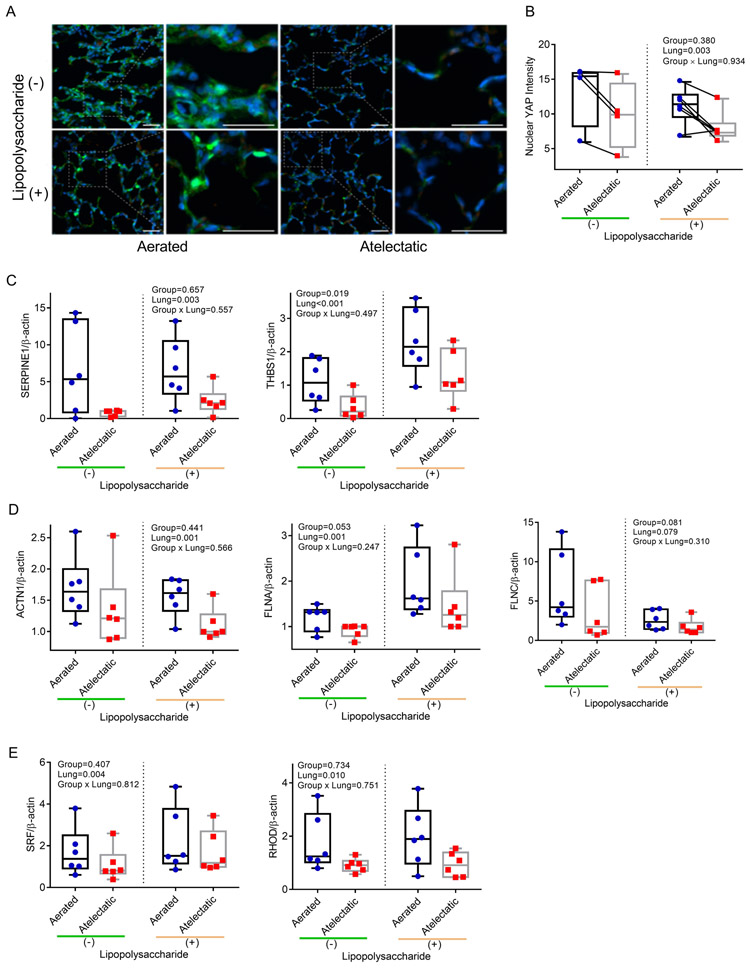
Therefore, we examined YAP and found that nuclear YAP distribution was markedly reduced in atelectatic vs aerated regions (Figure 9A, B). mRNA expression of YAP-responsive genes, such as SERPINE1 and THBS1, was also lower in atelectasis independent of LPS exposure (Figure 9C). Furthermore, lower protein levels of SERPINE1 (P=0.029) and numerically lower of THBS1 (P=0.113) were found in atelectatic vs aerated regions (Supplemental Digital Content 1, Figure S12D, E).
Figure 9. YAP is involved in atelectasis-associated barrier dysfunction.
(A) Representative images for YAP (green) from aerated and atelectatic regions in LPS(−) (n=6 animals) or LPS(+) (n=4 animals). Nuclei were stained with Hoechst in blue and autofluorescent blood cells appear in red; scale bars represent 20 μm. (B) Nuclear YAP intensity was markedly reduced in atelectatic vs aerated regions. Each connected dot pair represents atelectatic and aerated regions from a single animal. (C-E) Real-time polymerase chain reaction validation documenting lower mRNA expression for YAP-responsive genes SERPINE1 and THBS1 (C), cytoskeleton-associated proteins ACTN1, FLNA and FLNC (D) and actin dynamics associated factors SRF and RHOD (E) in atelectatic than in aerated regions (n=6 animals). P-values corresponding to analyzed effects (LPS exposure (Group), lung region (Lung), and their interaction (Group × Lung)) are indicated.
Because YAP’s role in barrier function relates to regulation and response to actin cytoskeleton dynamics29, we further validated the mRNA expression of genes involved in cytoskeleton organization (ACTN1, FLNA and FLNC). These were less expressed in atelectatic regions, even with LPS exposure (Figure 9D). Furthermore, SRF, associated with actin dynamics and YAP signaling, and RHOD, the regulator of actin cytoskeleton dynamics, presented lower mRNA expression in atelectatic vs aerated regions (Figure 9E).
Discussion
In a large animal model of unilateral lung collapse with global cardiopulmonary measurements consistent with clinical conditions, we documented transcriptomic changes in atelectasis indicative of dysregulated pulmonary immunity and alveolar-capillary barrier, two major processes in the development of acute lung injury. Exposure to systemic inflammation exacerbated lung injury in atelectatic tissue and enhanced its transcriptomic immune response, particularly leukocyte-related processes, which became larger than present in aerated lung and at least in part attributable to ISGs. Pulmonary barrier dysfunction was associated with defective distribution of YAP independent of systemic inflammation. Therefore, our study of injurious transcriptomic profiles in atelectatic lung regions provides new data to inform clinical controversies on tolerance of atelectasis in patients7-9 and targets for mechanistic studies on the biology and molecular management of atelectasis.
While atelectasis is a major pathophysiological process in clinical anesthesia, there is poor understanding of its biological and molecular mechanisms and their relationship to lung injury. Our experimental model aimed to mimic atelectasis and cardiopulmonary conditions comparable to those found in humans. Histological lung injury was mild in atelectatic and normally aerated regions and further exacerbated by LPS exposure. This is consistent with previous findings in atelectatic and aerated regions in large animals with either non-injurious17,30 or moderately injurious ventilation settings31,32. The used VT=10 ml/kg aimed to increase the normally-aerated lung33. Our measurements of regional lung aeration and strain confirmed the absence of overdistension in sampled areas.
Gene expression profiles in atelectatic tissue were dramatically distinct from those in aerated regions despite their comparable histological findings. Independent of systemic inflammation, more than half of the biological processes, including metabolic processes in RNA, lipid and protein, were negatively enriched in atelectatic tissue. Together with previous findings34 and our in vivo 18F-FDG measurement of regional lower metabolic activity (Kis), these findings indicate less cellular activation due to atelectasis.
In LPS-unexposed atelectasis, gene sets involved in cytokine production, leukocyte migration and cellular stress, all fundamental immune responses, were negatively enriched. Functional and transcription factor prediction analyses identified NF-κB signaling, the classic pathway in inflammation, immune and stress responses, as involved in such dysregulation. Additionally, NF-κB associated factors (CXCL8 and F2RL1) were less expressed in atelectatic than aerated regions. These findings support the concept of local immune dysregulation in atelectatic tissue likely through NF-κB signaling.
They are also consistent with previous findings of impaired alveolar macrophage function15 and less regional lung injury30,31 in atelectasis. However, they contrast with reports of atelectasis association with a proinflammatory state in rats11,12. The discrepancy might be explained by the different species, the duration of atelectasis and unknown sterility of rodent preparation. Our large animal model allowed for mimicking of clinical settings preserving sterility, and including alterations in regional lung blood volume, and exposure to a systemic inflammatory trigger and circulating inflammatory cells and mediators.
LPS exposure resulted in positively enriched transcriptomic patterns of immune responses, particularly leukocyte-related processes, in atelectatic vs aerated tissue. Such finding contrasted with their negative enrichment in LPS-unexposed atelectasis. Our genomic findings are likely due to changes in cellular transcriptomic activity in atelectatic tissue by LPS exposure, rather than to changes in local cellular composition. This is because neutrophilic infiltrates assessed by counts and myeloperoxidase were similar in atelectatic and aerated regions during LPS exposure, and the fraction of parenchymal cells in sampled tissues were likely similar in the acute condition. Expanding a previous report of vulnerability to infection in atelectatic lung exposed to Group B streptococci35, our findings suggest that the altered transcriptomic response of the atelectatic lung could contribute to its susceptibility to inflammatory stimuli.
ISGs are a broad gene family in immunity and host defenses,26 and with high expression in stimulated blood leukocytes36. For instance, CXCL9 and CXCL10 promote the recruitment of inflammatory cells (i.e., neutrophils, T-lymphocytes, monocytes and Natural killers)37. Our findings of upregulated ISGs in LPS-exposed atelectasis suggest their association with increased sensitivity of atelectasis to systemic inflammation. Such association was reinforced by functional analysis showing positively enriched leukocyte migration and by higher CXCL10 protein levels in LPS-exposed atelectasis. Recently, ISGs have been related to ARDS severity and prognosis38,39, further supporting their role in lung injury. Our data also suggest that ISGs in atelectatic tissue could be regulated through transcription factors IRFs and STATs25 and by regulators ZNF39527 and NOD228. Therefore, our results provide molecular support to the frequently stated but poorly demonstrated cause-effect relationship between atelectasis and pneumonia, ventilator-induced lung injury and postoperative pulmonary complications6. They simultaneously raise concerns on consequences of currently proposed management methods allowing for substantial lung collapse8-10, particularly in the presence of substantial inflammatory response.
We also found that atelectasis was associated with negatively enriched biological processes related to structural cells (endothelium and epithelium), structural elements (cytoskeleton and cell junction) and tissue repair function. Of note, such processes were still negatively enriched in LPS-exposed atelectasis, implying that atelectasis dysregulated barrier function transcriptomics independent of LPS exposure. These results contrast with the local transcriptomic response associated with immunity, which shifted from inhibited without to enhanced with LPS. While wet/dry ratios and Fees were lower in atelectatic than aerated regions, the numerically larger increase of Fees in atelectasis by LPS relative to aerated regions suggest higher susceptibility of atelectasis to permeability increase, consistent with transcriptomic findings. The LPS-exposed atelectatic sample with highest Fees clustered well with the transcriptomic data of other samples suggesting no undue effect on transcriptomics analysis.
Our findings pointed to reduced YAP signaling in atelectasis: less nuclear YAP distribution, low levels of YAP-responsive proteins SERPINE1 and THBS1, and less expression of cytoskeleton-associated genes (e.g., ACTN1, FLNA, FLNC, SRF and RHOD), independent of LPS exposure. YAP, the primary transcriptional effector of the Hippo pathway, is distributed in respiratory epithelial cells40. With roles in lung homeostasis and development41, YAP was increased after murine lung injury due to hemorrhage and LPS42, and recently associated with alveolar epithelium repair and regeneration following pneumonectomy43 or bacterial pneumonia44. It also contributes to barrier function by regulating and responding to actin cytoskeleton dynamics29. Lack of cyclic stretch45 and presence of cell contact46 during atelectasis could modulate the Hippo-YAP pathway. Thus, our data suggest that reduced YAP activity may participate in atelectasis-associated barrier dysfunction by compromising cytoskeleton organization and cell repair. Such findings constitute potential mechanisms for previous reports of increased permeability with pulmonary ultrastructural cell damage (i.e., microvascular disruption and epithelial damage)47,48 and vascular injury49 in atelectatic regions.
Our findings of reduced nuclear YAP in atelectasis, and protein levels consistent with the immune and alveolar-capillary barrier function provide plausibility to our transcriptomic data. The asynchrony between gene expression and protein synthesis is well documented, implying that transcriptomic changes are not necessarily simultaneous with corresponding protein levels50. Future studies will need to address the effects of the reported early regional transcriptional patterns on regional cellular function, protein levels, and their time courses.
Our use of image-guided tissue sampling not only allowed for better characterization of the tissue samples with their corresponding in vivo imaging endophenotypes, but also for paired statistics (aerated×atelectatic lung). This approach reduces the number of studied animals and may be considered in future studies in the field.
Our study has limitations. We studied only female sheep. While this reduces biological variability, it may limit the generalization of the results to males as it is unknown whether atelectasis-related lung injury presents sex dependence. Whereas non-dependent single-lung collapse with pneumothorax reflects conditions during thoracic surgery, associated regional intrapulmonary tissue pressures and perfusion patterns likely differ from those encountered in the more common clinical presentation of dependent atelectasis, and could determine different inflammatory responses in the distinct atelectasis conditions. Our animal model describes acute atelectasis (8h) as present in the first hours following intubation in critical care and surgical settings. Thus, results are expected to relate to mechanisms of lung injury in the early stages of atelectasis and did not address changes related to re-expansion. Our direct measurements for barrier damage were limited to wet/dry ratios. The exact cellular sources and functional roles of ISGs and YAP in atelectasis could not be established in this large animal study. Mechanistic studies with pharmacological modulation in models allowing for genetic manipulation will be required to establish the role of the identified pathways in atelectatic injury. While further investigation will be necessary to assess these topics, our study strongly indicates that atelectatic lung is associated with marked transcriptomic changes in immune and barrier dysfunction.
In conclusion, our study identified a local differential transcriptomic response consistent with impairment of the acute pulmonary immune function in atelectatic vs aerated lung in the absence of any additional inflammatory stimulus. In contrast, a distinct inflammatory pattern with augmentation of the transcriptomic immune response was present in atelectatic tissue exposed to systemic LPS. Locally increased ISGs were potentially associated with the atelectatic tissue response to systemic LPS. Our results also documented the dysregulation of transcriptomic responses related to alveolar-capillary barrier integrity in atelectatic tissue irrespective of systemic inflammation. This dysregulation was likely associated with YAP signaling. Therefore, modulators of these pathways might be novel candidate targets in atelectasis-associated lung injury.
Supplementary Material
6. Acknowledgements:
The authors thank Steve Weise, Department of Radiology (Nuclear Medicine and Molecular Imaging), Massachusetts General Hospital, Boston, Massachusetts, for the expert technical support with computed tomographic and positron emission tomographic imaging. We also thank Dana-Farber/Harvard Cancer Center in Boston, MA, for the use of the Specialized Histopathology Core, which provided histology and immunohistochemistry service. Dana-Farber/Harvard Cancer Center is supported in part by an NCI Cancer Center Support Grant # NIH 5 P30 CA06516.
10. Funding Statement: This work was supported by NIH-NHLBI grant RO1 HL121228.
Footnotes
Clinical trial number and registry URL: Not applicable.
Prior Presentations: Part of the work has been presented at the American Thoracic Society 2019 International Conference in Dallas, Texas, May 17-22, 2019.
Conflicts of Interest: Dr. Hutchinson received funding from the Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, National Institutes of Health Award UL 1TR002541), the Harvard Medical School Foundry, the NIEHS Center for Environmental Science, the Harvard Center for AIDS Research, Aztra-Zeneca and Boehringer-Ingelheim and Dr. Baron received funding from Merck for projects not related to the current work. The remaining authors declare no conflicts of interest.
References
- 1.Weiser TG, Regenbogen SE, Thompson KD, Haynes AB, Lipsitz SR, Berry WR, Gawande AA: An estimation of the global volume of surgery: a modelling strategy based on available data. The Lancet 2008; 372:139–44 [DOI] [PubMed] [Google Scholar]
- 2.Duggan M, Kavanagh BP: Pulmonary Atelectasis: A Pathogenic Perioperative Entity. Anesthesiology 2005; 102:838–54 [DOI] [PubMed] [Google Scholar]
- 3.Hedenstierna G, Edmark L: Mechanisms of atelectasis in the perioperative period. Best Pract Res Clin Anaesthesiol 2010; 24:157–69 [DOI] [PubMed] [Google Scholar]
- 4.Thompson BT, Chambers RC, Liu KD: Acute Respiratory Distress Syndrome N Engl J Med Edited by Drazen JM. 2017; 377:562–72 [DOI] [PubMed] [Google Scholar]
- 5.Muders T, Wrigge H: New insights into experimental evidence on atelectasis and causes of lung injury. Best Pract Res Clin Anaesthesiol 2010; 24:171–82 [DOI] [PubMed] [Google Scholar]
- 6.Tusman G, Böhm SH, Warner DO, Sprung J: Atelectasis and perioperative pulmonary complications in high-risk patients. Current Opinion in Anaesthesiology 2012; 25:1–10 [DOI] [PubMed] [Google Scholar]
- 7.Futier E, Constantin J-M, Paugam-Burtz C, Pascal J, Eurin M, Neuschwander A, Marret E, Beaussier M, Gutton C, Lefrant J-Y, Allaouchiche B, Verzilli D, Leone M, De Jong A, Bazin J-E, Pereira B, Jaber S: A Trial of Intraoperative Low-Tidal-Volume Ventilation in Abdominal Surgery. N Engl J Med 2013; 369:428–37 [DOI] [PubMed] [Google Scholar]
- 8.Writing Committee for the PROBESE Collaborative Group of the PROtective VEntilation Network (PROVEnet) for the Clinical Trial Network of the European Society of Anaesthesiology, Bluth T, Serpa Neto A, Schultz MJ, Pelosi P, Gama de Abreu M: Effect of Intraoperative High Positive End-Expiratory Pressure (PEEP) With Recruitment Maneuvers vs Low PEEP on Postoperative Pulmonary Complications in Obese Patients: A Randomized Clinical Trial. JAMA 2019; 321:2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.PROVE Network Investigators for the Clinical Trial Network of the European Society of Anaesthesiology, Hemmes SNT, Gama de Abreu M, Pelosi P, Schultz MJ: High versus low positive end-expiratory pressure during general anaesthesia for open abdominal surgery (PROVHILO trial): a multicentre randomised controlled trial. Lancet 2014; 384:495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marhong JD, Telesnicki T, Munshi L, Del Sorbo L, Detsky M, Fan E: Mechanical Ventilation during Extracorporeal Membrane Oxygenation. An International Survey. Annals ATS 2014; 11:956–61 [DOI] [PubMed] [Google Scholar]
- 11.Tojo K, Nagamine Y, Yazawa T, Mihara T, Baba Y, Ota S, Goto T, Kurahashi K: Atelectasis causes alveolar hypoxia-induced inflammation during uneven mechanical ventilation in rats. ICMx 2015; 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minamiya Y, Saito H, Takahashi N, Kawai H, Ito M, Hosono Y, Motoyama S, Ogawa J: Polymorphonuclear leukocytes are activated during atelectasis before lung reexpansion in rat. Shock 2008; 30:81–6 [DOI] [PubMed] [Google Scholar]
- 13.Chu EK, Whitehead T, Slutsky AS: Effects of cyclic opening and closing at low- and high-volume ventilation on bronchoalveolar lavage cytokines*. Critical Care Medicine 2004; 32:168–74 [DOI] [PubMed] [Google Scholar]
- 14.Nakamura M, Fujishima S, Sawafuji M, Ishizaka A, Oguma T, Soejima K, Matsubara H, Tasaka S, Kikuchi K, Kobayashi K, Ikeda E, Sadick M, Hebert CA, Aikawa N, Kanazawa M, Yamaguchi K: Importance of Interleukin-8 in the Development of Reexpansion Lung Injury in Rabbits. Am J Respir Crit Care Med 2000; 161:1030–6 [DOI] [PubMed] [Google Scholar]
- 15.Shennib H: The Effects of Pulmonary Atelectasis and Reexpansion on Lung Cellular Immune Defenses. Arch Surg 1984; 119:274. [DOI] [PubMed] [Google Scholar]
- 16.Birukov KG, Jacobson JR, Flores AA, Ye SQ, Birukova AA, Verin AD, Garcia JGN: Magnitude-dependent regulation of pulmonary endothelial cell barrier function by cyclic stretch. American Journal of Physiology-Lung Cellular and Molecular Physiology 2003; 285:L785–97 [DOI] [PubMed] [Google Scholar]
- 17.Motta-Ribeiro GC, Hashimoto S, Winkler T, Baron RM, Grogg K, Paula LFSC, Santos A, Zeng C, Hibbert K, Harris RS, Bajwa E, Vidal Melo MF: Deterioration of Regional Lung Strain and Inflammation during Early Lung Injury. Am J Respir Crit Care Med 2018; 198:891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prost de N, Feng Y, Wellman T, Tucci MR, Costa EL, Musch G, Winkler T, Harris RS, Venegas JG, Chao W, Vidal Melo MF: 18F-FDG kinetics parameters depend on the mechanism of injury in early experimental acute respiratory distress syndrome. J Nucl Med 2014; 55:1871–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder T, Vidal Melo MF, Musch G, Harris RS, Venegas JG, Winkler T: Modeling Pulmonary Kinetics of 2-Deoxy-2-[18F]fluoro-d-glucose During Acute Lung Injury. Academic Radiology 2008; 15:763–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinbaugh MJ, Pantano L, Kirchner RD, Barrera V, Chapman BA, Piper ME, Mistry M, Khetani RS, Rutherford KD, Hofmann O, Hutchinson JN, Ho Sui S: bcbioRNASeq: R package for bcbio RNA-seq analysis. F1000Res 2018; 6:1976 [Google Scholar]
- 22.Janky R, Verfaillie A, Imrichová H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, Svetlichnyy D, Kalender Atak Z, Fiers M, Marine J-C, Aerts S: iRegulon: From a Gene List to a Gene Regulatory Network Using Large Motif and Track Collections PLoS Comput Biol Edited by Bussemaker HJ. 2014; 10:e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schurch NJ, Schofield P, Gierliński M, Cole C, Sherstnev A, Singh V, Wrobel N, Gharbi K, Simpson GG, Owen-Hughes T, Blaxter M, Barton GJ: How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016; 22:839–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Protti A, Cressoni M, Santini A, Langer T, Mietto C, Febres D, Chierichetti M, Coppola S, Conte G, Gatti S, Leopardi O, Masson S, Lombardi L, Lazzerini M, Rampoldi E, Cadringher P, Gattinoni L: Lung Stress and Strain during Mechanical Ventilation: Any Safe Threshold? Am J Respir Crit Care Med 2011; 183:1354–62 [DOI] [PubMed] [Google Scholar]
- 25.Tamura T, Yanai H, Savitsky D, Taniguchi T: The IRF Family Transcription Factors in Immunity and Oncogenesis. Annu Rev Immunol 2008; 26:535–84 [DOI] [PubMed] [Google Scholar]
- 26.Schneider WM, Chevillotte MD, Rice CM: Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu Rev Immunol 2014; 32:513–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroeder L, Herwartz C, Jordanovski D, Steger G: ZNF395 Is an Activator of a Subset of IFN-Stimulated Genes. Mediators of Inflammation 2017; 2017:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura S, Davis KM, Weiser JN: Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J Clin Invest 2011; 121:3657–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Kim YH, Kim J, Park DY, Bae H, Lee D-H, Kim KH, Hong SP, Jang SP, Kubota Y, Kwon Y-G, Lim D-S, Koh GY: YAP/TAZ regulates sprouting angiogenesis and vascular barrier maturation. Journal of Clinical Investigation 2017; 127:3441–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tucci MR, Costa ELV, Wellman TJ, Musch G, Winkler T, Harris RS, Venegas JG, Amato MBP, Melo MFV: Regional Lung Derecruitment and Inflammation during 16 Hours of Mechanical Ventilation in Supine Healthy Sheep. Anesthesiology 2013; 119:156–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa ELV, Musch G, Winkler T, Schroeder T, Harris RS, Jones HA, Venegas JG, Vidal Melo MF: Mild Endotoxemia during Mechanical Ventilation Produces Spatially Heterogeneous Pulmonary Neutrophilic Inflammation in Sheep. Anesthesiology 2010; 112:658–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis JM, Dickerson B, Metlay L, Penney DP: Differential effects of oxygen and barotrauma on lung injury in the neonatal piglet. Pediatr Pulmonol 1991; 10:157–63 [DOI] [PubMed] [Google Scholar]
- 33.Kozian A, Schilling T, Schütze H, Senturk M, Hachenberg T, Hedenstierna G: Ventilatory protective strategies during thoracic surgery: effects of alveolar recruitment maneuver and low-tidal volume ventilation on lung density distribution. Anesthesiology 2011; 114:1025–35 [DOI] [PubMed] [Google Scholar]
- 34.Bellani G, Guerra L, Musch G, Zanella A, Patroniti N, Mauri T, Messa C, Pesenti A: Lung Regional Metabolic Activity and Gas Volume Changes Induced by Tidal Ventilation in Patients with Acute Lung Injury. Am J Respir Crit Care Med 2011; 183:1193–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaam van AH, Lachmann RA, Herting E, De Jaegere A, Iwaarden van F, Noorduyn LA, Kok JH, Haitsma JJ, Lachmann B: Reducing Atelectasis Attenuates Bacterial Growth and Translocation in Experimental Pneumonia. Am J Respir Crit Care Med 2004; 169:1046–53 [DOI] [PubMed] [Google Scholar]
- 36.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, Chaussabel D: Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 2007; 109:2066–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groom JR, Luster AD: CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 2011; 89:207–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nick JA, Caceres SM, Kret JE, Poch KR, Strand M, Faino AV, Nichols DP, Saavedra MT, Taylor-Cousar JL, Geraci MW, Burnham EL, Fessler MB, Suratt BT, Abraham E, Moss M, Malcolm KC: Extremes of Interferon-Stimulated Gene Expression Associate with Worse Outcomes in the Acute Respiratory Distress Syndrome PLoS ONE Edited by Rosenberger P. 2016; 11:e0162490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Englert JA, Cho MH, Lamb AE, Shumyatcher M, Barragan-Bradford D, Basil MC, Higuera A, Isabelle C, Vera MP, Dieffenbach PB, Fredenburgh LE, Kang JB, Bhatt AS, Antin JH, Ho VT, Soiffer RJ, Howrylak JA, Himes BE, Baron RM: Whole blood RNA sequencing reveals a unique transcriptomic profile in patients with ARDS following hematopoietic stem cell transplantation. Respir Res 2019; 20:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lange AW, Sridharan A, Xu Y, Stripp BR, Perl A-K, Whitsett JA: Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. Journal of Molecular Cell Biology 2015; 7:35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isago H, Mitani A, Mikami Y, Horie M, Urushiyama H, Hamamoto R, Terasaki Y, Nagase T: Epithelial Expression of YAP and TAZ Is Sequentially Required in Lung Development. Am J Respir Cell Mol Biol 2020; 62:256–66 [DOI] [PubMed] [Google Scholar]
- 42.Hu C, Sun J, Du J, Wen D, Lu H, Zhang H, Xue Y, Zhang A, Yang C, Zeng L, Jiang J: The Hippo–YAP pathway regulates the proliferation of alveolar epithelial progenitors after acute lung injury. Cell Biol Int 2019; 43:1174–83 [DOI] [PubMed] [Google Scholar]
- 43.Liu Z, Wu H, Jiang K, Wang Y, Zhang W, Chu Q, Li J, Huang H, Cai T, Ji H, Yang C, Tang N: MAPK-Mediated YAP Activation Controls Mechanical-Tension-Induced Pulmonary Alveolar Regeneration. Cell Reports 2016; 16:1810–9 [DOI] [PubMed] [Google Scholar]
- 44.LaCanna R, Liccardo D, Zhang P, Tragesser L, Wang Y, Cao T, Chapman HA, Morrisey EE, Shen H, Koch WJ, Kosmider B, Wolfson MR, Tian Y: Yap/Taz regulate alveolar regeneration and resolution of lung inflammation. Journal of Clinical Investigation 2019; 129:2107–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Codelia VA, Sun G, Irvine KD: Regulation of YAP by Mechanical Strain through Jnk and Hippo Signaling. Current Biology 2014; 24:2012–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gumbiner BM, Kim N-G: The Hippo-YAP signaling pathway and contact inhibition of growth. J Cell Sci 2014; 127:709–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fanelli V, Mascia L, Puntorieri V, Assenzio B, Elia V, Fornaro G, Martin EL, Bosco M, Delsedime L, Fiore T, Grasso S, Ranieri VM: Pulmonary atelectasis during low stretch ventilation: “Open lung” versus “lung rest” strategy*. Critical Care Medicine 2009; 37:1046–53 [DOI] [PubMed] [Google Scholar]
- 48.Duggan M, McCaul CL, McNamara PJ, Engelberts D, Ackerley C, Kavanagh BP: Atelectasis Causes Vascular Leak and Lethal Right Ventricular Failure in Uninjured Rat Lungs. Am J Respir Crit Care Med 2003; 167:1633–40 [DOI] [PubMed] [Google Scholar]
- 49.Duggan M, McNamara PJ, Engelberts D, Pace-Asciak C, Babyn P, Post M, Kavanagh BP: Oxygen Attenuates Atelectasis-induced Injury in the In Vivo Rat Lung. Anesthesiology 2005; 103:522–31 [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Beyer A, Aebersold R: On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016; 165:535–50 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.