Abstract
Environmental toxicants such as dioxins and polycyclic aromatic carbons are risk factors for pancreatitis and pancreatic cancer. These toxicants activate aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor, of which activation regulates many downstream biological events, including xenobiotic metabolism, inflammation, and cancer cell growth and transformation. Here, we identified that environmental toxicant-activated AHR increased expression of metastasis associated lung adenocarcinoma transcript 1 (MALAT1) in pancreatic cancer cells and pancreatic tissues. The MALAT1 is a long noncoding (lnc) RNA which interacts with Enhancer of Zeste 2 (EZH2), a histone methyltransferase with epigenetic silencer activity, and the MALAT1-EZH2 interaction increased its epigenetic silencing activity. In contrast, AHR antagonist, CH223191 or resveratrol, counteracted the AHR-mediated MALAT1 induction and MALAT1-enahnced EZH2 activity. Collectively, these results revealed a novel pathway of how environmental exposure leads to epigenetic alteration via activation of AHR-MALAT1-EZH2 signaling axis under pancreatic tissue- and cancer cell-context.
Keywords: Environmental toxicants, Aryl hydrocarbon receptor, MALAT1, EZH2, Epigenetic regulation, Pancreas
1. Introduction
Pancreatitis is a prerequisite for development of pancreatic ductal adenocarcinoma (PDA), which is one of the deadliest diseases with no effective prevention strategy currently available [1, 2]. Accumulated epidemiological studies showed a strong connection between cigarette smoking and PDA. Cigarette smoking is considered as a major risk factor for pancreatitis and PDA, which contains various environmental toxicants, including dioxins and benzo(a)pyrene (BaP), agonists of AHR [3-6].
The AHR is a ligand-activated transcription factor essential for mediating xenobiotic metabolism, immune responses, inflammation, differentiation as well as cancer cell growth and malignancy [7, 8]. It is activated by various environmental toxicants such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), BaP, and phytochemicals as well as tryptophan metabolites. Interestingly, depending on type of ligand, AHR can be either activated or inhibited and this selective feature makes AHR an important pharmacological target [9, 10].
The MALAT1 is a lncRNA which acts molecular scaffolds for various riboprotein complexes and functions as a transcriptional and epigenetic regulator [11]. MALAT1 interacted with EZH2, a histone methyltransferase that mediates gene silencing via tri-methylation of histone 3 lysing 27 (H3K27me3) [12, 13]. Indeed, EZH2 can interact with many lncRNAs other than MALAT1, including HOTAIR [14, 15], H19 [16], MEG3 [17] and XIST [18]. These interactions likely regulate the distribution, level, and activity of EZH2. Furthermore, it was reported that MALAT1 is highly expressed and associated with poor prognosis in pancreatic cancer [19, 20]. However, the underlying mechanisms for MALAT1 regulation by environmental toxicant exposure and MALAT1-directed regulation of EZH2 function in pancreatic cancer cell or tissue remain unclear.
In the present study, we showed that environmental toxicant-activated AHR induced MALAT1 and that MALAT1 interacted with EZH2 and increased its activity in pancreatic cancer cells and tissues. In contrast, treatment of AHR antagonist, CH223191[21] or resveratrol [22], inhibited the increase of MALAT1, EZH2 activity, and H3K27me3 levels. Taken together, our findings revealed a novel pathway that links environmental exposure to epigenetic regulation via activation of AHR-MALAT1-EZH2 signaling axis in pancreatic cancer cells and tissues.
2. Materials and Methods
2.1. Cell culture and Chemicals.
Panc-1 and AsPC-1 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and were maintained at 37°C in the presence of 5% CO2 in DMEM medium supplemented with 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin solution (Sigma Aldrich, St Louis). TCDD was purchased from AccuStandard, Inc. (New Heaven, CT); BaP was purchased from Sigma Aldrich (St. Louis, MO); CH223191 and Resveratrol were purchased from Cayman Chemical (Ann Arbor, MI). Small interfering RNAs (siRNAs) targeting MALAT1 (ID #: n511399 and n511398) and negative control siRNAs were purchased from Thermo Fisher Scientific. The siRNAs targeting AHR (siRNA ID: SASI_Hs02_00332181 and SASI_Hs01_00140198) were purchased from Sigma Aldrich.
2.2. Animal experiment.
C57BL/6J wild type mice (9-12 weeks old, 25-30 g body weight) were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were allocated for treatment with corn oil as control, TCDD dissolved in corn oil, or Resveratrol in corn oil. Each group contains 4-5 mice. Before any treatment, mice were fasted up to 12 hr. TCDD (30 μg/kg) intraperitoneally (i.p.) was injected into mice which was then anesthetized after 24 hr. In TCDD plus resveratrol treatment group, mice were pre-treated with resveratrol (20 mg/kg, i.p.) and after 90 min, mice were treated with TCDD (30 μg/kg, i.p.). After 24 hours, pancreatic tissue samples were collected for further analyses. The animal experiments were performed in compliance with the guidelines established by the Animal Care Committee of University of Cincinnati. The animals were acclimated to temperature- and humidity-controlled rooms with a 12-h light/dark cycle for 1 week prior to use.
2.3. Chromatin immunoprecipitation assay.
A chromatin immunoprecipitation (ChIP) assay was carried out with AHR (Enzo Life Sciences, Inc., Cat#: ALX-804-423-R100) and RNA polymerase II (Active Motif, Carlsbad, CA, Cat# 39097) antibodies using Panc-1 cell ChIP lysate treated with TCDD for 2 hour with the ChIP-IT Express Chromatin Immunoprecipitation Kit (Active Motif), according to the manufacturer’s protocol. The ChIP primer set that covers MALAT1 gene promoter proximal region (−442~−201): (forward): 5’-AGGAGAGAGGTGGGAAAGGAAG-3’ and (reverse) 5’-TGGTTCTAACCGGCTCTAGC-3’. All the ChIP-PCR reactions were carried out using a 7300HT Real-Time PCR system or a QuantStudio 3 Real Time PCR system with a 96-well block module (Applied Biosystems). The cycling conditions were 56°C for 30 min and 95°C for 10 min, followed by 48 cycles of 95°C for 25 s and 60°C for 60 s.
2.4. RNA immunoprecipitation (RIP) assay.
To detect the level of interaction between lncRNAs and EZH2, we performed the RIP assay as previously established [23]. Briefly, cells were fixed with formaldehyde and sheared by sonication. After DNase treatment, the fixed chromatins were immunoprecipitated with IgG or EZH2 antibody (Active Motif, Cat # 39933) with RNAse inhibitor. The purified lncRNAs and input RNA lysate were analyzed by qRT-PCR with the individual lncRNA primer set (Supplementary Table 1).
2.5. Real-time quantitative PCR (RT-qPCR).
The cDNAs were synthesized with the obtained RNA lysate from cells or tissues using a high-capacity cDNA Reverse Transcription Kit (Applied Biosystems) and RT-qPCR was performed. The obtained results were normalized to GAPDH or β-actin control. All the primer set sequences for detecting specific mouse or human genes by qRT-PCR are listed (Supplementary Table 1).
2.6. Histological analysis.
Paraffin-embedded liver sections were sectioned, deparaffinized in xylene, and rehydrated through a series of graded ethanol solutions. The morphological changes in pancreatic tissue sections stained with hematoxylin and eosin (H&E) were examined under a light microscope for histological analysis.
2.7. Histone extraction.
Histones were extracted from cells or tissues using EpiQuik total histone extraction kit according to the manufacturer’s protocol (EpiGentek, Farmingdale, NY).
2.8. Nuclear extraction and H3K27me3 enzymatic assay.
Nuclear extract from cells or pancreatic tissues were collected using EpiQuik Nuclear Extraction kit (Cat # OP-0002-1, EpiGentek, Inc., Farmingdale, NY). By using obtained nuclear extracts, we performed H3K27me3 enzymatic assay with EpiQuik Histone Methyltransferase Activity/Inhibition Assay Kit (EpiGentek, Inc., Cat # P3005). All the procedures were performed according to the manufacturer’s protocol.
2.9. Immunoblotting.
Total proteins extracted from either cells or minced pancreatic tissues (30-100 μg per lane) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. After being blocked with TBST or PBST buffer containing 5% non-fat milk, the membranes were incubated with proper primary antibody. Proteins of interest were detected with either anti-rabbit or anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology) and visualized with an enhanced chemiluminescence (ECL) detection kit using a C-DiGit Blot Scanner from LI-COR (Lincoln, NE).
2.10. Immunohistochemical staining.
After pancreatic tissue sections were deparaffinized, the sections were immersed in 0.01 mM sodium citrate (pH 6.0) and heated in a microwave oven (98°C) for antigenic retrieval. The deparaffinized sections were incubated with peroxidase-blocking reagent (Biogenex, CA, USA) to block endogenous peroxidase activity and then incubated with nonspecific staining blocking reagent (Vector Laboratories, Burlingame, CA, USA). The sections were incubated with primary antibodies at 4°C overnight. Anti-EZH2 antibody (Cell Signaling Technology, Danvers, MA, Cat # 5246) and anti-H3K27me3 antibody (Active motif, Cat #: 39155) were employed. The sections were subsequently incubated with peroxidase-conjugated secondary antibodies (Vector Laboratories) and 3, 3-diaminobenzine-tetrachloride (DAB; Vector Laboratories), according to the manufacturer’s instructions. The sections were counterstained with hematoxylin and observed under a microscope.
2.11. Statistical analysis.
Statistical significance was determined by the unpaired Student’s t-test with a two tailed distribution or one-way analysis of variance (ANOVA) with a post-hoc Tukey's test. Data are presented as the mean ± SD. Statistical analysis was performed with Graph Pad Prism 6.0 software. Statistical significance was set at a P-value of <0.05.
3. Results
3.1. Environmental toxicant induced MALAT1, a lncRNA that interacted with EZH2.
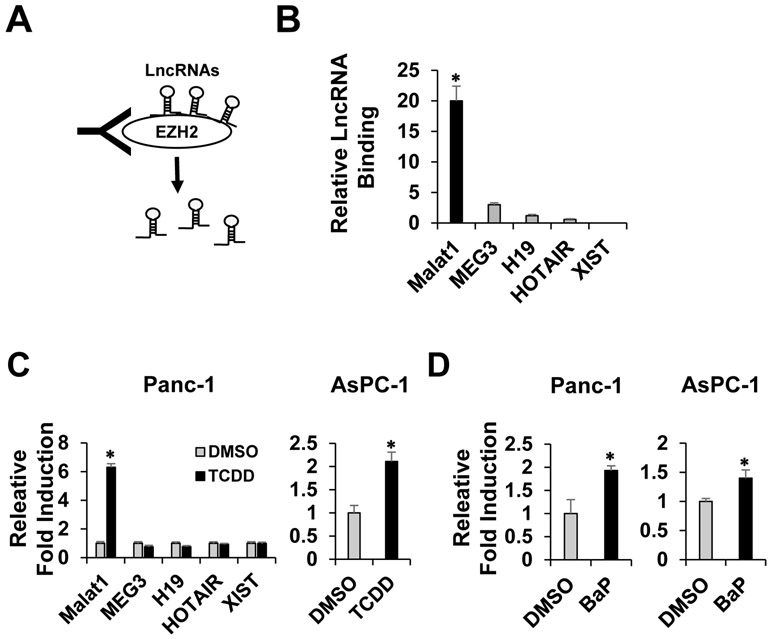
Many previous reports showed that various lncRNAs formed complexes with EZH2 and regulated activity or distribution of EZH2, an epigenetic writer and silencer with histone methyltransferase function [24, 25]. By employing RIP assay, we determined the types and levels of lncRNA-EZH2 interactions under pancreatic cancer cell context. After Panc-1 human pancreatic cancer cells were fixed with formaldehyde, EZH2-associated lncRNAs were pull down by EZH2 antibody (Fig 1A). The qRT-PCR was performed using the obtained RNA lysate with appropriate primer sets for determining the levels of binding interactions between EZH2 and these lncRNAs, including MALAT1, HOTAIR, MEG3, H19, and XIST, as previously reported as EZH2 interacting lncRNAs. Among lncRNAs, we identified that MALAT1 most strongly interacted with EZH2. MEG3, H19, and HOTAIR also interacted with EZH2 with different levels however no interaction was detected between XIST and EZH2 (Fig. 1B), indicating existence of various lncRNA-EZH2 complexes. The induction levels of CYP1A1, a representative AHR downstream target gene, by TCDD or BaP treatment in Panc-1 and Aspc-1 cells were presented (Supplementary Fig. 1). The primer sequences for all the lncRNAs were listed (Supplementary Table 1).
Figure 1. MALAT1-EZH2 interaction and environmental toxicant-induced MALAT1 induction.
A schematic diagram for RIP assay. EZH2-associated lncRNAs were pulled down with EZH2 antibody (A). To measure the strength of binding interactions between EZH2 and the pull-downed lncRNAs, a RT-qPCR assay was performed with each lncRNA primer set (B). TCDD-mediated lncRNA induction in Panc-1 and AsPC-1 human pancreatic cancer cells (C). BaP-mediated MALAT1 induction in Panc-1 and AsPC-1 cells (D). Values are represented as mean ± SD. *Significant difference or induction P < 0.05.
In parallel, we determined which lncRNA can be induced by environmental exposure. TCDD, a potent AHR agonist, was used as a model compound. We found out that only MALAT1 was significantly induced by TCDD treatment, not others (Fig. 1C, left). MALAT1 was also induced in AsPC-1 human pancreatic cancer cells (Fig. 1C. right). Furthermore, treatment of BaP, another AHR agonist and ubiquitous environmental toxicant, induced MALAT1 in both Panc-1 and AsPC-1 cells (Fig. 1D). These results together indicated that MALAT1 is an EZH2-interacting lncRNA which is significantly induced by environmental toxicant exposure in pancreatic cancer cells.
3.2. Role of AHR in the MALAT1 induction
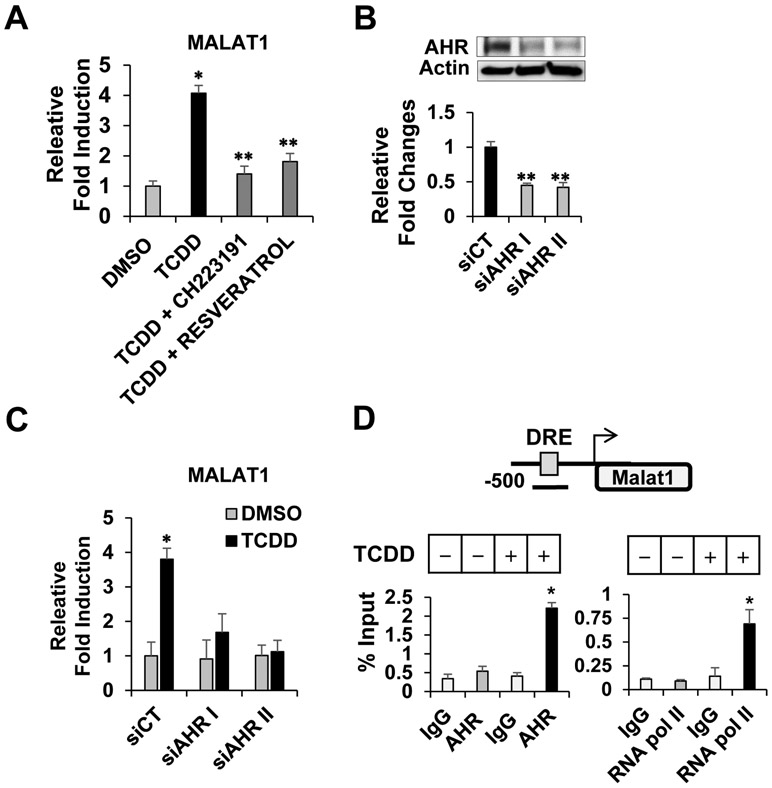
Since TCDD or BaP are AHR agonists, we examined a role of AHR in the MALAT1 induction. Cells were treated with TCDD alone or together with AHR antagonist either CH223191 or resveratrol. The induction of MALAT1 was markedly inhibited by AHR antagonist (Fig. 2A). To further determine whether AHR proteins is required for the MALAT1 induction, we depleted AHR proteins using small interfering RNA (siRNA) targeting AHR. Cells were transfected with two different siRNAs targeting different regions of AHR (siAHR I and II) or scrambled control siRNA (siCT). The knockdown of AHR was confirmed by immunoblotting and RT-qPCR. The reduced AHR mRNA and protein levels were observed (Fig. 2B). Furthermore, the AHR depletion using this interfering siRNA approach significantly decreased MALAT1 induction, confirming that AHR was essential for the MALAT1 induction (Fig. 2C). Next, we performed gene promoter analysis using PROMO program [26] whether there is any potential AHR binding sequence located in the MALAT1 gene promoter region. We identified a potential AHR binding site, namely Dioxin Response Element (DRE), in the proximal region of MALAT1 gene promoter (5’-GCGTGCGCAGTCACGC-3’) (Fig. 2D, top). To examine whether TCDD-activated AHR is actually recruited to the DRE, we performed chromatin immunoprecipitation (ChIP) assay. The recruitment of AHR to the DRE site was detected and in parallel, RNA polymerase II, a positive control for transcriptional activation, also interacted with the DRE, indicating that AHR plays a role in transcriptional activation of MALAT1 via the DRE site (Fig. 2D, bottom). These results demonstrated that AHR is essential for the TCDD-induced MALT1 induction.
Figure 2. AHR-mediated MALAT1 induction.
TCDD treatment induced MALAT1 while co-treatment of AHR antagonist, either CH223191 or resveratrol, inhibited the MALAT1 induction (A). Transfection of siRNAs targeting AHR (siAHR I and II) reduced AHR protein and mRNA levels but not with scrambled control siRNA (siCT) (B). AHR depletion with siAHR I and II significantly decreased the MALAT1 induction (C). A schematic diagram of human MALAT1 gene proximal promoter region. A DRE site is shown as square. Black bar indicated the location of ChIP primer in the MALAT1 gene promoter region (D, top). A ChIP assay showed that TCDD treatment facilitated recruitment of AHR and RNA polymerase II to the DRE site (D, bottom). *Significant induction or **repression P < 0.05.
3.3. Role of AHR-MALAT1 signaling in the regulation of EZH2 activity.
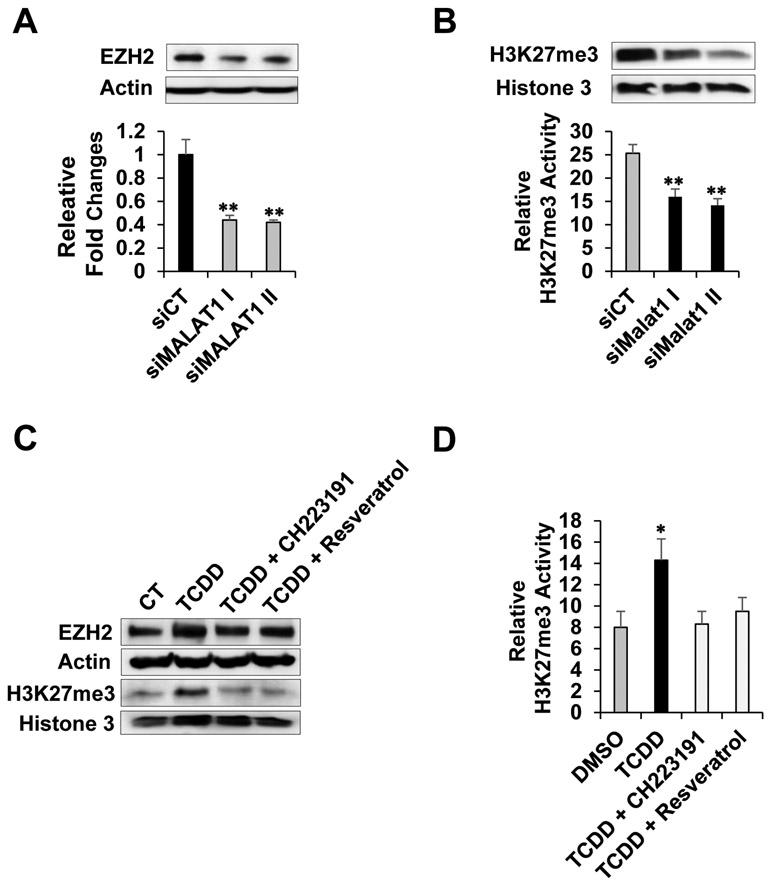
Previous report showed that MALAT1 interacted with EZH2 and regulated its function in pancreatic cancer cells [27, 28]. However, overarching effects of MALAT1 knockdown on EZH2 level and activity remain elusive. Therefore, we transfected cells with siRNAs targeting MALAT1 (siMalat1 I and II) or control siRNA (siCT) in Panc-1 cells and determined effects of MALAT1 depletion on EZH2 and H3K27me3 levels. We observed decreased levels of EZH2 and H3K27me3 (Fig. 3A, top). The MALAT1 depletion efficiency was determined by RT-qPCR (Fig 3A, bottom). Next, using nuclear extracts from the control and MALAT1-depleted cells, we performed the enzymatic assay for measuring H3K27 trimethylation activity since H3K27me3 is a substrate of EZH2 (EpiGenteck, Inc.). Consistently, there was greatly reduced H3K27me3 enzymatic activity observed with the MALAT1-depleted nuclear extracts (Fig. 3B).
Figure 3. MALAT1 depletion or AHR-mediated MALAT1 induction on EZH2 level and activity.
Cells were transfected with siRNAs targeting MALAT1 (siMALAT1 I & II) or control siRNA (siCT). Effects of MALAT1 depletion on the EZH2 protein levels (A, top). The MALAT1 depletion was confirmed by RT-qPCR (A, bottom). Effects of MALAT1 depletion on H3K27me3 marks (B, top) and H3K27me3 enzymatic activities (B, bottom). Effects of TCDD ± CH223191 (5 μM) or resveratrol (20 μM) for 24 hours on the levels of EZH2 protein and H3K27me3 (C) and H3K27me3 enzymatic activities (D).
Next, we examined effects of TCDD with or without AHR antagonist treatment on MALAT1 and EZH2 levels, and H3K27me3 enzymatic activities by employing the same approaches above. TCDD increased MALAT1, EZH2, and H3K27me3 levels whereas co-treatment of TCDD with AHR antagonist either CH223191 or resveratrol inhibited the increases (Fig. 3C, top and bottom). Similarly, the TCDD-enhanced H3K27me3 enzymatic activities and its inhibition by CH223191 or resveratrol were observed (Fig. 3D). Collectively, these results indicated that TCDD increased MALAT1, EZH2, and H3K27me3 levels as well as the enzymatic activity of EZH2.
3.4. Effects of TCDD on Malat1-EZH2 signaling axis in vivo
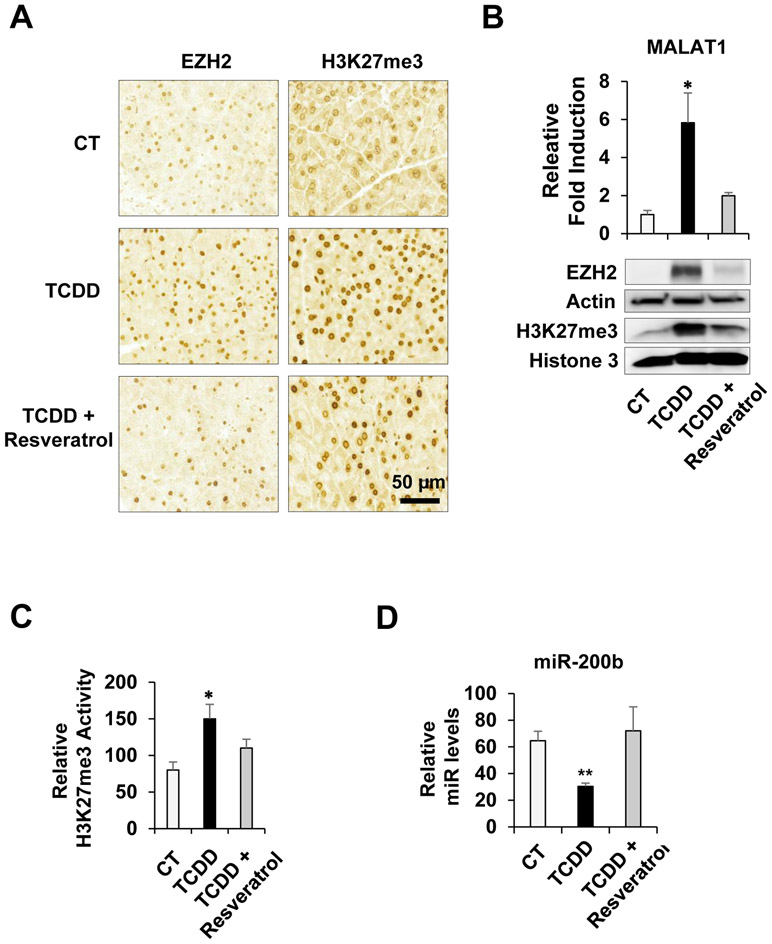
In order to determine effects of TCDD on the Malat1-EZH2 signaling activation in vivo, TCDD was administered to mice with or without pre-treatment of resveratrol. After 24 hours, pancreatic tissues were collected and then immunostaining with anti-EZH2 or anti-H3K27me3 antibody was performed. Significantly increased EZH2 and H3K27me3 staining were observed in the TCDD-treated pancreatic tissues while pre-treatment of resveratrol prevented them (Fig. 4A). The result from RT-qPCR analysis consistently showed that TCDD treatment markedly induced MALAT1 expression however pre-treatment of resveratrol inhibited it (Fig. 4B). Representative images of haemotoxylin and eosin (H&E) staining were shown (Supplementary Fig. 2A). The induction of CYP1A1 by TCDD and its inhibition by resveratrol in the pancreatic tissues were confirmed by RT-qPCR (Supplementary Fig. 2B). Similarly, results from immunoblotting showed that TCDD treatment increased EZH2 and H3K27me3 levels while resveratrol counteracted them (Fig. 4C). The H3K27me3 enzymatic assay was performed with tissue nuclear extracts. TCDD treatment enhanced H3K27me3 enzymatic activities whereas the pre-treatment of resveratrol prevented the enhancement (Fig. 4D). Lastly, to determine effects of the enhanced EZH2 activity on downstream target gene expression, we examined expression levels of miR-200b, a known EZH2 target gene [29, 30]. We found out that miR-200b was significantly downregulated by TCDD but restored its expression level by resveratrol, validating effects of TCDD-mediated MALAT1-EZH2 signaling activation on downstream target gene expression. Taken together, results clearly demonstrated that AHR-activated MALAT1-EZH2 signaling pathway in vivo.
Figure 4. Effects of TCDD-activated AHR on MALAT1-EZH2 signaling in vivo.
Immunostaining analysis of control-, TCDD-, and TCDD plus Resveratrol-treated pancreatic tissue sections (N = 4-5 per treatment) with EZH2 and H3K27me3 antibodies. Scale bar, 50 μm (A). TCDD-mediated induction of MALAT1 and its inhibition by resveratrol in pancreatic tissues (B, top). Immunoblotting analysis of pancreatic tissues treated with TCDD, TCDD plus Resveratrol, or control with EZH2 or H3K27me3 antibody. Beta-Actin (Actin) or histone 3 were used as controls (B, bottom). The H3K27me3 enzymatic assay was performed with the pancreatic tissue nuclear lysates treated with TCDD, TCDD plus Resveratrol, or control (C). The RT-qPCR with TCDD, TCDD plus Resveratrol, or control pancreatic tissue RNA lysates was performed to determine the miR-200b level changes (D).
4. Discussion
Accumulating epidemiological evidences have indicated that environmental exposure is a main risk factor in pancreatic inflammation and cancer. Our results in this study demonstrated that environmental exposure-activated AHR induced a lncRNA MALAT1 and this induction subsequently increased epigenetic silencing function of EZH2. Overarchingly, these results showed a novel molecular mechanism of how environmental exposure leads to epigenetic alteration in pancreatic cancer cell and tissue context, which, in part, occurs via AHR-MALAT1-EZH2 signaling activation.
Moreover, given that higher expression or dysregulated activities of EZH2 or MALAT1 played an important roles in cancer stem cell property and malignancy [31, 32], these results provide the possibility for a pro-oncogenic role of AHR in the epigenetic dysregulation. AHR can function as a linchpin molecule that connects environmental exposure to the EZH2-mediated epigenetic dysregulation via MALAT1 induction in pancreatitis and pancreatic cancer. In addition, as shown in Fig. 4D, low levels of miR-200b, one of downstream target gene of MALAT1-EZH2 signaling pathways, is highly correlated with epithelial-mesenchymal transition, cancer stem cell features, and drug resistance in pancreatic cancer [33, 34].
It is also of interest that AHR can selectively be activated or inhibited, depending on ligand-type. Our results demonstrated that either AHR antagonist or depletion inhibited the MALAT1 induction and prevented the increased EZH2 enzymatic activity in both in vitro and in vivo (Fig. 3 and 4), suggesting that AHR antagonism has a potential to be a novel prevention strategy. However, it is highly conceivable that depending on environmental toxicant- or ligand-type, AHR induced other lncRNAs that differentially regulates EZH2 function. Therefore, environmental toxicant- or ligand-dependent regulation of lncRNAs via AHR need to be further systematically investigated. Lastly, in line with our results as shown in this study, it is highly interesting to investigate a role of AHR-MALAT1-EZH2 signaling under experimental settings of pancreatitis and pancreatic cancer in the future.
Supplementary Material
Highlights.
TCDD increased MALAT1 expression via AHR activation.
AHR antagonists inhibited AHR-mediated MALAT1 induction.
MALAT1 interacted with EZH2 and increased its epigenetic silencing function.
The AHR-MALAT1-EZH2 signaling linked environmental exposure to epigenetic alteration.
Acknowledgements
This work was supported by NIEHS P30ES006096 and Steven Goldman Memorial Pancreatic Cancer Research Grant.
Footnotes
Conflict of interest
All the authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lowenfels AB, Maisonneuve P, Cavallini G, et al. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N Engl J Med. 328 (1993) 1433–1437, https://www.nejm.org/doi/full/10.1056/nejm199305203282001. [DOI] [PubMed] [Google Scholar]
- [2].Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 388 (2016) 73–85, 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- [3].Yuan C, Morales-Oyarvide V, Babic A, Clish CB, Kraft P, Bao Y, Qian ZR, Rubinson DA, Ng K, Giovannucci EL, Ogino S, Stampfer MJ, Gaziano JM, Sesso HD, Cochrane BB, Manson JE, Fuchs CS, Wolpin BM. Cigarette Smoking and Pancreatic Cancer Survival. J Clin Oncol. 35 (2017) 1822–1828, https://ascopubs.org/doi/10.1200/JCO.2016.71.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Muto H, Takizawa Y. Dioxins in cigarette smoke. Arch Environ Health. 44 (1989) 171–174, https://www.tandfonline.com/doi/abs/10.1080/00039896.1989.9935882. [DOI] [PubMed] [Google Scholar]
- [5].Stedman RL. The chemical composition of tobacco and tobacco smoke. Chem Rev. 68 (1968) 153–207, https://pubs.acs.org/doi/abs/10.1021/cr60252a002. [DOI] [PubMed] [Google Scholar]
- [6].Kasai A, Hiramatsu N, Hayakawa K, Yao J, Maeda S, Kitamura M. High levels of dioxin-like potential in cigarette smoke evidenced by in vitro and in vivo biosensing. Cancer Res. 66 (2006) 7143–7150, https://cancerres.aacrjournals.org/content/66/14/7143. [DOI] [PubMed] [Google Scholar]
- [7].Puga A, Ma C, Marlowe JL. The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem Pharmacol. 77 (2009): 713–22, 10.1016/j.bcp.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Safe S, Lee SO, Jin UH. Role of the aryl hydrocarbon receptor in carcinogenesis and potential as a drug target. Toxicol Sci. 135 (2013) 1–16, 10.1093/toxsci/kft128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer. 14 (2014) 801–14, 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kolluri SK, Jin UH, Safe S. Role of the aryl hydrocarbon receptor in carcinogenesis and potential as an anti-cancer drug target. Arch Toxicol. 91 (2017) 2497–2513, 10.1007/s00204-017-1981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and Pathophysiological functions. RNA Biol. 14 (2017)1705–1714, 10.1080/15476286.2017.1358347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 75 (2015) 1322–1331, 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li P, Zhang X, Wang H, et al. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol Cancer Ther. 16 (2017) 739–751, 10.1158/1535-7163.MCT-16-0591. [DOI] [PubMed] [Google Scholar]
- [14].Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone Modification complexes. Science. 329 (2010) 689–693, 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 32 (2013) 1616–1625, 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 333 (2013) 213–221, 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- [17].Kaneko S, Bonasio R, Saldaña-Meyer R, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 53 (2014) 290–300, 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 322 (2008) 750–756, 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 36 (2015) 2403–2407, 10.1007/s13277-014-2850-8 [DOI] [PubMed] [Google Scholar]
- [20].Li L, Chen H, Gao Y, et al. Long Noncoding RNA MALAT1 Promotes Aggressive Pancreatic Cancer Proliferation and Metastasis via the Stimulation of Autophagy. Mol Cancer Ther. 15 (2016) 2232–2243, 10.1158/1535-7163.MCT-16-0008. [DOI] [PubMed] [Google Scholar]
- [21].Zhao B, Degroot DE, Hayashi A, He G, Denison MS. CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci. 117 (2010) 393–403, 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Casper RF, Quesne M, Rogers IM, et al. Resveratrol has antagonist activity on the aryl hydrocarbon receptor: implications for prevention of dioxin toxicity. Mol Pharmacol. 56 (1999) 784–790, http://molpharm.aspetjournals.org/content/56/4/784.long. [PubMed] [Google Scholar]
- [23].Khanal T, Leung YK, Jiang W, Timchenko N, Ho SM, Kim K. NR2E3 is a key component in p53 activation by regulating a long noncoding RNA DINO in acute liver injuries. FASEB J. 33 (2019) 8335–8348, 10.1096/fj.201801881rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 1856 (2015) 151–164, 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang Y, Xie Y, Li L, et al. EZH2 RIP-seq Identifies Tissue-specific Long Non-coding RNAs. Curr Gene Ther. 18 (2018) 275–285, 10.2174/1566523218666181008125010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Farré D, Roset R, Huerta M, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31 (2003) 3651–3653, 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Han T, Jiao F, Hu H, Yuan C, Wang L, Jin Z, Song W, Wang L EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget. 7 (2016) 11194–11207, 10.18632/oncotarget.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ougolkov AV, Bilim VN, Billadeau DD. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin Cancer Res. 14 (2008) 6790–6796, 10.1158/1078-0432.ccr-08-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ning X, Shi Z, Liu X, et al. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 359 (2015) 198–205, 10.1016/j.canlet.2015.01.005. [DOI] [PubMed] [Google Scholar]
- [30].Bao B, Ali S, Banerjee S, et al. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 72 (2012) 335–345, 10.1158/0008-5472.CAN-11-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 106 (2012) 243–247, 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jiao F, Hu H, Yuan C, et al. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 32 (2014) 2485–2492, 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- [33].Li Y, VandenBoom TG 2nd, Kong D, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 69 (2009) 6704–6712, 10.1158/0008-5472.can-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bao B, Wang Z, Ali S, et al. Metformin inhibits cell proliferation, migration and invasion by attenuating CSC function mediated by deregulating miRNAs in pancreatic cancer cells. Cancer Prev Res (Phila). 5 (2012) 355–364, 10.1158/1940-6207.capr-11-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.