Abstract
Lysosomes transcend the role of degradation stations, acting as key nodes to inter-organelle crosstalk and signal transduction. Lysosomes communicate with the nucleus through physical proximity and functional interaction. In response to external and internal stimuli, lysosomes actively adjust their distribution between peripheral and perinuclear regions and modulate lysosome-to-nucleus signaling pathways, and the nucleus in turn fine-tunes lysosomal biogenesis and functions through transcriptional controls. Changes in coordination between these two essential organelles are associated with metabolic disorders, neurodegenerative diseases, and aging. This review addresses recent advances in lysosome-and-nucleus communication by multi-tiered regulatory mechanisms and discusses how these regulations couple metabolic inputs with organellar motility, cellular signaling, and transcriptional network.
Keywords: lysosome-to-nucleus signaling, lysosome positioning, lysosomal adaptation, lysosomal metabolites, transcription factors
Lysosomes as degradation and signaling hubs
Lysosomes were first discovered by the Nobel laureate Christian de Duve in 1955 as a “scientific serendipity” [1], which are later found in nearly all eukaryotic cells (vacuoles in plants and yeasts; lysosomes in animals) and have the reputation of being “waste-to-energy” incinerators inside the cell. Enclosed in a single phospholipid bilayer membrane, lysosomes maintain an acidic interior (pH 4.5~5.5) through pumping cytosolic protons into the internal lumen by vacuolar H+-ATPase, which is aided by other lysosomal ion channels [2]. The acidic lumen of lysosomes accommodates approximately 60 acidic hydrolases and provides them an optimal environment to break down various biological macromolecules [3, 4]. These biological macromolecules are delivered into lysosomes from extracellular or intracellular space through endocytosis (see Glossary) or autophagy (Box 1), respectively [5–7], and their degradation products, such as amino acids, fatty acids, monosaccharides and nucleosides, are exported to the cytosol for reuse via lysosomal transporters and/or vesicle trafficking [8–10]. In several specialized cell types, lysosomes are the critical part of the secretory pathway known as lysosomal exocytosis that is essential for plasma membrane repair and cholesterol homeostasis [11–14].
BOX 1: Lysosomes in autophagy.
In response to nutrient deprivation, autophagy is a highly regulated catabolic process that captures unwanted intracellular components, including macromolecules and even entire organelles and delivers them to lysosomes for bulk degradation and recycling. Three major types of autophagy have been identified in eukaryotes: chaperone-mediated autophagy (CMA), microautophagy, and macroautophagy. CMA and microautophagy both take place directly on lysosomes. In CMA, protein substrates with KFERQ-like motifs are recognized by HSC70 and co-chaperones and internalized into the lysosome with the aid of lysosome-associated membrane protein type 2A (LAMP2A) [114–116] . In microautophagy, cytosolic materials are directly captured into the lysosome via the invagination of lysosomal membrane [117]. On the other hand, macroautophagy, a major type of autophagy, requires extensive membrane remodeling and the formation of autophagosome, a specialized double membrane-bound vesicle that engulfs cytoplasmic contents and delivers these cargos to the lysosome through direct fusion (autolysosome) [118, 119]. Upon the fusion, the cargos are degraded inside the acidic lumen of autolysosome by various hydrolases for recycling. All three forms of autophagy depend on the proper function of lysosomes.
In addition to their well-appreciated housekeeping function as cellular degradative compartments, lysosomes now emerge as crucial signaling hubs to relay external and internal stimuli, such as growth factors, nutrient availability, amino acid, glucose and lipid metabolism, to signaling complexes like mammalian/mechanistic target of rapamycin (mTOR) and AMP activated protein kinase (AMPK) and undergo adaptation to meet cellular needs [15–17]. The adaption of lysosomes occurs at multiple layers, and the communication between the lysosome and the nucleus plays a crucial role in this process. Here, we overview three regulatory layers of lysosome-and-nucleus communication, including peripheral-to-perinuclear positioning of lysosomes, nuclear translocation of signaling factors from lysosomes, and transcriptional control of lysosomal adaptation by nuclear factors. We highlight what molecular players are involved in these regulations, how these regulations couple with metabolic and environmental changes, and how these discoveries have broadened our view of lysosomal contributions to health, diseases, and aging (Box 2).
BOX2: Lysosome dysfunction and diseases.
Dysfunction of lysosomes has been associated with multiple human diseases. Mutations in lysosomal hydrolase and/or transporter genes result in aberrant accumulation of a variety of metabolites within the lysosomal lumen, leading to metabolic diseases collectively called lysosomal storage diseases (LSDs) [120–122]. Dysfunction of lysosomal-autophagic pathways is detected in neurodegenerative diseases, such as Parkinson’s, Alzheimer’s, Huntington’s diseases and amyotrophic lateral sclerosis (ALS), that share a common feature with neuronal disposition of protein aggregates [123–126]. Defects in lysosome trafficking machinery are also involved in LSDs and neurodegenerative diseases, as reviewed in [127]. Overexpression of TFEB/TFE3 stimulates lysosomal exocytosis and promotes the clearance of undigested materials in several cell models of LSDs [67, 128, 129]. Overexpression of TFEB or pharmacological stimulation of its activity also offers protection against the pathology of Parkinson’s, Alzheimer’s and Huntington’s diseases in mouse models [65, 130–133]. In invertebrate model organisms, the nuclear retention of TFEB caused by the mutation of a nuclear export protein XPO1 enhances autophagy and protects ALS-afflicted fruit flies from developing neurodegeneration [134].
Despite the protection against LSDs and neurodegenerative diseases, the enhanced lysosomal-autophagic process caused by TEFB/TFE3 overexpression can drive tumor progression by recycling internal cellular constituents in cancer cells and compensating the shortage of external nutrients due to insufficient vascularization [135, 136] . For instance, pancreatic ductal adenocarcinoma (PDA) exhibits constitutive activation of MITF transcription factors to maintain the amino acid pool derived from autophagy [137] , and consequently the pathogenesis of PDA requires a high level of autophagy [138]. It is also known that TFE-fusion renal cell carcinomas (TFE-fusion RCCs) are resulted from chromosomal translocation and MITF gene fusion that lead to the overexpression of TFEB and TFE3 encoding genes [139, 140]. On the other hand, FOXO1 is shown to activate the autophagic process, which leads to cell death and a protection against human colon cancer [84, 140].
Moreover, the lysosomal-autophagic activity decreases with age. FOXO and TFEB transcription factors can co-regulate lysosome biogenesis and autophagy genes [85, 86], and their increased activities contribute to lifespan extension in diverse organisms [76, 141, 142] and protection against age-associated diseases like osteoarthritis [85].
Peripheral to perinuclear positioning of lysosomes
Lysosomes are broadly distributed in the cytoplasm and can change positioning in responding to different metabolic and signaling inputs [18]. In particular, the movement from the cell periphery toward the perinuclear region (retrograde) and from the perinuclear region toward the cell periphery (anterograde) contributes to lysosome-related cellular processes, including cell migration, autophagy and immune responses, as well as to the regulation of lysosomal signaling pathways [19]. The intracellular movement of lysosomes is governed by two families of microtubule-based motors, kinesins and dynein. Generally, in an unpolarized cell, kinesins move towards the microtubule plus end (towards the cell periphery) while dynein moves toward the minus end (towards the nucleus or microtubule organizing center) [20, 21]. Perinuclear movement of lysosomes has been observed in a variety of lysosomal storage disorder (LSD) diseases (Table 1) and can be triggered upon nutrient deprivation that is the key input to modulate lysosomal signaling. This movement is governed by multiple cooperative mechanisms, including the enhancement of coupling with dynein and its activator dynactin, the promotion of tethering between lysosomes and the perinuclear endoplasmic reticulum (ER)/Golgi complex, and the inhibition of anterograde transport involving coupling lysosomes to kinesins (Figure 1).
Table1:
Lysosome positioning changes in human disease models.
| Disease | Lysosome positioning * | Model system | Reference |
|---|---|---|---|
| Mucolipidosis type IV | Perinuclear | Primary mouse fibroblasts | [25] |
| Niemann-Pick type C | Perinuclear | Primary mouse cell isolations | [107] |
| Patient derived fibroblasts | [108] | ||
| Juvenile CLN3 | Perinuclear | HeLa cell | [109] |
| Huntington’s | Perinuclear | Mouse striatal derived cell line | [110] |
| Axonal/Peripheral 1 | Mouse primary DRG neurons | [111] | |
| Alzheimer’s | Axonal/Peripheral 2 | Mouse hippocampus | [112, 113] |
A broad definition of lysosomes, including lysosomes, autophagosomes1 (1) and amphisomes2 (fusion between autophagosomes and endosomes) is applied.
Figure 1. Molecular mechanism of lysosome positioning.
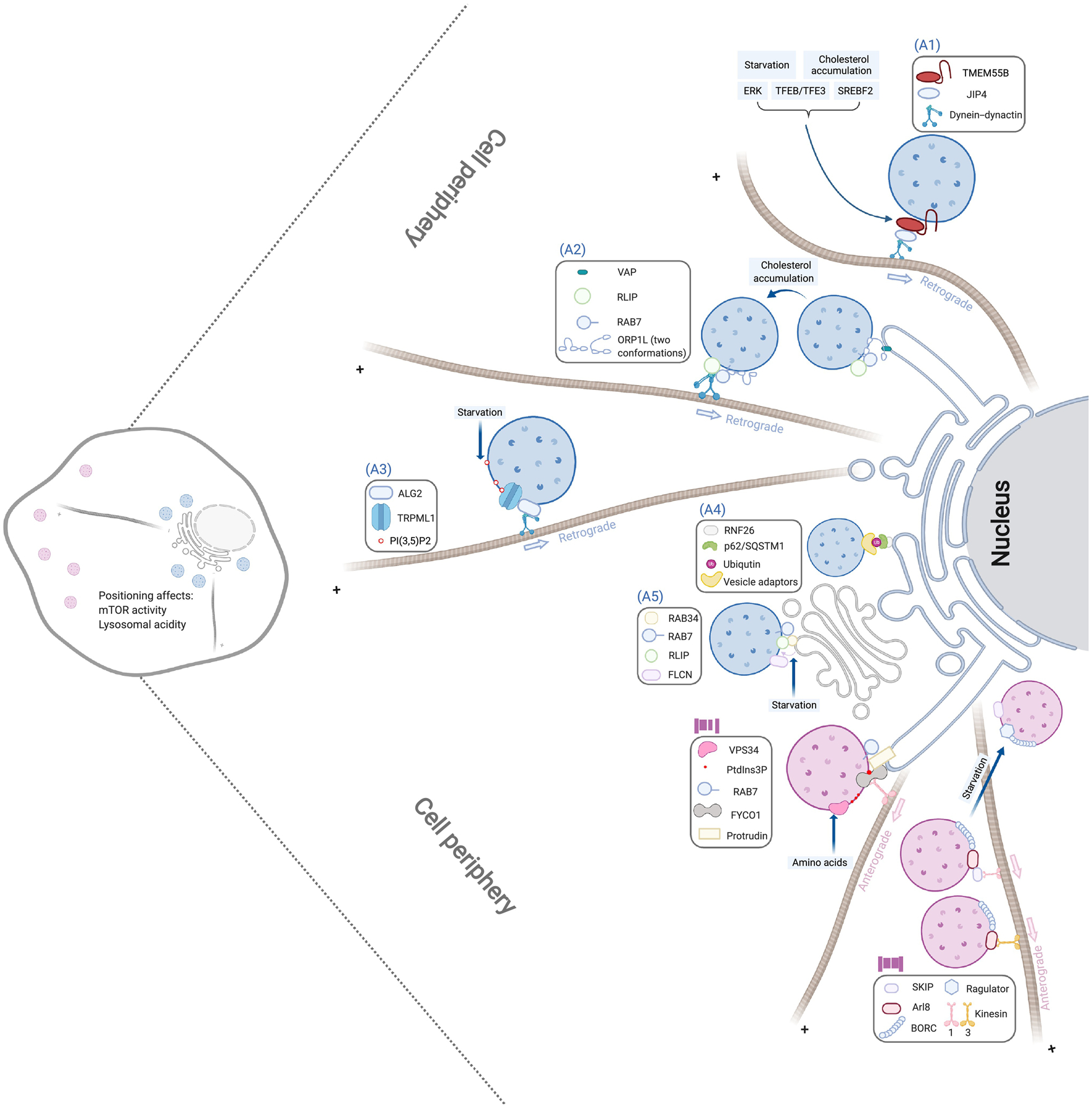
The perinuclear clustering of lysosomes (marked by blue) is driven by the following machineries: (A1) TMEM55B binding with the dynein adaptor JIP4 to recruit the dynein-dynactin complex; (A2) ORP1L adopting a conformation to facilitate the detachment of lysosomes from ER and the recruitment of dynein-dynactin by the Rab7-RILP complex; (A3) Lysosomal lipid PI(3,5)P2 activating the TRPML1 calcium channel to allow the recruitment of dynein-dynactin by the calcium sensor ALG2; (A4) ER-located RNF26 ubiquitinating p62/SQSTM1 and recruiting various vesicle adaptors through their ubiquitin-binding-domains; and (A5) Lysosome-associated FLCN promoting the interaction between lysosomal Rab7 and RILP and Golgi-localized Rab34. On the other hand, periphery positioning of lysosomes (marked in pink) is mediated by (B1) VPS34-induced PtdIns3P to promote the transfer of kinesin-1 from ER Protrudin to lysosomal FYCO1 and (B2) BORC recruitment of Arl8 to interact with different kinesins, which is blocked by its interaction with Ragulator. Each of these pathways can be regulated by different stimuli as indicated in light blue boxes, and lysosomal positioning regulates lysosomal acidity and mTOR activation. These pathways are shown as independent for presentation clarity, but multiple machineries can be present in a single lysosome.
Perinuclear retrograde movement associated with dynein-dynactin
Three independent mechanisms have been reported to couple lysosomes with the dynein-dynactin retrograde motor. First, the lysosome associated small GTPase Rab7 can interact with a homodimeric effector protein Rab7-Interacting Lysosomal Protein (RILP), which subsequently recruits the p150Glued subunit of the dynein–dynactin complex and drives lysosomal distribution toward the perinuclear region [22] (Figure 1, pathway A2). RILP is also in association with Oxysterol-binding protein-related protein 1 (ORP1L), which can adopt different conformations in response to the cholesterol level of the lysosome and in turn modulate the formation of lysosome-ER contact sites. In normal conditions, the ER protein VAP interacts with the Rab7-RILP complex at those contact sites and competes off p150Glued and dynein–dynactin [23]. Upon cholesterol accumulation induced by chemical or genetic intervention, ORP1L adopts a conformation that prevents the formation of the lysosome-ER contact and consequently the interaction between VAP and the Rab7-RILP complex, leading to the perinuclear movement of lysosomes [23].
Second, the lysosomal calcium channel TRPML1 can bind with the calcium senor ALG2 [24], which physically associates with dynein-dynactin and mediates the retrograde transport and perinuclear accumulation of lysosomes [25] (Figure 1, pathway A3). TRPML1 is the predominant calcium channel that controls calcium release in the lysosome [26] and can be activated by lysosome specific phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) [27] whose biosynthesis is regulated by both nutrient and growth factors [28]. In response to starvation, autophagy induction or cytosolic alkalization, lysosomes undergo perinuclear redistribution, which is blocked by acute TRPML1 inhibition or PI(3,5)P2 depletion [25]. Conversely, the activation of TRPML1 sufficiently induces perinuclear accumulation of lysosomes in a PI(3,5)P2 and ALG-2 dependent manner [25]. Notably, although mutations of the human TRPML1 gene are linked with mucolipidosis type IV (ML-IV) LSD [26], the TRPML1-ALG-2 mechanism does not contribute to the perinuclear accumulation of lysosomes in ML-IV, and neither does it contribute to perinuclear accumulation of lysosomes in Niemann-Pick disease type C1, another LSD characterized by cholesterol accumulation [25].
Third, lysosomal transmembrane protein TMEM55B can directly bind with the dynein scaffold protein JIP4 and recruit dynein-dynactin to lysosomes through JIP4, inducing lysosomal retrograde transport and perinuclear positioning [29] (Figure 1, pathway A1). It is known that TMEM55B overexpression causes the clustering of lysosomes around the nucleus, while its knockdown increases the concentration of lysosomes in the cell periphery [29]. Interestingly, starvation up-regulates the expression of TMEM55B through the transcription factor EB (TFEB)/transcription factor E3 (TFE3), the master regulator of lysosomal biogenesis [29, 30] (see the section “Transcriptional control of lysosomal adaptation”). The transcription of TMEM55B also responds to cholesterol levels, which is mediated by SREBF2 and TFEB/TFE3 [29]. As a result, the TMEM55B-JIP4 mechanism contributes to the perinuclear redistribution of lysosomes caused by chemical and genetic induction of lysosomal cholesterol accumulation [29]. In addition to the transcriptional control, the TMEM55B-JIP4 mechanism is also subject to the post-transcriptional regulation by ERK [29, 30].
Perinuclear retrograde movement via ER/Golgi interaction
Additionaly, lysosomal positioning close to the perinuclear region is mediated by the interaction of lysosomes with ER and Golgi. Rab34, a small GTPase primarily localized to the perinuclear Golgi complex, can interact with RILP [31] and Rab7 to promote lysosome-Golgi tethering and perinuclear retention of lysosomes [32] (Figure 1, pathway A5). Rab34 is required for starvation-induced perinuclear accumulation of lysosomes and is sufficient to promote lysosome movement toward the nucleus upon overexpression [32]. Lysosome associated Folliculin (FLCN) facilitates this regulatory mechanism through interacting with RILP and promoting the loading of active Rab34 onto RILP upon starvation [32].
At the same time, an ER-located ubiquitin ligase RNF26 can recruit and ubiquitinate p62/SQSTM1 (the ubiquitin-binding protein involved in autophagosome cargo recruitment), which further attracts ubiquitin-binding domains of various vesicle adaptors, including EPS15, TAX1BP1 and TOLLIP, and restricts their cognate vesicles in the perinuclear region [33] (Figure 1, pathway A4). This anchoring process is terminated upon deubiquitination of p62/SQSTM1 by the RNF26-associated deubiquitinating enzyme USP15, which facilitates the vesicle release toward the periphery [33]. Depletion of RNF26, or knocking down of p62/SQSTM1, EPS15, TAX1BP1 or TOLLIP leads to redistribution of lysosomes toward the peripheral region, while depletion of USP15 decreases the number of periphery-located lysosomes and overall endosome mobility [33]. It is currently unclear whether the activity of RNF-16 and/or UPS15 is modulated by the nutrient status of the cell.
Peripheral anterograde transport with kinesins
Furthermore, starvation inhibits anterograde transport of lysosomes toward the periphery. Anterograde transport can be mediated by a lysosomal eight-unit complex BORC, which recruits a small GTPase, Arl8, to the lysosomal membrane [20] (Figure 1, pathway B2). Arl8 in turn interacts with different kinesins to drive peripheral movement of lysosomes [34]. Arl8 requires an effector protein SKIP to establish the connection with kinesin-1 [35] but directly binds to kinesin-3 [20]. Knocking down of Arl8, SKIP, BORC, kinesin-1 or kinesin-3 causes perinuclear redistribution of lysosomes without starvation [20], [34, 35]. In the condition of starvation, BORC interaction with the Ragulator complex is enhanced, which prevents BORC-Arl8b binding and consequently leads to lysosomal accumulation around the nucleus [21]. However, overexpression of Arl8 or kinesin-1 can block this starvation-induced perinuclear accumulation [36].
At the same time, anterograde movement can be regulated by phosphatidylinositol 3-phosphate (PtdIns3P), which promotes lysosome-ER contact and the translocation of lysosomes to the cell periphery [37] (Figure 1, pathway B1). PtdIns3P on the lysosome recruits a PtdIns3P-binding protein FYCO1 to the lysosome membrane and another PtdIns3P-binding protein Protrudin on the ER membrane, which facilitates the transfer of kinesin-1 from protrudin to FYCO1 to initiate anterograde transportation [37, 38]. The production of PtdIns3P on the lysosome is catalyzed by the lipid kinase VPS34, which is stimulated by an increased level of amino acids [39]. Thus, upon starvation, this PtdIns3P-dependent anterograde transportation mechanism is comprised due to inactive VPS34.
Collectively, the emerging research on lysosome positioning starts to reveal the tip of the iceberg for this delicately balanced system with high dynamics. Positioning changes have been reported to affect lysosomal acidity and autophagic flux [36, 40, 41] as well as signal transduction (see the section “Lysosomal positioning & signaling activation”). Further understanding of its regulatory mechanisms will advance our current knowledge on the roles of lysosomes in cellular signaling, organismal development, pathogenesis and aging.
Lysosome-to-nucleus interaction in signaling transduction
Importantly, the interaction between lysosomes and the nucleus does not just occur at the morphological level. Dependent on their abilities to cluster at the perinuclear region, produce various metabolites and anchor signaling factors, lysosomes can communicate with the nucleus to regulate signal transduction and transcription responses. Three examples of such regulations are highlighted here.
Lysosomal positioning & signaling activation
As a master regulator of cellular growth and metabolism, mTOR complex 1 (mTORC1) plays crucial roles in cancer, neurodegeneration, metabolic disorders and aging. Recent studies reveal mTORC1 activation at the lysosomal surface, which is regulated by Ragulator, GATOR1 and FLCN in complex with FNIP (see reviews [42–44]), and also the interesting link between mTORC1 activation and lysosomal positioning [36]. On one hand, mTORC1 influences lysosomal positioning in response to nutrient and growth factors (see above and Figure 1, pathways B2 and A5). More specifically, Ragulator physically interacts with BORC and in turn comprises its ability to recruit kinesins, which is enhanced upon starvation or weakened by growth factors to drive the perinuclear or peripheral movement of lysosomes, respectively [21, 45]. Meanwhile, direct binding of FLCN with RILP, in response to starvation, promotes the loading of active Rab34, the tethering of lysosomes to the perinuclear Golgi complex, and the retaining of lysosomes around the nucleus [32]. On the other hand, lysosomal positioning can in turn influence mTORC1 activation. Overexpression of proteins mediating the anterograde transport of lysosomes, such as BORC, ARL8, Protrudin and FYCO1, enhances mTORC1 activity [36, 38]. Conversely, genetic knockdown or knockout of those anterograde transport mediators attenuates mTORC1 activation in response to nutrient and growth factors [36, 38, 46].
However, there are also studies that associate peripheral positioning of lysosomes with a reduction in mTORC1 activity [47, 48]. For example, cytosolic acidification resulted from hypoxia leads to peripheral distribution of lysosomes and mTORC1 inhibition [48]. This controversy might be related to specific mechanisms activating mTORC1 under different conditions. In response to growth factors, perinuclear clustering of lysosomes attenuates the activation of mTORC2 and AKT that can augment mTORC1 activation [46]; while in response to cytosolic acidification, peripheral redistribution of lysosomes drives lysosome-bound mTOR away from the perinuclear pool of RHEB that activates it [48].
Nuclear translocation of lysosomal FABPs
As a metabolic active site, lysosomes are responsible for the digestion and degradation of macromolecules and the production of various metabolites. A crucial group of bioactive metabolites comprise fatty acids (FAs) and FA derivatives that are sources of energy fuel, membrane building, and signal transduction [49]. Within the lysosomal lumen, acidic lipases and phospholipases can de-esterify neutral lipids and phospholipids, respectively, to generate FAs and FA derivatives, which may be exported via vesicular routes and/or lipid binding proteins and transporters [50].
In the cytoplasm, hydrophobic FAs and FA derivatives rely on fatty acid binding proteins (FABPs) to facilitate their transportation between organelles [51]. This family of proteins therefore act as lipid chaperones to mediate the metabolic and signaling effects of FAs and FA derivatives [51]. In particular, specific FABPs can selectively carry FAs to the nucleus and activate a family of nuclear hormone receptors known as peroxisome proliferator-activated receptors (PPARs) [52]. FABP proteins are conserved from Caenorhabditis elegans to humans, with nine homologs in each organism. One of C. elegans FABPs, LBP-8, is reported to be translocated from the lysosome to the nucleus upon lysosomal lipolysis induced by overexpression of a lysosomal acid lipase LIPL-4 [53]. The LBP-8 protein is able to bind a variety of FAs and FA derivatives [53, 54], and carries a structurally conserved nuclear localization signal [54]. Overexpression of LIPL-4 also leads to an increased level of oleoylethanolamide (OEA), a FA derivative that directly binds to LBP-8 and requires its nuclear translocation to activate the nuclear hormone receptor complex NHR-80 and NHR-49 (Figure 2) [53, 54]. This complex in turn upregulates lbp-8 gene to form a positive feedback loop [53] and mitochondrial genes to induce lysosome-mitochondria crosstalk [55]. The activation of this lysosome-to-nucleus retrograde lipid signaling pathway by overexpressing lipl-4 or lbp-8 or supplementing OEA promotes longevity, lipid catabolism and redox homeostasis [53, 55].
Figure 2. Lysosome and nucleus interaction in signaling and transcription.
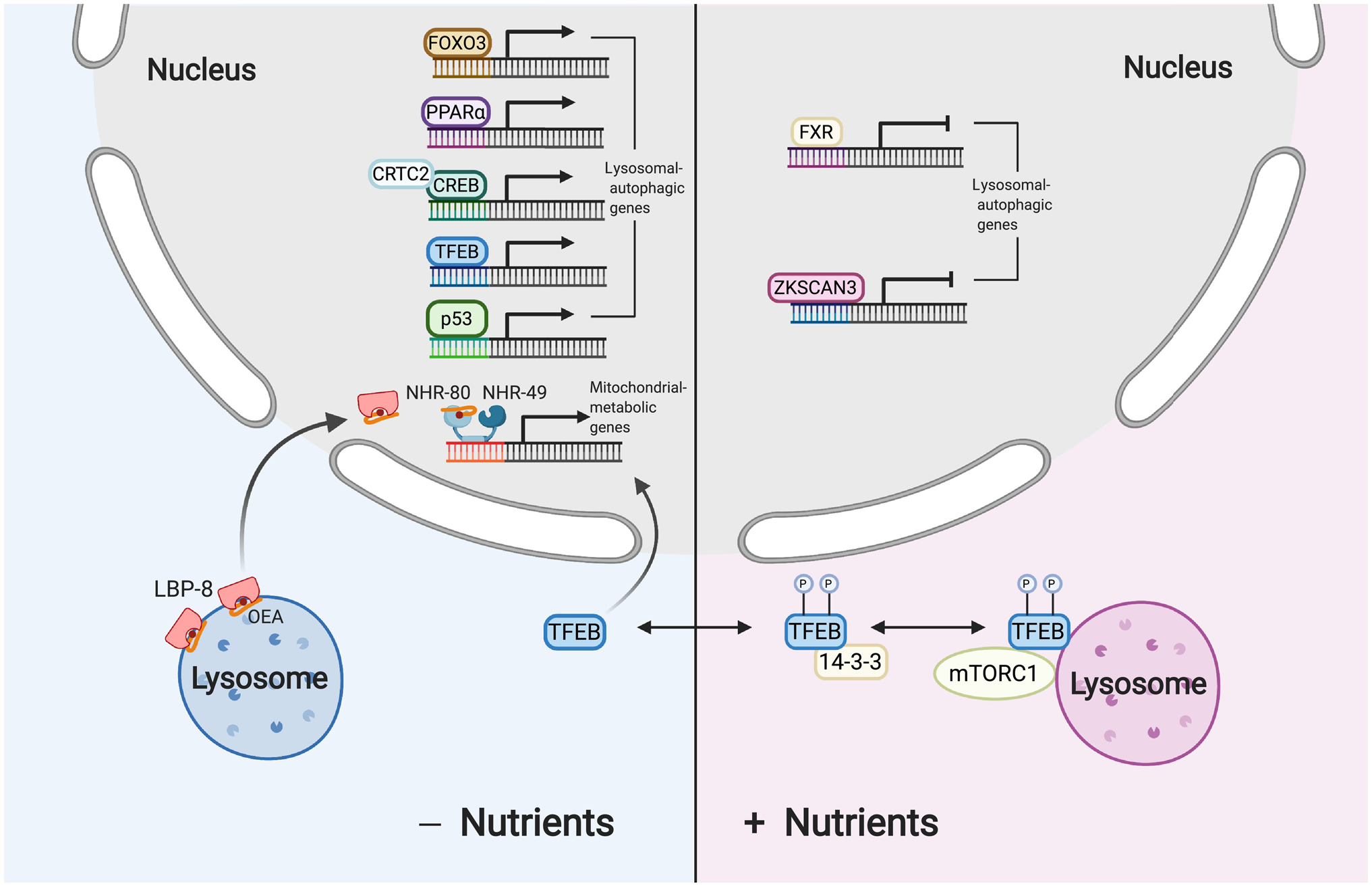
For the nucleus-to-lysosome interaction, the lysosomal-autophagic pathway is transcriptionally regulated responding to the nutrient status. Upon starvation, transcription factors, TFEB/TFE3 (shown as TFEB), FOXO3, CREB-CRTC2 PPARα, and p53 bind to the promoter region of lysosomal-autophagic genes and activate their expression. Meanwhile, ZKSCAN3 represses the expression of lysosomal-autophagic genes, so does FXR through competing with PPARα and dissociating the CREB-CRTC2 complex in well-fed conditions. For the lysosome-to-nucleus interaction, starvation can induce the nuclear translocation of fatty acid binding protein LBP-8 that binds oleoylethanolamine (OEA) and deliver it to the nuclear hormone receptor complex NHR-49-NHR-80 to induce mitochondrial metabolic genes. In contrast, nutrient repletion results in lysosomal docking of mTORC1 that phosphorylates TFEB/TFE3. The binding of phosphorylated TFEB/TFE3 with cytosolic 14-3-3 proteins prevents its nuclear translocation.
Lysosome-nucleus shuttling of TFEB and MITF-1
Unlike nuclear hormone receptors mostly residing in the nucleus, numerous transcription factors shuttle between the cytoplasm and the nucleus in response to different stimuli. It is a common mechanism that transcription factors are sequestered in the cytoplasm until specific cues trigger their translocation to the nucleus and regulate the expression of their target genes accordingly. In particular, transcription factors, TFEB and MITF-1, can be recruited to the lysosome through interacting with active Rag GTPase heterodimers, leading to mTORC1-dependent inhibition of their activities (Figure 2) [56]. TFEB and MITF-1 both belong to the MITF subfamily of helix-loop-helix leucine zipper transcription factors that also include TFE3 [57]. When nutrient is abundant, TFEB and MITF-1 are constantly cycling between the lysosome and the cytosol, and the lysosome-associated fraction can be phosphorylated by mTORC1 and released back to the cytosol and tethered with the 14-3-3 protein [56, 58–60]. The TFEB-14-3-3 tethering blocks the nuclear translocation of TFEB and consequently its activity.
Upon starvation or mTORC1 inactivation, dephosphorylated TFEB fails to tether with 14-3-3 and translocates into the nucleus and shows a stronger association with lysosomes as well [62, 65–67]. Similarly, inactivation of mTORC1 can also lead to the accumulation of MITF-1 into the nucleus and at the lysosome [56]. In the nucleus, TFEB activates a series of target genes that control lysosomal biogenesis and autophagy [15, 61] (see the section “TFEB/TFE3 transcription factors”). Besides mTORC1, the phosphorylation and nuclear localization of TFEB can be regulated by several other kinases and phosphatases, including ERK2, protein phosphatase 2A (PP2A), as well as calcineurin, a Ca2+ triggered serine-threonine phosphatase [62–64]. Interestingly, calcineurin is activated by Ca2+ release through the ion channel TRPML1 that is required for promoting Ca2+- dependent lysosomal movement towards the perinuclear region upon nutrient deprivation [25]. Thus, lysosomal positioning might also couple with the modulation of TFEB activity.
Nucleus-to-lysosome interaction in the control of lysosomal adaptation
In supporting cellular homeostasis, not only lysosomal signals actively influence nuclear transcription through altering lysosomal positioning and promoting nuclear translocation of chaperone proteins and transcription factors, but nuclear transcription also dynamically regulates genes involved in lysosomal biogenesis and functions and autophagy. Several transcription factors are implicated in this nuclear control of lysosomal adaptation, in order to ensure that lysosomes can adjust their values and activities in the context of environment fluctuation (Figure 2).
TFEB/TFE3 transcription factors
The discovery of the coordinated lysosomal expression and regulation (CLEAR) gene network and its responsible transcription factor TFEB suggests that the lysosomal-autophagic pathway is under a transcriptional control [15, 16]. Genes in the CLEAR network carry a CLEAR motif (TCACG) that is recognized and positively regulated by TFEB [15], and are extensively implicated in the lysosomal-autophagic process, such as autophagosome biogenesis, lysosome acidification, hydrolysis, ion homeostasis and exocytosis [29, 65, 66]. In addition to TFEB, TFE3 also induces the expression of lysosomal biogenesis and autophagy genes via binding with the CLEAR motif [67].
TFEB and TFE3 act independently to some extent, and are both regulated by mTORC1, which phosphorylates them at critical serine residues, leading to 14-3-3 tethering and cytoplasmic retention [67–69]. Nutrient deprivation inhibits the mTORC1-dependent phosphorylation, promotes calcineurin-mediated dephosphorylation following lysosomal Ca2+ release through TRPML1, and drives the translocation of TFEB/TFE3 into the nucleus [62]. Transcriptional induction of TFEB/TFE3 target genes enhances lysosome biogenesis and autophagic flux, which enable cells and organisms to meet the catabolic need required for survival under starvation. Other kinases responsive to metabolic cues and growth factors, such as MAP4K3, ERK, and GSK3, can also phosphorylate TFEB/TFE3 and inhibit their nuclear localization [61, 70–73].
As a counterpart of TFEB, a zinc-finger transcription factor harboring KRAB and SCAN domains (ZKSCAN3) has been reported to translocate into the nucleus when nutrient is available and repress a large set of genes related to the lysosomal-autophagic pathway [74]. Knockdown of ZKSCAN3 increases lysosome biogenesis and promotes autophagy in cells [74]. However, studies from ZKSCAN3 knockout mice show the dispensable role of this transcription factor in regulating lysosomal or autophagic gene expression [75]. It remains unclear what might cause this discrepancy, and one possible explanation is that ZKSCAN3 knockout over long time leads to transcriptional adaptation that hinders its effects.
FOXO transcription factors and p53
The class O of forkhead transcription factors (FOXO) are involved in regulating numerous cellular processes including cell metabolism, cell cycle and proliferation, oxidative stress response, and longevity [76–78]. C. elegans and Drosophila melanogaster each have one FOXO, but mammals carry four members, FOXO1, FOXO3, FOXO4 and FOXO6 [76, 79, 80]. AKT is the primary regulator of FOXO, which phosphorylates FOXO and causes its tethering with 14-3-3 and retention in the cytoplasm [81]. In muscle cells, FOXO3 is required for the transcriptional induction of autophagy-related genes [82]. More recently, analysis of ChIP-seq data shows that FOXO3 directly binds to and regulates a network of autophagy-related genes in adult neural stem and progenitor cells, where the activation of FOXO3 promotes autophagic flux [83]. In addition to FOXO3, FOXO1 has been shown to induce autophagy upon oxidative stress or serum starvation in human colon cancer cell line HCT116 and human cervical carcinoma cell line HeLa, which is however independent of its transcription activity [84]. Moreover, the expression of autophagy-related genes is reduced in the cartilage-specific triple knockout mice of FOXO1, 3 and 4, but induced in chondrocytes overexpressing the constitutively active form of FOXO1 [85].
For FOXO3, although ChIP-seq analysis reveals its binding to many autophagy-related genes, it has been noted that a number of those genes are not transcriptionally regulated by FOXO3, suggesting a co-factor function of FOXO3. In supporting this idea, a more recent study in C. elegans shows that TFEB/HLH-30 directly interacts with FOXO/DAF-16 by forming a combinatorial transcription factor complex [86]. Further transcriptomic analysis indicates that these two transcription factors jointly regulate many target genes involved in the longevity pathway [86]. Given the vital function of FOXO in multiple stress and metabolic processes, its coordination with TFEB adds another layer to the regulation of lysosomal adaption.
In addition, FOXO3a itself is subject to the transcriptional activation by p53, a key tumor suppressor [87, 88]. p53 family members also up-regulate the expression of other genes involved the lysosomal-autophagic process, ranging from upstream factors mediating AMPK activation and mTOR inhibition, core autophagy components to lysosomal proteins [88–91]. On the other hand, basal cytoplasmic p53 has been shown to repress autophagy in different cellular contexts likely through transcription-independent mechanisms [90]. Thus, p53 regulates the lysosomal-autophagic process in a dual fashion, acting as both an inducer and a suppressor, which demands further investigation.
Nuclear Hormone Receptors
Nuclear hormone receptors are a family of ligand-inducible transcription factors that are commonly activated by various lipid signals, and some of them have been linked with the transcriptional regulation of the lysosomal-autophagic process. In hepatic cells, farnesoid X receptor (FXR), a well-known bile acid receptor, is activated by feeding-associated increase in bile acids and transcriptionally repress lysosomal and autophagic genes. In one way, this repression is mediated through the interaction between FXR and CREB, which hinders the binding of CREB with its coactivator CRTC2. Upon starvation, FXR-mediated inhibition of the CREB-CRTC2 complex is removed, allowing the transcriptional induction of lysosomal and autophagic genes [92].
In another way, this repression is mediated through the competition between FXR and PPARα on the same binding motif in the promoter regions of lysosomal and autophagic genes. Fasting activates PPARα to induce the transcription of lysosomal and autophagic gene in the liver, while feeding activates FXR to repress the transcription of the same group of genes [93]. Interestingly, TFEB itself is a transcription target of FXR and CREB [92], which in turn transcriptionally regulates PPARα and its coactivator PGC1α [61]. Thus, these two mechanisms might coordinate to enhance lysosomal adaption in response to metabolic changes. At the same time, specific FABP can directly bind with PPARα and regulate its transcriptional activity [94], and OEA is a well-known endogenous agonist of PPARα [95], which may suggest the role of lysosomal FABP and lipid signals in the transcriptional regulation of lysosomal adaptation.
Conclusion Remarks
Lysosomes, traditionally viewed as terminal degradative organelles in catabolic processes, their fundamental roles in nutrient sensing and metabolic signal transduction have been emerged and recognized in the past decade. A variety of sensors and transducers within the cell refine metabolic cues from lysosomes and launch signaling cascades to promote appropriate cellular responses. The crosstalk between the lysosome and the nucleus is a crucial part of this signaling orchestra and occurs at multiple layers, which include the modulation of lysosomal motility and positioning, the shuttling of signaling factors between the two compartments, and the transcriptional control of lysosomal adaption, as reviewed in this article.
Moreover, accumulating evidence suggest that lysosomes widely interact and cooperate with other organelles in addition to the nucleus. For example, lysosome-ER contact plays crucial roles in regulating lysosomal positioning, Ca2+ homeostasis and mTOR signaling [38, 96, 97]. Both ER and lysosomes are intracellular Ca2+ stores. The release of lysosomal Ca2+ is implicated in endocytic membrane trafficking and fusion, autophagy, lysosome exocytosis and plasma membrane repair [98, 99]. A recent study shows that the inositol 1,4,5-trisphosphate (IP3) receptor is activated by IP3 populated at the contact site between ER and lysosomes and regulates Ca2+ exchange between them [100]. In addition, lysosome-ER contact is vital to monitor and regulate cholesterol levels, and OSBP can function at lysosome-ER contact sites and transport cholesterol across the contact to activate mTORC1 [97]. Physical contact between lysosomes and mitochondria is also reported in healthy untreated Hela cells, which is mediated by Rab7 [101]. Upon the recruitment of the Rab7 GTPase-activating protein TBCF1D15 to mitochondria by FIS1, increased GTP hydrolysis releases the contact between lysosomes and mitochondria [101]. Unraveling how lysosomes are able to coordinately interact with different organelles and the physiological relevance underlying these interactions will be a challenging and promising direction for future research. Cutting edge imaging techniques, like lattice light-sheet microscopy, cryogenic super-resolution fluorescence microscopy coupled with focused ion beam scanning electron microscopy [102–106], is shedding new light on this area.
Most of our knowledge regarding the signaling role of lysosomes comes from studies in cell culture systems. In an organism, LSDs usually show a wide range of cell and organ specificity [120]. Although this specificity may be due to different cellular/metabolic demands in different cells and organs, the heterogeneity of lysosomes across different cell types could contribute to different pathologies of distinct organs associated with LSDs [36, 40, 41]. Therefore, it will be crucial to harness the power of tissue/cell type specific proteomics and metabolomics of lysosomes and high-resolution, high-dynamics microscopic analyses of lysosomes to investigate their heterogeneity, complex interaction with other organelles, and cell-autonomous and non-autonomous regulatory mechanisms at the organism level. These systemic studies will provide new ways for improving lysosomal functions in various tissues and deepen our current knowledge about the functional roles of lysosomes in health and diseases.
Outstanding questions:
Other than nutrient deprivation, are there environmental cues that modulate lysosome motility and positioning?
How does lysosomal positioning regulate lysosomal signaling in addition to mTOR, and do lysosomes recruit different proteins to the surface at peripheral and perinuclear regions for these regulations?
What is the interrelationship between lysosomal positioning and functional heterogeneity and signaling transduction, and how can we apply our knowledge of lysosomal positioning from cell culture systems to in vivo organisms?
Do the retrograde movement of lysosomes facilitate lysosome-to-nucleus signaling communication and the transcriptional activation of lysosomal functions?
Do changes in lysosomal positioning associated with diseases contribute to alternations in lysosome-to-nucleus communication and the transcriptional control of lysosomal adaptation?
How can we improve current imaging methods for better in vivo visualization of lysosomal heterogeneity, dynamics and interactions with other organelles?
How does different transcription factors coordinate to regulate lysosomal adaptation in response to different external and internal stimuli?
Is the lysosome-to-nucleus communication mechanism mediated by FABPs conserved in mammals? What factors recruit FABPs to the lysosome and facilitate its translocation into the nucleus? What lysosome transporters coordinate with FABPs to export signaling lipids, and the specific nuclear hormone receptors acting downstream of lysosomal FABPs?
Are there other lipid chaperone proteins and lipid signals mediating lysosome-to-nucleus retrograde signaling? Are there lipid chaperone proteins mediating lipid-related inter-organelle communication beyond the lysosome and the nucleus?
Are there a variety of transcription factors that can be shuttled between the cytosol, the lysosome and the nucleus in response to different stimuli, and what are the molecular mechanisms underlying this dynamic regulation?
What roles do epigenetic factors play in the fine tuning of lysosomal adaptation to various environmental cues, and how do they cooperate with various transcription factors?
At the organismal level, how heterogenous are lysosomes across different cell types and tissues? What is the physiological and functional relevance of lysosomal heterogeneity, and how is it related to different pathologies of organs associated with lysosomal storage disorders, neurodegenerative diseases and aging?
Highlights:
Lysosomal and nuclear communication coordinates lysosomal positioning, metabolism and signaling with nuclear transcription to support fundamental cellular activities.
Lysosomal positioning between peripheral and perinuclear regions is subject to cooperative regulation in response to metabolic stimuli and contributes to lysosomal functions and signaling.
Retrograde transport of chaperone proteins and transcription factors from the lysosome to the nucleus is actively regulated by metabolic signaling.
Nuclear transcriptional networks comprise a variety of different transcription factors that act independently or coordinately to fine tune lysosomal-autophagic gene expression.
Acknowledgment
We thank NIH grants R01AG045183 (M.C.W.), R01AT009050 (M.C.W.), R01AG062257 (M.C.W.), DP1DK113644 (M.C.W.), March of Dimes Foundation (M.C.W.), Welch Foundation (M.C.W.). We apologize to colleagues whose work we were not able to cite owing to space limitations.
Glossary
- AKT
also known as protein kinase B, is a serine/threonine kinase and main regulator of metabolism
- ARL8
ADP ribosylation factor-like 8, is small G protein and regulator of lysosomal anterograde trafficking
- BORC
BLOC-1-related complex, is a multi-subunit complex that regulates lysosome positioning
- CREB
cAMP response element-binding protein, is a transcription factor
- Endocytosis
a process by which cells internalize materials surrounding the cell membrane through membrane invagination and formation of vesicles
- ERK
known as extra-cellular signal regulated protein kinase, is a member of mitogen activated protein kinases
- EPS15
epidermal growth factor receptor substrate 15, is an endocytic adaptor protein interacting with RNF26/SQSTM1
- Exocytosis
an energy-dependent secretory process that transports cellular materials confined within vesicles out of the cell membrane into the extracellular space
- FIS1
mitochondria fission 1 protein, is a regulator of mitochondrial fragmentation
- FYCO1
FYVE and coiled-coil domain-containing protein 1, is a regulator of lysosomal retrograde transportation
- GSK3
glycogen synthase kinase 3, is a serine/threonine protein kinase
- Lipolysis
a biochemical pathway through which ester bonds in neutral lipids, triglycerides are hydrolyzed, resulting in the generation of fatty acids and glycerol
- Lysosome adaptation
Lysosomes actively adjust their biosynthesis and activities in response to intracellular and environmental inputs, which is transcriptionally coordinated by multiple factors.
- MITF
Microphthalmia-associated transcription factor in the family of basic helix-loop-helix leucine zipper transcription factors
- Oxysterol-binding protein-related protein 1(ORP1L)
is a member of the human oxysterol-binding protein (OSBP) family
- OSBP
oxysterol-binding protein, is a receptor for a variety of oxysterols
- PGC1α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha, is a transcriptional co-activator for several nuclear receptors
- RNF26
Ring finger protein 26, is ER-associated E3 ubiquitin ligase
- SKIP
SifA-kinesin interacting protein, also known as PLEKHM2, is an adaptor protein between kinesin motor protein and ARL-8
- TAX1BP1
Tax1-binding protein 1, is an autophagy receptor and vesicle adaptor protein interacting with RNF26/SQSTM1
- TBCF1D15
TBC1 domain family member 15, is a GTPase activation protein for Rab7
- TOLLIP
Toll interacting protein, is a negative regulator of toll-like receptor-mediated signaling as well as a vesicle adaptor protein interacting with RNF26/SQSTM1
- TRPML1
transient receptor potential, mucolipin subfamily, member 1, is a cation-permeable channel that is localized on both late endosomes and lysosomes
- USP15
ubiquitin carboxyl-terminal hydrolase 15, is a deubiquitinating enzyme
- VPS34
vacuolar protein sorting 34, is a class 3 phosphatidylinositol (PtdIns) 3-kinase and an endosomal trafficking regulator
- XPO1
exportin 1, is a eukaryotic nuclear export protein
- 14-3-3
is a family of structural similar intracellular phosphor-serine/threonine binding proteins that are found in all eukaryotes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Appelmans F, Wattiaux R, and De Duve C, Tissue fractionation studies. 5. The association of acid phosphatase with a special class of cytoplasmic granules in rat liver. Biochem J, 1955. 59(3): p. 438–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mindell JA, Lysosomal acidification mechanisms. Annu Rev Physiol, 2012. 74: p. 69–86. [DOI] [PubMed] [Google Scholar]
- 3.Saftig P, Schroder B, and Blanz J, Lysosomal membrane proteins: life between acid and neutral conditions. Biochem Soc Trans, 2010. 38(6): p. 1420–3. [DOI] [PubMed] [Google Scholar]
- 4.Chapel A, et al. , An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol Cell Proteomics, 2013. 12(6): p. 1572–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tooze SA, Abada A, and Elazar Z, Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb Perspect Biol, 2014. 6(5): p. a018358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabinowitz JD and White E, Autophagy and metabolism. Science, 2010. 330(6009): p. 1344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorkin A and von Zastrow M, Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol, 2009. 10(9): p. 609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saftig P and Klumperman J, Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol, 2009. 10(9): p. 623–35. [DOI] [PubMed] [Google Scholar]
- 9.Lloyd JB, Metabolite efflux and influx across the lysosome membrane. Subcell Biochem, 1996. 27: p. 361–86. [DOI] [PubMed] [Google Scholar]
- 10.Liu B, et al. , LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science, 2012. 337(6092): p. 351–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blott EJ and Griffiths GM, Secretory lysosomes. Nat Rev Mol Cell Biol, 2002. 3(2): p. 122–31. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez A, et al. , Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J Cell Biol, 1997. 137(1): p. 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaiswal JK, Andrews NW, and Simon SM, Membrane proximal lysosomes are the major vesicles responsible for calcium-dependent exocytosis in nonsecretory cells. J Cell Biol, 2002. 159(4): p. 625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chieregatti E and Meldolesi J, Regulated exocytosis: new organelles for non-secretory purposes. Nat Rev Mol Cell Biol, 2005. 6(2): p. 181–7. [DOI] [PubMed] [Google Scholar]
- 15.Sardiello M, et al. , A gene network regulating lysosomal biogenesis and function. Science, 2009. 325(5939): p. 473–7. [DOI] [PubMed] [Google Scholar]
- 16.Perera RM and Zoncu R, The Lysosome as a Regulatory Hub. Annual Review of Cell and Developmental Biology, 2016. [DOI] [PMC free article] [PubMed]
- 17.Ballabio A and Bonifacino JS, Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol, 2020. 21(2): p. 101–118. [DOI] [PubMed] [Google Scholar]
- 18.van Bergeijk P, Hoogenraad CC, and Kapitein LC, Right Time, Right Place: Probing the Functions of Organelle Positioning. Trends Cell Biol, 2016. 26(2): p. 121–134. [DOI] [PubMed] [Google Scholar]
- 19.Pu J, et al. , Mechanisms and functions of lysosome positioning. J Cell Sci, 2016. 129(23): p. 4329–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pu J, et al. , BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell, 2015. 33(2): p. 176–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pu J, Keren-Kaplan T, and Bonifacino JS, A Ragulator-BORC interaction controls lysosome positioning in response to amino acid availability. J Cell Biol, 2017. 216(12): p. 4183–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordens I, et al. , The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol, 2001. 11(21): p. 1680–5. [DOI] [PubMed] [Google Scholar]
- 23.Rocha N, et al. , Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol, 2009. 185(7): p. 1209–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vergarajauregui S, Martina JA, and Puertollano R, Identification of the penta-EF-hand protein ALG-2 as a Ca2+-dependent interactor of mucolipin-1. J Biol Chem, 2009. 284(52): p. 36357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, et al. , A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol, 2016. 18(4): p. 404–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong XP, et al. , The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature, 2008. 455(7215): p. 992–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong XP, et al. , PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun, 2010. 1: p. 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bridges D, et al. , Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell, 2012. 23(15): p. 2955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willett R, et al. , TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat Commun, 2017. 8(1): p. 1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takemasu S, et al. , Phosphorylation of TMEM55B by Erk/MAPK regulates lysosomal positioning. J Biochem, 2019. 166(2): p. 175–185. [DOI] [PubMed] [Google Scholar]
- 31.Wang T and Hong W, Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol Biol Cell, 2002. 13(12): p. 4317–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starling GP, et al. , Folliculin directs the formation of a Rab34-RILP complex to control the nutrient-dependent dynamic distribution of lysosomes. EMBO Rep, 2016. 17(6): p. 823–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jongsma ML, et al. , An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell, 2016. 166(1): p. 152–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guardia CM, et al. , BORC Functions Upstream of Kinesins 1 and 3 to Coordinate Regional Movement of Lysosomes along Different Microtubule Tracks. Cell Rep, 2016. 17(8): p. 1950–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosa-Ferreira C and Munro S, Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell, 2011. 21(6): p. 1171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korolchuk VI, et al. , Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol, 2011. 13(4): p. 453–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raiborg C, et al. , Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature, 2015. 520(7546): p. 234–8. [DOI] [PubMed] [Google Scholar]
- 38.Hong Z, et al. , PtdIns3P controls mTORC1 signaling through lysosomal positioning. J Cell Biol, 2017. 216(12): p. 4217–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nobukuni T, et al. , Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A, 2005. 102(40): p. 14238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson DE, et al. , The position of lysosomes within the cell determines their luminal pH. J Cell Biol, 2016. 212(6): p. 677–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gowrishankar S and Ferguson SM, Lysosomes relax in the cellular suburbs. J Cell Biol, 2016. 212(6): p. 617–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J and Guan KL, mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol, 2019. 21(1): p. 63–71. [DOI] [PubMed] [Google Scholar]
- 43.Savini M, Zhao Q, and Wang MC, Lysosomes: Signaling Hubs for Metabolic Sensing and Longevity. Trends Cell Biol, 2019. 29(11): p. 876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu GY and Sabatini DM, mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol, 2020. [DOI] [PMC free article] [PubMed]
- 45.Filipek PA, et al. , LAMTOR/Ragulator is a negative regulator of Arl8b- and BORC- dependent late endosomal positioning. J Cell Biol, 2017. 216(12): p. 4199–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia R and Bonifacino JS, Lysosome Positioning Influences mTORC2 and AKT Signaling. Mol Cell, 2019. 75(1): p. 26–38 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clippinger AJ and Alwine JC, Dynein mediates the localization and activation of mTOR in normal and human cytomegalovirus-infected cells. Genes Dev, 2012. 26(18): p. 2015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walton ZE, et al. , Acid Suspends the Circadian Clock in Hypoxia through Inhibition of mTOR. Cell, 2018. 174(1): p. 72–87 e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duffy J, Mutlu AS, and Wang MC, Lipid Metabolism, Lipid Signalling and Longevity, in Ageing: Lessons from C. elegans, Olsen A and Gill MS, Editors. 2017, Springer International Publishing: Cham: p. 307–329. [Google Scholar]
- 50.Thelen AM and Zoncu R, Emerging Roles for the Lysosome in Lipid Metabolism. Trends Cell Biol, 2017. 27(11): p. 833–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furuhashi M and Hotamisligil GS, Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov, 2008. 7(6): p. 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan NS, et al. , Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol, 2002. 22(14): p. 5114–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Folick A, et al. , Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science, 2015. 347: p. 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tillman MC, et al. , Structural characterization of life-extending Caenorhabditis elegans Lipid Binding Protein 8. Sci Rep, 2019. 9(1): p. 9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramachandran PV, et al. , Lysosomal Signaling Promotes Longevity by Adjusting Mitochondrial Activity. Dev Cell, 2019. 48(5): p. 685–696 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martina JA and Puertollano R, Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol, 2013. 200(4): p. 475–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rehli M, et al. , Cloning and characterization of the murine genes for bHlH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics, 1999. [DOI] [PubMed]
- 58.Bronisz A, et al. , Microphthalmia-associated transcription factor interactions with 14-3- 3 modulate differentiation of committed myeloid precursors. Mol Biol Cell, 2006. 17(9): p. 3897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Settembre C, et al. , A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J, 2012. 31(5): p. 1095–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roczniak-Ferguson A, et al. , The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal, 2012. 5(228): p. ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Settembre C, et al. , TFEB links autophagy to lysosomal biogenesis. Science, 2011. 332(6036): p. 1429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Medina DL, et al. , Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol, 2015. 17(3): p. 288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tong Y and Song F, Intracellular calcium signaling regulates autophagy via calcineurin-mediated TFEB dephosphorylation. Autophagy, 2015. 11(7): p. 1192–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W, et al. , Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc Natl Acad Sci U S A, 2015. 112(11): p. E1373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Medina DL, et al. , Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell, 2011. 21(3): p. 421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sbano L, et al. , TFEB-mediated increase in peripheral lysosomes regulates store-operated calcium entry. Sci Rep, 2017. 7: p. 40797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martina JA, et al. , The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal, 2014. 7(309): p. ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu Y, et al. , YWHA/14-3-3 proteins recognize phosphorylated TFEB by a noncanonical mode for controlling TFEB cytoplasmic localization. Autophagy, 2019. 15(6): p. 1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martina JA, et al. , MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy, 2012. 8(6): p. 903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marchand B, et al. , Glycogen synthase kinase-3 (GSK3) inhibition induces prosurvival autophagic signals in human pancreatic cancer cells. J Biol Chem, 2015. 290(9): p. 5592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu CL, et al. , MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat Commun, 2018. 9(1): p. 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, et al. , Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol, 2016. 18(10): p. 1065–77. [DOI] [PubMed] [Google Scholar]
- 73.Li L, et al. , A TFEB nuclear export signal integrates amino acid supply and glucose availability. Nat Commun, 2018. 9(1): p. 2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chauhan S, et al. , ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell, 2013. 50(1): p. 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan H, et al. , The role of ZKSCAN3 in the transcriptional regulation of autophagy. Autophagy, 2017. 13(7): p. 1235–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Webb AE and Brunet A, FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci, 2014. 39(4): p. 159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klotz LO, et al. , Redox regulation of FoxO transcription factors. Redox Biol, 2015. 6: p. 51–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng Z, The FoxO-Autophagy Axis in Health and Disease. Trends Endocrinol Metab, 2019. 30(9): p. 658–671. [DOI] [PubMed] [Google Scholar]
- 79.Tissenbaum HA, DAF-16: FOXO in the Context of C. elegans. Curr Top Dev Biol, 2018. 127: p. 1–21. [DOI] [PubMed] [Google Scholar]
- 80.Webb AE, Kundaje A, and Brunet A, Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell, 2016. 15(4): p. 673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tzivion G, Dobson M, and Ramakrishnan G, FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta, 2011. 1813(11): p. 1938–45. [DOI] [PubMed] [Google Scholar]
- 82.Mammucari C, et al. , FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab, 2007. 6(6): p. 458–71. [DOI] [PubMed] [Google Scholar]
- 83.Audesse AJ, et al. , FOXO3 directly regulates an autophagy network to functionally regulate proteostasis in adult neural stem cells. PLoS Genet, 2019. 15(4): p. e1008097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Y, et al. , Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol, 2010. 12(7): p. 665–75. [DOI] [PubMed] [Google Scholar]
- 85.Matsuzaki T, et al. , FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med, 2018. 10(428). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin XX, et al. , DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity. Nat Commun, 2018. 9(1): p. 4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Renault VM, et al. , The pro-longevity gene FoxO3 is a direct target of the p53 tumor suppressor. Oncogene, 2011. 30(29): p. 3207–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kenzelmann Broz D, et al. , Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev, 2013. 27(9): p. 1016–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crighton D, et al. , DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell, 2006. 126(1): p. 121–34. [DOI] [PubMed] [Google Scholar]
- 90.Tasdemir E, et al. , Regulation of autophagy by cytoplasmic p53. Nat Cell Biol, 2008. 10(6): p. 676–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brady OA, et al. , The transcription factors TFE3 and TFEB amplify p53 dependent transcriptional programs in response to DNA damage. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seok S, et al. , Transcriptional regulation of autophagy by an FXR-CREB axis. Nature, 2014. 516(7529): p. 108–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee JM, et al. , Nutrient-sensing nuclear receptors coordinate autophagy. Nature, 2014. 516(7529): p. 112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hostetler HA, et al. , L-FABP directly interacts with PPARalpha in cultured primary hepatocytes. J Lipid Res, 2009. 50(8): p. 1663–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fu J, et al. , Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature, 2003. 425(6953): p. 90–3. [DOI] [PubMed] [Google Scholar]
- 96.Garrity AG, et al. , The endoplasmic reticulum, not the pH gradient, drives calcium refilling of lysosomes. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lim CY, et al. , ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat Cell Biol, 2019. 21(10): p. 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li P, Gu M, and Xu H, Lysosomal Ion Channels as Decoders of Cellular Signals. Trends Biochem Sci, 2019. 44(2): p. 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu H and Ren D, Lysosomal physiology. Annu Rev Physiol, 2015. 77: p. 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Atakpa P, et al. , IP3 Receptors Preferentially Associate with ER-Lysosome Contact Sites and Selectively Deliver Ca2+ to Lysosomes. Cell Reports, 2018. [DOI] [PMC free article] [PubMed]
- 101.Wong YC, Ysselstein D, and Krainc D, Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature, 2018. 554(7692): p. 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen BC, et al. , Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science, 2014. 346(6208): p. 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo Y, et al. , Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell, 2018. 175(5): p. 1430–1442 e17. [DOI] [PubMed] [Google Scholar]
- 104.Liu Z, Lavis LD, and Betzig E, Imaging live-cell dynamics and structure at the single-molecule level. Mol Cell, 2015. 58(4): p. 644–59. [DOI] [PubMed] [Google Scholar]
- 105.Valm AM, et al. , Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature, 2017. 546(7656): p. 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hoffman DP, et al. , Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science, 2020. 367(6475). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reid PC, Sugii S, and Chang TY, Trafficking defects in endogenously synthesized cholesterol in fibroblasts, macrophages, hepatocytes, and glial cells from Niemann-Pick type C1 mice. J Lipid Res, 2003. 44(5): p. 1010–9. [DOI] [PubMed] [Google Scholar]
- 108.Kwiatkowska K, et al. , Visualization of cholesterol deposits in lysosomes of Niemann-Pick type C fibroblasts using recombinant perfringolysin O. Orphanet J Rare Dis, 2014. 9: p. 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Uusi-Rauva K, et al. , Neuronal ceroid lipofuscinosis protein CLN3 interacts with motor proteins and modifies location of late endosomal compartments. Cell Mol Life Sci, 2012. 69(12): p. 2075–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Erie C, et al. , Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington’s disease. Eur J Neurosci, 2015. 42(3): p. 1941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wong YC and Holzbaur EL, The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci, 2014. 34(4): p. 1293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tammineni P, et al. , Impaired retrograde transport of axonal autophagosomes contributes to autophagic stress in Alzheimer’s disease neurons. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gowrishankar S, et al. , Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci U S A, 2015. 112(28): p. E3699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cuervo AM and Dice JF, A receptor for the selective uptake and degradation of proteins by lysosomes. Science, 1996. 273(5274): p. 501–3. [DOI] [PubMed] [Google Scholar]
- 115.Chiang HL, et al. , A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science, 1989. 246(4928): p. 382–5. [DOI] [PubMed] [Google Scholar]
- 116.Dice JF, Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci, 1990. 15(8): p. 305–9. [DOI] [PubMed] [Google Scholar]
- 117.Oku M and Sakai Y, Three Distinct Types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. Bioessays, 2018. 40(6): p. e1800008. [DOI] [PubMed] [Google Scholar]
- 118.Zhao YG and Zhang H, Autophagosome maturation: An epic journey from the ER to lysosomes. J Cell Biol, 2019. 218(3): p. 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakatogawa H, Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol, 2020. [DOI] [PubMed]
- 120.Schultz ML, et al. , Clarifying lysosomal storage diseases. Trends Neurosci, 2011. 34(8): p. 401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Parenti G, Andria G, and Ballabio A, Lysosomal storage diseases: from pathophysiology to therapy. Annu Rev Med, 2015. 66: p. 471–86. [DOI] [PubMed] [Google Scholar]
- 122.Marques ARA and Saftig P, Lysosomal storage disorders - challenges, concepts and avenues for therapy: beyond rare diseases. J Cell Sci, 2019. 132(2). [DOI] [PubMed] [Google Scholar]
- 123.Dehay B, et al. , Lysosomal impairment in Parkinson’s disease. Mov Disord, 2013. 28(6): p. 725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Uddin MS, et al. , Autophagic dysfunction in Alzheimer’s disease: Cellular and molecular mechanistic approaches to halt Alzheimer’s pathogenesis. J Cell Physiol, 2019. 234(6): p. 8094–8112. [DOI] [PubMed] [Google Scholar]
- 125.Martin DD, et al. , Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci, 2015. 38(1): p. 26–35. [DOI] [PubMed] [Google Scholar]
- 126.Sharma A, et al. , Cerebrospinal Fluid from Sporadic Amyotrophic Lateral Sclerosis Patients Induces Mitochondrial and Lysosomal Dysfunction. Neurochem Res, 2016. 41(5): p. 965–84. [DOI] [PubMed] [Google Scholar]
- 127.Lie PPY and Nixon RA, Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol Dis, 2019. 122: p. 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Song W, et al. , TFEB regulates lysosomal proteostasis. Hum Mol Genet, 2013. 22(10): p. 1994–2009. [DOI] [PubMed] [Google Scholar]
- 129.Spampanato C, et al. , Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med, 2013. 5(5): p. 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsunemi T, et al. , PGC-1alpha rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med, 2012. 4(142): p. 142ra97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Martini-Stoica H, et al. , The Autophagy-Lysosomal Pathway in Neurodegeneration: A TFEB Perspective. Trends Neurosci, 2016. 39(4): p. 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Martini-Stoica H, et al. , TFEB enhances astroglial uptake of extracellular tau species and reduces tau spreading. J Exp Med, 2018. 215(9): p. 2355–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Polito VA, et al. , Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med, 2014. 6(9): p. 1142–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Silvestrini MJ, et al. , Nuclear Export Inhibition Enhances HLH-30/TFEB Activity, Autophagy, and Lifespan. Cell Rep, 2018. 23(7): p. 1915–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kimmelman AC and White E, Autophagy and Tumor Metabolism. Cell Metab, 2017. 25(5): p. 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Perera RM, Di Malta C, and Ballabio A, MiT/TFE Family of Transcription Factors, Lysosomes, and Cancer. Annu Rev Cancer Biol, 2019. 3: p. 203–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Perera RM, et al. , Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature, 2015. 524(7565): p. 361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang S, et al. , Pancreatic cancers require autophagy for tumor growth. Genes Dev, 2011. 25(7): p. 717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kauffman EC, et al. , Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol, 2014. 11(8): p. 465–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Calcagni A, et al. , Modelling TFE renal cell carcinoma in mice reveals a critical role of WNT signaling. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Martins R, Lithgow GJ, and Link W, Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell, 2016. 15(2): p. 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lapierre LR, et al. , The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun, 2013. 4: p. 2267. [DOI] [PMC free article] [PubMed] [Google Scholar]


