Abstract
Background
CDK12 genomic alterations occur in several tumor types, but little is known about their oncogenic role and clinical significance.
Objective
To describe the landscape of CDK12 alterations across solid cancers and the clinical features of CDK12-altered prostate cancer.
Design, setting, and participants
A single-center retrospective study of 26743 patients across 25 solid tumor types who underwent tumor sequencing was performed. Clinicopathologic features and outcomes were assessed in prostate cancer.
Outcome measurements and statistical analysis
CDK12 alterations and their association with genomic characteristics are described. For prostate cancer patients, overall survival and time to castration resistance were assessed using univariable and multivariable Cox regression analysis.
Results and limitations
CDK12 alterations were identified in 404/26743 patients (1.5%) overall, but were most frequent in prostate (100/1875, 5.3%) and ovarian cancer (43/1034, 4.2%), in which they were associated with a high prevalence of truncating variants and biallelic inactivation. CDK12 alterations defined a genomic subtype of prostate cancer with a unique copy-number alteration profile and involvement of distinct oncogenic pathway alterations, including cell-cycle pathway genes. CDK12-altered prostate cancer was associated with somewhat more aggressive clinical features and shorter overall survival (median 64.4 vs 74.9 mo; p = 0.032) independent of standard clinical factors and tumor copy-number alteration burden (adjusted hazard ratio 1.80, 95% confidence interval 1.12–2.89; p = 0.024). The study is limited by its retrospective nature.
Conclusions
CDK12 alteration is a rare event across solid cancers but defines a clinically distinct molecular subtype of prostate cancer associated with unique genomic alterations and slightly more aggressive clinical features.
Patient summary
CDK12 gene alterations occur rarely across tumor types, but more frequently in prostate cancer, where they are associated with genomic instability, cell-cycle pathway gene alterations, and somewhat worse clinical outcomes, warranting further investigation of therapeutic targeting of this disease subset.
Keywords: CDK12, Clinical sequencing, Genomics, Prostate cancer, Tumor biology
1. Introduction
Genomic alterations in CDK12 have been observed in various cancer types [1–4] but their oncogenic function and clinical relevance are poorly understood. CDK12 encodes a cyclin-dependent serine/threonine kinase involved in the regulation of the cell cycle and of DNA repair by homologous recombination (HR). Specifically, it has been shown that CDK12 loss of function suppresses the expression of several HR genes, at least in part, via intronic polyadenylation [5–8]. Growing evidence suggests that biallelic CDK12 loss determines a distinct phenotype of ovarian and prostate cancer characterized by high genomic instability and tandem duplications [9–11].
It has been reported that CDK12 alterations occur in 4–11 % of prostate cancer cases and are more frequent in the metastatic castration-resistant setting (mCRPC) [3,9,12]. Despite preclinical work suggesting that CDK12 loss or inhibition could impair genes in the HR repair pathway, recent evidence has shown that PARP inhibitors have limited efficacy in mCRPC patients harboring a CDK12 alteration [13,14]. However, Wu and colleagues [9] recently demonstrated that CDK12 loss determines a novel molecular subtype of prostate cancer associated with a higher proportion of fusion neoantigens and potentially high immune infiltration and response to anti–PD1/PD-L1 agents.
In this study we sought to survey the landscape of CDK12 mutations across different cancer types. We identified prostate cancer as the cancer type with the highest prevalence of CDK12 oncogenic alterations and the largest proportion of truncating variants, with biallelic inactivation occurring frequently, and characterized the genomic architecture of CDK12-altered prostate carcinomas. We describe clinicopathologic characteristics and clinical outcomes for patients with prostate cancers harboring CDK12 alterations. Our work adds to recent publications [15,16] by integrating pan-cancer genomic analysis from a clinical sequencing assay, identifying distinct genomic alterations associated with CDK12-altered prostate cancer, and describing clinical features of the largest set of prostate cancer patients with CDK12 alterations identified using a single assay.
2. Patients and methods
2.1. Study design and patients
All patients in the study underwent tumor sequencing performed using the Memorial Sloan Kettering Integrated Molecular Profiling of Actionable Cancer Targets (MSK-IMPACT) clinical sequencing assay, a hybridization capture-based, next-generation sequencing platform, with 341, 410, or 468 genes (all panels included CDK12) [17,18]. After excluding 1965 cases with high microsatellite instability (MSI) cases, defined as an MSI sensor [19] score >10, and/or high tumor mutation burden (TMB) cases, defined as TMB ≥ 20 mutations/Mb, because of the high likelihood of passenger alterations in CDK12, a total of 26743 patients across 25 solid tumor types (minimum number of patients 100 per cancer type) who had MSK-IMPACT sequencing between July 2014 and April 12, 2019 were assessed for the presence of CDK12 alterations. For patients with multiple sequenced samples, one sample was selected according to the following hierarchy: tumor purity (highest tumor purity as assessed by pathologist), gene panels (most recent), and coverage (highest sample sequencing coverage). Chart review was performed to extract clinical and pathologic data for prostate cancer patients. Data collection and retrospective analysis were performed with approval from the MSKCC institutional review board.
2.2. Genomic analysis
TMB was calculated as the total number of nonsynonymous mutations excluding CDK12, divided by the number of bases sequenced. Recurrent oncogenic alterations were defined as oncogenic according to OncoKB [20] (version August 28, 2019) and present in at least 1% in the whole cohort. Canonical oncogenic pathway-level alterations were computed using curated pathway templates [21]. MutSigCV v.1.4 was used to determine significantly mutated genes (false discovery rate [FDR] <0.1) [22]. Segmented copy-number data were processed using the CNtools package v1.4. The fraction of genome altered (FGA) was calculated for each sample as the percentage of the genome with log2 copy ratios >0.2 or <−0.2. Thresholds for copy-number alteration gain and loss were set at log2 copy ratios of >0.2 and <−0.2, respectively. CDK12 biallelic (CDK12-Bi) inactivation was defined as either deep deletion, two or more deleterious mutations in the same tumor sample, one mutation with concurrent heterozygous loss of the wild-type (WT) allele, or one mutation with concurrent copy-neutral loss-of-heterozygosity (CN-LOH) as computed using the FACETS algorithm [23]. FACETS is an allele-specific copy-number analysis pipeline providing accurate, purity- and ploidy-corrected, integer DNA copy-number calls and cancer cell fraction (CCF) from sequencing data. Integer copy-number calls were used to determine the presence of CDK12-Bi inactivation due to heterozygous loss of the WT allele or CN-LOH. FACETS was also used to determine clonality. Mutations were defined as clonal if the upper bound of the 95% confidence interval (CI) for CCF was ≥0.8, and all other mutations were called subclonal. Of note, samples with CDK12 rearrangement events were excluded in the biallelic analysis.
2.3. Prostate cancer outcomes
Baseline clinical characteristics and outcomes were available for a subset of patients with histologically confirmed prostate cancer who underwent tumor genomic profiling. Outcomes assessed were overall survival (OS), time to castration resistance, and time on treatment with first-line abiraterone acetate or enzalutamide for castration-resistant disease (Supplementary Fig. 1). For OS, follow-up started at the time of first metastasis and ended with patient death, with censorship occurring at last patient contact. For time to castration-resistance, follow-up started at the time of start of continuous androgen deprivation therapy (ADT) and ended at the development of castration-resistant disease as documented in the medical record, with censorship occurring at last patient assessment. For time-on-treatment analysis, follow-up started at the time of starting the agent as first-line therapy for CRPC and ended with discontinuation of the agent, with censorship occurring at last patient assessment. Median follow-up time was calculated for patients without the event only.
2.4. Statistical analysis
The Kruskal-Wallis test was used to evaluate differences in TMB, FGA, number of breakpoints, median segment size, tumor purity, and sequencing coverage among the three groups defined by CDK12 allelic status (CDK12-WT vs CDK12 monoallelic [CDK12-Mono] vs CDK12-Bi). We used a pairwise post-hoc Mann-Whitney test corrected for multiple comparisons using the FDR method. Fisher’s exact test (adjusted for multiple comparisons) was used to evaluate differences in the prevalence of copy-number gain and copy-number loss, and the prevalence of oncogenic pathway alterations based on CDK12 allelic status. We used a binomial test to determine whether the ratio of missense mutations present in the kinase domain of CDK12 differed significantly from the proportion of nucleic acids present in the kinase domain of CDK12. We used the Wilcoxon signed-rank test to determine whether TMB, FGA, and the number of breakpoints differed significantly between paired samples collected at two different time points. Time-to-event outcomes were analyzed using the Kaplan-Meier method and compared via the log-rank test. The association between CDK12 alteration and survival was evaluated using univariable and multivariable Cox proportional-hazards regression models. Multivariable analysis was adjusted for standard clinical prognostic factors: age at diagnosis (continuous), Gleason score at diagnosis (<8 vs ≥8), prostate-specific antigen (PSA) at diagnosis (continuous), visceral metastasis at diagnosis (presence vs absence), de novo metastatic status at diagnosis (presence vs absence), and FGA. Multivariable tests were performed using analysis of variance to compare the models with and without the extra term. The p values reported are two-tailed. When applicable, multiple testing correction was performed using the FDR method and FDR < 0.05 was considered significant. All analyses were performed using R v3.5.2 (www.R-project.org) and Bioconductor v3.4.
3. Results
3.1. Landscape of CDK12 alterations across cancer types
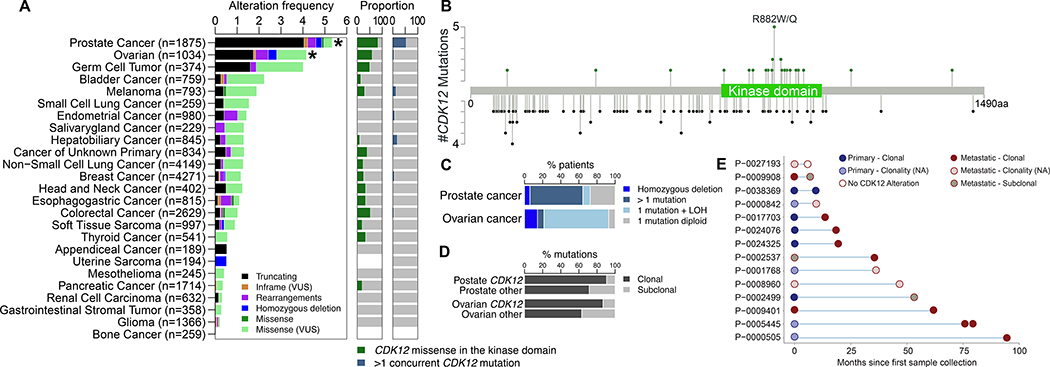
We assessed the prevalence of CDK12 alterations among 26743 patients across 25 solid cancer types profiled using the MSK-IMPACT clinical sequencing test. A total of 404 patients (1.5%) had tumors with at least one CDK12 alteration. Prostate (n = 100/1875, 5.3%) and ovarian (n = 43/1034, 4.2%) cancers were the types with the highest frequencies of CDK12 oncogenic alterations (Fig. 1A), including somatic point mutations, rearrangements, and deep deletions. Using MutSigCV [22], we found that CDK12 was significantly recurrently mutated only in prostate and ovarian cancers compared with background mutation rates (FDR < 0.001 and FDR = 0.056, respectively; Fig. 1A). The majority of CDK12 alterations in prostate cancer were truncating mutations, followed by missense mutations, rearrangements, and deep deletions (Supplementary Fig. 2A,B). Most truncating mutations were located within or upstream of the kinase domain (101/105, 96%), whereas missense mutations were more frequently clustered within the kinase domain (25/30, 83%), which was higher than expected by chance (p < 0.001; Fig. 1B), suggesting that these mutations probably have a deleterious effect on CDK12 function. Moreover, in prostate cancer the majority of patients with missense mutations in the kinase domain of CDK12 (18/24, 75%) had a concurrent truncating mutation suggestive of CDK12-Bi inactivation, which was not observed in other cancer types. In total, we identified 61/84 (73%) and 26/28 patients (93%) with putative CDK12-Bi inactivation in prostate and ovarian cancer, respectively (Fig. 1C). The principal genomic event responsible for CDK12-Bi inactivation in prostate cancer was multiple concurrent mutations of CDK12 (49/61, 75%), whereas in ovarian cancer it was LOH (20/26, 77%; Fig. 1C). We observed a higher prevalence of clonal CDK12 mutations than non-CDK12 mutations in prostate and ovarian cancer (105/116 [91%] vs 140/196 [71%]; p < 0.001; and 26/30 [87%] vs 82/130 [63%]; p = 0.016, respectively; Fig. 1D, Supplementary Fig. 2C).
Fig. 1 –
Landscape of somatic CDK12 mutations across 25 cancer types in the MSK-IMPACT cohort. (A) Somatic CDK12 alteration prevalence (left panel), proportion of kinase domain mutations among all missense mutations (middle panel), and proportion of cases with >1 CDK12 mutation in an individual tumor (right panel) across 26743 tumors from 25 different cancer types. CDK12 is significantly mutated in prostate and ovarian cancer only. * Significantly mutated according to MutSigCV. (B) Distribution of missense (upper green lollipop) and truncating mutations (lower black lollipop) in CDK12 in prostate cancer. (C) Prevalence and type of biallelic inactivation of CDK12 in prostate and ovarian cancer. (D) Prevalence of CDK12 clonal mutations compared to all other mutations in prostate and ovarian cancer. (E) CDK12 alterations in matched tumors in the prostate CDK12 alteration cohort, including localized primaries and later metastases and other matched tumors from the same patients. LOH = loss of heterozygosity; NA = not evaluable.
Of 14 prostate cancer patients with more than one tumor sequenced longitudinally, 13 showed shared CDK12 alterations among all matched tumors, with one patient having samples collected up to 7.8 yr apart (Fig. 1E). Of note, the only patient who did not have shared CDK12 alterations in their matched tumors had a CDK12 rearrangement known to be prone to false-negative results in hybrid capture DNA sequencing. FGA and the number of breakpoints increased over time, whereas TMB did not change significantly between paired samples (Supplementary Fig. 2D). The frequency of CDK12-Bi inactivation did not differ between primary (34/48, 70%) and metastatic tumors (27/36, 75%) in which zygosity could be determined. These analyses suggest that CDK12 alteration is an early event in the genomic makeup of CDK12-altered metastatic prostate cancer.
3.2. Genomic architecture of CDK12-altered prostate cancer
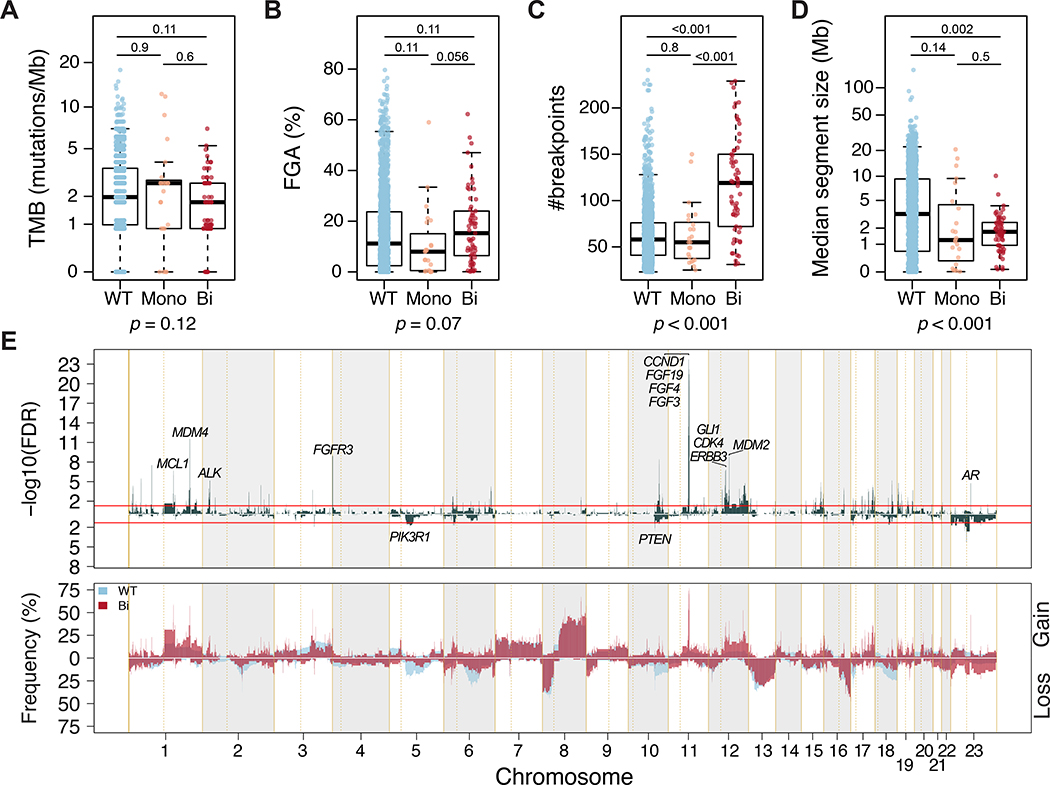
Of 84/100 CDK12-altered prostate cancers for which zygosity could be determined, 23 had CDK12 monoallelic inactivation (CDK12-Mono) and 61 had biallelic inactivation (CDK12-Bi). There was no difference in tumor purity or sequencing coverage by CDK12 alteration status (Supplementary Fig. 3A,B). TMB and overall FGA were similar between CDK12 -WT, CDK12-Mono, and CDK12-Bi (Fig. 2A, B), but we observed a higher fraction of genome gain in CDK12-Bi prostate cancer (Supplementary Fig. 3C). Of note, CDK12-Bi prostate cancer was associated with a higher number of breakpoints and smaller median size of copy-number–altered segments (Fig. 2C,D and Supplementary Fig. 3E,F). We compared copy-number alteration patterns and found that 13% of the evaluated genome was differentially affected between CDK12-WT and CDK12-Bi cases (Fig. 2E and Supplementary Table 1). This highly fragmented genome with a pattern of narrow spikes in copy-number–altered segments distributed along the genome (Fig. 2E) is suggestive of a tandem duplicator phenotype [9]. Chromosome 11q13.3, a locus that includes CCND1, FGF19, FGF4, and FGF3, was more frequently affected by copy-number gain in CDK12-Bi than in CDK12-WT tumors. Overall, these results suggest that CDK12-Bi inactivation may shape the genomic architecture of prostate cancer.
Fig. 2 –
Genomic architecture of CDK12-altered prostate cancer. (A–D) Comparison of (A) tumor mutational burden (TMB), (B) fraction of genome altered (FGA), (C) number of breakpoints, and (D) median size of copy number alterations in prostate cancers with wild-type CDK12 (WT, blue), CDK12 with monoallelic inactivation (Mono, orange), or CDK12 with biallelic inactivation (-Bi). The p values are derived from the Kruskal-Wallis H test. The q value (adjusted for multiple comparison) derived from the pairwise Mann-Whitney test is shown above each comparison. (E) Prevalence plot (bottom) and corresponding -log10(FDR) value derived from Fisher’s exact test (top) comparing copy-number gains and losses between CDK12-WT (blue) and CDK12-Bi (red) prostate cancer. Selected oncogenic genes within affected chromosomal regions are shown. Red lines indicate FDR = 0.05. FDR = false discovery rate.
3.3. Oncogenic alterations associated with CDK12-altered prostate cancer
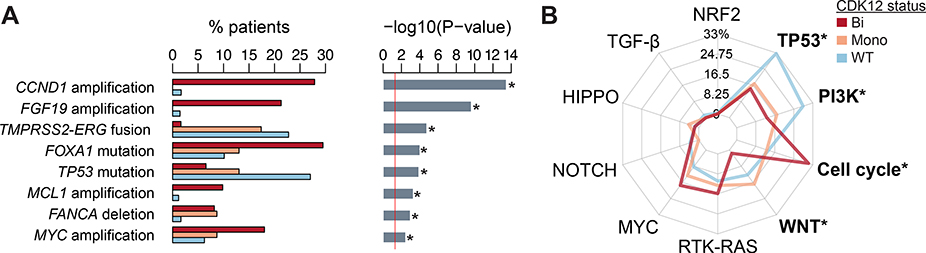
We investigated differences in the prevalence of 53 recurrent oncogenic alterations (Methods 2.2and Supplementary Table 2) by CDK12 allelic status. We found eight recurrent oncogenic alterations that differed in prevalence by CDK12 status (FDR < 0.05; Fig. 3A and Supplementary Table 2): CCND1, MCL1, and MYC were more frequently amplified, FANCA was more frequently lost, and FOXA1 was more frequently mutated in the CDK12-Bi group. TP53 mutations and TMPRSS2-ERG fusions were less frequently observed in the CDK12-Bi group. Next, we interrogated the alteration prevalence of ten canonical oncogenic signaling pathways: cell cycle, Hippo, Myc, Notch, Nrf2, PI-3-Kinase/Akt, RTK-RAS, TGFβ signaling, p53, and β-catenin/Wnt [21]. The cell cycle pathway was more frequently altered in CDK12-Bi than in CDK12-WT tumors, whereas alterations in the p53, PI-3–Kinase/Akt, and β-catenin/Wnt pathways were less frequently observed in CDK12-Bi than in CDK12-WT tumors (Fig. 3B).
Fig. 3 –
Oncogenic alterations associated with CDK12-altered prostate cancer. (A) Prevalence of recurrent oncogenic alteration in prostate cancer with CDK12 biallelic (Bi, red) compared with CDK12 monoallelic (Mono, orange) inactivation or CDK12 wild-type tumors (WT, blue). The p values are derived from Fisher’s exact test (* FDR < 0.05). (B) Radar plots showing the percentage of patients with alterations in the corresponding canonical oncogenic signaling pathways according to CDK12 alterations. Statistically significant pathways associated with CDK12 alteration are indicated with an asterisk (* FDR < 0.05). FDR = false discovery rate.
3.4. Clinicopathologic features and outcomes for CDK12-altered prostate cancer
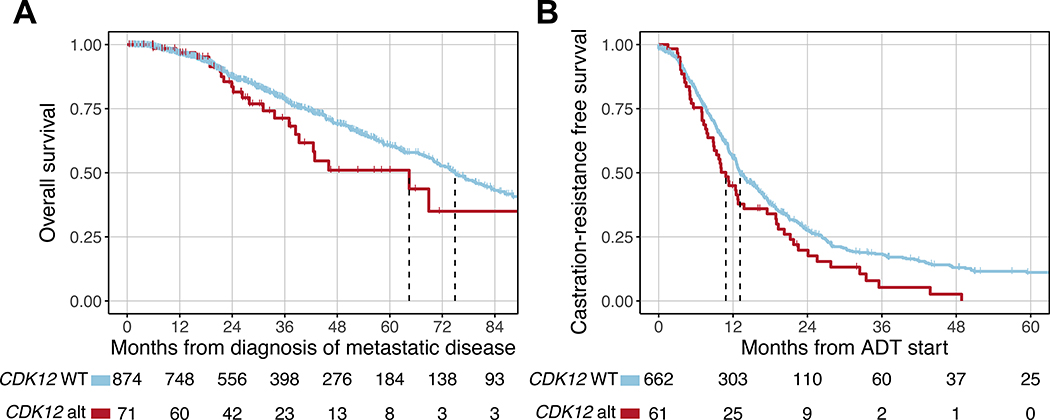
A subset of 1465 patients with prostate cancer were evaluable for clinical outcomes, of whom 100 had CDK12 alterations and 1365 were CDK12-WT (Supplementary Fig. 1). Baseline clinicopathologic characteristics are shown in Table 1. Patients with CDK12-altered tumors had clinical features associated with worse prognosis, including a higher prevalence of de novo metastatic disease (40% vs 26%), higher PSA at diagnosis (median 14.8 vs 9.0 ng/mL), and a higher prevalence of Gleason score ≥8 (80% vs 57%). Patients with CDK12-Bi inactivation had the worst baseline characteristics (Supplementary Table 4). Clinical status at the time of tissue collection for molecular profiling is summarized in Supplementary Table S4 and shown in cBioPortal [26] (www.cbioportal.org/study?id=prad_cdk12_mskcc_2020). We interrogated the association between CDK12 status and OS. There were a total of 315 deaths (CDK12-WT: 290/874 evaluable patients; CDK12-altered: 25/71) and the median follow-up time for censored patients was 30.2 mo. Compared to CDK12-WT patients, patients in the CDK12-altered group had shorter OS from diagnosis of metastatic disease (median 64.4 vs 74.9 mo; Fig. 4A). This difference was maintained after adjustment for known prognostic factors, including FGA [24] (adjusted hazard ratio [aHR] 1.80, 95% CI 1.12–2.89; p = 0.024). We also examined time to castration resistance from the start of ADT. There were a total of 522 events (CDK12-WT: 468/662 evaluable patients; CDK12-altered: 54/61) and the median follow-up time for censored patients was 12.2 mo. Patients in the CDK12-altered group had a shorter time to the development of castration-resistant disease (median 10.8 vs 13.1 mo; aHR 1.49, 95% CI 1.09–2.03; p = 0.017; Fig. 4B). Finally, among patients who received first-line abiraterone or enzalutamide for castration-resistant disease, those with CDK12-altered tumors had a similar time on treatment compared to CDK12-WT patients (median 9.7 vs 8.7 mo; aHR 1.08, 95% CI 0.57–1.51; p = 0.8; Supplementary Fig. 4). Collectively, these results suggest that CDK12 oncogenic alterations define a subset of somewhat more aggressive prostate cancer that progresses slightly more rapidly to castration resistance and is associated with slightly worse survival.
Table 1:
Clinical and pathological characteristics of CDK12-altered and CDK12-WT prostate cancer patients
| CDK12-altered | CDK12-WT | |
|---|---|---|
| Number of patients | 100 | 1365 |
| Age at diagnosis - years | ||
| Median (IQR) | 61 (54–67) | 63 (56–68) |
| Histology at diagnosis - n (% of known cases) | ||
| Adenocarcinoma | 94 (97) | 1216 (99) |
| Adenocarcinoma with neuroendocrine features | 2 (2) | 12 (0.9) |
| Other | 1 (1) | 6 (0.4) |
| Unknown | 3 | 131 |
| PSA at diagnosis - ng/mL | ||
| Median (IQR) | 14.8 (7.7–77.8) | 9.0 (5.3–24.1) |
| Gleason - n (% of known cases) | ||
| 6 | 7 (8) | 108 (9) |
| 7 | 10 (12) | 435 (34) |
| ≥8 | 68 (80) | 727 (57) |
| Not evaluable | 15 | 95 |
| Metastatic at diagnosis - n (% of known cases) | 39 (40) | 346 (26) |
| Unknown | 2 | 23 |
| Sites of metastasis at tissue collection - n (% of total metastatic patients) | ||
| Bone | 48 (60) | 528 (57) |
| Nonregional lymph nodes | 44 (55) | 380 (41) |
| Liver | 5 (6.3) | 93 (10) |
| Lung | 4 (5) | 101 (11) |
| Visceral | 7 (8.8) | 165 (18) |
Fig. 4 –
Clinical outcomes for CDK12-altered prostate cancer. (A) Overall survival from diagnosis of metastatic disease. (B) Time to development of castration-resistant prostate cancer from ADT initiation. ADT = androgen deprivation therapy.
4. Discussion
We analyzed the prevalence and types of somatic CDK12 alterations across >26000 tumors spanning 25 cancer types and found that prostate and ovarian cancers were the malignancies with the highest prevalence of CDK12 alterations, similar to a recent report [15]. The overall prevalence of CDK12 alterations in prostate cancer was 5.3%, which falls within the previously reported range [3,15]. The majority of CDK12 alterations in these cancers were oncogenic and clonal and resulted in biallelic inactivation, suggesting that CDK12 loss of function has an important biological role in a subset of prostate and ovarian cancers. In prostate cancer in particular, the main event presumed to be responsible for CDK12-Bi inactivation is the co-occurrence of two deleterious mutations, whereas in CDK12-altered ovarian cancer it is an LOH event, which can span a larger chromosomal segment and may not be specific to CDK12. While we were not able to definitively demonstrate the existence of CDK12 homozygous inactivation via copy-number alteration only, we included CDK12 deep deletion in the definition of the CDK12-altered group. Future studies are needed to determine whether complete loss of CDK12 can occur through deletion alone in prostate cancer. CDK12-Bi inactivation in prostate cancer was associated with a higher prevalence of small-fragment copy-number alterations and a higher number of breakpoints across the genome, consistent with the previously described tandem duplicator phenotype [9,25], as tandem duplications are difficult to characterize directly using targeted panel sequencing assays. We found that CDK12 mutations occur clonally in metastatic prostate cancer, and were identified in primary matched tumors from patients who developed metastatic disease, suggesting that CDK12 loss of function is an early event in the pathogenesis of CDK12-altered metastatic prostate cancer. Our study differs from the pan-cancer analysis by Sokol et al. [15] in that we identified early occurrence of CDK12 alterations in patients with matched tumors, and include a description of clinicopathologic features and clinical outcomes for prostate cancer patients, all within the same data set. Furthermore, our analysis of copy-number alterations in prostate tumors with CDK12-Bi inactivation compared with CDK12-WT tumors revealed numerous differentially altered regions. This includes amplification of 11q13.3, a region that encompasses the oncogene CCND1, and enrichment of alterations in cell-cycle pathway genes in patients with CDK12 loss of function. We did not identify a significant co-occurrence of alterations in CDK12 with DNA-repair gene alterations.
Identifying vulnerabilities of CDK12-altered prostate cancer is particularly important given our finding of somewhat more aggressive clinicopathologic features associated with CDK12 alteration, including more aggressive disease at diagnosis, slightly shorter OS, and shorter time to castration resistance. A study in an independent cohort of 46 patients with CDK12-altered prostate cancer by Reimers et al. [16] recently revealed similar outcomes with regard to time to castration resistance and time on treatment with first-line abiraterone and enzalutamide. Our larger study differs in that it also showed shorter OS for patients with CDK12-altered cancer, and differences in outcomes were maintained after adjusting for common clinical prognostic variables, probably facilitated by the larger sample size, all within a single institutional data set using a single clinical tumor-sequencing assay. We noted a similar rate of Gleason 8–10 prostate cancer at diagnosis for patients with CDK12-altered disease in comparison to the Reimers study (80% vs 88%). One vulnerability of CDK12-altered prostate cancer may be a higher sensitivity to immune checkpoint blockade owing to the higher neoantigen burden resulting from the tandem duplicator phenotype associated with CDK12 loss of function [9], a hypothesis that is currently being tested in a multi-institutional phase 2 study combining nivolumab with ipilimumab (NCT03570619). Of note, the slightly worse clinical outcomes we identified for CDK12-altered prostate cancer were unaffected by adjustment for FGA, suggesting that CDK12 loss of function impacts disease behavior through a mechanism independent of broad copy-number changes, possibly by promoting genomic alterations in specific pathways, or by impacting gene expression through increased intronic polyadenylation [7]. Limitations of our study include its retrospective nature, the use of a targeted panel assay for genomic profiling, which can miss genomic rearrangements, and potential selection bias arising from the inclusion of patients only from a tertiary referral center.
5. Conclusions
In summary, our results suggest that CDK12 loss of function defines a distinct subset of disease associated with unique genomic alterations and somewhat worse clinical outcomes, and highlights the need for novel therapies targeting CDK12-altered prostate cancer.
Supplementary Material
Acknowledgments
Financial disclosures: Wassim Abida certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Howard I. Scher is an uncompensated consultant/advisory board member for Amgen, ESSA Pharma Inc., Janssen Research & Development LLC, Janssen Biotech Inc., and Menarini Silicon; a compensated consultant/advisory board member for Ambry Genetics Corporation, Konica Minolta Inc., OncLive Insights, Physicians Education Resource, and WCG Oncology; has received research funding from Epic Sciences, Illumina Inc., Janssen, Menarini Silicon, and ThermoFisher; and has received travel/accommodation expenses from Amgen, Clovis Oncology, ESSA Pharma Inc., Menarini Silicon, OncLive Insights, Physician Education Resource, Prostate Cancer Foundation, and WCG Oncology. Philip W. Kantoff holds investments in Context Therapeutics LLC, DRGT, Placon, Seer Biosciences, and Tarveda Therapeutics; is a company board member for Context Therapeutics LLC; is a consultant/scientific advisory board member for Bavarian Nordic Immunotherapeutics, DRGT, GE Healthcare, Janssen, New England Research Institutes Inc., OncoCellMDX, Progenity, Sanofi, Seer Biosciences, Tarveda Therapeutics, and Thermo Fisher; and sits on data safety monitoring boards for Genentech/Roche and Merck. Michael J. Morris is an unpaid consultant for Astellas, Bayer, and Endocyte; is a paid consultant for Advanced Accelerator Applications, Blue Earth Diagnostics, Tokai, Tolmar, and Oric; and has received institutional research funding from Bayer, Sanofi, Endocyte, Progenics, Corcept, and Roche/Genentech. Dmitriy Zamarin has received consulting fees from Western Oncolytics, Agenus Inc., Tesaro, and Merck, and has received research funding from Merck. David B. Solit has received consulting fees or honoraria from Pfizer, Loxo Oncology, Lilly Oncology, Illumina, and Vivideon Therapeutics. Wassim Abida has received consulting fees from Clovis Oncology, Janssen Pharmaceutica, MORE Health, and ORIC Pharmaceuticals; research funding from AstraZeneca, Zenith Epigenetics, Clovis Oncology, and GlaxoSmithKline; and travel expenses from Clovis Oncology, GlaxoSmithKline, ORIC Pharmaceuticals, and the Prostate Cancer Foundation. Dana Rathkopf has received research funding from Janssen, Celgene, Ferring, Novartis, Taiho, Tracon, AstraZeneca, and Genentech/Roche, and has been an uncompensated consultant for Janssen, Tracon, AstraZeneca, Genentech, and Bayer. The remaining authors have nothing to disclose.
Funding/Support and role of the sponsor: This study was supported by Young Investigator and Challenge Awards from the Prostate Cancer Foundation (W.A., N.S., K.H.S.); Department of Defense Prostate Cancer Research Program Physician Research AwardW81XWH-17-1-0124 (W.A.) and Early Investigator Research AwardW81XWH-18-1-0330 (K.H.S.); and National Cancer Institute (NCI) cancer center support grant P30CA008748, and NCI prostate cancer SPORE grant P50CA092629. The sponsors played no direct role in the study.
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.eururo.2020.03.024.
References
- [1].Bell D, Berchuck A, Birrer M, et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Abeshouse A, Ahn J, Akbani R, et al. The tancer. Cell 2015;163:1011–25. 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. 10.1016/j.cell.2015.05.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Armenia J, Wankowicz SAM, Liu D, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50:645–51. 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ekumi KM, Paculova H, Lenasi T, et al. Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the Cdk12/CycK complex. Nucleic Acids Res 2015;43:2575–89. 10.1093/nar/gkv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Blazek D, Kohoutek J, Bartholomeeusen K, et al. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 2011;25:2158–72. 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dubbury SJ, Boutz PL, Sharp PA. CDK12 activates HRR by suppressing intronic polyadenylation, 2018. Nature 2018;564:141–5. 10.1038/s41586-018-0758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Krajewska M, Dries R, Grassetti AV, et al. CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat Commun 2019;10:1757 10.1038/s41467-019-09703-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu Y-M, Cie M, Lonigro RJ, et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer article inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 2018;173:1770–82. 10.1016/j.cell.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Popova T, Manié E, Boeva V, et al. Ovarian cancers harboring inactivating mutations in CDK12 display a distinct genomic instability pattern characterized by large tandem duplications. Cancer Res 2016;76:1882–91 10.1158/0008-5472.CAN-15-2128. [DOI] [PubMed] [Google Scholar]
- [11].Quigley DA, Dang HX, Zhao SG, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018;174:758–769.e9. 10.1016/j.cell.2018.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Abida W, Armenia J, Gopalan A, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol 2017;1:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Non-BRCA DNA Damage Repair Gene Alterations and Response to the PARP Inhibitor Rucaparib in Metastatic Castration-Resistant Prostate Cancer: analysis from the phase 2 TRITON2 study Abida Wassim, Campbell David, Patnaik Akash, Shapiro Jeremy D, Sautois Brieuc, Vogelzang Nicholas J, Voog Eric G, Bryce Alan H., McDermott Ray, Ricci Francesco, Rowe Julie, Zhang Jingsong, Piulats Josep Maria, Fizazi Karim, Merseburger Axel S, Higano Celestia S., Krieger Laurence E., Ryan Charles J, Feng Felix Y., Simmons Andrew D, Loehr Andrea, Despain Darrin, Dowson Melanie, Green Foad, Watkins Simon P., Golsorkhi Tony and Chowdhury Simon Clin Cancer Res February 21 2020. 10.1158/1078-0432.CCR-20-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mateo J, Porta N, McGovern UB, et al. TOPARP-B: a phase II randomized trial of the poly(ADP)-ribose polymerase (PARP) inhibitor olaparib for metastatic castration resistant prostate cancers (mCRPC) with DNA damage repair (DDR) alterations. J Clin Oncol 2019;37(15 Suppl):5005 10.1200/jco.2019.37.15_suppl.5005. [DOI] [Google Scholar]
- [15].Sokol ES, Pavlick D, Frampton GM, et al. Pan-cancer analysis of CDK12 loss-of-function alterations and their association with the focal tandem-duplicator phenotype. Oncologist 2019;24:1526–33. 10.1634/theoncologist.2019-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reimers MA, Yip SM, Zhang L, et al. Clinical outcomes in cyclin-dependent kinase 12 mutant advanced prostate cancer. Eur Urol 2020;77:333–41. 10.1016/j.eururo.2019.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cheng DT, Mitchell T, Zehir A, et al. MSK-IMPACT: a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30:1015–6 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chakravarty D, Gao J, Phillips S, Kundra R. OncoKB: a precision oncology knowledge base. Precis Oncol 2017;1:1–16. 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 2018;173:321–337. e10. 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:1–9. 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hieronymus H, Schultz N, Gopalan A, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A 2014;111:11139–44. 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Viswanathan SR, Ha G, Hoff AM, et al. Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell 2018;174:433–447.e19. 10.1016/j.cell.2018.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–4. 10.1158/2159-8290.CD-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.