Abstract
The products of most secondary metabolite biosynthetic gene clusters (BGCs) have yet to be discovered, in part due to low expression levels in laboratory cultures. Reporter-guided mutant selection (RGMS) has recently been developed for this purpose: a mutant library is generated and screened, using genetic reporters to a chosen BGC, to select transcriptionally active mutants that then enable the characterization of the ‘cryptic’ metabolite. The requirement for genetic reporters limits the approach to a single pathway within genetically tractable microorganisms. Herein, we utilize untargeted metabolomics in conjunction with transposon mutagenesis to provide a global read-out of secondary metabolism across large numbers of mutants. We employ self-organizing map analytics and imaging mass spectrometry to identify and characterize seven cryptic metabolites from mutant libraries of two different Burkholderia species. Applications of the methodologies reported can expand our understanding of the products and regulation of cryptic BGCs across phylogenetically diverse bacteria.
Introduction
Natural products have been indispensable as a starting point for drug discovery and a source of inspiration for researchers across multiple disciplines.1,2 It is by now well-established that many natural product biosynthetic gene clusters (BGCs) are silent under standard laboratory conditions, meaning they are at best sparingly expressed, so that the cognate products do not accumulate to detectable levels.3–6 The natural products discovered thus far, therefore, are based on a fraction of constitutively expressed biosynthetic genes. Products of silent BGCs, by contrast, have been largely overlooked and provide an underexplored source of microbial metabolites. Recently, several approaches have been developed for activating silent BGCs, including co-culture assays, heterologous expression, insertion of active promoters, ribosome engineering, high-throughput elicitor screening (HiTES), and reporter-guided mutant selection (RGMS).7–14 While these methods have collectively provided new ‘cryptic’ metabolites, insights into the regulation of the corresponding BGCs are only starting to emerge.15
RGMS provides a forward genetic strategy for activating silent BGCs and is, along with HiTES, especially useful in deciphering the underlying regulatory circuits.16–18 In RGMS, a reporter gene is inserted into the silent cluster as a facile read-out of expression and mutants are then generated and selected based on their ability to activate the desired BGC. RGMS was first applied to silent BGCs in Streptomyces sp. PGA64, where a new glycosylated gaudimycin was identified.17 Additional applications followed in Streptomyces coelicolor and Streptomyces albus.19,20 Two technical aspects are critical when considering RGMS: the source of mutants and the expression read-out. So far UV-induced DNA damage and transposon (Tn) mutagenesis have been used to create libraries of mutants.16–20 Tn mutagenesis is desirable as it allows for rapid identification of the gene, which when interrupted via Tn insertion, results in activation of the chosen cluster. Both visual inspection of pigmented molecules and cluster-specific reporter genes have been used to monitor BGC expression, with the latter approach presenting a much more general option as it can be applied to diverse natural product structural classes.
We recently extended RGMS to Burkholderia thailandensis, a saprophytic β-proteobacterium that is genetically tractable and contains more than a dozen silent BGCs.15,21–23 In one study, we mutagenized wild-type B. thailandensis and inspected the resulting mutants for phenotypic changes, ultimately leading to the characterization of three new cryptic antibiotics, thailandenes A-C.22 In another study, we used transcriptional and translational fluorescent protein fusions as cluster-specific reporters and identified disruption of nucleotide biogenesis as a trigger for induction of the silent malleicyprol BGC.23–26 One drawback of this approach is that two rounds of genetic manipulations are necessary, the first to generate the reporter constructs and the second for creation of the library of Tn mutants. In the current work, we explore the idea of replacing genetic reporters with mass spectrometry-based methods. Specifically, we report the use of HPLC-MS-coupled self-organizing map (SOM) analytics with a small number of mutants, as well as rapid imaging mass spectrometry (IMS) with a larger number of mutants for the identification of cryptic metabolites. We have utilized these strategies in two different Burkholderia strains and disclose seven cryptic metabolites in three different compound classes. In the mutants that overproduced these cryptic metabolites, the site of Tn insertion is identified as well. Together, our results show that the combination of Tn mutagenesis and MS-based metabolomic approaches provide a useful platform for engineering metabolically activated strains and thereby deciphering both the products and the underlying regulation of silent BGCs.
Results and Discussion
High-throughput detection of cryptic metabolites.
We began with the goal of rapidly surveying the secondary metabolomes of bacterial mutant libraries in a high-throughput fashion using MS-based methods. One of the key advantages of an MS read-out is that genetic manipulations are not necessary and, rather than monitor the expression of a single BGC, the global secondary metabolome can be surveyed, as detected by MS. We considered self-organizing map (SOM) analytics based on HPLC-MS data as well as rapid imaging mass spectrometry (IMS), both of which have recently been used for detection of natural products in multidimensional metabolomic datasets.27–32 The first approach facilitates simultaneous analysis of a small number of samples (<100) to identify differentially synthesized natural products within a treatment condition (in this case Tn mutants). The second allows for rapid screening of a much larger number of mutants (>500) but is less comprehensive due to the lack of chromatographic resolution resulting in increased ion suppression. For these purposes, we selected Burkholderia plantarii and Burkholderia gladioli (Table S1), plant pathogens in the phytogenic group of the Burkholderia genus. Bioinformatic analyses by antiSMASH reveals at least 25 and 19 BGCs in B. plantarii and B. gladioli, respectively (Tables S2, S3).33 The products for many of these BGCs are unknown, making the strains ideal targets for genome mining. For mutagenesis, we selected the EZ-Tn5 method,34 which has been applied broadly to diverse Gram-positive and Gram-negative bacteria. B. gladioli has previously been mutagenized with the Himar1 Tn;35 to the best of our knowledge, Tn mutants of B. plantarii have not yet been reported.
HPLC-MS and SOM analysis with B. plantarii.
EZ-Tn5 mutagenesis with a kanamycin resistance marker led to a mutation frequency of 2 × 10−7 (B. plantarii) and 6 × 10−7 (B. gladioli), which was sufficient for our application. After growth on selective media, ~600 B. plantarii and ~1800 B. gladioli mutants were arrayed and stored in 96-well plates. Because of the smaller number of mutants, we decided to apply the HPLC-MS-coupled SOM approach, which is inherently lower-throughput compared to IMS, to B. plantarii: 72 randomly selected Tn mutants from our library were used to inoculate 20 mL flask cultures, which were grown overnight. The cell-free supernatants were subsequently analyzed by HPLC-Qtof-MS. The unmutagenized wild-type (wt) was treated in a similar fashion in four replicates. Untargeted feature extraction - each feature consists of a specific m/z, intensity, and retention time - was used to align ~2200 unique ions across the 72 mutants and the wt samples of B. plantarii. The multidimensional data then underwent Kohonen map analysis, in which a competitive learning algorithm was used to sort the mass features by their abundance profiles across all mutants; these were then organized in a square grid with each pixel representing a unique collection of metabolites with highly covarying responses to transposon mutagenesis.27,28 The four wt maps were averaged and three-fold of this wt composite was subtracted from each Tn profile, resulting in 72 distinct differential heatmaps (Figure S1). Regions of the self-organized maps were colored based on m/z intensity, with yellow indicative of >3-fold abundance in the Tn mutant relative to wt.
Visual inspection of all 72 heatmaps showed discernible regions of abundant features for most Tn mutants, indicating that a significant (>3-fold) impact on the metabolome had occurred (Figure S1). Specifically, four regions of interest (ROI) were identified; the features from these regions are tabulated (Figure 1A, Table S4). ROI-1 consisted of a broad range of ions, many of these in the m/z range of 300–600. ROI-2 contained a single ion with a neutral mass of 602.3. ROI-3 consisted of several groups of ions, some of these with HR-MS that matched the 4-hydroxy-3-methylquinoline (HMQ) group of compounds, which are known to be produced by Burkholderia spp.36–38 The corresponding hmq cluster is present in the B. plantarii genome. ROI-4 contained several high molecular weight features. Candidate metabolites for further study were identified in each of the ROIs (Table S4). We focused our efforts on ROI-4 as the enhanced production of high molecular weight compounds, which did not match any reported natural products in various databases, optimized the possibility of finding new metabolites. Specifically, we focused on two compounds with m/z 809.4068 and 1021.5221 (Figure 1A). Detection of these features in all 72 mutants showed that their production levels were tightly coupled, suggesting either that they are derived from the same biosynthetic pathway or that their biogenesis is strictly co-regulated from disparate BGCs (Figure 1B).
Figure 1.
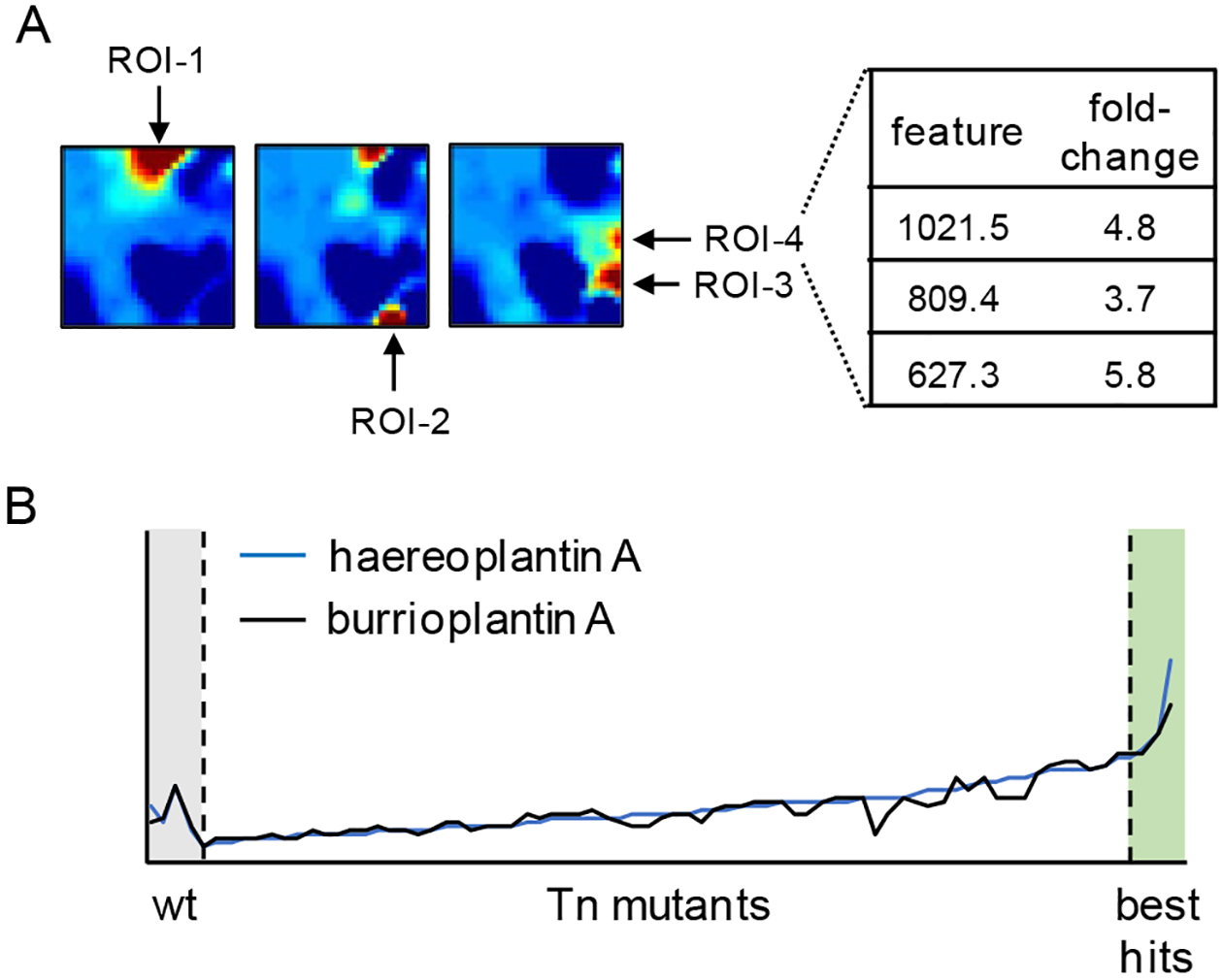
Analysis of B. plantarii Tn mutants using HPLC-MS-based SOM analytics. (A) Three representative differential maps are shown. As described in SI methods, these were obtained by extracting features from HPLC-Qtof-MS data for each Tn mutant, arraying the features in a grid using self-organizing map software, and subtracting three-times the wt map from each mutant to yield the differential maps shown (Figure S1). Four major ROIs, which emerged from this analysis, are marked. Select metabolites in ROI-4 are highlighted. (B) Production of two compounds haereoplantin A (m/z 809.4) and burrioplantin A (m/z 1021.5) are correlated across all Tn mutants. One of these, in the ‘best hits’ zone, was selected for further study.
IMS analysis with B. gladioli.
To implement the second detection approach, we used laser ablation electrospray ionization mass spectrometry (LAESI-MS), a method that requires minimal sample preparation and provides broad molecular coverage with μM detection sensitivity for many metabolites.39–41 960 B. gladioli Tn mutants were cultured in 96-well plates, subjected to solid-phase extraction in the same format, and directly analyzed by LAESI-MS. Ions observed above an abundance threshold were then binned by mass for each Tn mutant. These were subtracted from twice the average value detected for that bin in the wt samples. Positive values of the resulting difference matrix were then displayed in a 3D plot, which shows the m/z and intensity for all metabolites as a function of each the 960 Tn mutants (Figure 2, top). All metabolites that appear in the 3D map are overproduced in the Tn mutant and are present at low levels, or not at all, in the wt strain. Here again, we could easily observe significant metabolomic changes resulting from Tn mutagenesis.
Figure 2.
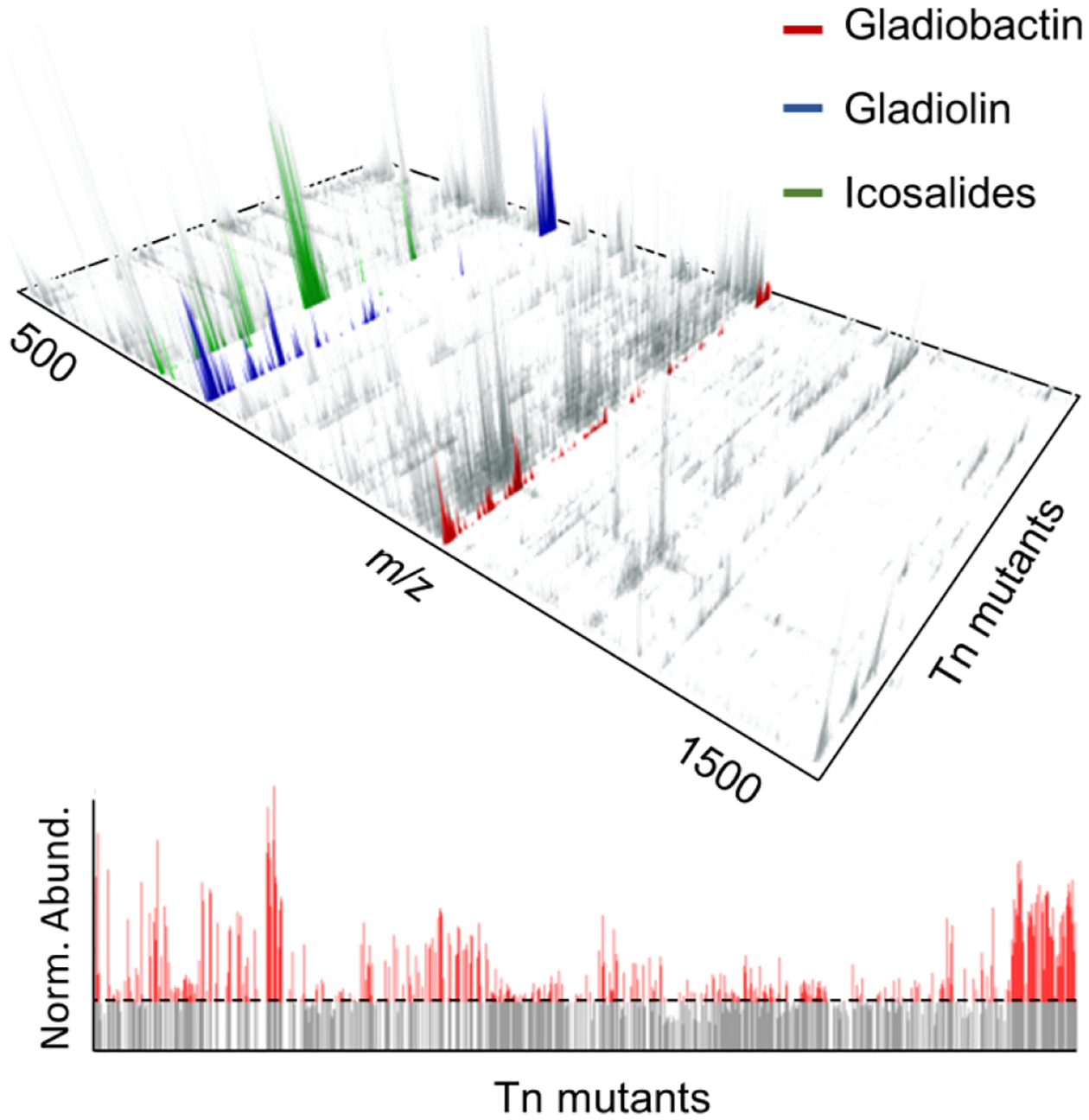
Analysis of B. gladioli Tn mutants using imaging mass spectrometry. (Top) 3D-Map of the global secondary metabolome of B. gladioli detected by LAESI-MS. The map shows the m/z and intensity of detected metabolites as a function of 960 Tn mutants. It was generated by analyzing the metabolome of each Tn mutant with LAESI-MS and extracting all ions detected for each Tn mutant with the appropriate software (see SI methods). Two-fold of the intensity observed in the wt sample was subtracted from the corresponding metabolite in the Tn mutant to give the 3D difference plot. Note that ions with intensities lower than 2-fold that of the wt are not shown. Peaks corresponding to gladiolin, icosalide, and a compound investigated further, gladiobactin, are highlighted. (Bottom) 2D-Slice out of the 3D plot focusing on gladiobactin intensity as a function of 960 Tn mutant. The dashed line indicates the level of gladiobactin synthesis by wt B. gladioli.
MS-guided dereplication identified gladiolin and icosalide A and B in our 3D map (Figure 2, top); HPLC-Qtof-MS data collected on the supernatants of the relevant Tn mutants were consistent with these assignments (Table S5).42–46 Gladiolin belongs to the etnangien group of natural products; its production from B. gladioli was recently reported.42,43 In our 3D map, gladiolin synthesis was enhanced in ~20% of Tn mutants. Icosalides were initially identified from two fungal species;44 more recently they were reported from B. gladioli as well.45,46 Among the remaining metabolites, which could not be matched to known ones, we focused on a compound with m/z 1105.5884. A 2D slice from the 3D plot shows the production of this metabolite across the 960 Tn mutants and facilitated identification of the best overproducer (Figure 2, bottom).
Both SOM-based HPLC-MS analysis and IMS provided candidate metabolites for further investigation, which are described below. More generally, the results show that Tn mutagenesis in combination with MS-based metabolomics is an effective method of eliciting and detecting cryptic metabolites in a medium- to high-throughput manner from bacterial extracts.
Discovery of cryptic metabolites from B. plantarii.
We focused on two compounds with m/z 809.4068 and 1021.5221 to verify that they represented new cryptic metabolites. Production cultures of the Tn mutant that exhibited optimal titers for these two compounds were generated and the metabolites isolated in yields of 0.25–0.5 mg/L, leading to five variants of the first compound and a single member of the second; these have been named haereoplantin A-E and burrioplantin A, respectively. The structure of haereoplantin A was determined using multidimensional nuclear magnetic resonance (NMR) spectroscopy. Analysis of 1H, COSY, and HSQC data revealed six amide protons (Figure S2). The peptide was N-terminally acylated and consisted of six amino acids, five of these non-canonical. Together with analysis of HMBC and NOESY data, the amino acids were identified as two successive (E)-2,3-didehydrobutyrine (E-Dhb), β-hydroxyleucine (β-OH-Leu), 4-hydroxyphenylglycine (Hpg), an unusual p-aminobenzoate, followed by threonine (Thr). The sequence of the amino acids was determined by extensive HMBC and NOESY spectral analysis (Figure 3A, Table S6). The acyl group was found to be a saturated decanoyl chain. Marfey’s analysis was subsequently used to assign the absolute configuration of the α-carbons and the β-hydroxy group,47 thus completing the structure of haereoplantin A (Figure 3B, Table S7). The structure was consistent with HR-MS data ([M+H]+obs 809.4068, [M+H]+calc 809.4080, Δppm = 1.5). We similarly determined the structures of haereoplantin B-E, which varied in the N-terminal acyl group (variant B), carried D-Leu in place of β-OH-D-Leu (variant C), L-Leu in place of L-Hpg (variant D), or lacked the C-terminal L-Thr (variant E) (Figure 3C, Tables S8–S11). Haereoplantins are new members of the ‘haereo’ compound family; they are similar to haereogladins and haereoglumins, which were recently reported from Burkholderia gladioli and Burkholderia glumae, respectively, but contain a different amino acid sequence and/or different acyl group.48 Haereogladins were identified in a mushroom infection model; the haereoglumin BGC was silent in the native host and its product was characterized after heterologous expression of the BGC in E. coli.48
Figure 3.
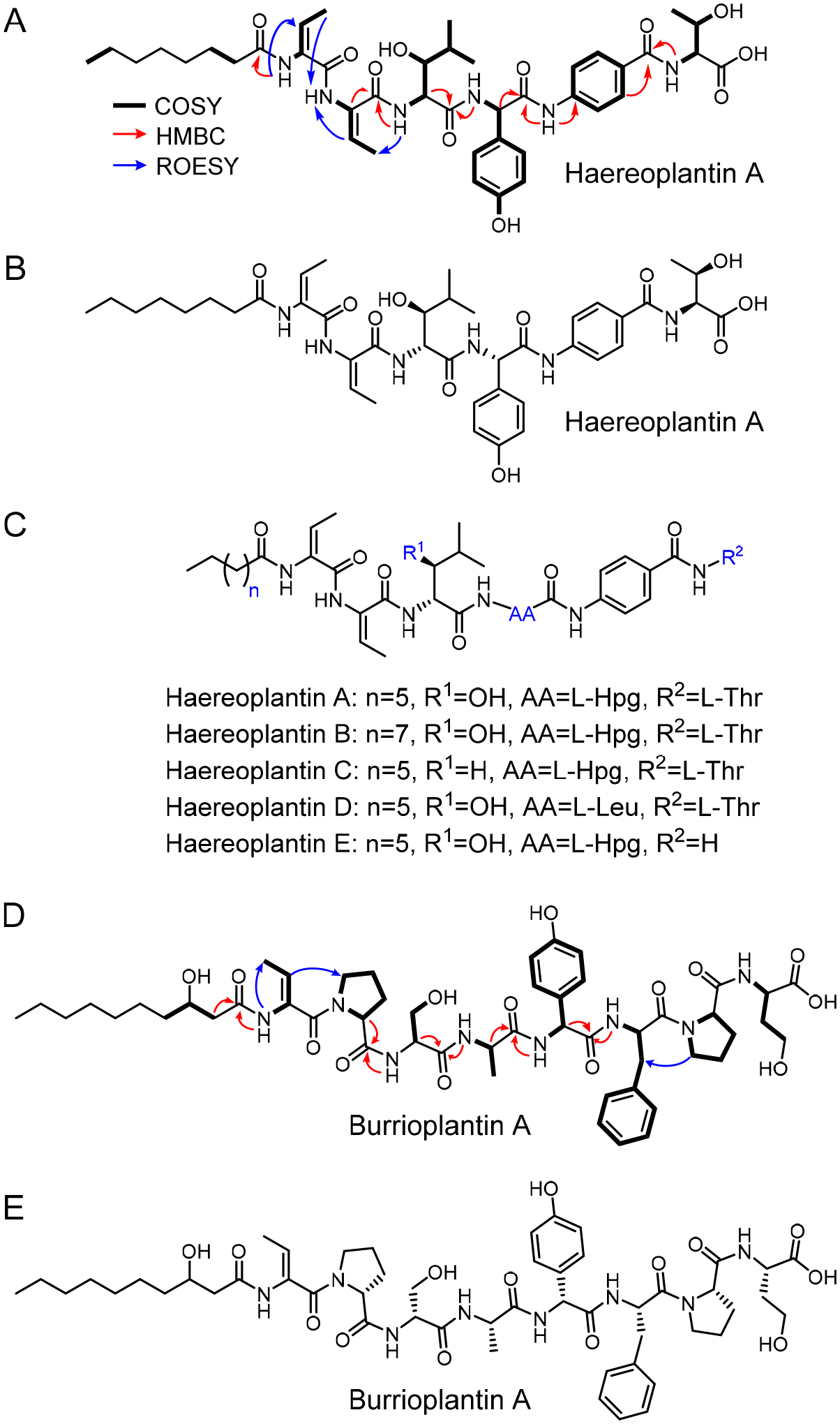
Identification of haereoplantins and burrioplantin from B. plantarii. (A) Relevant NMR correlations used to solve the structure of haereoplantin A. (B) Structure of haereoplantin A, including absolute configuration of chiral centers. (C) Structures of haereoplantin A–E. (D) Relevant NMR correlations used to solve the structure of burrioplantin A. (E) Structure of burrioplantin A, including absolute configuration of α-carbons.
The structure of burrioplantin A was similarly elucidated using NMR. It too consisted of an acylated peptide, in this case a β-hydroxydecanoyl group, with eight amino acid building blocks: Z-Dhb, proline (Pro, x2), serine (Ser), alanine (Ala), Hpg, phenylalanine (Phe), and homoserine (Figure 3D, Figure S3). The identity and sequence of these was determined via NMR and the absolute configurations by Marfey’s analysis to complete the structure, a linear acylated octapeptide with several non-canonical amino acids (Figure 3E, Tables S7, S12). HR-MS data gave a molecular formula of C51H72N8O14 ([M+H]+obs 1021.5221, [M+H]+calc 1021.5241, Δppm =2.0) and was consistent with the proposed structure. Burrioplantin A is a new relative of burrioglumin A, previously identified from B. glumae; Hpg and homoserine in burrioplantin are substituted by valine (Val) and Thr, respectively in burrioglumin.48
Identification of hpt and bpt BGCs.
Both haereoplantins and burrioplantin A appear to be products of non-ribosomal peptide synthetase (NRPS) gene clusters. Inspection of the genome of B. plantarii revealed two NRPS BGCs with adenylation domain specificities that approximately matched the sequences of haereoplantins and burrioplantin A. They are homologous to the corresponding haereoglumin and burrioglumin BGCs in B. glumae. We verified the involvement of these BGCs, which we have name hpt (haereoplantins) and bpt (burrioplantin A, Figure 4A, B), by generating a gene inactivation mutant of the large NRPS within each gene cluster. Specifically, hptC and bptE were replaced with a tetracycline resistance marker (tet) in the genome of the Tn mutant that led to enhanced haereoplantin/burrioplantin A production. Analysis of the hptC::tet strain showed that production of haereoplantin A was abolished (Figure 4C); burrioplantin A could still be observed in this mutant. Likewise, analysis of the extracts of the bptE::tet Tn mutant showed that burrioplantin A synthesis was abrogated, whereas haereoplantins were still detected (Figure 4C). A biosynthetic model for these compounds can be proposed largely based on previous work with haereoglumin and burrioglumin (Figure S4, Tables S13, S14).48 Speculatively, the co-regulation of the hpt and bpt BGCs can be rationalized on grounds of the biological activity determined for these compounds, namely as biosurfactants produced simultaneously to enable motility to new host niches. In sum, six new metabolites could be identified from B. plantarii using Tn mutagenesis in conjunction with HPLC-MS and SOM analytics. Numerous additional compounds remain to be explored (Table S4), thus providing motivation for further downstream investigations of the Tn mutant library.
Figure 4.
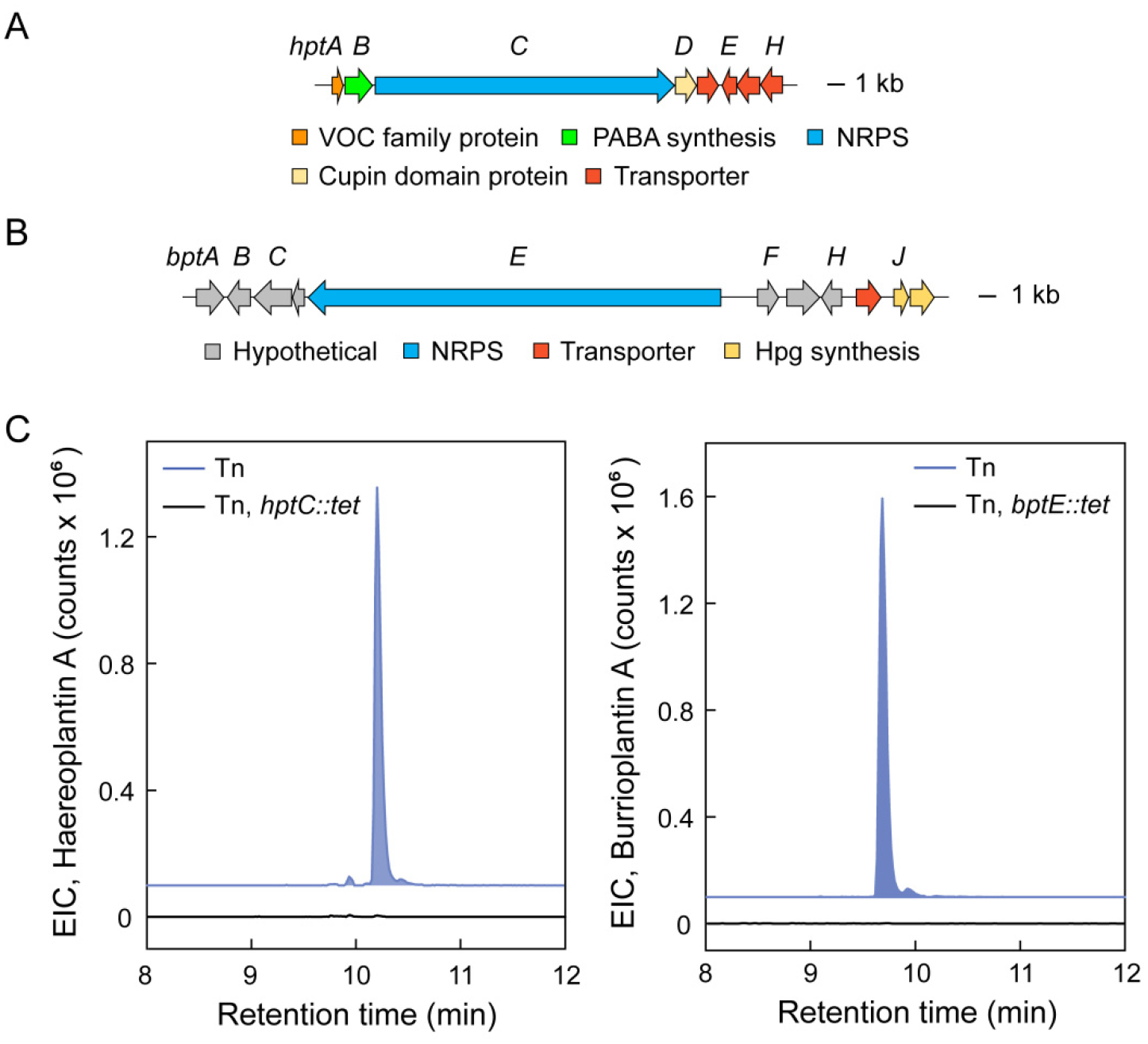
Validation of the haereoplantin and burrioplantin BGCs. (A, B) The hpt and bpt biosynthetic loci. Genes are color coded as indicated. (C) (Left) Extracted ion chromatogram for haereoplantin A in the overproducing Tn mutant (blue trace) as well as the overproducing Tn mutant in which hptC is replaced with tet by insertional mutagenesis (black trace). (Right) Extracted ion chromatogram for burrioplantin A in the overproducing Tn mutant (blue trace) as well as the overproducing Tn mutant in which bptE is removed by insertional mutagenesis (black trace).
Discovery of a cryptic metabolite from B. gladioli 3D maps.
We focused on the compound with m/z 1105.5884, termed gladiobactin A, to verify that it represented a new cryptic metabolite. The Tn mutant overproducer was used to prepare large-scale cultures and gladiobactin was subsequently isolated in ~3 mg yield (from 4 L). Analysis of 1D/2D NMR data allowed us to solve the structure of gladiobactin A. 1H, COSY and TOCSY data revealed eight amino acids, which we identified as Ser (x2), β-hydroxyaspartate (β-OH-Asp), modified ornithines (Orn, x3), lysine (Lys), and glycine (Gly). NOESY and HMBC data showed that the Lys, Gly and the modified Orn residues form a macrocycle, with the key macrolactam bond generated through the amino group of an Orn side-chain and the carboxylic acid of another Orn residue (Figure 5A, Figure S5). NOESY and HMBC data provided the sequence of amino acids within this macrocycle. They also showed that the two remaining Orn carried an unusual modification, which we identified as diazonium diolates, as identified by both 1H and 13C NMR chemical shift patterns. The occurrence of this modification of Orn, an amino acid referred to as graminine, has recently been elegantly demonstrated by the Hertweck group.49,50 HR-MS/MS data were consistent with the presence of two graminine residues, as they showed the characteristic loss of NO during MS fragmentation (Figure S6). Analysis of NMR data also facilitated structural elucidation of the linear portion of gladiobactin, consisting of a saturated dodecanoyl group carried on a β-OH-Asp-Ser-Ser tripeptide, the latter residue linked to the macrocyclic Orn. Absolute configurations were subsequently resolved by Marfey’s analysis, thus completing the structure of gladiobactin A (Figure 5B, Tables S7, S15). Gladiobactin is a siderophore in the gramibactin class, recently identified from B. glumae. It had been detected by Hermenau et al. from B. gladioli, but sufficient material was not isolated for NMR analysis as the wt produces negligible amounts under standard growth conditions.50 Its structure was therefore suggested on the basis of HR-MS/MS data and knowledge regarding products of similar BGCs in other Burkholderia; the structure reported is identical to the one we derived from NMR and stereochemical analyses (Figure 5B). A biosynthetic pathway for gladiobactin can be proposed based on prior work with gramibactin by the Hertweck group (Figure S7, Table S16).49,50 The module and domain arrangements of the corresponding gladiobactin BGC (Figure 5C, gld) give rise to a co-linear biosynthetic pathway and agree with the absolute Cα configurations determined experimentally for gladiobactin A.
Figure 5.
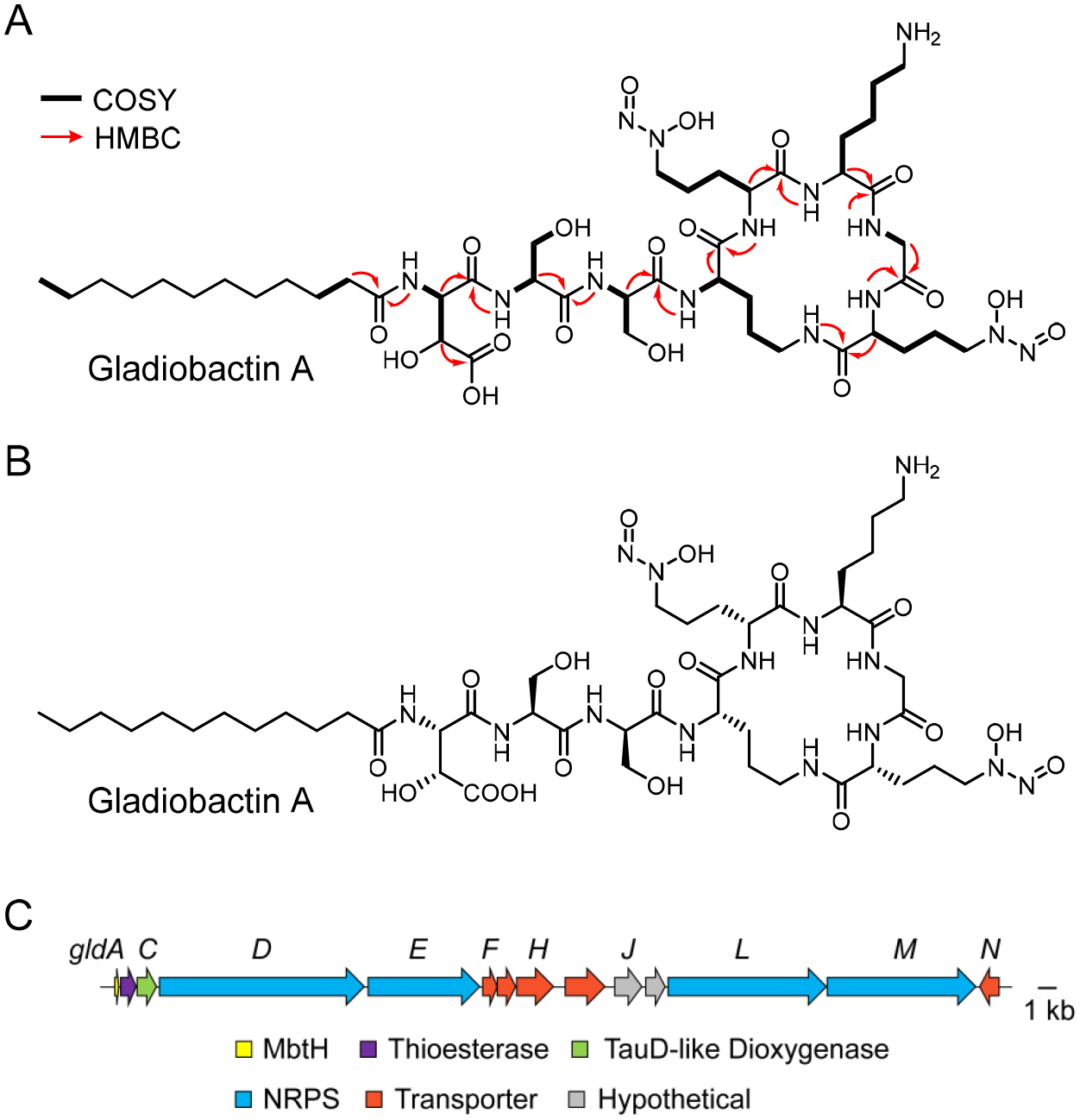
Gladiobactin structure and BGC. (A) Relevant NMR correlations used to solve the structure of gladiobactin A. (B) Structure of gladiobactin A, including absolute configuration of chiral centers. (C) Gladiobactin BGC; genes are color coded as indicated.
Identification of the sites of Tn insertion.
With several new natural products characterized, we next sought to identify the location of Tn insertion as a first step toward deciphering the mechanism by which secondary metabolite synthesis was enhanced. Using arbitrary PCR and nested primer sets, we identified Tn insertion into a superfamily II DNA/RNA helicase in the B. plantarii mutant, which synthesized elevated levels of haereoplantins and burrioplantin A (Figure 6A). Sequence analyses showed that the helicase likely belongs to the DEAD box subfamily of helicases, which are primarily involved in unwinding double-stranded RNA and generally contribute toward RNA metabolism and turnover.51 The insertion occurred on the 3’-end of the gene (between basepairs 1257 and 1258 of 1593 basepairs total) (Figure 6A). It is also possible that polar effects emanating from Tn insertion are responsible for enhanced biosynthetic activities. To differentiate between these possibilities, we knocked out the helicase in the wt background via insertion of tet. The resulting helicase::tet strain recapitulated the haereoplantin and burrioplantin overproduction phenotype, verifying that disruption of the helicase leads to enhanced synthesis of these compounds (Figure 6B). This gene has previously not been linked with the induction of silent BGCs, and further studies are necessary to elucidate how its inactivation leads to secondary metabolite synthesis.
Figure 6.
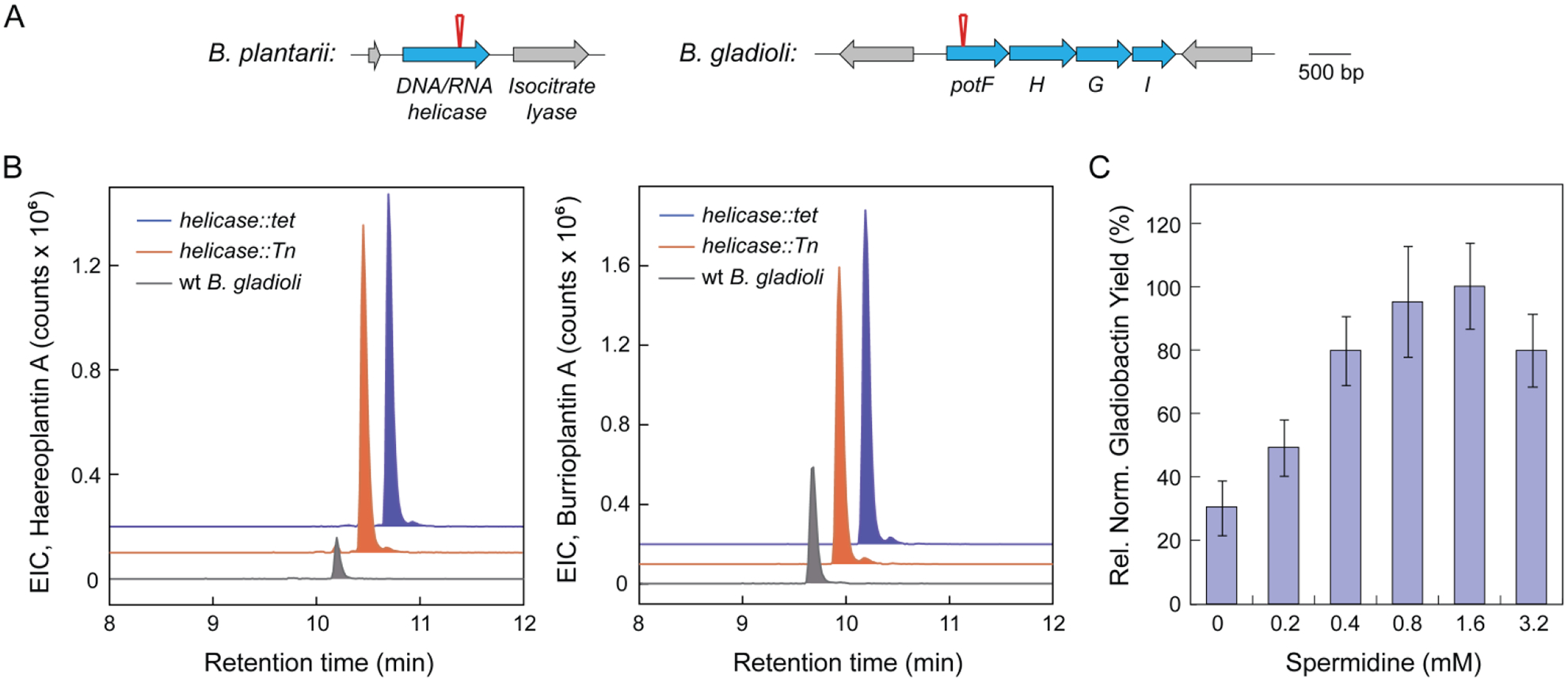
Identification of Tn insertion sites in the haereoplantin, burrioplantin, and gladiobactin overproducers. (A) Tn insertion in B. plantarii occurs in a DNA/RNA helicase, as indicated by the red pin. In B. gladioli, Tn insertion occurred into potF (red pin) in the pot operon. (B) Disruption of the DNA/RNA helicase recapitulates the effect of Tn insertion. (Left) Extracted ion chromatograms of haereoplantin A in wt B. plantarii (gray trace), the helicase::Tn mutant (red), and the helicase deletion mutant (helicase::tet, blue). (Right) Extracted ion chromatograms of burrioplantin A in wt B. plantarii (gray), Tn mutant (red), and helicase deletion mutant (blue). (C) Induction of gladiobactin synthesis by spermidine. Shown is cell density-normalized relative quantification of gladiobactin A as a function of exogenous spermidine concentrations. The relative levels of gladiobactin in each culture supernatant were determined from integration of the gladiobactin peak observed in extracted-ion chromatograms. The average from two independent measurements is reported. Error bars represent standard error.
In the best gladiobactin overproducing mutant, we observed insertion into a potF homolog, a putrescine transport protein.52,53 Tn insertion occurred between basepairs 334 and 335 (1095 basepairs total) (Figure 6A). Intracellular levels of polyamines, such as spermidine and putrescine, are regulated by several mechanisms, including transport/uptake. The pot operon is responsible for putrescine uptake, whereas the spd operon imports spermidine.52,53 Interestingly, the structures of these two transport complexes are highly analogous, with the specificity provided by a periplasmic binding protein that interacts with putrescine (PotF) or with spermidine (SpdD). We envisioned two mechanisms by which Tn insertion into potF could induce gladiobactin production: (1) the Pot complex could still be operative, but in the absence of PotF, it may instead import spermidine as the analogous membrane-spanning components may be able to interact with SpdD; (2) alternatively, the Pot complex may be entirely inactive, thereby abolishing putrescine import. In both cases, the putrescine-spermidine balance would be altered in favor of spermidine. We tested whether increased spermidine levels would induce gladiobactin production by supplementing cultures of B. gladioli with varying concentrations of the polyamine. At 1.6 mM, we observed a ~3.5-fold overproduction of gladiobactin, relative to untreated cultures (Figure 6C). Moreover, a dose-response analysis showed a parabolic dependence of gladiobactin levels on spermidine titers, with an increase in siderophore yield between 0.2 and 1.6 mM spermidine.
We therefore propose that Tn insertion into potF alters the spermidine balance inside the cell, which is a trigger for gladiobactin synthesis. Why elevated spermidine elicits this response remains to be determined. One possibility is oxidative stress from higher-than-normal concentrations of the polyamine, as this connection has previously been made in E. coli.54–58 The correlation between gladiobactin yields and spermidine in the low-concentration region may therefore indicate activation of a stress response pathway, which could in turn induce the gld BGC. Consistent with this idea, is the decrease in gladiobactin beyond 1.6 mM spermidine, an effect likely associated with toxic effects of spermidine (Figure 6C).54,55 Indeed, we determined a minimal inhibitory concentration of ~5 mM for spermidine against B. gladioli. While further work is necessary, the results suggest that spermidine exhibits hormetic properties, stimulating metabolism at low concentrations, but inhibiting growth at high titers.10,59,60 A similar phenomenon has been observed with other small molecule elicitors of silent BGCs.10,13
Conclusion
Many natural product BGCs are insufficiently expressed in the laboratory under standard growth conditions, and new approaches have therefore been needed to enhance microbial secondary metabolite output. RGMS is a forward genetic method that allows for selection of mutant strains, in which expression of silent BGCs is activated. Because the endogenous producer is used and artificial regulatory elements are avoided, RGMS is useful in investigating both the product and regulation of silent BGCs. A down-side of RGMS has been that two rounds of genetic manipulations are necessary, one for creating a reporter strain and a second for the library of mutants. In the current work, we show that next-generation mass spectrometry approaches can effectively monitor and map the secondary metabolomes of Tn mutant libraries. Therefore, the prospects of a genetics-free RGMS method are provided: UV-induced or chemical mutagenesis can generate a mutant library, which could subsequently be screened using the medium- to high-throughput metabolomics workflows described herein. This combination can be applied to any culturable bacterium, notably rare actinomycetes that are rich in natural product BGCs but difficult to manipulate genetically.3,6 With the advances in next-generation DNA sequencing, the genome sequence of the selected mutant could be determined to pinpoint the mutation(s) that lead to enhanced secondary metabolism.
Both MS-based detection methods employed in this study revealed obvious secondary metabolic responses to Tn mutagenesis, and multiple mutants were found to enhance production of a given natural product. While we did not determine the site of Tn insertion for every mutant, it is statistically very likely that Tn insertions in different loci led to activation the hpt, bpt, and gld BGCs. This notion is in line with previous work, which has shown that disruption of multiple genes can lead to enhanced expression of a given BGC. In a comprehensive study in S. coelicolor, 724 Tn insertions (from a library of 51,443 Tn mutants) were found to increase undecylprodigiosin production, a frequency of 1.4%.19 In B. thailandensis, 17 Tn insertions from a total of 960 Tn mutants were found to induce three different silent BGCs, a frequency of 1.8%.23 The 1–2% frequency from these studies, with which Tn mutants give rise to metabolically active strains, suggests that even sub-saturating Tn libraries will be sufficient for inducing silent BGCs. Moreover, as phenotypic changes are often linked and some mutations can globally enhance secondary metabolism, both SOM and IMS are ideal methods for surveying secondary metabolomes on a global level and detecting linked metabolic changes, as in the case of haereoplantin and burrioplantin.
Further assessment of the potF::Tn mutant surprisingly uncovered a link between spermidine levels and gladiobactin synthesis. Importantly, spermidine was growth-inhibitory at elevated concentrations, indicative of a stress response that may underpin enhanced gladiobactin biosynthesis. Production of siderophores to sequester toxic metals is often a tell-tale sign of the oxidative stress response, and high spermidine titers have been reported to trigger oxidative stress in E. coli.54,55 These links provide future avenues for examining the biological mechanisms that lead to increased gladiobactin biosynthesis in response to spermidine. Moreover, it will be interesting to investigate whether polyamine supplementation is a general method for inducing cryptic metabolite biosynthesis in other Burkholderia and beyond.
Supplementary Material
Acknowledgments
We are grateful to the National Institutes of Health (DP2-AI-124786 to M.R.S.) and the Princeton Catalysis Institute for funding this work. A.Y. was supported by the Uehara Memorial Foundation and the Kyoto University Foundation. C.Z. acknowledges funding from the Edward C. Taylor 3rd Year Fellowship in Chemistry. A. L. was supported by a graduate fellowship from the China Scholarship Council.
Footnotes
The Supporting Information is available free of charge at:
Detailed description of materials and methods, including transposon mutagenesis, HPLC-MS-based SOM analysis, imaging mass spectrometry-based 3D map analysis, purification and structural elucidation of cryptic metabolites, creation of site-specific B. plantarii inactivation mutants, spermidine supplementation experiments.
The authors declare no competing financial interests.
References
- 1.Newman DJ, and Cragg GM (2020) Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod 83, 770–803. [DOI] [PubMed] [Google Scholar]
- 2.Nicolaou KC, Sorensen EJ, and Winssinger N (1998) The art and science of organic and natural products synthesis. J. Chem. Educ 75, 1226–1258. [Google Scholar]
- 3.Nett M, Ikeda H, and Moore BS (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep 26, 1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensen PR, Chavarria KL, Fenical W, Moore BS, and Ziemert N (2014) Challenges and triumphs to genomics-based natural product discovery. J. Ind. Microbiol. Biotechnol 41, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, and Cheng YQ (2014) Genome-guided discovery of diverse natural products from Burkholderia sp. J. Ind. Microbiol. Biotechnol 41, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baltz RH (2017) Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol 44, 573–588. [DOI] [PubMed] [Google Scholar]
- 7.Ochi K, and Hosaka T (2013,) New strategies for drug discovery: Activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol 97, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H, Sandiford SK, and Van Wezel GP (2014) Triggers and cues that activate antibiotic production by actinomycetes. J. Ind. Microbiol. Biotechnol 41, 371–386. [DOI] [PubMed] [Google Scholar]
- 9.Rutledge PJ, and Challis GL (2015) Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol 13, 509–523. [DOI] [PubMed] [Google Scholar]
- 10.Okada BK, and Seyedsayamdost MR (2017) Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev 41, 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren H, Wang B, and Zhao H (2017) Breaking the silence: new strategies for discovering novel natural products. Curr. Opin. Biotechnol 48, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi SS, Katsuyama Y, Bai L, Deng Z, Ohnishi Y, and Kim ES (2018) Genome engineering for microbial natural product discovery. Curr. Opin. Microbiol 45, 53–60. [DOI] [PubMed] [Google Scholar]
- 13.Mao D, Okada BK, Wu Y, Xu F, and Seyedsayamdost MR (2018) Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol 45, 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Hindra, and Elliot MA (2019) Unlocking the trove of metabolic treasures: activating silent biosynthetic gene clusters in bacteria and fungi. Curr. Opin. Microbiol 51, 9–15. [DOI] [PubMed] [Google Scholar]
- 15.Li A, Mao D, Yoshimura A, Rosen PC, Martin WL, Gallant É, Wühr M, and Seyedsayamdost MR (2020) Multi-omic analyses provide links between low-dose antibiotic treatment and induction of secondary metabolism in Burkholderia thailandensis. MBio 11, e03210–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang SH, Li J, Yin H, Zheng JT, Yang X, Wang H Bin, Luo JL, Bai H, and Yang KQ (2009) Application of a double-reporter-guided mutant selection method to improve clavulanic acid production in Streptomyces clavuligerus. Metab. Eng 11, 310–318. [DOI] [PubMed] [Google Scholar]
- 17.Guo F, Xiang S, Li L, Wang B, Rajasärkkä J, Gröndahl-Yli-Hannuksela K, Ai G, Metsä-Ketelä M, and Yang K (2015) Targeted activation of silent natural product biosynthesis pathways by reporter-guided mutant selection. Metab. Eng 28, 134–142. [DOI] [PubMed] [Google Scholar]
- 18.Baral B, Akhgari A, and Metsä-Ketelä M (2018) Activation of microbial secondary metabolic pathways: Avenues and challenges. Synth. Syst. Biotechnol 3, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z, Wang Y, Chater KF, Ou HY, Xu HH, Deng Z, and Tao M (2017) Large-scale transposition mutagenesis of Streptomyces coelicolor identifies hundreds of genes influencing antibiotic biosynthesis. Appl. Environ. Microbiol 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed Y, Rebets Y, Tokovenko B, Brötz E, and Luzhetskyy A (2017) Identification of butenolide regulatory system controlling secondary metabolism in Streptomyces albus J1074. Sci. Rep 7, 9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brett PJ, DeShazer D, and Woods DE (1998) Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol 48, 317–320. [DOI] [PubMed] [Google Scholar]
- 22.Park J-D, Moon K, Miller C, Rose J, Xu F, Ebmeier CC, Jacobsen JR, Mao D, Old WM, DeShazer D, and Seyedsayamdost MR (2019) Thailandenes, Cryptic Polyene Natural Products Isolated from Burkholderia thailandensis Using Phenotype-Guided Transposon Mutagenesis. ACS Chem. Biol 15, 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao D, Yoshimura A, Wang R, and Seyedsayamdost MR (2020) Reporter-Guided Transposon Mutant Selection for Activation of Silent Gene Clusters in Burkholderia thailandensis. ChemBioChem, In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trottmann F, Franke J, Richter I, Ishida K, Cyrulies M, Dahse HM, Regestein L, and Hertweck C (2019) Cyclopropanol Warhead in Malleicyprol Confers Virulence of Human- and Animal-Pathogenic Burkholderia Species. Angew. Chem. Int. Ed 58, 14129–14133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke J, Ishida K, and Hertweck C (2012) Genomics-Driven Discovery of Burkholderic Acid, a Noncanonical, Cryptic Polyketide from Human Pathogenic Burkholderia Species. Angew. Chem. Int. Ed 124, 11779–11783. [DOI] [PubMed] [Google Scholar]
- 26.Biggins JB, Ternei MA, and Brady SF (2012) Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J. Am. Chem. Soc 134, 13192–13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodwin CR, Sherrod SD, Marasco CC, Bachmann BO, Schramm-Sapyta N, Wikswo JP, and McLean JA (2014) Phenotypic mapping of metabolic profiles using self-organizing maps of high-dimensional mass spectrometry data. Anal. Chem 86, 6563–6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodwin CR, Covington BC, Derewacz DK, McNees CR, Wikswo JP, McLean JA, and Bachmann BO (2015) Structuring microbial metabolic responses to multiplexed stimuli via self-organizing metabolomics maps. Chem. Biol 22, 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derewacz DK, Covington BC, McLean JA, and Bachmann BO (2015) Mapping Microbial Response Metabolomes for Induced Natural Product Discovery. ACS Chem. Biol 10, 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu F, Wu Y, Zhang C, Davis KM, Moon K, Bushin LB, and Seyedsayamdost MR (2019) A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat. Chem. Biol 15, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watrous JD, and Dorrestein PC (2011) Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol 9, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esquenazi E, Yang YL, Watrous J, Gerwick WH, and Dorrestein PC (2009) Imaging mass spectrometry of natural products. Nat. Prod. Rep 26, 1521–1534. [DOI] [PubMed] [Google Scholar]
- 33.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, De Los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, and Medema MH (2017) AntiSMASH 4.0 - improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goryshin IY, and Reznikoff WS (1998) Tn5 in vitro transposition. J. Biol. Chem 273, 7367–7374. [DOI] [PubMed] [Google Scholar]
- 35.Somprasong N, McMillan I, Karkhoff-Schweizer RR, Mongkolsuk S, and Schweizer HP (2010) Methods for genetic manipulation of Burkholderia gladioli pathovar cocovenenans. BMC Res. Notes 3, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vial L, Lépine F, Milot S, Groleau M-C, Dikempe V, Woods DE, and Déziel E (2008) Burkholderia pseudomallei, B. thailandensis, and B. ambifaria produce 4-hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum sensing regulation. J. Bacteriol 190, 5339–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada BK, Wu Y, Mao D, Bushin LB, and Seyedsayamdost MR (2016) Mapping the trimethoprim-induced secondary metabolome of Burkholderia thailandensis. ACS Chem. Biol 11, 2124–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, and Seyedsayamdost MR (2017) Synergy and target promiscuity drive structural divergence in bacterial alkylquinoline biosynthesis. Cell Chem. Biol 24, 1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemes P, and Vertes A (2007) Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal. Chem 79, 8098–8106. [DOI] [PubMed] [Google Scholar]
- 40.Fincher JA, Korte AR, Reschke B, Morris NJ, Powell MJ, and Vertes A (2017) Enhanced sensitivity and metabolite coverage with remote laser ablation electrospray ionization-mass spectrometry aided by coaxial plume and gas dynamics. Analyst 142, 3157–3164. [DOI] [PubMed] [Google Scholar]
- 41.Li H, and Vertes A (2017) Solvent gradient electrospray for laser ablation electrospray ionization mass spectrometry. Analyst 142, 2921–2927. [DOI] [PubMed] [Google Scholar]
- 42.Song L, Jenner M, Masschelein J, Jones C, Bull MJ, Harris SR, Hartkoorn RC, Vocat A, Romero-Canelon I, Coupland P, Webster G, Dunn M, Weiser R, Paisey C, Cole ST, Parkhill J, Mahenthiralingam E, and Challis GL (2017) Discovery and Biosynthesis of Gladiolin: A Burkholderia gladioli Antibiotic with Promising Activity against Mycobacterium tuberculosis. J. Am. Chem. Soc 139, 7974–7981. [DOI] [PubMed] [Google Scholar]
- 43.Irschik H, Schummer D, Höfle G, Reichenbach H, Steinmetz H, and Jansen R (2007) Etnangien, a macrolide-polyene antibiotic from Sorangium cellulosum that inhibits nucleic acid polymerases. J. Nat. Prod 70, 1060–1063. [DOI] [PubMed] [Google Scholar]
- 44.Boros C, Smith CJ, Vasina Y, Che Y, Dix AB, Darveaux B, and Pearce C (2006) Isolation and identification of the icosalides - Cyclic peptolides with selective antibiotic and cytotoxic activities. J. Antibiot. (Tokyo) 59, 486–494. [DOI] [PubMed] [Google Scholar]
- 45.Dose B, Niehs SP, Scherlach K, Flórez LV, Kaltenpoth M, and Hertweck C (2018) Unexpected Bacterial Origin of the Antibiotic Icosalide: Two-Tailed Depsipeptide Assembly in Multifarious Burkholderia Symbionts. ACS Chem. Biol 13, 2414–2420. [DOI] [PubMed] [Google Scholar]
- 46.Jenner M, Jian X, Dashti Y, Masschelein J, Hobson C, Roberts DM, Jones C, Harris S, Parkhill J, Raja HA, Oberlies NH, Pearce CJ, Mahenthiralingam E, and Challis GL (2019) An unusual: Burkholderia gladioli double chain-initiating nonribosomal peptide synthetase assembles “fungal” icosalide antibiotics. Chem. Sci 10, 5489–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marfey P (1984) Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun 49, 591–596. [Google Scholar]
- 48.Thongkongkaew T, Ding W, Bratovanov E, Oueis E, Garcĺa-Altares M, Zaburannyi N, Harmrolfs K, Zhang Y, Scherlach K, Müller R, and Hertweck C (2018) Two Types of Threonine-Tagged Lipopeptides Synergize in Host Colonization by Pathogenic Burkholderia Species. ACS Chem. Biol 13, 1370–1379. [DOI] [PubMed] [Google Scholar]
- 49.Hermenau R, Ishida K, Gama S, Hoffmann B, Pfeifer-Leeg M, Plass W, Mohr JF, Wichard T, Saluz HP, and Hertweck C (2018) Gramibactin is a bacterial siderophore with a diazeniumdiolate ligand system. Nat. Chem. Biol 14, 841–843. [DOI] [PubMed] [Google Scholar]
- 50.Hermenau R, Mehl JL, Ishida K, Dose B, Pidot SJ, Stinear TP, and Hertweck C (2019) Genomics-Driven Discovery of NO-Donating Diazeniumdiolate Siderophores in Diverse Plant-Associated Bacteria. Angew. Chem. Int. Ed 58, 13024–13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byrd AK, and Raney KD (2012) Superfamily 2 helicases. Front. Biosci 17, 2070–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Igarashi K, and Kashiwagi K (1999) Polyamine transport in bacteria and yeast. Biochem. J 344, 633–642. [PMC free article] [PubMed] [Google Scholar]
- 53.Igarashi K, Ito K, and Kashiwagi K (2001) Polyamine uptake systems in Escherichia coli. Res. Microbiol 152, 271–278. [DOI] [PubMed] [Google Scholar]
- 54.Stewart TM, Dunston TT, Woster PM, and Casero RA (2018) Polyamine catabolism and oxidative damage. J. Biol. Chem 293, 18736–18745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tkachenko AG, and Nesterova L. Yu. (2003) Polyamines as modulators of gene expression under oxidative stress in Escherichia coli. Biochemistry (Moscow) 68, 850–856. [DOI] [PubMed] [Google Scholar]
- 56.Lae Jung I, and Gyu Kim I (2003) Transcription of ahpC, katG, and katE genes in Escherichia coli is regulated by polyamines: polyamine-deficient mutant sensitive to H2O2-induced oxidative damage. Biochem. Biophys. Res. Comm 301, 915–922. [DOI] [PubMed] [Google Scholar]
- 57.Chagneau CV, Garcie C, Bossuet-Greif N, Tronnet S, Brachmann AO, Piel J, Nougayrède J-P, Martin P, and Oswald E (2019) The Polyamine Spermidine Modulates the Production of the Bacterial Genotoxin Colibactin. mSphere 4, e00414–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peralta DR, Adler C, Corbalán NS, Paz Garciá EC, Pomares MF, and Vincent PA (2016) Enterobactin as part of the oxidative stress response repertoire. PLoS One 11, e0157799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies J, Spiegelman GB, and Yim G (2006) The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol 9, 445–453. [DOI] [PubMed] [Google Scholar]
- 60.Romero D, Traxler MF, López D, and Kolter R (2011) Antibiotics as signal molecules. Chem. Rev 111, 5492–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


