Abstract
Despite the crucial roles of lipids in metabolism, we are still at the early stages of comprehensively annotating lipid species and their genetic basis. Mass spectrometry(MS)-based discovery lipidomics offers the potential to globally survey lipids and their relative abundances in various biological samples. To discover the genetics of lipid features obtained through high resolution LC-MS/MS, we analyzed liver and plasma from 384 Diversity Outbred (DO) mice, and quantified 3,283 molecular features. These features were mapped to 5,622 lipid quantitative trait loci (QTL) and compiled into a public web-resource termed LipidGenie. This data is cross-referenced to the human genome and offers a bridge between genetic associations in humans and mice. Harnessing this resource, we used genome-lipid association data as an additional aid to identify a number of lipids, for example gangliosides through their association with B4galnt1, and found evidence for a group of sex-specific phosphatidylcholines through their shared locus. Finally, LipidGenie’s ability to query either mass or gene-centric terms, suggests acyl chain-specific functions for proteins of the ABHD family.
Beyond their roles in energy storage and membrane structure, lipids are central actors of myriad metabolic functions and molecular signaling.1,2 As our understanding of these diverse lipid functions grows, so too does our appreciation for the complexity of the lipidome of mammalian systems.3 Mass spectrometry has emerged as the central tool to dissect and quantify the myriad of lipid species beyond traditional clinical measures.4–6 Specifically, using liquid chromatography (LC) coupled with high resolution tandem mass spectrometry (MS/MS), over one thousand unique lipid features from a complex mixture can be quantified in under an hour.7 A chromatographic feature in this context is defined by a unique combination of mass and retention time. From these features, hundreds of individual lipids are routinely identified; however, the majority of the features remain unannotated.8,9 The result is that more often than not, the majority of mass spectrometry data are not leveraged.8,10,11
One strategy for lipid feature identification is to group compounds likely to be related. For example, members of a lipid class often (i) appear within a defined chromatographic retention time, (ii) occupy a characteristic mass range, and (iii) exhibit similar dissociation patterns when subjected to fragmentation.12 Most efforts to improve lipid identification rates exploit one or all of these steps3,13–15 - including our laboratories recent description of a software suite that constructs tailored MS/MS libraries for automated lipid spectral identification.16,17 Others have sought to build on this information by adding external complementary data, such as measurement of collisional cross section,10 or labile hydrogen counting, among others.18 All of these methods show great promise but share in the common theme that they incorporate lipid chemical properties into their identification inference.
In the field of shotgun proteomics, genome sequence data are used to identify experimental peptide tandem mass spectra. Given the success of this field we wondered whether genomic information could be leveraged similarly in the field of lipidomics. Unfortunately, it is not possible to directly predict lipid identities from genomic data; however, shared genetic regulation among lipids could provide key information to facilitate identification. For example, a recent large-scale multi-omic study of a knock-out yeast library demonstrated dramatic regulation of the lipidome,19 nuclear magnetic resonance (NMR)-based untargeted metabolomics identified disease-associated metabolites and genomic regions via quantitative trait loci (QTL) mapping,20,21 and in humans, data from genome wide association studies (GWAS) were used to assist in small molecule identifications from both MS and NMR data.22–25 We thus aimed to create a global genome-lipid association map that would offer genetic location as a fourth (iv), orthogonal dimension of data to assist in lipid feature identification.
To construct a global genome-lipid association map we measured plasma and liver lipids from a mouse population using LC-MS/MS and performed QTL mapping.26–28 This study is carried out using Diversity Outbred (DO) mice, a multiparent population (MPP) derived from 8 highly diverse founder strains (Figure 1a).24,25,26 A key advantage of the MPP is that we can identify the additive genetic effects contributed by each founder strain at a quantitative trait locus (QTL). Unlike standard bi-parental crosses where the founder haplotype effects are either increasing or decreasing, the haplotype effects in a MPP are complex and enable us to distinguish chance co-localization from pleiotropic effects. In addition, we can compare founder haplotype effects across different studies using DO mice to identify shared effects on traits that were not directly measured in only one group of DO mice. Specifically, we were able to use liver gene expression from a previously published study to propose candidate genes for the lipid features in this study.30 Further, each of the eight inbred DO founder mouse strains (129, A/J, B6, CAST, NOD, NZO, PWK, WSB) contributes to generate distinct allele effect patterns at each locus, thus providing an additional criterion for gene identification.31 Finally, careful control of external sources of variation such as diet and environmental conditions, allows the extraordinary phenotypic diversity of the DO32 to be directly attributed to genetic diversity. The DO population has already been used extensively to map clinical traits,33 transcripts,32 proteins,34 gut microbiota and bile acids35 providing a wealth of existing data to integrate with global genome-lipid associations.
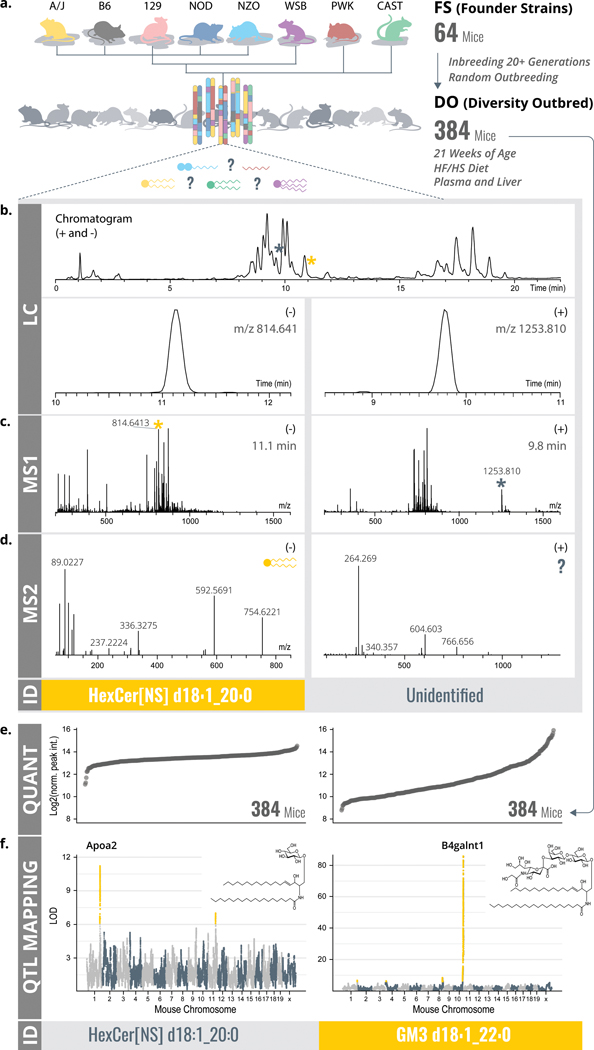
Figure 1. LC-MS/MSL lipidomics and QTL mapping as ways to lipid identification.
a, A modified MTBE lipid extraction40 was performed on plasma and liver from 64 FS and 384 DO mice. b-d, Lipid extracts were analyzed by LC-MS/MS. Identifications were obtained through LipiDex16 based on retention time window (b), exact mass (c), retention time window and tandem mass fragmentation (d). e, Quantitative values over large dynamic ranges for both identified and unidentified features were obtained. f, All lipidomic features (identified and unidentified) were then mapped onto the mouse genome via QTL mapping, revealing genomic position and founder strain allele effect pattern as results for each QTL. This additional information enabled identification of otherwise unidentified features. Abbreviations: MTBE (methyl-tert-butyl ether), FS (founder strains), DO (diversity outbred), LC-MS/MS (liquid chromatography - tandem mass spectrometry), QTL (quantitative trait loci), m/z (mass-to-charge), RT (retention time), LOD (logarithm of odds).
Here we describe the first discovery lipidomics analysis on a cohort of DO mice. In doing so, we utilize QTL position as an independent piece of information to guide lipid identification and apply it to define unidentified mass spectral features. We demonstrate the utility of genome-lipid associations to assign identification or function in independent studies through a web-based resource - LipidGenie (http://lipidgenie.com/).
Results
QTL mapping connects lipids to their genetic regulators
To explore how global association of mass spectrometry data to genomic coordinates could assist lipid identification, we collected whole lipidome profiles of plasma and liver tissue from 64 FS and 384 DO mice using high resolution LC-MS/MS (Figure 1a). Altogether, we performed 894 LC-MS/MS discovery lipidomics experiments from which we extracted approximately 4,500,000 tandem mass spectra (Figure 1d). From the full scan mass spectrometry data (Figure 1c), we detected and quantified 19,636 molecular features; 12,429 in plasma and 7,207 in liver (Figure 1b). Next, we applied the LipiDex algorithm16 to (1) match the tandem mass spectra to their respective features, (2) eliminate features derived from adduction, dimerization, in-source fragmentation, etc., and (3) to assign molecular identities when possible (Figure 1d). From the 3,283 distinct molecular features that remained, we identified 594 lipids (from 1,721 features) in plasma and 584 lipids (from 1,562 features) in liver (see Methods: Lipidomics Data Analysis for details). This discovery approach allows for broad, untargeted lipidome coverage. However, we note that some lipid classes, including PS, LysoPS, PA, LysoPA, LysoPG, cholesterol, are either partially or wholly, missed by the current method. For instance, , RPLC-ESI-MS/MS poorly retains PA and PS species. Further, cholesterols and other sterols have poor electrospray ionization efficiencies.5,9,36,37 Extended Data Figure S2a–b and Table 1 provide an overview of the identified lipids that span roughly 30 lipid subclasses from five of the major classes: fatty acyls, glycerolipids, phospholipids, sphingolipids, and sterol lipids.38 For ~70% of these identifications we find MS/MS evidence to detail fatty acid composition, otherwise we report sum composition.
Table 1.
Breakdown of lipid identifications in plasma and liver samples by one of 31 classes. “Molecular Level” refers to lipids identified with individual fatty acid rather than as a sum composition.
| Plasma |
Liver |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lipid Category | Lipid Class | Abbreviation(s) | Count | % of IDd | Molecular Level | % of Class | Count | % of IDd | Molecular Level | % of Class |
| Fatty Acyl | 29 | 4.6% | 29 | 100.0% | 37 | 5.9% | 37 | 100.0% | ||
| Acyl Carnitine | AC | 2 | 0.3% | 2 | 100.0% | 6 | 1.0% | 6 | 100.0% | |
| Fatty Acid* | FA | 27 | 4.3% | 27 | 100.0% | 31 | 5.0% | 31 | 100.0% | |
| Glycerolipid | 210 | 33.1% | 165 | 78.6% | 185 | 29.7% | 111 | 60.0% | ||
| Diglyceride | DG, Alkenyl-DG | 2 | 0.3% | 2 | 100.0% | 17 | 2.7% | 15 | 88.2% | |
| Triglyceride | TG, Alkanyl-TG, Alkenyl-TG | 208 | 32.8% | 163 | 78.4% | 168 | 27.0% | 96 | 57.1% | |
| Phospholipid | 287 | 45.3% | 194 | 67.6% | 303 | 48.6% | 238 | 78.5% | ||
| Cardiolipin | CL | 0 | 0.0% | 9 | 1.4% | 8 | 88.9% | |||
| Lyso-Phosphocholine | Lyso-PC | 27 | 4.3% | 27 | 100.0% | 18 | 2.9% | 18 | 100.0% | |
| Lyso-Phosphoethanolamine | Lyso-PE | 5 | 0.8% | 5 | 100.0% | 10 | 1.6% | 10 | 100.0% | |
| Lyso-Phosphoinositol | Lyso-PI | 1 | 0.2% | 1 | 100.0% | 3 | 0.5% | 3 | 100.0% | |
| Phosphocholine | PC | 129 | 20.3% | 69 | 53.5% | 88 | 14.1% | 62 | 70.5% | |
| Phosphoethanolamine | PE, PE-NMe2 | 24 | 3.8% | 22 | 91.7% | 80 | 12.8% | 57 | 71.3% | |
| Phosphoglycerol | PG | 2 | 0.3% | 2 | 100.0% | 26 | 4.2% | 26 | 100.0% | |
| Phosphoinositol | PI | 23 | 3.6% | 19 | 82.6% | 23 | 3.7% | 21 | 91.3% | |
| Plasmalogen | Plasmanyl-PC, Plasmenyl-PE, Plasmanyl-PE, Plasmenyl-PC | 76 | 12.0% | 49 | 64.5% | 46 | 7.4% | 33 | 71.7% | |
| Sphingolipid | 102 | 16.1% | 41 | 40.2% | 96 | 15.4% | 46 | 47.9% | ||
| Ceramide | Cer[NS], HexCer[NS], HexCer[AP], Cer[NP], Cer[AS], Cer[AP] | 40 | 6.3% | 34 | 85.0% | 58 | 9.3% | 39 | 67.2% | |
| Ganglioside* | GM2/GM3 | 13 | 2.1% | 7 | 53.8% | 8 | 1.3% | 6 | 75.0% | |
| Sphingomyelin | SM | 49 | 7.7% | 0 | 0.0% | 29 | 4.7% | 0 | 0.0% | |
| Sphingosine | SP | 0 | 0.0% | 1 | 0.2% | 1 | 100.0% | |||
| Sterol Lipid | 6 | 0.9% | 6 | 100.0% | 2 | 0.3% | 2 | 100.0% | ||
| Cholesteryl Ester | CE | 6 | 0.9% | 6 | 100.0% | 2 | 0.3% | 2 | 100.0% | |
| * hand-identified | Count | % of Total | Molecular Level | % of IDd | Count | % of Total | Molecular Level | % of IDd | ||
| Identified | 634 | 36.8% | 435 | 68.6% | 623 | 39.9% | 434 | 69.7% | ||
| Unidentified | 1087 | 63.2% | 939 | 60.1% | ||||||
| Total | 1721 | 100.0% | 1562 | 100.0% | ||||||
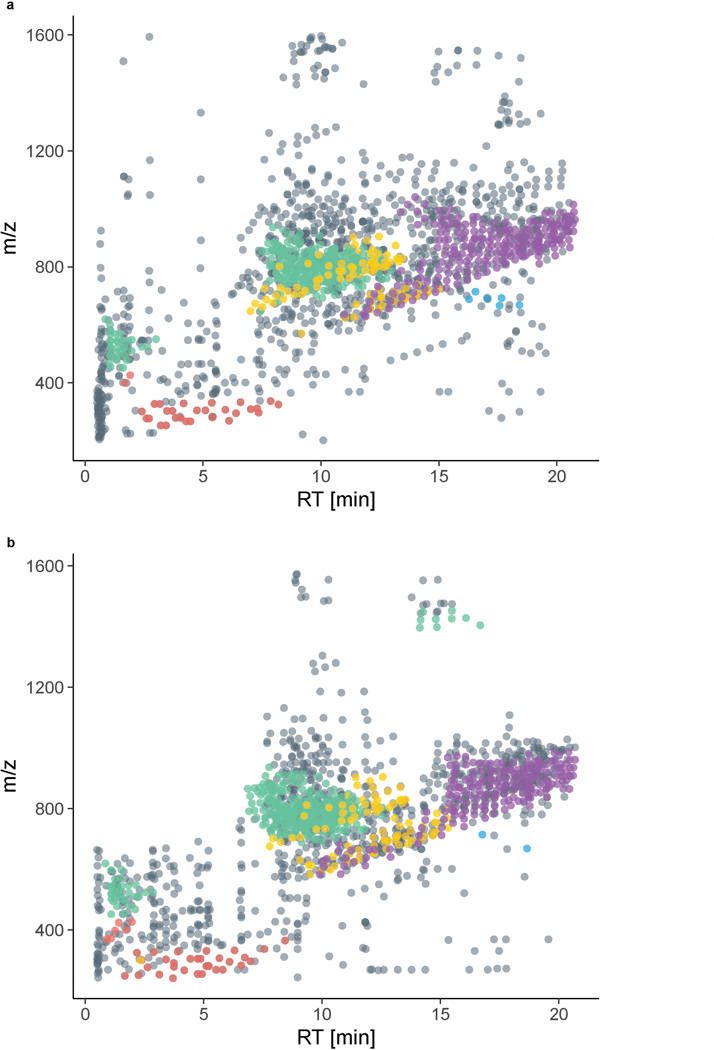
Figure S1a and b present a bird’s-eye view of these plasma and liver lipidomes. Here each distinct molecular feature is plotted as a function of its m/z and chromatographic retention time. Identified lipids are colored by class; we note members of individual lipid classes group well, adding confidence to their identification. Triglycerides (TG),39 for example, as hydrophobic lipids with three fatty acids can be found at high m/z and late chromatographic retention. From this perspective, we observe that the unidentified molecular features, ⅔ of all detected species, are either clustered around identified lipid classes or exist on m/z and retention islands. We conclude that these data can be further interrogated to (1) expand existing lipid class coverage and (2) reveal the presence of additional lipid classes.
Next, we extracted quantitative information from all detected molecular features across all 384 animals, creating a molecular trait for each feature. Figure 1e displays the quantitative values of two examples of individual molecular traits from plasma; one identified as a sphingolipid and one unidentified. Plasma HexCer[NS] d18:1_20:0 has a relative abundance dynamic range of ~15-fold across all 384 animals. For comparison, we plot the abundance of a molecular feature with a mass of 1252.8028 Da. Here we see an even greater dynamic range of ~75-fold, however, the feature was unidentified using our traditional data processing. Correlation to a candidate gene region ultimately led to the identification of the feature. To correlate these MS-derived lipid quantitative phenotypes (vide supra) with genomic variation we performed quantitative trait locus (QTL) mapping using R/qtl2.27.
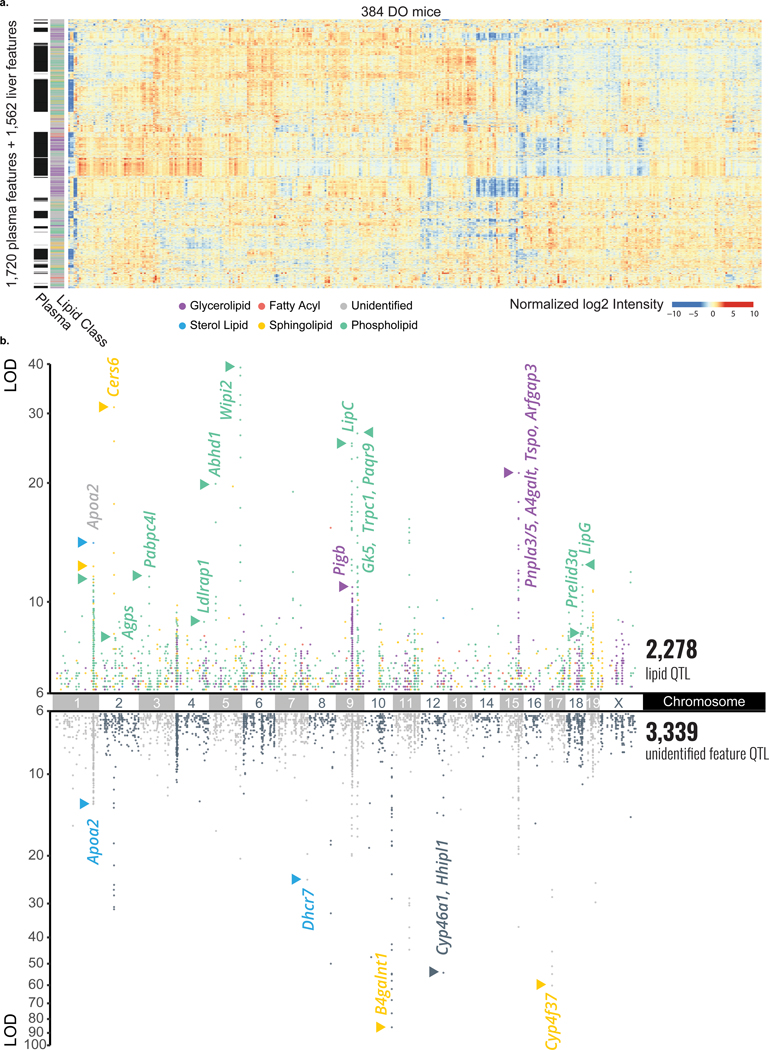
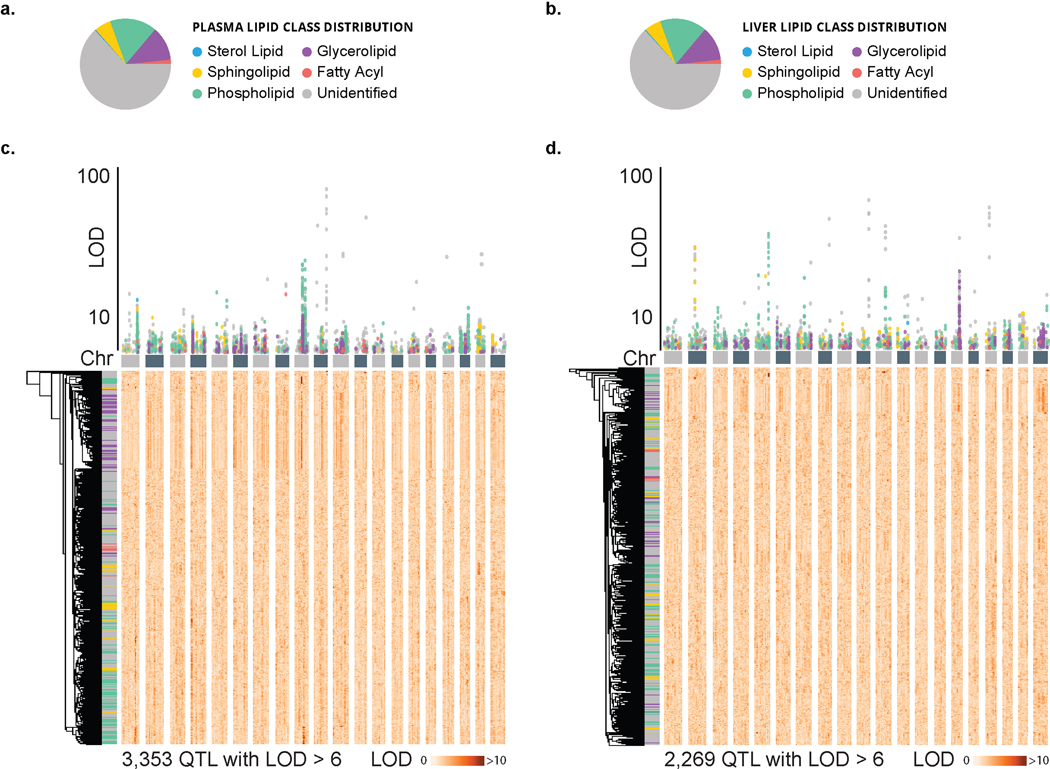
Figure 2a displays a hierarchically clustered heatmap of these quantitative results for all measured molecular traits (1,721 and 1,562 for plasma and liver, respectively) across all 384 animals. Notably, we observe considerable clustering by lipid class, even across tissue type (y-axis). We detected 3,348 plasma lipid QTL for 1,405 of the 1,721 (81.6 %) traits (logarithm of odds (LOD) score > 6). 1,351 of these were from identified lipids, while 1,997 were from unidentified features. Similarly, in liver, we detected 2,269 lipid QTL for 1,190 of the 1,562 (76.2 %) traits, of which 927 were from identified lipids while 1,342 were from unidentified features Figure 2b and Extended Data Figure S2c–d present the genetic correlations for this entire collection of significant QTL extracted in a Manhattan plot. We note that the unidentified molecular traits cluster among the various identified lipid classes, which provides further evidence that these features are of biological origin and amenable for further interrogation. Secondly, the unidentified features occupy additional distinct loci, implicating previously unidentified lipid classes.
Figure 2. Large scale lipid quantitative profiling and subsequent QTL mapping reveals hotspots of associated lipids.
a, In plasma, we quantified 1,721 lipidomic features, 621 of which were identified, and in liver, we quantified 1,562 lipidomic features, 615 of which were identified. Hierarchical clustering of all 3,283 lipidomic features’ intensities by the 384 DO mice resulted in distinct clustering by lipid class, notably across tissue type. b, When mapped onto the mouse genome, 1,405 plasma and 1,190 liver features showed at least one QTL with an LOD > 6 as displayed in a Manhattan plot (n = 3,353 + 2,269 = 5,622 total QTL). A number of lipid hotspots are shared by identified lipids and unidentified features (e. g. at Apoa2), while others only appear among the unidentified features (e. g. at B4galnt1). Abbreviations: MTBE (methyl-tert-butyl ether), Chr (chromosome), DO (diversity outbred), ESI (electrospray ionization), LC-MS/MS (liquid chromatography - tandem mass spectrometry), QTL (quantitative trait loci), m/z (mass-to-charge), RT (retention time), LOD (logarithm of odds).
QTL map recapitulates APOA2 biology and informs cholesteryl ester lipid identifications
Several genetic loci are strongly associated with lipids and appear as hotspots - locations on the genome where multiple lipid QTL co-map (Figure 2b and Extended Data Figure S2b). To better explore these regions, we asked whether these co-mapping lipid QTL shared a common genetic relationship to segregating alleles at the locus. One advantage of the DO mice is that shared founder strain allele effect patterns can be indicative of a common genetic regulator.28 Thus, we define a lipid QTL hotspot as multiple lipid QTL co-mapping (± 2 Mbp) with a shared founder strain allele effect pattern. We identified a number of hotspots; many of these are detailed in Supplementary Table 1 with their respective lipid class and likely candidate gene drivers. To garner additional support for founder strain specific genetic effects on lipid abundance, we profiled plasma and liver lipids for each of the founder strains (4 males, 4 females, Supplementary Tables 10 and 11).
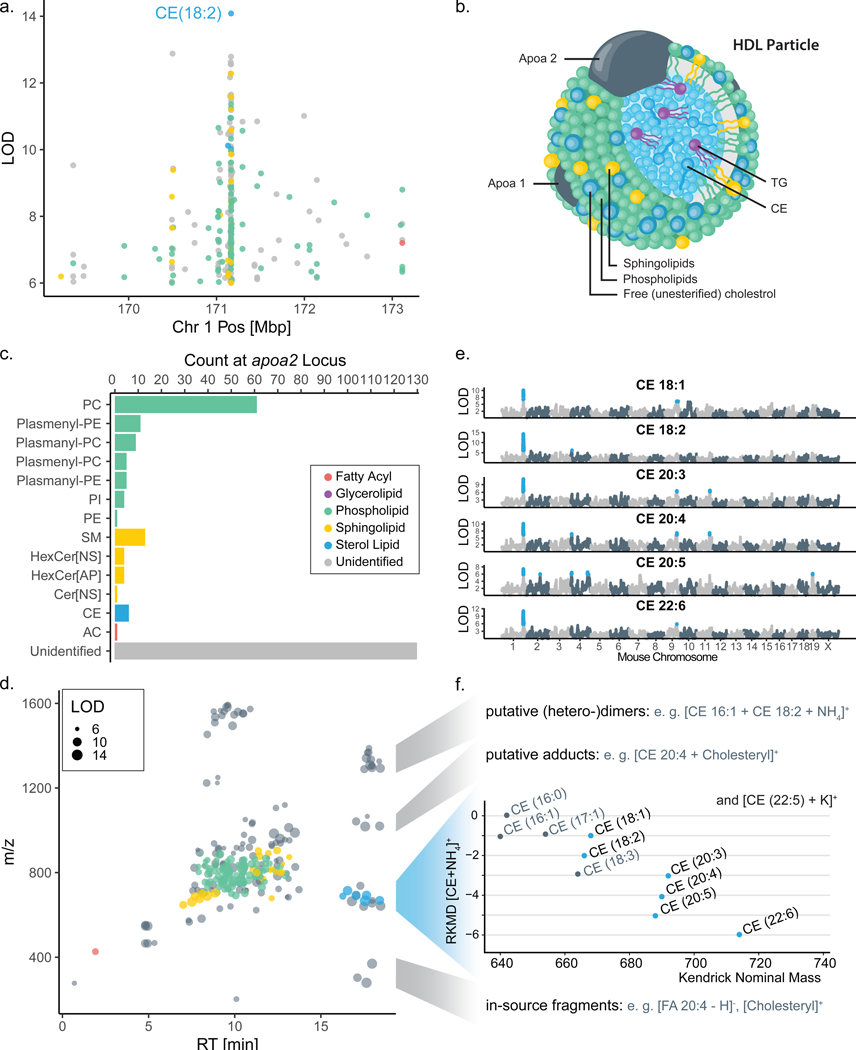
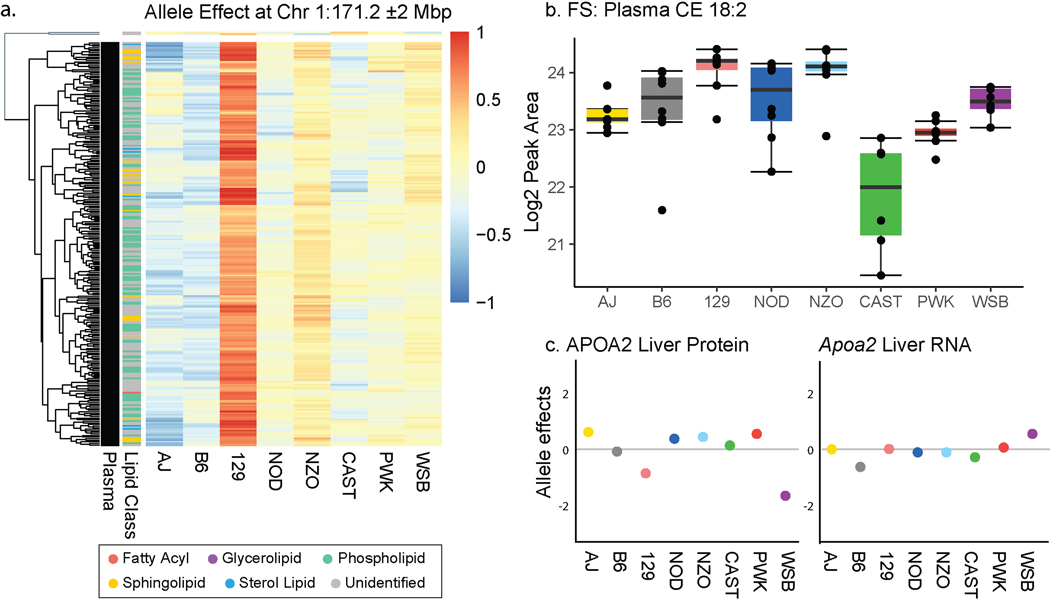
Figure 3a highlights a lipid QTL hotspot on chromosome 1:171 Mbp. Here, 255 lipid traits, all from plasma, co-localize with a shared allele effect pattern of upregulation associated with alleles derived from the founder strain 129 (Extended Data Figure S3a). The lipid with the highest LOD at this locus was a cholesteryl ester (CE 18:2), which was also elevated in founder strain 129 plasma (Extended Data Figure S3b). At 171 Mbp on chromosome 1, strain 129 possesses a missense SNP in Apoa2 gene (rs8258226), resulting in a 61Ala >Val substitution in the protein apolipoprotein-II.41 A prior DO study identified APOA2 protein and mRNA expression QTL in liver tissue but these displayed different allele effects than plasma lipid QTL, suggesting that the causal variants that modulate their respective levels differ (Extended Data Figure S3c).34 Notably, APOA2 protein is a major component of high density lipoprotein (HDL) particles in plasma, corroborated by human HDL traits mapping to APOA2 in GWAS,42 and is considered a principal genetic regulator of plasma HDL levels in mice.43,44,45,46 The other major components of HDL particles are phospholipids (35–50%) and cholesteryl esters (30–40%) (Figure 3b).47 Consistent with this composition, seven sub-types of phospholipids and various cholesteryl esters (CE) map to the Apoa2 locus (Figure 3c). Sphingolipids, a minor component of HDL particles, map in four different sub-classes to this Apoa2 locus. We conclude that this hotspot illuminates the molecular composition of HDL particles in mice, while also linking an additional 130 unidentified lipid features to this locus.
Figure 3. Co-mapping of lipids at the Apoa2 locus facilitated identification of additional cholesteryl esters.
a, One lipid hotspot on chromosome 1 at 171 Mbp is shared by 255 plasma lipid features co-mapping with a common 129 high allele effect (Extended Data Figure S3a). b, The candidate gene at this locus is Apoa2, which encodes for apolipoprotein II, which is carried on HDL cholesterol particles along with c, a variety of lipid classes, mostly phospho- and sphingolipids, which mapped to the locus. d, When plotting all 255 Apoa2-specific lipid features in the m/z-RT plane, a group of unidentified features sharing the RT region with CEs stood out. e, Notably, all six CEs show their primary QTL at this locus, as visible from their individual LOD plots. f, Subsets of the unidentified features could subsequently be identified as CE-related features, including heterodimers, cholesterol-adducts and in-source fragments. Abbreviations: HDL (high-density lipoprotein), CE (cholesteryl ester), QTL (quantitative trait loci), LOD (logarithm of odds), m/z (mass-to-charge), RT (retention time), FA (fatty acid), PC (phosphatidylcholine), PE (phosphatidylethanolamine), PI (phosphatidylinositol), SM (sphingomyelin), Cer (ceramide), AC (acylcarnitine), RKMD (referenced Kendrick mass defect).49
To test if a shared QTL would enable identification of additional lipids, we plotted the 255 lipid traits that map to the Apoa2 locus as a function of chromatographic retention time, mass, and identification status (Figure 3d). A cluster of unidentified lipid features shared retention time with CEs, a class of lipids that are often devoid of informative fragments.48 All CEs showed their major QTL at the Apoa2 locus (Figure 3e) providing greater confidence in their identification. The shared genetic regulation further allowed us to predict a CE identity for the cluster of unidentified co-mapping features. Examination of their total masses49 and tandem mass spectra supported the annotation of five additional CEs (Figure 3f), while another 18 lipid features’ m/z and RT were consistent with technical artifacts of CEs: eleven heterodimers, four cholesterol adducts, and three in-source fragments.
QTL map provides an orthogonal tool for lipid identification
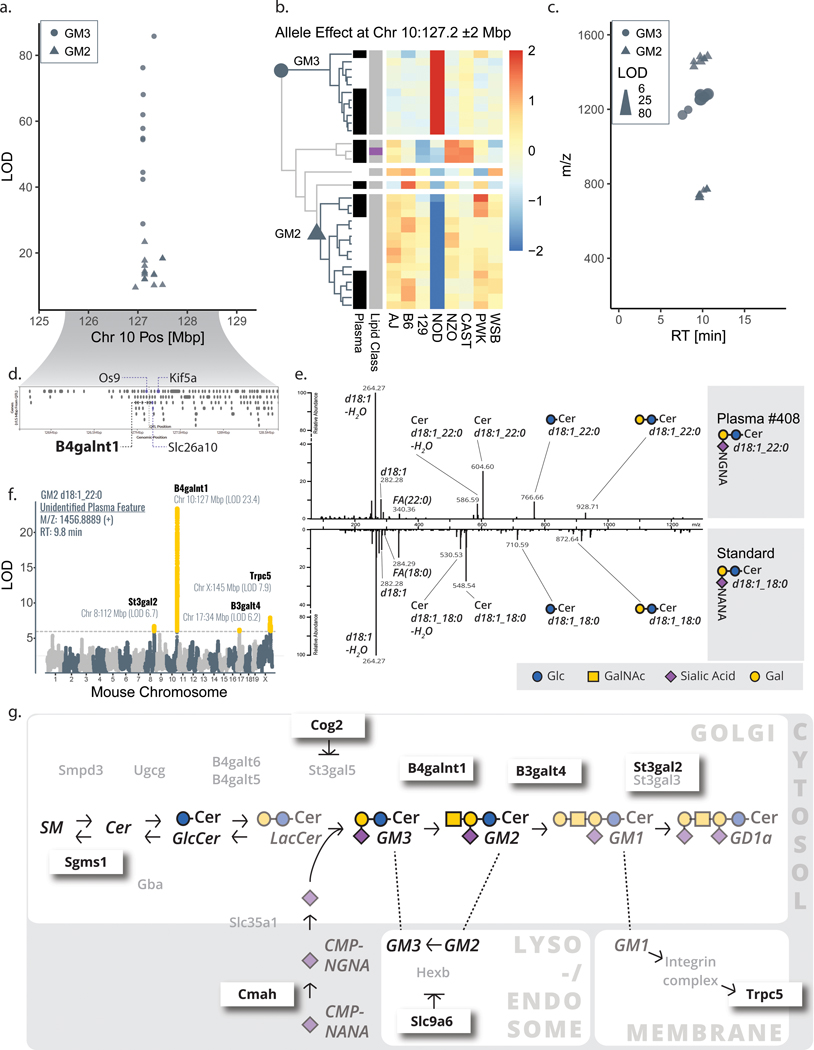
On chromosome 10, at 127 Mbp we observed a significant lipid QTL hotspot. At this site, over twenty-five plasma and liver lipid features mapped with the highest overall significance (Figure 4a). These features also shared a common allele dependence; i.e., NOD-driven and split between NOD high vs. low effect (Figure 4b). None of these lipid features were identified following our conventional data analysis strategy, which leverages retention time, mass, and tandem mass spectra. The features were observed in two distinct clusters based on m/z and RT, suggesting they could derive from two distinct lipid classes (Figure 4c). Given that these unidentified features (1) appeared as two defined lipid classes and (2) were high scoring at a genetic locus with opposite allele effects, we reasoned that identification of the causal gene may enable their identification.
Figure 4. Lipid features mapping to B4galnt1 lead to identification of GM2 and GM3 gangliosides.
a, A hotspot of solely unidentified features with exceptionally strong correlation was composed of 11 liver and 15 plasma features mapping to chromosome 10:127 Mbp with b, a similar NOD-driver allele pattern (two main clusters from hierarchical clustering, row-scaled, Euclidean cutoff of h=2.5). Two groups of lipid features (circles vs. triangles) emerged as distinct in strength of LOD (a), directionality of allele effect (b), and m/z space (c). d, The candidate gene B4galnt1 pointed us to the putative identifications of GM3 (circles) and GM2 (triangles) gangliosides, which were confirmed by e, spectral matching with a human GM3 standard. f, Secondary QTL for these gangliosides, as exemplary shown for GM2 d18:1_22:0, mapped to eight additional candidate genes within 4 Mbp of the 15 total ganglioside hotpots that were previously linked to ganglioside metabolism. g, The various candidate genes influencing GM3 and GM2 levels span well-known enzymes (e. g. B3galt4) but also include indirect affectors including Cog2 and Slc9a6. Abbreviations: NOD (non-obese diabetic mouse strain NOD/ShiLtJ), Mbp (megabase pair), LOD (logarithm of odds), QTL (quantitative trait loci), Chr (chromosome), m/z (mass-to-charge), RT (retention time), Glc (glucose), GalNAc (N-acetylgalactosamine), Gal (galactose), SM (sphingomyelin), Cer (ceramide), NGNA (N-glycolylneuraminic acid), NANA (N-acetylneuraminic acid).
The genetic effects of SNPs and other genomic variants can influence lipid abundance. For example, SNPs in coding regions can affect protein product function. In the extreme, a missense variant in proteins involved in lipid metabolism could very likely affect lipid abundance. SNPs in non-coding regions, such as promoters and enhancers, can alter gene expression. To identify candidate SNPs, we analyzed the SNPs associated with the lipids by identifying those with the founder strain SNP database at each QTL (R/qtl2; scan1snps()27) and subsequently causing missense, frameshift, stop lost/gained, incomplete terminal codons, in-frame deletions/insertions, altering 3’ or 5’ UTR sequences, splice acceptor/donor/region, predicated to cause nonsense-mediated decay, initiator codon or mature miRNA variants (according to the Sequence Ontology (SO) consortium)50. At the chromosome 10 hotspot we identified several candidate genes with potentially causal mutations (Figure 4d). We included in our analysis, but did not focus on genes with synonymous, stop retained, up-/downstream, intergenic, intron and non-coding transcript (exon) variants (which represent ~97% of all SNPs in the database).
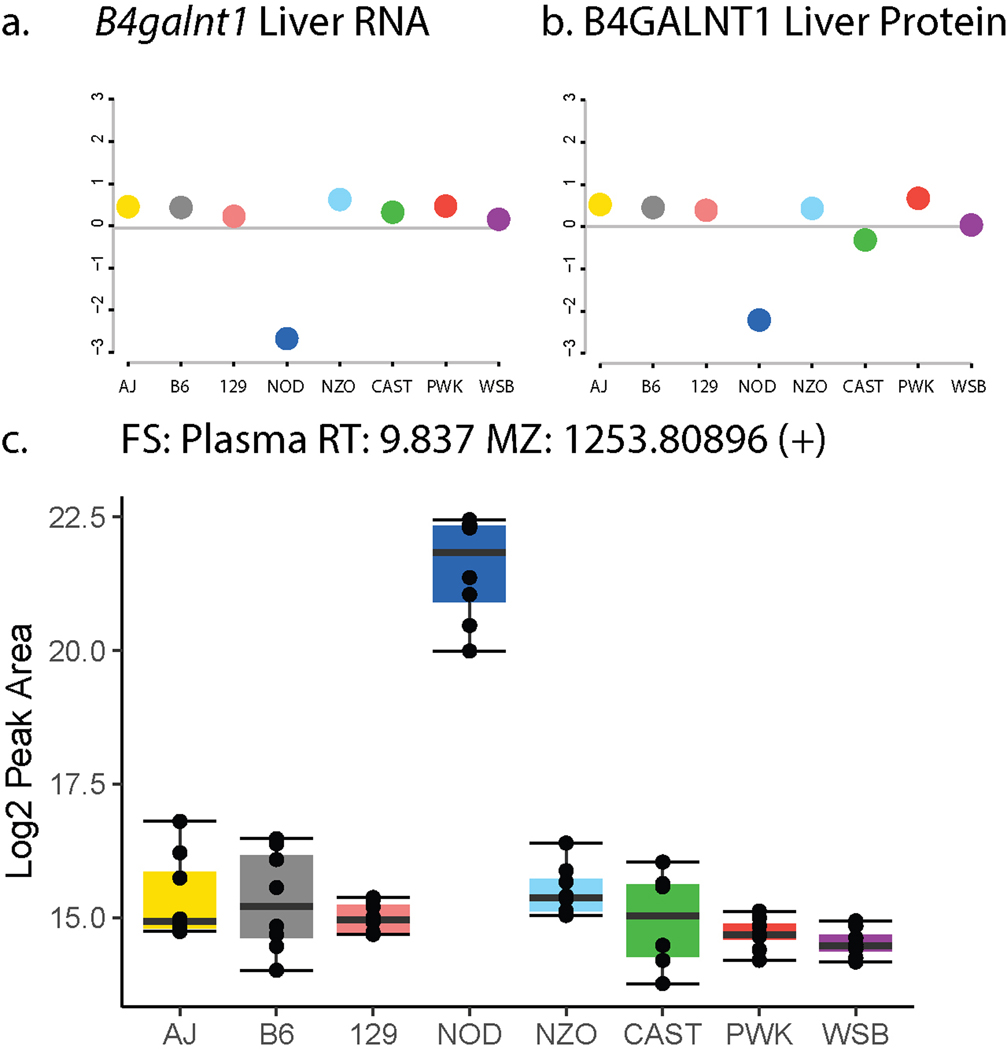
In cases of altered gene expression, we further narrowed down the list of candidates by directly assessing transcriptomics data. While we did not profile hepatic gene expression in the DO cohort used for lipid QTL analysis, we surveyed a recently published hepatic QTL data set to match allele effects of mRNA expression and protein QTL that are within the location of the candidate gene (cis-eQTL and pQTL, respectively).34 We asked if any transcripts or proteins presented a similar NOD-driven allele effect at the lipid locus on chromosome 10. Of the protein coding genes within ±2 Mbp of the lipid QTL, 55 showed a cis-eQTL (Supplementary Table 2). However, the only cis-eQTL that was strongly and uniquely driven by NOD alleles was B4galnt1 (Extended Data Figure S4a). Furthermore, 16 cis-pQTL were identified for genes within this region, including B4GALNT1 (Supplementary Table 3). Similar to the cis-eQTL, the only pQTL that showed an NOD-driven allele effect pattern was for B4GALNT1 (Extended Data Figure S4b). Consequently, the 3’ UTR variant in B4galnt1 SNP rs13462597 was our strongest candidate as the genetic regulation of hepatic B4galnt1 transcript and protein expression matches that of the unidentified lipids.
B4galnt1 encodes for β−1,4 N-acetylgalactosaminyltransferase 1, an enzyme that catalyzes the conversion of GM3 to GM2 gangliosides.51 With this candidate gene in mind, we investigated whether the unidentified lipids could be classified as gangliosides. Their precursor m/z and tandem mass spectra were consistent with monosialic gangliosides, which we further confirmed by comparison with a GM3 ganglioside standard (Figure 4e). In total, we confidently identified 26 lipid features as six unique GM2 and seven unique GM3 species (Supplementary Table 4). Consistent with an NOD-driven effect, NOD mice have higher abundance of GM3 gangliosides in pancreas,52 and we confirmed NOD had higher abundance of GM3 in plasma in an independent lipidomic analysis of founder strain mice (Extended Data Figure S4c).
By identifying the features mapping to chromosome 10:127 Mbp as gangliosides, we recognized that ganglioside abundances, like the levels of most lipid species, were polygenic, that is regulated by multiple loci (Figure 4f). From the 26 identified ganglioside features we gain a total of 62 QTL annotations, describing more than 15 unique loci (at least two ganglioside features with LOD > 6.0) on 10 chromosomes (Supplementary Table 5). Interestingly, these newly annotated ganglioside QTL mapped to candidate genes of the ganglioside pathway (Sgms153, B3galt4, St3gal2, Cmah54), even more distant regulators of ganglioside metabolism (Slc9a655, Cog256, Trcp557, Cdh13), and regions of the genome with yet undescribed ganglioside regulation (Figure 4g).
LipidGenie identifies candidate genetic regulators for lipid features
To make these genome-lipid associations accessible to the community we created a web-based resource; LipidGenie (http://lipidgenie.com). With LipidGenie, lipid features can be searched by m/z, lipid identifier, or lipid class. The search returns QTL of the matching features and allows the user to explore the genetic region, founder strain allele effects, and associated SNPs. In addition, with LipidGenie individual genes or gene regions can be queried for lipid associations.
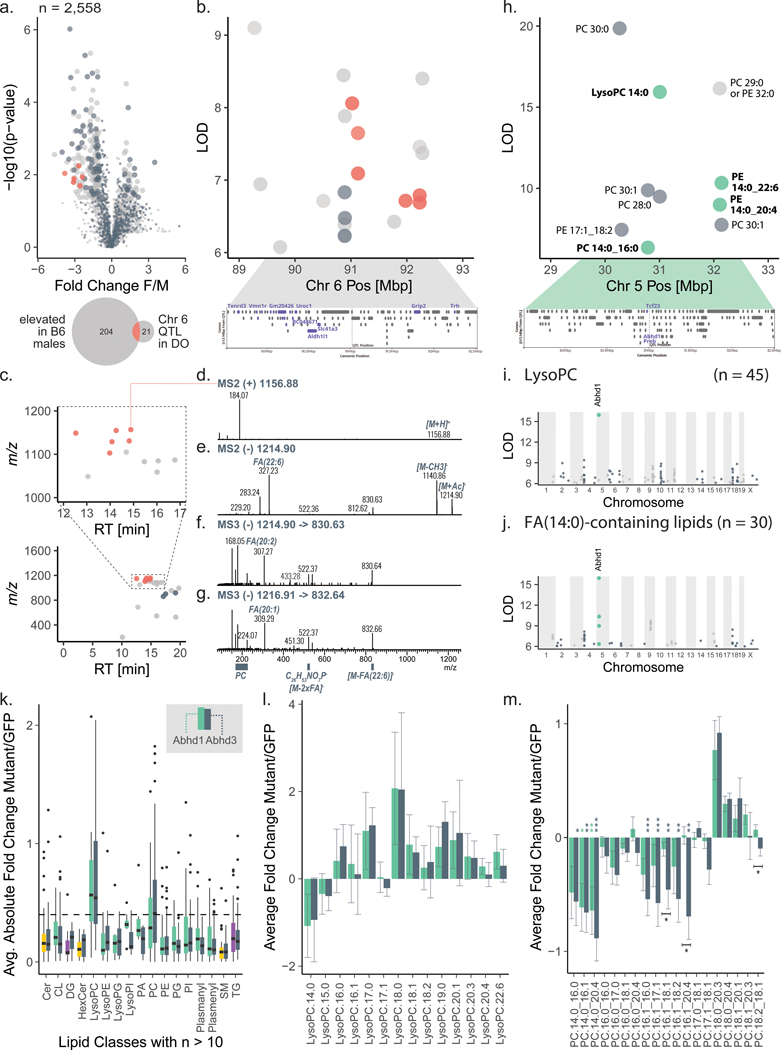
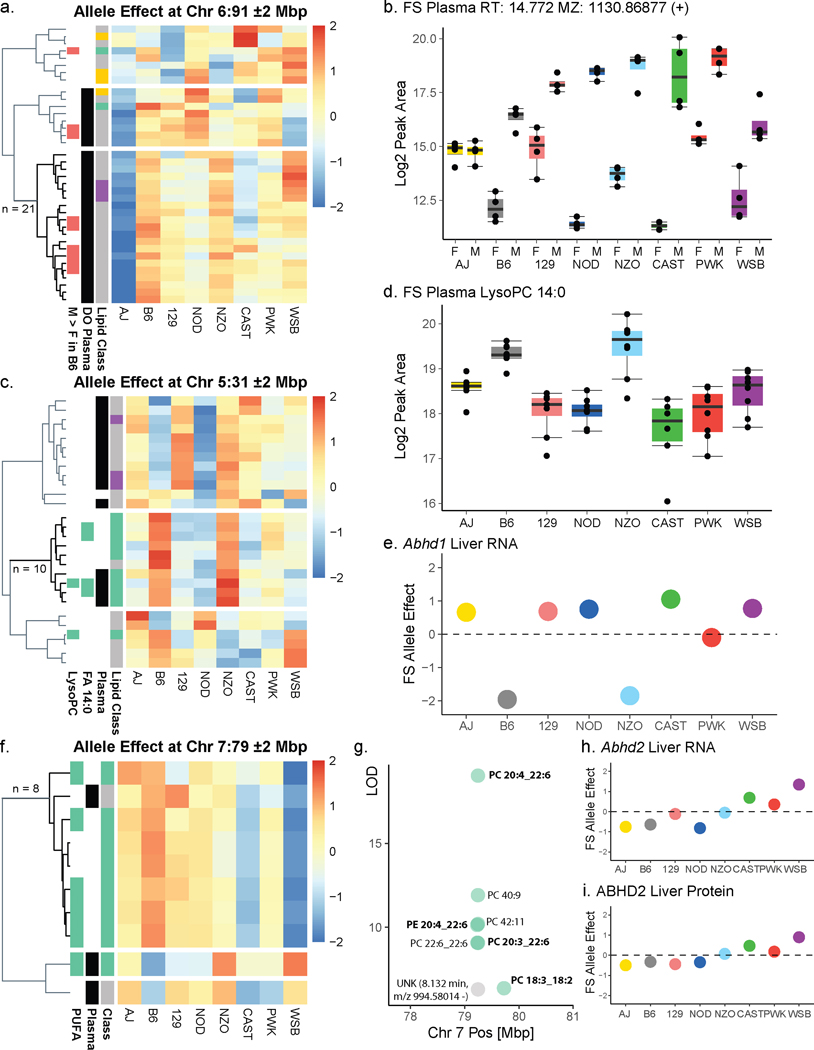
To validate LipidGenie we explored sex-associated lipid features that were observed within the B6 founder strain. In this study we quantified 2,558 lipid features in B6 plasma and found 254 features that showed significantly different levels by sex (Figure 5a). As is common in LC-MS lipidomics, most of these sex-specific features were unidentified after the database search (n = 197). Utilizing LipidGenie’s m/z search parameter and a 10 ppm m/z window, we found significant genome-lipid associations for 127 of the sex-specific features, of which 79 were unidentified. Strikingly, a group of six unidentified lipids mapped to the same genetic locus on chromosome 6 at 91 Mbp (Figure 5b, Supplementary Table 6); all had similar allele effect patterns (Extended Data Figure S5a) and were elevated in males (Figure 5a and Extended Data Figure S5b). At the locus, a total of 12 out of 21 co-mapping features shared a lipid class-like behavior, i.e., clustered in m/z-RT space (Figure 5c). To further characterize these lipids, we collected additional tandem mass spectra in both positive and negative mode (Figure 5d–g).
Figure 5. Web resource LipidGenie guides exploration of genome-lipid connections.
a, We quantified 2,558 features in B6 plasma (n=4 for each sex). 254 features were sex-specific (FC > 1.0, p < 0.05, non-paired, two-sided Student’s t-test). Precursor m/z (±10 ppm) matching to our DO database provided genetic information for ⅓ of the otherwise unidentified features. b, Six male-specific unidentified features (red) share a QTL on Chr 6:91 Mbp with a common A/J down effect (Extended Data Figure S5a). c, The features further clustered in m/z-RT space. d-g, Targeted fragmentation spectra (exemplary spectra for two species ([M+H]+ m/z 1156 and 1158) in positive (MS2) and negative (MS2 and MS3) mode) exhibited signals consistent with a lipid class built of a PC headgroup and three FAs. h, The DO database further confirmed LysoPC 14:0 mapping to Abhd160 with j, an enrichment of FA 14:0 containing lipids k, We compare Hepa1-6 overexpressing ABHD1 and ABHD3 versus a control overexpressing GFP (n=12 for each, 4 biological x 3 technical replicates, boxplots are defined with first and third quartiles for lower and upper hinges, 1.5x interquartile range for the length of the whiskers, center line at median). The boxplots show absolute FC of each mutant over GFP by lipid class; the dashed line is at FC=0.4 l, The lowest (negative) FC for both is observed for LysoPC 14:0; isomers are summed. m, All 14:0 containing PCs exhibit a negative FC for ABHD1 and ABHD3 mutants consistently, while 18:0 containing species are showing opposing positive FC. Plotted are sum-normalized, log2-transformed FC means with error bars representing 95% confidence interval, significance indicated by * (p < 0.05), ** (p < 0.01), *** (p < 0.001) of non-paired two-sided Student’s t-test, equal variance, n=12 for each, details in source data. Abbreviations: DO (diversity outbred), FC (fold change), m/z (mass-to-charge), QTL (quantitative trait loci), Chr (chromosome), Mbp (megabase pair), SNP (single nucleotide polymorphism), PC (phosphatidylcholine), F/M (female-to-male), RT (retention time), FA (fatty acid), PE (phosphatidylethanolamine), GFP (green fluorescent protein)
The spectra showed shared fragmentation patterns consistent with a phosphatidylcholine (PC) class identity. Strikingly, one fatty acid seemed to be either FA 22:6 (Figure 5e) or FA 16:0, but only MS3 spectra showed the presence of a second acyl chain expected for PCs (Figure 5f–g). These features also shared an m/z 522 fragment that matched the formula of LysoPC 19:0, C26H53NO7P−.
We next leveraged the LipidGenie associations to generate hypotheses about the nature of this lipid class. At chromosome 6 at 91 Mbp, we found SNPs with matching allele effects in several genes including Txnrd3, Vmn1r, Uroc1, Aldh1l1, Slc41a3, Grip2, and Trh (Figure 5b). One possible candidate on chromosome 6 is Vmn1r, encoding for vomeronasal receptors, the organs that sense pheromones. Not only could this gene explain the observed sex difference, it also points us to PC estolides as a potential class identity. Estolides are lipids containing fatty acid esters of hydroxy fatty acids (FAHFAs). Consistent with the observed 16:0 or 22:6 fragments in MS2 spectra of the unidentified lipids, 16:0 and 22:6 can be esterified to hydroxy fatty acids to form FAHFAs.58 This hypothesis is further supported by accounts of FAHFAs as pheromones in spiders and TG estolides in mammalian scent glands.59 The potential estolide identity is intriguing, but definitive identification will require follow-up studies. Further evidence is likely contained in the genetic associations. Similar to our earlier example with gangliosides, we observed co-mapping of these 12 lipids at other loci (e. g., Chr 10, at 84 Mbp, Chr 12, at 84 Mbp), thereby offering potential pathway information (Supplementary Table 7) and highlighting the power of genome-lipid associations obtained with LipidGenie.
We next explored whether LipidGenie would also offer insights when querying for identified lipid features. Recently, Parker et al. found an association between LysoPC 14:0 and chromosome 5 at 31 Mbp using multi-omic QTL mapping of the hybrid mouse diversity panel60. From these data, they postulated that the protein encoded by candidate gene Abhd1 (alpha beta hydrolase domain containing 1) regulates plasma levels of LysoPCs. Note, ABHD1 has no annotated function. LipidGenie’s lipid search provides a direct means to test this putative functional annotation of ABHD1.
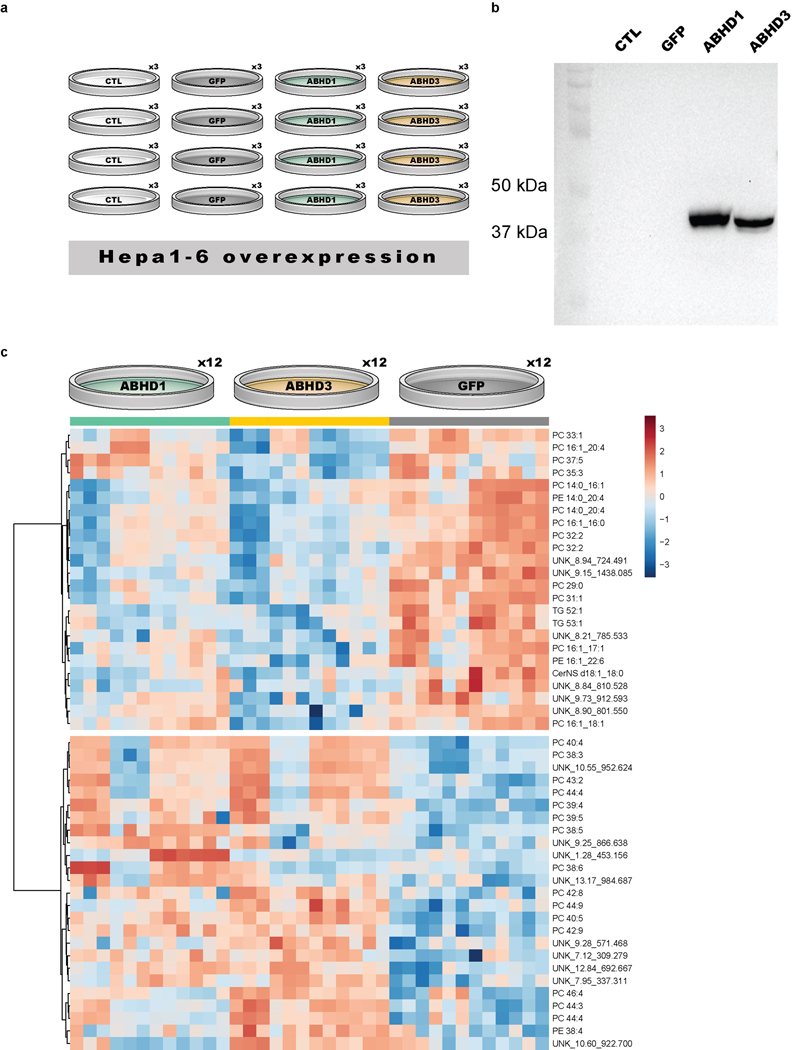
LipidGenie confirmed that plasma LysoPC 14:0 has a strong QTL at the abhd1 locus (Figure 5h), and further found the B6 and NZO high allele effect consistent with the 3’ UTR variant rs29681817 (Extended Data Figure S5c). This observation is further supported by an independent measure of the founder strain mice and a hepatic cis-eQTL in Abhd1 with matching opposite allele effects (Extended Data Figure S5d–e). To connect the function of ABHD1 protein to LysoPCs, we asked whether other LysoPCs (n = 45) mapped to this gene region. However, we did not find general mapping of LysoPCs to this locus (Figure 5i), but instead found other lipids co-mapping on chromosome 5, at 31 Mbp, including PC 14:0_16:0, PE 14:0_20:4, PE 14:0_22:6, PC 28:0, PC 30:0, and PC 30:1 (Figure 5h). These fatty acid signatures suggest a myristic acid (14:0) specific association. Given the high degree of lipid structural resolution contained within LipidGenie, we demonstrate that 14:0-containing lipids (n = 30) have an enriched hotspot at the Abhd1 locus (Figure 5j). With these data we propose that ABHD1 is a phospholipase for myristic acid containing phospholipids; consistent with the function of a related and highly homologous gene, abhd3.61,62 14:0-containing phospholipids have also been mapped to ABHD3 in human GWAS.63 To validate this hypothesis, we overexpressed ABHD1 and ABHD3 in Hepa1-6 cells (Extended Data Figure S6a–b) and measured their lipidome with respect to cells overexpressing GFP as control (Supplementary Table 12). Hierarchical clustering of the top 49 features showed two clusters, one with increased levels in the mutants over control and the other one decreased (Extended Data Figure S6c). We noticed a majority of identified lipids among the most significantly different features, and when plotting the average fold change by lipid class, LysoPC and PC phospholipids stood out (Figure 5k). Upon closer look, we could confirm the predicted fatty acid dependency for both LysoPC and PC lipids, particularly prominent in 14:0 containing phospholipids (Figure 5l–m). While ABHD1 and ABHD3 mutants exhibited largely similar lipidomic profiles, differences as in PC 16:1_20:4 that was only decreased in the Abhd3 mutant, may also point to differential functions. The 14:0 specificity could be relevant to human health as plasma LysoPC 14:0 is a predictor of diabetes risk in humans. Finally, our proposed function might provide a clue to understanding why ABHD1 is associated with oxidative stress, a prominent hallmark of metabolic diseases. 61,64,65
Having documented the diverse utility of LipidGenie for lipid queries, we lastly tested its use for gene-based queries. ABHD2, another member of the alpha beta hydrolase domain protein family, acts on arachidonylglycerol, among other substrates.66,67 A LipidGenie query of Abhd2 does indeed provide evidence for this polyunsaturated fatty acid pathway specificity. Specifically, within 2 Mbp of Abhd2, LipiGenie returned ten liver phospholipids. Eight of these lipids shared an allele effect pattern, and contained poly-unsaturated fatty acids - i.e., 18:2, 18:3, 20:3, 20:4, and 22:6 (Extended Data Figure S5f–g). Further, ABHD2 showed matching opposite WSB and CAST effects in both liver cis-eQTL and pQTL (Extended Data Figure S5h–i).34
Discussion
Discovery lipidomics presently relies on measurement of various chemical properties for lipid identification. These properties are most often hydrophobicity, mass, and fragmentation pattern. Unfortunately, application of only these strategies to complex mammalian lipid mixtures results in many unidentified lipid features. Here we investigated the power of genome-lipid associations to facilitate lipid identification.
To construct a large-scale map of genome-lipid associations, we performed QTL analysis for over 5,000 plasma and liver lipid QTL, of which over 60% stem from unidentified spectral features. To our knowledge, this QTL map is the broadest in scope and depth of lipids analyzed and QTL identified in mice.60,68,69 With these data, we first tested our hypothesis by analyzing one of several QTL hotspots; the Apoa2 locus. The identified lipids mapping to this locus belonged to 11 different classes and, together with APOA2, constitute the known components of HDL particles. With this association, 23 unidentified lipid features could be classified as cholesteryl esters and related features.
To further test the concept, we selected a second hotspot containing only unidentified lipid features (10:127 Mbp). Genetic mapping to B4galnt1 enabled their identification as GM3 and GM2 gangliosides. In fact, the identification allowed for a comprehensive investigation of their complex polygenic regulation. We identified a total of eight candidate genes that likely contribute different functions in the pathway, including three (Slc9a6, Cog2, Trpc5) that exert indirect effects on ganglioside biosynthetic enzymes.
Having confirmed the value of genome-lipid associations for lipid mass spectral data annotation, we built an interactive, query-able resource - LipidGenie. Using the lipid query function, we demonstrated LipidGenie’s ability to facilitate lipid identification and in one instance revealed a potentially new sub-class of PC lipids (PC-estolides). Beyond assisting lipid identification, LipidGenie can provide evidence for gene function, and when queried for either lipid ID or gene ID, LipidGenie revealed acyl-chain specificity for ABHD1 and ABHD2, respectively. We confirmed the putative phospholipase function of ABHD1 in cells overexpressing the mouse protein while comparing to ABHD3.
We envision the genome-lipid associations contained within LipidGenie to be a valuable resource for researchers across multiple fields. We anticipate it will be immediately useful for directed analysis of key unidentified features in exploratory lipidomics analyses and lead to recovery of more data for biological studies. A limitation of the approach is that lipid identification remains a manual process and this tool does not remove the requirement for expert knowledge and care in spectral interpretation for its use. With all this said, we hope it will garner excitement for potentially novel genetic regulation of lipid metabolism. Finally, through integration with other large data resources, e.g., protein-protein interactions, pathway tools, tissue-specific QTL, GWAS data, etc., these genome-lipid associations will allow more global integration of lipid data into current knowledge bases. Especially the integration with human loci will allow for cross-validation to inform human health and disease.70,71
Methods
Animal Husbandry and Sample Collection.
All experiments involving mice were preapproved by an AAALAC-accredited Institutional Animal Care and Use Committee of the College of Agricultural Life Sciences (CALS) at the University of Wisconsin-Madison. The CALS Animal Care and Use Protocol number associated with the study is A005821, A.D. Attie, Principal Investigator. Equal numbers of male and female Diversity Outbred (DO) mice and the eight founder strains (C57BL/6J (B6), A/J, 129S1/SvImJ (129), NOD/ShiLtJ (NOD), NZO/HILtJ (NZO), PWK/PhJ (PWK), WSB/EiJ (WSB), and CAST/EiJ (CAST)) were all obtained from the Jackson Labs and have been previously described.32,33,72 Briefly, all mice were housed within the vivarium at the Biochemistry Department, University of Wisconsin-Madison, and maintained on a Western-style high-fat/high-sucrose (HF/HS) diet (44.6% kcal fat, 34% carbohydrate and 17.3% protein) from Envigo Teklad (TD.08811) for 16 weeks. All mice were maintained in a temperature and humidity-controlled room on a 12 hr light/dark cycle (lights on at 6AM and off at 6PM), and provided water ad libitum. At ~22 weeks of age, mice were sacrificed following a 4 hr fast. Plasma and liver were collected from each mouse and flash frozen in liquid nitrogen. One sample from each tissue per mouse was used for lipidomic analyses.
Mouse Genotyping and Haplotype Reconstruction.
We collected tail biopsies for DNA extraction28 at 4 to 6 weeks of age when animals arrived at the University of Wisconsin and were assigned to single-housed pens. We shipped DNA to Neogen (Lincoln, NE) for genotyping using the Mouse Universal Genotyping Array (GigaMUGA; 143,259 markers). Genotype calls were subject to quality control as described in Broman et al.73 Genotypes were used to reconstruct the 8-founder haplotype mosaic of each DO mouse using the hidden Markov model in the R/qtl2 software package.27,32 The haplotype-reconstruction uses information at each genetic markers and its neighbors to assign an eight-state haplotype probability that accounts for both heterozygosity and uncertainty in haplotype assignments.26 We interpolated the founder haplotype probabilities onto an evenly spaced grid of 69,005 pseudo-markers for mapping analysis. Sample mix-ups (one pair of samples) were resolved using islet gene expression data as described in Keller et al. 2018.74
Plasmids and Cell Culture Expression.
Mouse Abhd1 (CMV6 promoter, Myc-DDK-tagged, MR206471) and mouse Abhd3 (CMV6 promoter, Myc-DDK-tagged, MR206458) plasmids were obtained from Origene. Manufacturer’s sequencing primers were used to confirm plasmid insert. His-tagged CMV6-GFP plasmid was a gift from J. Simcox. All plasmids were transformed into E. coli (ThermoFisher Scientific, 18258012). Plasmids were maxiprepped according to manufacturer’s instructions (Qiagen, 12362).
5x105 Hepa1-6 cells (ATCC® CRL-1830) were seeded in 6-well plates with DMEM (ThermoFisher Scientific, 12100061). After 16 hours, cells were reconditioned with fresh media for 2 hours. Cells were transfected in triplicate with Lipofectamine2000 (ThermoFisher Scientific, 11668019) according to manufacturer’s instructions. Transfection efficiency was confirmed by visualizing GFP. After 24 hours, media was replaced. 48 hours after transfection, cells were washed in cold 1X PBS and scraped to be released from the plate. Released cells were pelleted by centrifugation and snap-frozen in liquid nitrogen. The frozen cell pellets were stored at −80 °C until lysis. Hepa1-6 cells were a gift from J. Simcox.
For Western Blots, cell pellets were lysed in 2x SDS-PAGE loading buffer and boiled at 95C for 5 min. Samples were run on a 10% SDS-PAGE gel for 1.5 h at 120V, standard is Precision Plus Dual Color Protein Standards (Bio-Rad, 1610394). Samples were wet-transferred onto PVDF membrane (Bio-Rad, 1620177) for 1.5 h at 100 V. Following transfer, membrane was blocked in 5% milk in TBST for 1 h at room temperature. Membrane was incubated overnight at 4C with 1:2000 rabbit anti-MYC antibody (CST, 2278 clone 71D10) in blocking buffer. Primary antibody was removed by washing 3X with 1X TBST. Membrane was incubated with 1:2000 goat anti-rabbit-HRP conjugated antibody (CST, 7074S) in blocking buffer. Samples were visualized with Clarity Western ECL Substrate (Bio-Rad, 1705060) on a ThermoFisher iBright FL1500 Imaging System. Uncropped and unprocessed scans are supplied in the Source Data file.
Lipidomics Sample Preparation. Plasma.
40 μL (30 μL for founder strains, FS) of plasma and 10 μL SPLASH Lipidomix internal standard mixture (Avanti Polar Lipids, Inc.) were aliquoted into a tube. Protein was precipitated by addition of 215 μL MeOH. Control samples comprised an aliquot of mixed male and female B6 plasma (Chow diet), extracted with each batch. After the mixture was vortexed for 10 s, 750 μL methyl tert-butyl ether (MTBE) were added as extraction solvent and the mixture was vortexed for 10 s and mixed on an orbital shaker for 6 min. Phase separation was induced by adding 187.5 μL of water followed by 20 s of vortexing. All steps were performed at 4 °C on ice. Finally, the mixture was centrifuged for 4 min at 14,000 x g at 4 °C and 150 μL of the lipophilic upper layer were transferred to glass vials and dried by vacuum centrifuge for 60 min. The dried extracts were re-suspended in 100 μL MeOH/Toluene (9:1, v/v).
Lipidomics Sample Preparation. Liver.
20 (± 2) mg liver tissue, frozen in liquid nitrogen along with 20 μL SPLASH Lipidomix internal standard mixture were aliquoted into a tube with a metal bead and 1150 μL of MTBE/MeOH (10:3, v/v) were added for protein precipitation. Control samples for DO comprised aliquots of sample pooled from FS, extracted with each batch. All steps were performed at 4 °C on ice. The mixture was homogenized by bead beating for 4 min at 25 Hz and shaking on an orbital shaker for 6 min. After bead removal, 225 μL of water were added to each tube and the mixture was vortexed for 20 s. Finally, the mixture was centrifuged for 20 min at 13,000 x g at 4 °C after which 200 μL of the lipophilic upper layer were transferred to glass vials and dried by vacuum centrifuge for 60 min. The dried lipophilic extracts were re-suspended in 100 μL MeOH/Toluene (9:1, v/v).
Lipidomics Sample Preparation. Cells.
Hepa1-6 cells were scraped off of six well plates and transferred to 1.5 ml Eppendorf tubes. Cell pellets were kept frozen (less than −20 °C) until extraction. Cells were lysed and protein was precipitated by addition of 225 μL MeOH. 750 μL methyl tert-butyl ether (MTBE) were added as extraction solvent. The mixture was homogenized by vortexing for 10 s and shaking on an orbital shaker for 6 min. Phase separation was induced by adding 187.5 μL of water followed by 20 s of vortexing. All steps were performed at 4 °C on ice. Finally, the mixture was centrifuged for 8 min at 14,000 x g at 4 °C and 200 μL of the lipophilic upper layer were transferred to glass vials and dried by vacuum centrifuge for 60+ min. The dried extracts were re-suspended in 100 μL MeOH/Toluene (9:1, v/v).
LC-MS/MS.
Sample analysis by LC-MS/MS, running data-dependent acquisition (DDA) with dynamic exclusion and polarity switching, was performed in randomized order on an Acquity CSH C18 column held at 50 °C (2.1 mm x 100 mm x 1.7 μm particle diameter; Waters) using an Ultimate 3000 RSLC Binary Pump (400 μL/min flow rate; Thermo Scientific) for plasma, while for the liver and cell samples a Vanquish Binary Pump (400 μL/min flow rate; Thermo Scientific) was used. Mobile phase A consisted of 10 mM ammonium acetate in ACN/H2O (70:30, v/v) containing 250 μL/L acetic acid. Mobile phase B consisted of 10 mM ammonium acetate in IPA/ACN (90:10, v/v) with the same additives. Mobile phase B was initially held at 2% for 2 min and then increased to 30% over 3 min. Mobile phase B was further increased to 50% over 1 min and 85% over 14 min and then raised to 95% over 1 min and held for 7 min. The column was re-equilibrated for 2 min before the next injection.
Plasma.
Ten microliters of lipid extract were injected through SII for Xcalibur by an Ultimate 3000 RSLC autosampler (Thermo Scientific). The LC system was coupled to a Q Exactive Focus mass spectrometer run by Tune software version 2.5.0.2042 (Thermo Scientific) by a HESI II heated ESI source kept at 300 °C (Thermo Scientific). The inlet capillary was kept at 300 °C, sheath gas was set to 25 units, auxiliary gas to 10 units, and the spray voltage was set to 5,000 V (+) and 4,000 V (−), respectively. The MS was operated in polarity switching mode acquiring positive and negative mode MS1 and MS2 spectra (Top2) during the same separation. MS acquisition parameters were 17,500 resolving power, 1 × 106 automatic gain control (AGC) target for MS1 and 1 × 105 AGC target for MS2 scans, 100-ms MS1 and 50-ms MS2 ion accumulation time, 200- to 1,600-Th MS1 and 200- to 2,000-Th MS2 scan range, 1-Th isolation width for fragmentation, stepped HCD collision energy (20, 30, 40 units), 1.0% under fill ratio, and 10-s dynamic exclusion.
Liver.
One microliter of lipid extract was injected through SII for Xcalibur (Thermo Scientific) by a Vanquish Split Sampler HT autosampler (Thermo Scientific). The LC system was coupled to a Q Exactive HF mass spectrometer run by Tune software version 2.8.0.2688 (Thermo Scientific) by a HESI II heated ESI source kept at 300 °C (Thermo Scientific). The inlet capillary was kept at 300 °C, sheath gas was set to 25 units, auxiliary gas to 10 units, and the spray voltage was set to 4,000 V (+) and 3,500 V (−), respectively. The MS was operated in polarity switching dd-MS2 mode acquiring positive and negative mode MS1 and MS2 spectra (Top2 for positive, Top3 for negative mode) during the same separation. MS acquisition parameters were 60,000 resolution and 3 × 106 automatic gain control (AGC) target for MS1 and 15,000 resolution and 5 × 105 AGC target for MS2 scans, 100-ms MS1 and 35-ms MS2 ion accumulation time, 240 to 1,200-Th MS1 scan range for positive and to 1,600-Th for negative mode, and 200- to 2,000-Th MS2 scan range, 1.4-Th isolation width for fragmentation, stepped HCD collision energy (20, 25 units for positive, 20,30 units for negative mode), and 10-s dynamic exclusion.
Cells.
Ten microliters of lipid extract were injected through SII for Xcalibur (Thermo Scientific) by a Vanquish Split Sampler HT autosampler (Thermo Scientific). The LC system was coupled to a Q Exactive HF mass spectrometer run by Tune software version 2.9.3.2948 (Thermo Scientific) by a HESI II heated ESI source kept at 300 °C (Thermo Scientific). The inlet capillary was kept at 300 °C, sheath gas was set to 25 units, auxiliary gas to 10 units, and the spray voltage was set to 4,000 V (+) and 3,500 V (−), respectively. The MS was operated in polarity switching mode acquiring positive and negative mode MS1 and MS2 spectra (Top2) during the same separation. MS acquisition parameters were 30,000 resolving power, 1 × 106 automatic gain control (AGC) target for MS1 and 1 × 105 AGC target for MS2 scans, 100-ms MS1 and 50-ms MS2 ion accumulation time, 200- to 1,600-Th MS1 scan range, 1-Th isolation width for fragmentation, stepped HCD collision energy (20, 30, 40 units), 1.0% under fill ratio, and 10-s dynamic exclusion.
Lipidomics Data Analysis.
The resulting LC-MS lipidomics raw files were converted to mgf files via MSConvertGUI (ProteoWizard, Dr. Parag Mallick, Stanford University75) and processed using Compound Discoverer 2.0 (Thermo Fisher Scientific) and an in-house developed open-source software suite, LipiDex16. Briefly, these software tools use area under the peak intensity calculations to generate relative quantitation of each spectral feature as is typical for metabolomic analyses. All raw files were loaded into Compound Discoverer with blanks marked as such to generate two result files using the following Workflow Processing Nodes: Input Files, Select Spectra, Align Retention Times, Detect Unknown Compounds, Group Unknown Compounds, Fill Gaps and Mark Background Compounds for the so called “Aligned” result and solely Input Files, Select Spectra, and Detect Unknown Compounds for an “Unaligned” Result. Under Select Spectra, the retention time limits were set between 0.4 and 21 min, MS order as well as unrecognized MS order replacements were set to MS1. Under Align Retention Times the mass tolerance was set to 10 ppm and the maximum shift according to the dataset to 0.5 min. Under Detect Unknown Compounds, the mass tolerance was also set to 10 ppm, with an S/N threshold of 3, and a minimum peak intensity of 5E5 (DO) or 1E5 (FS, Cells). Further, [M+H]+1 and [M-H]-1 were selected as ions ([M+H]+1 and [M-H+TFA]-1 for cells) and a maximum peak width of 0.75 min as well as a minimum number of scans per peak equaling 5 were set. Lastly, for Group Unknown Compounds as well as Fill Gaps, mass tolerance was set to 10 ppm and retention time tolerance to 0.2 minutes. For best compound selection rules #1 and #2 were set to unspecified, while MS1 was selected for preferred MS order and [M+H]+1 as the preferred ion. For everything else, the default settings were used. Resulting peak tables were exported as excel files in three levels of Compounds, Compound per File and Features (just Features for the “Unaligned”) and later saved as csvs. In LipiDex’ Spectrum Searcher “LipiDex_HCD_Acetate”, “LipiDex_HCD_Plants”, “LipiDex_Splash_ISTD_Acetate”, “LipiDex_HCD_ULCFA”, and “Ganglioside_20171205” were selected as libraries for the DO while “LipidBlast2_Reformatted_CoonLab”, “LB_cleaned” and “Lipid_Spectral_Library_20170523” were selected for the FS. For the cells “LipiDex_HCD_Acetate”, “LipiDex_HCD_Plants”, “LipiDex_HCD_ULCFA”, “FAHFA”, and “Ganglioside_20200206” were selected. Extended Data Figure S7 details the lipid classes searched for in these databases with their respective adducts. We further kept the defaults of 0.01-Th for MS1 and MS2 search tolerances, a maximum of 1 returned search result, and an MS2 low mass cutoff of 61-Th. Under the Peak Finder tab, Compound Discoverer was chosen as peak table type, and its “Aligned” and “Unaligned” results, as well as the MS/MS results from Spectrum Researcher uploaded. Features had to be identified in a minimum of 1 file (4 files for the FS), however, the average lipid ID was based on a much higher average of 344 features found in plasma and 310 features in the liver dataset. We kept the defaults of a minimum of 75% of lipid spectral purity, an MS2 search dot product of at least 500 and reverse dot product of at least 700, as well as a multiplier of 2.0 (3.0 for FS) for FWHM window, a maximum 15 ppm mass difference, adduct/dimer and in-source fragment filtering, and a maximum RT M.A.D Factor of 3.5. As post-processing all features that were only found in 1 file and had no ID were deleted, and artifactual duplicates deleted.
For the FS liver dataset, peak areas were normalized to the 15:0–18:1(d7)-PC internal standard by dividing each peak area by the internal standards’ peak area of that sample and multiplying the result with the median of all internal standard peak areas. The quantification of the internal standard was obtained through TraceFinder 4.0 (Thermo Fisher Scientific). FS plasma results were normalized by dividing each peak area by the feature’s average batch control and multiplying with the median feature’s peak area over average batch controls. Reported is the log2 of all normalized values. Note that there is no data available for two CAST females as one animal died before sacrifice (CAST-4) and for another there was not enough plasma (CAST-3). For the cell experiments, peak areas were normalized to the sum of identified lipids, and log2-transformed.
QTL Mapping.
Note while the much smaller FS dataset was normalized to the internal PC standard; for the entire DO dataset, where many more LC-MS runs were collected, we used a batch correction approach (ComBat) to achieve normalization. In short, the ComBat method provided superior performance, especially in the case of the liver dataset. Specifically, the batch effects that occurred were easily and effectively corrected by application of the ComBat adjustment.
Prior to mapping analysis, the lipid metabolite data were adjusted for batch effects using the Combat algorithm76 as implemented in the R/sva software package.77 Batches correspond to sets of ~32 samples each that were run on the same day on the mass spectrometer. Effectiveness of batch correction was confirmed by visualization of the first few principal components. We note that batch adjustment substantially increased the yield of QTL, even though no genotype information is used in the correction process.
QTL mapping involves “scanning” the genome and testing for association between the 8-state haplotype probabilities and the batch corrected MS feature levels. The genome scans were performed for each lipid metabolite feature using the scan1() function in R/qtl2.23 This software fits a linear mixed with sex and DO breeding generation as additive covariates and random effect to account for the kinship structure of the DO mice and computes a log10 likelihood ratio statistic (LOD) to evaluate the significance of the genetic effect at each pseudo-marker locus. Sex-specific genetic associations were identified using a separate set of genome scans that included a sex x genotype interaction in the linear mixed model. We identified suggestive QTL at LOD > 6.0 and significant QTL at LOD > 7.4. These threshold values were estimated by permutation analysis to obtain a family-wise error correction for genome-wide QTL search.68 The family-wise error rate ensures that the maximum LOD score across the genome-wide search when applied to a trait with no QTL (i.e., a permuted trait) will exceed the threshold with a fixed probability. For the lenient threshold 6.0, the genome-wide probability of false QTL detection is 0.20. For the stringent threshold 7.4, the genome-wide error rate is controlled at 0.05. The lenient threshold is used to identify the almost-significant associations that co-localize on the genome in hotspots.
Data analysis and plotting.
Data analysis was largely performed using R79 in RStudio80. Data formatting was performed utilizing R/dplyr_0.8.381, R/tidyr_1.0.082 and R/reshape2_1.4.383 and visualizations were created using R/ggplot2_3.2.184, R/RColorBrewer_1.1–285, and for exploratory analysis, R/plotly_4.9.086. Heatmaps were generated using R/pheatmap_1.0.1287 and manhattan plots were generated based on code accessible via the R graph gallery.88 All boxplots were generated by ggplot2:geom_boxplot with the first and third quartiles (25th and 75th percentile) for lower and upper hinges, 1.5x interquartile range for the length of the whiskers, center line at median (50% quantile), and all raw data points, including outliers shown. Statistical t-test calculations were performed with MetaboAnalyst 4.0.89 A 95% Bayesian confidence interval (CI) for each QTL was calculated using the function find_peaks() in R/qtl2.27 Human Mouse homologues were obtained from the MGI homology database (available here: http://www.informatics.jax.org/downloads/reports/HOM_MouseHumanSequence.rpt).
Allele effects for each QTL were generated using the scan1blup() function of R/qtl2.27 SNP associations were performed using the scan1snps() function in R/qtl2_0.2027 accessing variants from the database cc_variants.sqlite (available here: https://ndownloader.figshare.com/files/18533342) and genes from mouse_genes_mgi.sqlite (available here: https://ndownloader.figshare.com/files/17609252) via R/RSQLite_2.1.2.90
To nominate candidate gene drivers at lipid-associated QTL, we integrated the lipid data collected in the present study, with hepatic gene expression data previously obtained from a separate cohort of DO mice.34 We reasoned a locus that demonstrated a hepatic cis-eQTL and a lipid-associated QTL with a similar allele effect patterns is likely to be driving the two phenotypes. We focused on cis-eQTL, as these are expression traits responding to local genetic variation. We computed the Pearson’s correlation between the allele effect patterns for all cis-eQTL at a locus to which one or more lipids co-mapped. We performed the same calculation for hepatic cis-pQTL identified in the previous study.34 For example, at the Chr 10 locus, we identified >25 QTL of unknown lipids in plasma and liver, all of which showed a strong NOD-driven allele effect pattern. About half of these QTL showed NOD as the high allele and half showed NOD as the low allele. We first computed the average allele effect pattern for the NOD-high lipids and the NOD-low lipids. We then identified 55 cis-eQTL and 16 cis-pQTL that were within ±2 Mbp of the lipid QTL at ~127 Mbp on Chr 10, and calculated the correlation between their allele effect patterns and the NOD-high and NOD-low lipid QTL. One gene showed a very strong correlation; B4galnt1. The overall correlation between the allele effects of the lipid QTL and the cis-eQTL or cis-pQTL was very low (e.g., 0), suggesting that the vast majority expression traits are responding to genetic variants different than the lipid traits. However, the correlation between the lipid traits and either the expression or protein level for B4galnt1 was >|0.97|. As B4galnt1 is a known gangliosidase, we then asked if the MS fragmentation pattern for the unknown lipids is consistent with gangliosides. It is worth noting that GM3 ganglioside standard (Cayman Chemicals, Ann Arbor, MI, Item No. 15587) contained N-acetyl-neuraminidate (NANA) - the only sialic acid made by humans. All gangliosides observed in the DO samples contain N-glycolyl-neuraminidate (NGNA), a major sialic acid in mice.91,92 This powerful approach enabled us to combine the lipid data from one DO study with the gene expression and proteomic data of another DO study to nominate one candidate gene.
Reporting summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability.
Genotypes and additional phenotype data associated with the DO mouse population have been deposited with Dryad (doi:10.5061/dryad.pj105; data files: Attie Islet eQTL data) (see Keller et al. 2018 for details).32 In addition, the data reported here are available for download and interactive web-based analysis at https://churchilllab.jax.org/qtlviewer/attie/islets. Genotyping used the Mouse Universal Genotyping Array (GigaMUGA; 143,259 markers).
Mass spectrometry data have been deposited in Chorus (http://chorusproject.org/) under ID 1610 (direct links to cell experiments https://chorusproject.org/anonymous/download/experiment/4984245205453479277, DO liver https://chorusproject.org/anonymous/download/experiment/a639bcc5602c441c9a1df94f4340d626, DO plasma https://chorusproject.org/anonymous/download/experiment/f8b273d222364f2a9d92cfdd0eb601b6, FS liver https://chorusproject.org/anonymous/download/experiment/c930cd419eb34dfebda7f53508c6969e, and FS plasma https://chorusproject.org/anonymous/download/experiment/9d4d025df0114687924d4075f3c927ca). Human Mouse homologues were obtained from the MGI homology database (available here: http://www.informatics.jax.org/downloads/reports/HOM_MouseHumanSequence.rpt). SNP associations were performed accessing variants from the database cc_variants.sqlite (available here: https://ndownloader.figshare.com/files/18533342) and genes from mouse_genes_mgi.sqlite (available here: https://ndownloader.figshare.com/files/17609252).Figures 1, 2, 3, 4, 5 and Extended Data Figures S1, S2, S3, S4, S5 and S6 have associated raw data.
Code Availability.
The data preparation and QTL mapping analysis are reproducibly documented in UNIX shell and R scripts posted on github (https://github.com/dmgatti/AttieMetabolomics). Code for data analysis and plotting is available at https://github.com/vanilink/DOLipids/ with input from Supplementary Tables 8 and 9. The genome-lipid associations are also accessible through an interactive web-based analysis tool that will allow users to replicate the analyses reported here (http://lipidgenie.com/). The source code for this resource can be found at https://github.com/coongroup/LipidGenie.
Extended Data
Extended Data Fig. 1. Identified lipids and unidentified features occupy characteristic regions in the m/z vs. RT space.
a, In plasma, we quantified 1,721 lipidomic features, 621 of which were identified, and b, In liver, we quantified 1,562 lipidomic features, 615 of which were identified. Abbreviations: m/z (mass-to-charge), RT (retention time).
Extended Data Fig. 2. Lipid profiling and subsequent QTL mapping reveals clusters of associated lipids.
a, Lipid class distribution of all 1,721 plasma and b, 1,562 liver lipidomic features. c, 1,405 plasma and d, 1,190 lipid features showed at least one QTL with an LOD > 6 as displayed in a Manhattan plot (n = 3,353 and 2,269 total QTL, respectively). Hierarchical clustering of these features against the 69,005 markers on the mouse genome, resulted in clustering of lipid class based on hotspots at the genetic level. Abbreviations: Chr (chromosome), DO (diversity outbred), QTL (quantitative trait loci), LOD (logarithm of odds).
Extended Data Fig. 3. Apoa2 as the candidate gene at the largest lipid hotspot.
a, 255 plasma (black) features mapping to the apoa2 locus on chromosome 1 share an allele effect pattern with upregulation in the 129 allele, while 2 mapping liver features (white) do not share the pattern (based on hierarchical clustering on allele effects, with a Euclidean distance cutoff of h = 1.5). b, The allele effect is exemplary replicated in an independent experiment of founder strain plasma CE(18:2) levels (n = 4 for each sex and strain, boxplots are defined with the first and third quartiles (25th and 75th percentile) for lower and upper hinges, 1.5x interquartile range for the length of the whiskers, center line at median (50% quantile)). c, The same pattern was not visible in previously reported34 Apoa2 liver protein and RNA allele effects. Abbreviations: CE (cholesteryl ester), FS (founder strain).
Extended Data Fig. 4. B4galnt1 as the candidate gene at the hotspot with the largest LOD.
a, The selection of B4galnt1 as the candidate gene for the chromosome 10:127 Mbp locus was corroborated by NOD-specific allele effects in previously reported liver eQTL and b, pQTL.34 c, The allele effect patterns of the later as gangliosides identified features mapping to the B4galnt1 locus could further be validated in an independent experiment of founder strain mice (exemplar GM3 pattern, n = 4 for each sex and strain, boxplots are defined with the first and third quartiles (25th and 75th percentile) for lower and upper hinges, 1.5x interquartile range for the length of the whiskers, center line at median (50% quantile)). Abbreviations: FS (founder strain), Mbp (megabase pair).
Extended Data Fig. 5. Allele effects characterize genome-lipid hotspots.
a, Hierarchical clustering of allele effects at Chr 6:91 Mbp resulted in 21 features with matching A/J down effect (main cluster featuring the six B6 male specific features (red) after row-scaling and Ward clustering, cutoff at h=5). b, Consistently, the pattern of male >> female was observed for each of the FS except for A/J as visible in the example for m/z 1130 (n = 4 for each sex and strain, boxplots are defined with the first and third quartiles (25th and 75th percentile) for lower and upper hinges, 1.5x interquartile range for the length of the whiskers, center line at median (50% quantile).) c, Hierarchical clustering of allele effects at Chr 5:31 Mbp locus resulted in 10 features with matching B6 and NZO up effect (main cluster featuring LysoPC 14:0 (turquoise) after row-scaling and Ward clustering, cutoff at h=8). d, This pattern could be replicated in the FS (n = 4 for each sex and strain, boxplots are defined with the first and third quartiles (25th and 75th percentile) for lower and upper hinges, 1.5x interquartile range for the length of the whiskers, center line at median (50% quantile)), as shown for LysoPC 14:0, as well as e, in opposite directionality in a liver eQTL.34 f, Hierarchical clustering of allele effects at Chr 7:79 Mbp locus resulted in 8 features with matching WSB down effect (main cluster featuring PUFA-containing phospholipids (turquoise) after row-scaling and Ward clustering, cutoff at h=2.5). g, The mapping phospholipids contained polyunsaturated fatty acids such as 20:4 and 22:6. h-i, Abhd2 liver RNA and protein allele effects34 matched with an opposite WSB high effect. Abbreviations: DO (diversity outbred), FS (founder strain), Chr (chromosome), Mbp (megabase pair), PC (phosphatidylcholine), PUFA (polyunsaturated fatty acid).
Extended Data Fig. 6. Overexpressing ABHD1 and ABHD3 results in distinct phospholipid signature.
a, Experimental design of the validation experiment featuring three technical and four biological replicates of Hepa1-6 cells either untransfected (CTL), transfected with a His-tag GFP control (GFP), or transfected with MYC-tagged ABHD1 or ABHD3. b, Western blot of Hepa1-6 overexpression of ABHD1 and ABHD3. Shown is an overlay of membrane and ECL blot for MYC-tag. c, Heatmap of top 49 features from discovery lipidomics experiment with p < 0.05 (ANOVA, Fisher’s LSD post-hoc). Features were sum-normalized and log2-transformed. Hierarchical clustering (Ward clustering, Euclidean distance) shows two clusters with opposite fold changes distinguishing between ABHD1 and ABHD3 and the GFP control.
Extended Data Fig. 7. Lipid class abbreviations and identifications with respective adduct types.
As searched for in LipiDex databases (see Methods).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants P41 GM108538 and 2R01DK101573. This research was performed using the compute resources and assistance of the UW-Madison Center For High Throughput Computing (CHTC) in the Department of Computer Sciences. The CHTC is supported by UW-Madison, the Advanced Computing Initiative, the Wisconsin Alumni Research Foundation, the Wisconsin Institutes for Discovery, and the National Science Foundation, and is an active member of the Open Science Grid, which is supported by the National Science Foundation and the U.S. Department of Energy’s Office of Science. We thank B. Paulson for help with sample preparation and D. Hwang for help with figures. We thank J. Simcox (Department of Biochemistry, University of Wisconsin-Madison, Madison, Wisconsin, USA) for providing the His-tagged CMV6-GFP plasmid and Hepa1-6 cells.
Footnotes
Competing Interest Statement
The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Han X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol 12, 668–679 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Yang L. et al. Recent advances in lipidomics for disease research. J. Sep. Sci 39, 38–50 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Kind T. et al. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 10, 755–758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross RW & Han X. Lipidomics at the Interface of Structure and Function in Systems Biology. Chemistry & Biology vol. 18 284–291 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cajka T & Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt. Chem 61, 192–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabassum R. et al. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat. Commun 10, 4329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiyonami R, Peake DA, Liu X & Huang Y Large-Scale Lipid Profiling of a Human Serum Lipidome Using a High-Resolution, Accurate-Mass LC/MS/MS Approach. in LIPID MAPS Annual Meeting 12–13 (pdfs.semanticscholar.org, 2015). [Google Scholar]
- 8.Slatter DA et al. Mapping the Human Platelet Lipidome Reveals Cytosolic Phospholipase A2 as a Regulator of Mitochondrial Bioenergetics during Activation. Cell Metab. 23, 930–944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Contrepois K. et al. Cross-Platform Comparison of Untargeted and Targeted Lipidomics Approaches on Aging Mouse Plasma. Sci. Rep 8, 17747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaženović I. et al. Increasing Compound Identification Rates in Untargeted Lipidomics Research with Liquid Chromatography Drift Time-Ion Mobility Mass Spectrometry. Anal. Chem 90, 10758–10764 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Mahieu NG & Patti GJ Systems-Level Annotation of a Metabolomics Data Set Reduces 25 000 Features to Fewer than 1000 Unique Metabolites. Analytical Chemistry vol. 89 10397–10406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaženović I, Kind T, Ji J & Fiehn O. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross RW The evolution of lipidomics through space and time. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids vol. 1862 731–739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koelmel JP et al. LipidMatch: an automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics 18, 331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartler J. et al. Deciphering lipid structures based on platform-independent decision rules. Nature Methods vol. 14 1171–1174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchins PD, Russell JD & Coon JJ LipiDex: An Integrated Software Package for High-Confidence Lipid Identification. Cell Syst 6, 621–625.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchins PD, Russell JD & Coon JJ Mapping Lipid Fragmentation for Tailored Mass Spectral Libraries. J. Am. Soc. Mass Spectrom 30, 659–668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kostyukevich Y. et al. Hydrogen/Deuterium Exchange Aiding Compound Identification for LC-MS and MALDI Imaging Lipidomics. Analytical Chemistry (2019) doi: 10.1021/acs.analchem.9b02461. [DOI] [PubMed] [Google Scholar]
- 19.Stefely JA et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nature Biotechnology vol. 34 1191–1197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumas M-E et al. Topological analysis of metabolic networks integrating co-segregating transcriptomes and metabolomes in type 2 diabetic rat congenic series. Genome Med. 8, 101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cazier J-B et al. Untargeted metabolome quantitative trait locus mapping associates variation in urine glycerate to mutant glycerate kinase. J. Proteome Res 11, 631–642 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Krumsiek J. et al. Mining the Unknown: A Systems Approach to Metabolite Identification Combining Genetic and Metabolic Information. PLoS Genet. 8, e1003005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin S-Y et al. An atlas of genetic influences on human blood metabolites. Nat. Genet 46, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rueedi R. et al. Metabomatching: Using genetic association to identify metabolites in proton NMR spectroscopy. PLOS Computational Biology vol. 13 e1005839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raffler J. et al. Identification and MS-assisted interpretation of genetically influenced NMR signals in human plasma. Genome Med. 5, 13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatti DM et al. Quantitative trait locus mapping methods for diversity outbred mice. G3: Genes, Genomes, Genetics 4, 1623–1633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broman KW et al. R/qtl2: Software for Mapping Quantitative Trait Loci with High-Dimensional Data and Multiparent Populations. Genetics 211, 495–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svenson KL et al. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190, 437–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chesler EJ et al. Diversity Outbred Mice at 21: Maintaining Allelic Variation in the Face of Selection. G3 6, 3893–3902 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer R. et al. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 6, 44 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aylor DL et al. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 21, 1213–1222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller MP et al. Genetic Drivers of Pancreatic Islet Function. Genetics 209, 335–356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller MP et al. Gene loci associated with insulin secretion in islets from nondiabetic mice. Journal of Clinical Investigation vol. 129 4419–4432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chick JM et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature 534, 500–505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kemis JH et al. Genetic determinants of gut microbiota composition and bile acid profiles in mice. doi: 10.1101/571075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallego SF, Højlund K & Ejsing CS Easy, Fast, and Reproducible Quantification of Cholesterol and Other Lipids in Human Plasma by Combined High Resolution MSX and FTMS Analysis. Journal of The American Society for Mass Spectrometry vol. 29 34–41 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Ogiso H, Suzuki T & Taguchi R. Development of a reverse-phase liquid chromatography electrospray ionization mass spectrometry method for lipidomics, improving detection of phosphatidic acid and phosphatidylserine. Analytical Biochemistry vol. 375 124–131 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Fahy E. et al. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res 50, S9–S14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liebisch G. et al. Shorthand notation for lipid structures derived from mass spectrometry. Journal of Lipid Research vol. 54 1523–1530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A & Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res 49, 1137–1146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su Z. et al. Genetic basis of HDL variation in 129/SvImJ and C57BL/6J mice: importance of testing candidate genes in targeted mutant mice. J. Lipid Res 50, 116–125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kettunen J. et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun 7, 11122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W. et al. Genome-wide association mapping of quantitative traits in outbred mice. G3 2, 167–174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pamir N. et al. Genetic control of the HDL proteome. Systems Biology (2018). [Google Scholar]
- 45.Wang X, Korstanje R, Higgins D & Paigen B. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Res. 14, 1767–1772 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blanco-Vaca F, Escolà-Gil JC, Martín-Campos JM & Julve J. Role of apoA-II in lipid metabolism and atherosclerosis: advances in the study of an enigmatic protein. J. Lipid Res 42, 1727–1739 (2001). [PubMed] [Google Scholar]
- 47.Kontush A, Lhomme M & Chapman MJ Unraveling the complexities of the HDL lipidome. J. Lipid Res 54, 2950–2963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy RC, Leiker TJ & Barkley RM Glycerolipid and Cholesterol Ester Analyses in Biological Samples by Mass Spectrometry. Biochim. Biophys. Acta 1811, 776 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lerno LA Jr, German JB & Lebrilla CB Method for the identification of lipid classes based on referenced Kendrick mass analyis. Anal. Chem 82, 4236–4245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eilbeck K. et al. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 6, R44 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagata Y. et al. Expression cloning of beta 1,4 N-acetylgalactosaminyltransferase cDNAs that determine the expression of GM2 and GD2 gangliosides. J. Biol. Chem 267, 12082–12089 (1992). [PubMed] [Google Scholar]
- 52.Dotta F. et al. Pancreatic islet ganglioside expression in nonobese diabetic mice: comparison with C57BL/10 mice and changes after autoimmune beta-cell destruction. Endocrinology 130, 37–42 (1992). [DOI] [PubMed] [Google Scholar]
- 53.Li Z. et al. Impact of sphingomyelin synthase 1 deficiency on sphingolipid metabolism and atherosclerosis in mice. Arterioscler. Thromb. Vasc. Biol 32, 1577–1584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergfeld AK et al. -glycolyl groups of nonhuman chondroitin sulfates survive in ancient fossils. Proc. Natl. Acad. Sci. U. S. A 114, E8155–E8164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strømme P. et al. X-linked Angelman-like syndrome caused by Slc9a6 knockout in mice exhibits evidence of endosomal–lysosomal dysfunction. Brain 134, 3369–3383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spessott W, Uliana A & Maccioni HJF Defective GM3 Synthesis in Cog2 Null Mutant CHO Cells Associates to Mislocalization of Lactosylceramide Sialyltransferase in the Golgi Complex. Neurochem. Res 35, 2161–2167 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Ledeen RW & Wu G. The multi-tasked life of GM1 ganglioside, a true factotum of nature. Trends in Biochemical Sciences vol. 40 407–418 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Yore MM et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159, 318–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McLean S, Davies NW, Nichols DS & Mcleod BJ Triacylglycerol estolides, a new class of mammalian lipids, in the paracloacal gland of the brushtail possum (Trichosurus vulpecula). Lipids 50, 591–604 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Parker BL et al. An integrative systems genetic analysis of mammalian lipid metabolism. Nature (2019) doi: 10.1038/s41586-019-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lord CC, Thomas G & Brown JM Mammalian alpha beta hydrolase domain (ABHD) proteins: Lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim. Biophys. Acta 1831, 792–802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Long JZ et al. Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nat. Chem. Biol 7, 763–765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Draisma HHM et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat. Commun 6, 7208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ha CY et al. The association of specific metabolites of lipid metabolism with markers of oxidative stress, inflammation and arterial stiffness in men with newly diagnosed type 2 diabetes. Clin. Endocrinol 76, 674–682 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Demirkan A. et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 8, e1002490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller MR et al. Unconventional endocannabinoid signaling governs sperm activation via the sex hormone progesterone. Science 352, 555–559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baggelaar MP, Maccarrone M & van der Stelt M. 2-Arachidonoylglycerol: A signaling lipid with manifold actions in the brain. Progress in Lipid Research vol. 71 1–17 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Jha P. et al. Systems Analyses Reveal Physiological Roles and Genetic Regulators of Liver Lipid Species. Cell Syst 6, 722–733.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jha P. et al. Genetic Regulation of Plasma Lipid Species and Their Association with Metabolic Phenotypes. Cell Syst 6, 709–721.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stacey D. et al. ProGeM: a framework for the prioritization of candidate causal genes at molecular quantitative trait loci. Nucleic Acids Res. 47, e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kastenmüller G, Raffler J, Gieger C & Suhre K. Genetics of human metabolism: an update. Hum. Mol. Genet 24, R93–R101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitok KA et al. Islet proteomics reveals genetic variation in dopamine production resulting in altered insulin secretion. Journal of Biological Chemistry vol. 293 5860–5877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Broman KW, Gatti DM, Svenson KL, Sen Ś & Churchill GA Cleaning Genotype Data from Diversity Outbred Mice. G3 9, 1571–1579 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi K. ‘kb’ & Choi, K. B. churchill-lab/gbrs: v01.5. (2017). doi: 10.5281/zenodo.291787. [DOI] [Google Scholar]
- 75.Adusumilli R & Mallick P. Data Conversion with ProteoWizard msConvert. Methods Mol. Biol 1550, 339–368 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Johnson WE, Evan Johnson W, Li C & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics vol. 8 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Leek JT, Johnson WE, Parker HS, Jaffe AE & Storey JD The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Churchill GA & Doerge RW Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Core Team. R: A Language and Environment for Statistical Computing. (2019). [Google Scholar]
- 80.Team RStudio. RStudio: Integrated Development Environment for R. (2016). [Google Scholar]
- 81.Wickham H, François R, Henry L & Müller K dplyr: A Grammar of Data Manipulation. (2019). [Google Scholar]
- 82.Wickham H & Henry L tidyr: Tidy Messy Data. (2019). [Google Scholar]
- 83.Wickham H & Others. Reshaping data with the reshape package. J. Stat. Softw. 21, 1–20 (2007). [Google Scholar]
- 84.Wickham H ggplot2: Elegant Graphics for Data Analysis. (Springer, 2016). [Google Scholar]
- 85.Neuwirth E RColorBrewer: ColorBrewer Palettes. (2014). [Google Scholar]
- 86.Sievert C plotly for R. (2018). [Google Scholar]
- 87.Kolde R pheatmap: Pretty Heatmaps. (2019). [Google Scholar]
- 88.Holtz Y Manhattan plot in R: a review. R graph gallery https://www.r-graph-gallery.com/101_Manhattan_plot.html. [Google Scholar]
- 89.Chong J. et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Research vol. 46 W486–W494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Müller K, Wickham H, James DA & Falcon S RSQLite: ‘SQLite’ Interface for R. (2019). [Google Scholar]
- 91.Kavaler S. et al. Pancreatic beta-cell failure in obese mice with human-like CMP-Neu5Ac hydroxylase deficiency. FASEB J. 25, 1887–1893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Salama A. et al. Neu5Gc and α1–3 GAL Xenoantigen Knockout Does Not Affect Glycemia Homeostasis and Insulin Secretion in Pigs. Diabetes 66, 987–993 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Bowden JA, Ulmer CZ, Jones CM, Koelmel JP & Yost RA NIST lipidomics workflow questionnaire: an assessment of community-wide methodologies and perspectives. Metabolomics 14, 53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotypes and additional phenotype data associated with the DO mouse population have been deposited with Dryad (doi:10.5061/dryad.pj105; data files: Attie Islet eQTL data) (see Keller et al. 2018 for details).32 In addition, the data reported here are available for download and interactive web-based analysis at https://churchilllab.jax.org/qtlviewer/attie/islets. Genotyping used the Mouse Universal Genotyping Array (GigaMUGA; 143,259 markers).
Mass spectrometry data have been deposited in Chorus (http://chorusproject.org/) under ID 1610 (direct links to cell experiments https://chorusproject.org/anonymous/download/experiment/4984245205453479277, DO liver https://chorusproject.org/anonymous/download/experiment/a639bcc5602c441c9a1df94f4340d626, DO plasma https://chorusproject.org/anonymous/download/experiment/f8b273d222364f2a9d92cfdd0eb601b6, FS liver https://chorusproject.org/anonymous/download/experiment/c930cd419eb34dfebda7f53508c6969e, and FS plasma https://chorusproject.org/anonymous/download/experiment/9d4d025df0114687924d4075f3c927ca). Human Mouse homologues were obtained from the MGI homology database (available here: http://www.informatics.jax.org/downloads/reports/HOM_MouseHumanSequence.rpt). SNP associations were performed accessing variants from the database cc_variants.sqlite (available here: https://ndownloader.figshare.com/files/18533342) and genes from mouse_genes_mgi.sqlite (available here: https://ndownloader.figshare.com/files/17609252).Figures 1, 2, 3, 4, 5 and Extended Data Figures S1, S2, S3, S4, S5 and S6 have associated raw data.