Abstract
Protective associations of fruits, vegetables, and fiber intake with colorectal cancer (CRC) risk have been shown in many, but not all epidemiological studies. One possible reason for study heterogeneity is that dietary factors may have distinct effects by CRC molecular subtypes. Here we investigate the association of fruit, vegetables, and fiber intake with four well-established CRC molecular subtypes separately and in combination. Nine observational studies including 9,592 cases with molecular subtypes for microsatellite instability (MSI), CpG island methylator phenotype (CIMP), and somatic mutations in BRAF and KRAS genes, and 7,869 controls were analyzed. Both case-only logistic regression analyses and polytomous logistic regression analyses (with one control set and multiple case groups) were used. Higher fruit intake was associated with a trend towards decreased risk of BRAF-mutated tumors [Odds ratio 4th vs. 1st quartile = 0.82 (95% confidence interval = 0.65-1.04)] but not BRAF-wildtype tumors [1.09 (0.97-1.22); P-difference as shown in case-only analysis = 0.02]. This difference was observed in case-control studies and not in cohort studies. Compared with controls, higher fiber intake showed negative association with CRC risk for cases with microsatellite stable (MSS)/MSI-low, CIMP-negative, BRAF-wildtype, and KRAS-wildtype tumors (Ptrend range from 0.03 to 3.4e-03), which is consistent with the traditional adenoma-CRC pathway. These negative associations were stronger compared with MSI-high, CIMP-positive, BRAF-mutated, or KRAS-mutated tumors, but the differences were not statistically significant. These inverse associations for fruit and fiber intake may explain, in part, inconsistent findings between fruit or fiber intake and CRC risk that have previously been reported.
Keywords: Fruit, Vegetables, Fiber, Colorectal cancer risk, Pooled analysis
INTRODUCTION
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide, and the number of cases is predicted to increase to 2.2 million new cases per year by 2030 (1). To date, various dietary factors have been investigated in relation to CRC risk in many epidemiological studies. Meta-analyses report that higher intake of fruit (2), vegetables (2), and fiber (3) is associated with decreased CRC risk. However, the World Cancer Research Fund and the American Institute for Cancer Research (WCRF/AICR) concluded that there is no “convincing strong evidence” for these dietary factors. High intake of foods containing dietary fiber only have “probable strong evidence” for decreasing the risk of CRC, and low intake of fruit and vegetables only have “limited-suggestive evidence” for increasing risk (https://www.wcrf.org/dietandcancer/colorectal-cancer) (4) because these preventive associations have been shown in many, but not all epidemiological studies (5–7). One possible reason for study heterogeneity is differential effects of diet on distinct CRC molecular subtypes.
Four molecular markers in particular have been well-studied with regard to CRC heterogeneity: microsatellite instability (MSI), the CpG island methylator phenotype (CIMP), and oncogenic mutations in BRAF and KRAS genes (8). Increasing evidence showed that some lifestyle risk factors can be differentially associated with these molecular markers (9). Probably the evidence is currently strongest for smoking which is more strongly associated with MSI-high, CIMP-positive and BRAF-mutated tumors (10,11). Furthermore, there is some evidence that body mass index and hormone replacement therapy are associated with MSI status (12). These findings provide a strong rationale to investigate if dietary risk factors may also be differentially associated with CRC molecular markers. Although several epidemiological studies have evaluated the association of fruit (13–19), vegetables (13–20), and fiber (13,16–22) intake with risk of CRC molecular subtypes, results from these studies have been inconsistent. Additionally, there has been no study to this point that has investigated all four characteristic molecular markers together despite the fact that these can point to differential pathways. Specifically, three different pathways to CRC development may be affected by the combination of molecular subtypes: a) a serrated pathway, b) an alternate pathway, and c) a traditional pathway (23,24). Only one meta-analysis reported the association of fiber intake with MSI status of CRC (12). However, this meta-analysis of three case-control studies used heterogeneous fiber definitions across studies (12). The direction of the association between fiber intake and MSI status in each study was different because one study found higher fiber intake was associated with decreased risk of MSS tumors (13), but another study reported decreased risk of both MSI-high and MSS/MSI-low tumors (16). On the contrary, one study found increased risk of MSS/MSI-low tumors with low fiber intake (17). To better address the hypothesis that these dietary factors may impact risk of CRC molecular subtypes differently, here we pooled 9 population-based studies with individual-level data harmonized in a consistent manner.
We analyzed data from 9,592 CRC cases and 7,869 controls within the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) and the Colon Cancer Family Registry (CCFR) to assess the association of fruit, vegetables, and fiber intake with CRC risk by molecular subtypes using data on MSI, CIMP, BRAF mutation, and KRAS mutation status. Identification of associations with specific molecular subtypes may point to specific diet-CRC associations and help inform underlying biological mechanisms relevant to CRC risk.
MATERIALS AND METHODS
Study Participants
This study population consisted of 8,783 CRC cases and 7,869 controls from 9 observational studies within GECCO and the CCFR with available tumor marker, fruit, fiber, vegetables, and total energy intake data. Additionally, 809 population-based CRC cases from the Mayo Clinic CCFR, which did not enroll population-based controls, were included in case-only analyses. Among study participants, 3,258 CRC cases and 3,984 controls were from cohort studies, and 6,334 CRC cases and 3,885 controls were from case-control studies. Descriptive characteristics of each study and mean intake of quartile cut points of fruits, vegetables, and fiber in each study are shown in Supplementary Tables S1 and S2. Additionally, details regarding each observational study are described in Supplementary Text. All CRC cases were defined by colorectal adenocarcinoma and confirmed by pathological records, medical records, and/or death certificate information. All study participants provided written informed consent, and each study was approved by their relevant research ethics committee or institutional review board.
Definition of Tumor Subtypes
Testing for MSI, CIMP, and mutations in the BRAF and KRAS gene was conducted previously by each study and according to individual study protocols. Details regarding marker testing in each study are described in Supplementary Text. Especially, Supplementary Table S3 shows study-specific markers used to assess MSI and definition of MSI status. Additionally, Supplementary Table S4 also shows study-specific panels used to assess CIMP status. We defined marker combinations for molecular subtypes, consistent with previously suggested classifications (23,25): Types 1-10. Details of the defined marker combinations are given in Figure 1 and Supplementary Text. Subtype classifications with fewer than 50 cases were excluded from analyses. With regard to colorectal carcinogenic pathways, previous studies reported that three different pathways to CRC development may be affected by the combination of molecular subtypes: a) a serrated pathway [Type 1: MSI-high, CIMP-positive, BRAF-mutated, KRAS-wildtype and Type 2: MSS/MSI-low, CIMP-positive, BRAF-mutated, KRAS-wildtype], b) an alternate pathway [Type 3: MSS/MSI-low, CIMP-negative, BRAF-wildtype, KRAS-mutated], and c) a traditional pathway [Type 4: MSS/MSI-low, CIMP-negative, BRAF-wildtype, KRAS-wildtype and Type 5: MSI-high, CIMP-negative, BRAF-wildtype, KRAS-wildtype] (23,24).
Exposure Data
In each study, demographic and lifestyle risk factor information was assessed via in-person interviews or structured self-administered questionnaires, as described in the Supplementary Text. Dietary variables were ascertained using food frequency questionnaires (FFQ) or diet history. Data were collected at study entry, at blood draw, or one to two years prior to sample ascertainment. A multistep, iterative data harmonization procedure was applied, reconciling each study’s unique protocols and data collection instruments (26). Multiple quality-control checks were performed, and outlying values of variables were truncated to the minimum or maximum value of an established range for each variable. Variables were combined into a single dataset using common definition, standardized coding, and standardized permissible values.
In our study analysis, we selected dietary intake variables for fruit (servings/day), vegetables (servings/day), and fiber (measured as g/day). Sex- and study-specific quartiles for dietary variables were created based on the distribution in the controls. Data harmonization was performed using SAS and T-SQL.
Statistical Analyses
We used the Chi-square and Mann-Whitney t tests to compare baseline characteristics between cases and controls. To test if the association of fruit, vegetables, and fiber intake with each molecular subtype differed, we conducted case-only logistic regression analysis. We also estimated the strength of the association between the dietary variables and risk for the specific molecular subtypes using polytomous logistic regression models with two case groups and one control set. The odds ratios (ORs) and their corresponding 95% confidential intervals (CIs) for the quartiles of dietary variables were compared with the lowest intake category as a reference. P-values for linear trends were calculated by treating quartiles of intake as a continuous variable. Participants with missing values of each dietary intake for fruit (n=1,933), vegetables (n=1,775), and fiber (n=5,422) were excluded from the analyses. In our primary analyses, minimally adjusted models (minimally adjusted OR) included study site, age at diagnosis, total energy consumption (kcal/day), and sex as covariates. To determine confounding factors for multivariate adjusted models (multivariate adjusted OR), we evaluated the association of the following CRC-related parameters: tobacco smoking, alcohol, body mass index, physical activity, history of diabetes mellitus, red meat intake, processed meat intake, and aspirin/non-steroidal anti-inflammatory drugs use, with dietary factors on the risk of CRC overall. We included those factors that changed the beta estimate of the dietary factors by more than 10% when compared with the minimally adjusted models (27). Based on this we included tobacco smoking, red meat intake, and processed meat intake in the multivariate adjusted models. Missing covariates were imputed by sex-specific mean for each study for age at diagnosis (n=8 missing), total energy consumption (n=5,473 missing), tobacco smoking (n=1,375 missing), red meat intake (n=1,709 missing), and processed meat intake (n=6,565 missing). Processed meat was not imputed for NFCCR because it was missing for all subjects (n=1,045 missing). We conducted a sub-group analysis stratified by study design (case-control or cohort study) as sensitivity analyses to evaluate differences in effects between study designs.
All P-values are two-sided. A P-value <0.05 was considered statistically significant for association with CRC risk. For assessing the heterogeneous association with molecular subtypes a Bonferroni corrected P-value < 0.05/10 was considered statistically significant to account for the 10 subtypes being tested. All statistical analyses were performed using R version 3.6.0.
RESULTS
Cases were more likely to be men; younger; past or current smokers; have a higher intake of red meat, processed meat, and energy; and have a lower intake of fruits and vegetables (Table 1). In a case-control analysis of all CRC cases combined, we observed an inverse association between fiber intake and overall CRC risk [multivariate adjusted OR 4th vs 1st quartile = 0.85 (95% CI = 0.76-0.97); Ptrend = 6.2e-03], but no statistically significant association between CRC risk with fruit intake [multivariate adjusted OR 4th vs 1st quartile = 1.04 (95% CI = 0.93-1.15); Ptrend = 0.99] and vegetable intake [multivariate adjusted OR 4th vs 1st quartile = 0.92 (95% CI = 0.82-1.03); Ptrend = 0.09] (Supplementary Table S5). Among CRC cases, 15.6% were MSI-high, 18.9% CIMP-positive, 12.8% BRAF-mutated, and 32.8% KRAS-mutated.
Table 1.
Baseline characteristics of cases and controls.
| Characteristics | Casesa | Controls | P valueb |
|---|---|---|---|
| n | 9,592 | 7,869 | |
| Age, mean (SD)c | 58.6 (11.5) | 60.9 (10.3) | <2.2e-16 |
| Sex (%) | |||
| Men | 4,883 (50.9) | 3,713 (47.2) | |
| Women | 4,709 (49.1) | 4,156 (52.8) | 9.9e-07 |
| Tobacco smoking (%)d | |||
| Never smoker | 3,790 (41.8) | 3,292 (46.9) | |
| Past or current smoker | 5,273 (58.2) | 3,731 (53.1) | 1.6e-10 |
| Dietary intake | |||
| Red meat, servings/day, mean (SD)e | 0.77 (0.68) | 0.73 (0.65) | 5.0e-06 |
| processed meat, servings/day, mean (SD)f | 0.29 (0.34) | 0.25 (0.29) | 1.8e-08 |
| Total energy, kcal/day, mean (SD)g | 2118.3 (851.1) | 2024.2 (768.0) | 5.4e-07 |
| Fruits, servings/day, mean (SD)h | 1.82 (1.63) | 2.09 (1.75) | <2.2e-16 |
| Vegetables, servings/day, mean (SD)i | 2.51 (1.98) | 2.95 (2.15) | <2.2e-16 |
| Fiber, g/day, mean (SD)j | 22.9 (10.7) | 23.1 (10.4) | 0.30 |
| Location of colorectal cancer (%) | |||
| Proximal | 3.670 (38.3) | - | |
| Distal | 2,975 (31.0) | - | |
| Rectum | 2,526 (26.4) | - | |
| Missing | 421 (4.4) | - | - |
| Colorectal cancer stage (%) | |||
| I | 2,038 (21.2) | - | |
| II | 1,780 (18.6) | - | |
| III | 1,847 (19.3) | - | |
| IV | 817 (8.5) | - | |
| Missing | 3,110 (32.4) | - | - |
| Microsatellite instability (%) | |||
| High | 1,417 (15.6) | - | |
| Stable/low | 7,639 (84.4) | - | - |
| CpG island methylator phenotype (%) | |||
| High | 1,298 (18.9) | - | |
| Low | 5,569 (81.1) | - | - |
| BRAF (%) | |||
| Mutated | 1,109 (12.8) | - | |
| Wildtype | 7,566 (87.2) | - | - |
| KRAS (%) | |||
| Mutated | 2,355 (32.8) | - | |
| Wildtype | 4,831 (67.2) | - | - |
The number of cases includes 809 colorectal cancer cases from the Mayo Clinic CCFR, which is not included controls.
Based on Chi-square test or Mann-Whitney t test.
Age is missing for 4 cases and 4 controls.
Tobacco smoking is missing for 529 cases and 846 controls.
Red meat intake is missing for 743 cases and 966 controls.
Processed meat intake is missing for 4,606 cases and 1,959 controls.
Total energy intake is missing for 4,067 cases and 1,406 controls.
Fruit intake is missing for 850 cases and 1,083 controls.
Vegetables intake is missing for 731 cases and 1,044 controls.
Fiber intake is missing for 4,038 cases and 1,384 controls.
Abbreviations: SD: standard deviation.
When we analyzed the association of dietary variables with risk of each molecular CRC subtype in a case-only analysis (Table 2), we observed that fruit intake was differentially associated with BRAF-mutated compared with BRAF-wildtype tumors [multivariate adjusted OR 4th vs 1st quartile = 0.75 (95% CI = 0.60-0.94), P-difference = 0.02]. Additionally, a polytomous logistic regression analysis (Table 3) comparing BRAF-mutated and BRAF-wildtype tumors with controls showed that a trend towards inverse association between fruit intake and CRC risk was limited to BRAF-mutated tumors [multivariate adjusted OR 4th vs 1st quartile = 0.82 (95% CI = 0.65-1.04), Ptrend =0.06] as compared with controls, while there was no association between fruit intake and BRAF-wildtype tumors [multivariate adjusted OR 4th vs 1st quartile = 1.09 (95% CI = 0.97-1.22), Ptrend = 0.54] as compared with controls. In sub-group analyses, these associations were found statistically significant in case-control studies, but not in cohort studies (Table 4 and Supplementary Table S6). Fruit intake did not show a statistically significant differential association with other molecular subtypes. Neither vegetable nor fiber intake showed significant differential associations for any of the molecular markers as shown by the case-only analysis (Table 2). However, in the sub-group analyses, we found that fiber intake was differentially associated with CIMP-negative tumors as compared with CIMP-positive tumors in cohort studies (Table 4).
Table 2.
ORs and 95% CIs for the association of fruits, vegetables, and fiber intake with the risk of molecular subtypes of CRC in case-only analysis.
| Quartile | |||||
|---|---|---|---|---|---|
| Lowest (Q1) | Second (Q2) | Third (Q3) | Highest (Q4) | Ptrend | |
| Fruits (servings/day) | |||||
| MSI-high vs MSS/MSI-low | |||||
| High (n)/Stable/low cases (n) | 354/1844 | 383/2076 | 322/1701 | 238/1338 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.12 (0.94-1.33) | 0.99 (0.84-1.17) | 0.88 (0.73-1.07) | 0.14 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.14 (0.96-1.35) | 1.02 (0.86-1.21) | 0.91 (0.75-1.10) | 0.27 |
| CIMP-positive vs CIMP-negative | |||||
| High (n)/Low cases (n) | 274/1293 | 334/1559 | 274/1278 | 232/1066 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.09 (0.90-1.33) | 0.97 (0.80-1.19) | 0.95 (0.77-1.18) | 0.43 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.10 (0.91-1.34) | 1.00 (0.82-1.23) | 0.99 (0.80-1.23) | 0.73 |
| BRAF-mutated vs BRAF-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 284/1780 | 277/2161 | 254/1691 | 169/1336 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.91 (0.75-1.11) | 0.87 (0.72-1.06) | 0.71 (0.57-0.88) | 2.9e-03 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.93 (0.76-1.13) | 0.91 (0.75-1.10) | 0.75 (0.60-0.94) | 0.02 |
| KRAS-mutated vs KRAS-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 502/1093 | 703/1410 | 544/1070 | 430/853 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.16 (1.00-1.36) | 1.12 (0.96-1.30) | 1.06 (0.90-1.25) | 0.55 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.16 (0.99-1.35) | 1.11 (0.95-1.29) | 1.04 (0.88-1.22) | 0.75 |
| Vegetables (servings/day) | |||||
| MSI-high vs MSS/MSI-low | |||||
| High (n)/Stable/low cases (n) | 267/1438 | 470/2535 | 377/1976 | 204/1101 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.02 (0.86-1.21) | 1.03 (0.86-1.23) | 0.97 (0.79-1.20) | 0.87 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.03 (0.87-1.22) | 1.05 (0.87-1.25) | 0.98 (0.80-1.21) | 0.99 |
| CIMP-positive vs CIMP-negative | |||||
| High (n)/Low cases (n) | 261/1074 | 351/1715 | 294/1529 | 214/921 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.99 (0.82-1.21) | 1.02 (0.83-1.25) | 1.08 (0.86-1.35) | 0.47 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.00 (0.82-1.22) | 1.05 (0.86-1.29) | 1.10 (0.88-1.38) | 0.33 |
| BRAF-mutated vs BRAF-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 203/1442 | 362/2545 | 276/1982 | 150/1104 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.12 (0.92-1.36) | 1.08 (0.88-1.33) | 0.99 (0.78-1.26) | 0.89 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.13 (0.93-1.38) | 1.12 (0.91-1.38) | 1.01 (0.79-1.29) | 0.90 |
| KRAS-mutated vs KRAS-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 444/974 | 785/1531 | 598/1196 | 378/756 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.12 (0.97-1.30) | 1.07 (0.92-1.25) | 1.06 (0.89-1.26) | 0.68 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.12 (0.97-1.30) | 1.06 (0.91-1.24) | 1.06 (0.89-1.26) | 0.75 |
| Fiber (g/day) | |||||
| MSI-high vs MSS/MSI-low | |||||
| High (n)/Stable/low cases (n) | 199/1092 | 202/1094 | 191/1095 | 235/1101 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.98 (0.78-1.21) | 0.91 (0.72-1.14) | 1.11 (0.87-1.41) | 0.51 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.99 (0.79-1.23) | 0.94 (0.74-1.18) | 1.15 (0.90-1.49) | 0.35 |
| CIMP-positive vs CIMP-negative | |||||
| High (n)/Low cases (n) | 195/822 | 231/849 | 218/826 | 235/863 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.21 (0.96-1.52) | 1.15 (0.90-1.46) | 1.18 (0.92-1.52) | 0.30 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.23 (0.98-1.54) | 1.18 (0.93-1.50) | 1.22 (0.94-1.57) | 0.20 |
| BRAF-mutated vs BRAF-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 166/1070 | 171/1056 | 175/1049 | 183/1072 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.98 (0.77-1.24) | 0.98 (0.77-1.26) | 0.99 (0.76-1.29) | 0.97 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.99 (0.78-1.26) | 1.02 (0.80-1.31) | 1.05 (0.80-1.37) | 0.69 |
| KRAS-mutated vs KRAS-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 375/735 | 375/743 | 383/723 | 402/735 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.98 (0.82-1.17) | 1.04 (0.86-1.25) | 1.09 (0.89-1.32) | 0.34 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.97 (0.81-1.17) | 1.02 (0.84-1.23) | 1.04 (0.85-1.27) | 0.60 |
Minimally adjusted OR was adjusted for age, sex, study, and total energy (continuous).
In addition to minimally adjusted OR, multivariate adjusted OR was further adjusted for tobacco smoking (never-, past and current smoker <25, 25-<50, 50-<75, ≥75 pack-years), red meat intake (study- and sex-specific quartiles as continuous), and processed meat intake (study- and sex-specific quartiles as continuous).
Abbreviations: CI: confidence interval; OR: odds ratio.
Table 3.
ORs and 95% CIs for the association of fruits, vegetables, and fiber intake with the risk of molecular subtypes of CRC in case-control analysis.
| Quartile | ||||||
|---|---|---|---|---|---|---|
| Lowest (Q1) | Second (Q2) | Third (Q3) | Highest (Q4) | Ptrend | Pdiffa | |
| Fruits (servings/day) | ||||||
| MSI-high vs Controls | ||||||
| Cases (n)/Controls (n) | 275/1938 | 404/1736 | 297/1682 | 199/1430 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.27 (1.06-1.52) | 1.01 (0.84-1.22) | 0.85 (0.69-1.04) | 0.047 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.32 (1.10-1.58) | 1.09 (0.91-1.32) | 0.95 (0.77-1.16) | 0.40 | |
| MSS/MSI-low vs Controls | ||||||
| Cases (n)/Controls (n) | 1447/1938 | 2199/1736 | 1556/1682 | 1128/1430 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.22 (1.10-1.35) | 1.02 (0.92-1.13) | 0.98 (0.87-1.09) | 0.24 | 0.19 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.25 (1.13-1.39) | 1.07 (0.96-1.19) | 1.06 (0.94-1.18) | 0.83 | 0.37 |
| CIMP-positive vs Controls | ||||||
| Cases (n)/Controls (n) | 242/1938 | 360/1736 | 268/1682 | 205/1430 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.18 (0.97-1.42) | 0.95 (0.78-1.15) | 0.91 (0.73-1.12) | 0.14 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.21 (1.00-1.47) | 1.02 (0.84-1.25) | 1.02 (0.82-1.26) | 0.77 | |
| CIMP-negative vs Controls | ||||||
| Cases (n)/Controls (n) | 1124/1938 | 1656/1736 | 1250/1682 | 933/1430 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.26 (1.13-1.41) | 1.06 (0.95-1.19) | 0.94 (0.83-1.06) | 0.11 | 0.59 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.30 (1.16-1.46) | 1.12 (0.99-1.25) | 1.02 (0.90-1.16) | 0.82 | 0.87 |
| BRAF-mutated vs Controls | ||||||
| Cases (n)/Controls (n) | 233/1938 | 321/1736 | 235/1682 | 145/1430 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.09 (0.89-1.32) | 0.86 (0.70-1.06) | 0.72 (0.57-0.91) | 1.7e-03 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.13 (0.93-1.38) | 0.94 (0.77-1.16) | 0.82 (0.65-1.04) | 0.06 | |
| BRAF-wildtype vs Controls | ||||||
| Cases (n)/Controls (n) | 1407/1938 | 2298/1736 | 1577/1682 | 1117/1430 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.27 (1.14-1.41) | 1.05 (0.94-1.17) | 1.01 (0.90-1.13) | 0.43 | 6.5e-03 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.31 (1.18-1.45) | 1.10 (0.99-1.23) | 1.09 (0.97-1.22) | 0.54 | 0.03 |
| KRAS-mutated vs Controls | ||||||
| Cases (n)/Controls (n) | 438/1938 | 755/1736 | 548/1682 | 369/1430 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.39 (1.20-1.61) | 1.16 (0.99-1.34) | 1.04 (0.88-1.22) | 0.93 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.43 (1.23-1.66) | 1.21 (1.04-1.41) | 1.12 (0.95-1.33) | 0.38 | |
| KRAS-wildtype vs Controls | ||||||
| Cases (n)/Controls (n) | 883/1938 | 1577/1736 | 1070/1682 | 738/1430 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.31 (1.17-1.47) | 1.06 (0.94-1.20) | 1.05 (0.93-1.20) | 0.84 | 0.94 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.36 (1.21-1.53) | 1.13 (1.00-1.28) | 1.16 (1.02-1.32) | 0.18 | 0.63 |
| Vegetables (servings/day) | ||||||
| MSI-high vs Controls | ||||||
| Cases (n)/Controls (n) | 255/1772 | 441/1987 | 319/1828 | 178/1238 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.08 (0.90-1.29) | 0.98 (0.81-1.19) | 0.87 (0.70-1.09) | 0.17 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.10 (0.92-1.31) | 1.03 (0.85-1.25) | 0.91 (0.73-1.13) | 0.35 | |
| MSS/MSI-low vs Controls | ||||||
| Cases (n)/Controls (n) | 1397/1772 | 2301/1987 | 1779/1828 | 929/1238 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.03 (0.93-1.14) | 0.97 (0.87-1.08) | 0.89 (0.79-1.00) | 0.046 | 0.81 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.03 (0.93-1.15) | 0.99 (0.89-1.11) | 0.91 (0.81-1.03) | 0.13 | 0.93 |
| CIMP-positive vs Controls | ||||||
| Cases (n)/Controls (n) | 261/1772 | 380/1987 | 246/1828 | 191/1238 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.15 (0.95-1.38) | 1.00 (0.82-1.22) | 1.05 (0.84-1.31) | 0.96 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.17 (0.97-1.41) | 1.05 (0.86-1.28) | 1.09 (0.88-1.36) | 0.67 | |
| CIMP-negative vs Controls | ||||||
| Cases (n)/Controls (n) | 1060/1772 | 1711/1987 | 1451/1828 | 784/1238 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.05 (0.94-1.18) | 0.98 (0.87-1.11) | 0.90 (0.79-1.03) | 0.09 | 0.35 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.06 (0.95-1.19) | 1.00 (0.89-1.13) | 0.93 (0.81-1.06) | 0.22 | 0.26 |
| BRAF-mutated vs Controls | ||||||
| Cases (n)/Controls (n) | 207/1772 | 370/1987 | 228/1828 | 135/1238 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.17 (0.96-1.42) | 0.99 (0.80-1.22) | 0.90 (0.71-1.15) | 0.23 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.19 (0.98-1.45) | 1.04 (0.84-1.28) | 0.94 (0.74-1.20) | 0.45 | |
| BRAF-wildtype vs Controls | ||||||
| Cases (n)/Controls (n) | 1408/1772 | 2374/1987 | 1793/1828 | 914/1238 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.01 (0.91-1.12) | 0.93 (0.84-1.05) | 0.87 (0.77-0.98) | 0.01 | 0.94 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.02 (0.92-1.13) | 0.96 (0.86-1.07) | 0.89 (0.79-1.01) | 0.04 | 0.77 |
| KRAS-mutated vs Controls | ||||||
| Cases (n)/Controls (n) | 448/1772 | 812/1987 | 561/1828 | 315/1238 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.10 (0.95-1.27) | 1.01 (0.87-1.18) | 0.95 (0.80-1.13) | 0.40 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.11 (0.96-1.29) | 1.04 (0.89-1.21) | 0.98 (0.82-1.16) | 0.65 | |
| KRAS-wildtype vs Controls | ||||||
| Cases (n)/Controls (n) | 968/1772 | 1616/1987 | 1104/1828 | 611/1238 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.00 (0.89-1.13) | 0.96 (0.85-1.09) | 0.90 (0.79-1.04) | 0.12 | 0.71 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.02 (0.90-1.14) | 0.99 (0.88-1.12) | 0.93 (0.81-1.07) | 0.33 | 0.76 |
| Fiber (g/day) | ||||||
| MSI-high vs Controls | ||||||
| Cases (n)/Controls (n) | 214/1606 | 200/1621 | 184/1624 | 230/1634 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 0.88 (0.71-1.09) | 0.74 (0.59-0.92) | 0.86 (0.68-1.08) | 0.11 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 0.91 (0.73-1.12) | 0.79 (0.63-0.99) | 0.95 (0.75-1.20) | 0.47 | |
| MSS/MSI-low vs Controls | ||||||
| Cases (n)/Controls (n) | 1161/1606 | 1079/1621 | 1050/1624 | 1092/1634 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 0.88 (0.79-0.99) | 0.81 (0.72-0.91) | 0.78 (0.69-0.88) | 4.3e-05 | 0.56 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 0.90 (0.80-1.01) | 0.84 (0.74-0.94) | 0.83 (0.73-0.95) | 3.4e-03 | 0.40 |
| CIMP-positive vs Controls | ||||||
| Cases (n)/Controls (n) | 212/1606 | 226/1621 | 209/1624 | 233/1634 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 1.09 (0.88-1.34) | 0.93 (0.75-1.16) | 1.01 (0.80-1.27) | 0.69 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 1.12 (0.91-1.38) | 0.99 (0.80-1.24) | 1.11 (0.88-1.41) | 0.63 | |
| CIMP-negative vs Controls | ||||||
| Cases (n)/Controls (n) | 899/1606 | 836/1621 | 786/1624 | 840/1634 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 0.90 (0.80-1.02) | 0.79 (0.69-0.90) | 0.79 (0.69-0.91) | 1.8e-04 | 0.08 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 0.92 (0.81-1.04) | 0.82 (0.72-0.94) | 0.85 (0.74-0.98) | 9.0e-03 | 0.049 |
| BRAF-mutated vs Controls | ||||||
| Cases (n)/Controls (n) | 178/1606 | 170/1621 | 167/1624 | 181/1634 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 0.89 (0.71-1.12) | 0.79 (0.62-1.00) | 0.80 (0.62-1.03) | 0.053 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 0.92 (0.73-1.16) | 0.86 (0.67-1.09) | 0.90 (0.70-1.16) | 0.36 | |
| BRAF-wildtype vs Controls | ||||||
| Cases (n)/Controls (n) | 1139/1606 | 1038/1621 | 1012/1624 | 1059/1634 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 0.87 (0.78-0.97) | 0.80 (0.71-0.90) | 0.78 (0.68-0.88) | 4.0e-05 | 0.88 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 0.88 (0.79-0.99) | 0.83 (0.74-0.94) | 0.83 (0.73-0.95) | 3.5e-03 | 0.58 |
| KRAS-mutated vs Controls | ||||||
| Cases (n)/Controls (n) | 403/1606 | 370/1621 | 372/1624 | 391/1634 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 0.89 (0.76-1.05) | 0.84 (0.71-0.99) | 0.84 (0.71-1.01) | 0.047 | |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 0.90 (0.77-1.06) | 0.87 (0.73-1.03) | 0.89 (0.74-1.07) | 0.18 | |
| KRAS-wildtype vs Controls | ||||||
| Cases (n)/Controls (n) | 791/1606 | 733/1621 | 674/1624 | 739/1634 | ||
| Minimally adjusted OR (95%CI)b | 1.00 (Reference) | 0.91 (0.80-1.03) | 0.78 (0.68-0.89) | 0.80 (0.70-0.93) | 4.7e-04 | 0.44 |
| Multivariate adjusted OR (95%CI)c | 1.00 (Reference) | 0.93 (0.82-1.06) | 0.82 (0.72-0.94) | 0.88 (0.76-1.02) | 0.03 | 0.72 |
Pdiff test the difference of the dietary risk factor and colorectal cancer association for the two cancer subtypes, such as MSI-high vs MSS/MSI-low. This is based on the case-only analysis testing the difference in the trend across the 4 quartiles.
Minimally adjusted OR was adjusted for age, sex, study, and total energy (continuous).
In addition to minimally adjusted OR, multivariate adjusted OR was further adjusted for tobacco smoking (never-, past and current smoker <25, 25-<50, 50-<75, ≥75 pack-years), red meat intake (study- and sex-specific quartiles as continuous), and processed meat intake (study- and sex-specific quartiles as continuous).
Abbreviations: CI: confidence interval; OR: odds ratio.
Table 4.
ORs and 95% CIs for the association of fruits, vegetables, and fiber intake with the risk of molecular subtypes of CRC in case-only analysis, stratified by study design.
| Quartile | |||||
|---|---|---|---|---|---|
| Lowest (Q1) | Second (Q2) | Third (Q3) | Highest (Q4) | Ptrend | |
| Cohort studies | |||||
| Fruits (servings/day) | |||||
| MSI-high vs MSS/MSI-low | |||||
| High (n)/Stable/low cases (n) | 114/591 | 94/500 | 103/505 | 94/502 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.92 (0.67-1.25) | 0.93 (0.69-1.26) | 0.83 (0.60-1.15) | 0.31 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.92 (0.68-1.26) | 0.94 (0.69-1.27) | 0.83 (0.60-1.15) | 0.31 |
| CIMP-positive vs CIMP-negative | |||||
| High (n)/Low cases (n) | 126/575 | 129/479 | 112/512 | 119/495 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.15 (0.86-1.55) | 0.90 (0.66-1.22) | 1.04 (0.76-1.43) | 0.81 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.15 (0.85-1.54) | 0.90 (0.66-1.22) | 1.03 (0.75-1.41) | 0.74 |
| BRAF-mutated vs BRAF-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 109/586 | 86/508 | 100/510 | 90/501 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.88 (0.65-1.22) | 0.99 (0.73-1.35) | 0.96 (0.69-1.34) | 0.97 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.88 (0.64-1.22) | 0.99 (0.72-1.35) | 0.94 (0.67-1.32) | 0.89 |
| KRAS-mutated vs KRAS-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 205/457 | 219/349 | 214/366 | 194/375 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.32 (1.03-1.68) | 1.24 (0.97-1.58) | 1.05 (0.81-1.35) | 0.79 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.31 (1.03-1.66) | 1.22 (0.96-1.56) | 1.02 (0.79-1.32) | 0.93 |
| Vegetables (servings/day) | |||||
| MSI-high vs MSS/MSI-low | |||||
| High (n)/Stable/low cases (n) | 123/603 | 98/516 | 91/538 | 93/440 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.88 (0.65-1.19) | 0.78 (0.57-1.07) | 0.95 (0.69-1.32) | 0.55 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.90 (0.66-1.22) | 0.80 (0.59-1.09) | 0.96 (0.70-1.33) | 0.59 |
| CIMP-positive vs CIMP-negative | |||||
| High (n)/Low cases (n) | 146/573 | 126/505 | 105/528 | 109/454 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.13 (0.84-1.51) | 0.93 (0.68-1.26) | 1.12 (0.82-1.54) | 0.79 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.16 (0.86-1.55) | 0.94 (0.69-1.28) | 1.14 (0.83-1.57) | 0.75 |
| BRAF-mutated vs BRAF-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 114/611 | 106/501 | 86/539 | 79/454 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.24 (0.91-1.68) | 0.94 (0.68-1.29) | 1.00 (0.71-1.41) | 0.65 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.27 (0.94-1.73) | 0.96 (0.69-1.32) | 1.01 (0.71-1.42) | 0.67 |
| KRAS-mutated vs KRAS-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 215/484 | 227/348 | 219/369 | 171/345 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.33 (1.05-1.69) | 1.23 (0.97-1.57) | 1.01 (0.78-1.31) | 0.94 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.34 (1.05-1.70) | 1.23 (0.97-1.57) | 1.00 (0.77-1.30) | 0.99 |
| Fiber (g/day) | |||||
| MSI-high vs MSS/MSI-low | |||||
| High (n)/Stable/low cases (n) | 95/533 | 95/542 | 106/531 | 125/528 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.91 (0.66-1.25) | 1.01 (0.73-1.38) | 1.15 (0.84-1.58) | 0.28 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.92 (0.67-1.27) | 1.05 (0.76-1.44) | 1.19 (0.86-1.65) | 0.20 |
| CIMP-positive vs CIMP-negative | |||||
| High (n)/Low cases (n) | 112/516 | 125/525 | 137/507 | 153/519 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.12 (0.83-1.52) | 1.29 (0.95-1.76) | 1.35 (0.99-1.85) | 0.04 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.13 (0.83-1.53) | 1.33 (0.98-1.81) | 1.35 (0.98-1.85) | 0.04 |
| BRAF-mutated vs BRAF-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 95/544 | 99/535 | 105/527 | 109/536 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.06 (0.77-1.46) | 1.15 (0.83-1.58) | 1.17 (0.84-1.62) | 0.31 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.05 (0.77-1.45) | 1.17 (0.85-1.61) | 1.15 (0.82-1.61) | 0.34 |
| KRAS-mutated vs KRAS-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 200/409 | 206/399 | 200/395 | 228/404 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.03 (0.81-1.32) | 1.00 (0.78-1.29) | 1.14 (0.89-1.47) | 0.35 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.02 (0.80-1.30) | 0.98 (0.76-1.26) | 1.08 (0.84-1.40) | 0.64 |
| Case-control studies | |||||
| Fruits (servings/day) | |||||
| MSI-high vs MSS/MSI-low | |||||
| High (n)/Stable/low cases (n) | 240/1253 | 289/1576 | 219/1196 | 144/836 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.12 (0.94-1.33) | 0.99 (0.84-1.17) | 0.88 (0.73-1.07) | 0.58 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.24 (1.01-1.54) | 1.05 (0.85-1.28) | 0.93 (0.73-1.17) | 0.42 |
| CIMP-positive vs CIMP-negative | |||||
| High (n)/Low cases (n) | 148/718 | 205/1080 | 162/766 | 113/571 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.06 (0.81-1.39) | 1.03 (0.80-1.34) | 0.87 (0.65-1.17) | 0.41 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.08 (0.83-1.42) | 1.10 (0.84-1.43) | 0.96 (0.71-1.29) | 0.89 |
| BRAF-mutated vs BRAF-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 175/1194 | 191/1653 | 154/1181 | 79/835 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.92 (0.71-1.19) | 0.83 (0.65-1.06) | 0.58 (0.43-0.77) | 3.5e-04 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.94 (0.73-1.22) | 0.88 (0.69-1.12) | 0.64 (0.47-0.86) | 4.9e-04 |
| KRAS-mutated vs KRAS-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 297/636 | 484/1061 | 330/704 | 236/478 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.06 (0.87-1.29) | 1.04 (0.85-1.26) | 1.07 (0.87-1.33) | 0.58 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.05 (0.86-1.28) | 1.02 (0.84-1.24) | 1.05 (0.84-1.30) | 0.76 |
| Vegetables (servings/day) | |||||
| MSI-high vs MSS/MSI-low | |||||
| High (n)/Stable/low cases (n) | 144/835 | 372/2019 | 286/1438 | 111/661 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.12 (0.90-1.39) | 1.17 (0.93-1.46) | 0.97 (0.74-1.28) | 0.87 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.12 (0.91-1.39) | 1.19 (0.95-1.50) | 0.98 (0.75-1.30) | 0.75 |
| CIMP-positive vs CIMP-negative | |||||
| High (n)/Low cases (n) | 115/501 | 225/1210 | 189/1001 | 105/467 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.92 (0.70-1.20) | 1.07 (0.81-1.41) | 1.04 (0.75-1.42) | 0.46 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.91 (0.70-1.19) | 1.11 (0.84-1.46) | 1.06 (0.77-1.46) | 0.31 |
| BRAF-mutated vs BRAF-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 89/831 | 256/2044 | 190/1443 | 71/650 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.07 (0.82-1.40) | 1.16 (0.88-1.54) | 0.98 (0.69-1.38) | 0.77 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.07 (0.82-1.40) | 1.21 (0.91-1.60) | 1.00 (0.71-1.41) | 0.58 |
| KRAS-mutated vs KRAS-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 229/490 | 558/1183 | 379/827 | 207/411 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.01 (0.83-1.22) | 0.96 (0.78-1.18) | 1.10 (0.87-1.39) | 0.64 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.00 (0.83-1.21) | 0.95 (0.78-1.17) | 1.09 (0.86-1.38) | 0.70 |
| Fiber (g/day) | |||||
| MSI-high vs MSS/MSI-low | |||||
| High (n)/Stable/low cases (n) | 104/559 | 107/552 | 85/564 | 110/573 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.01 (0.75-1.38) | 0.77 (0.55-1.10) | 0.99 (0.68-1.45) | 0.65 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.03 (0.75-1.39) | 0.80 (0.56-1.13) | 1.02 (0.70-1.51) | 0.79 |
| CIMP-positive vs CIMP-negative | |||||
| High (n)/Low cases (n) | 83/306 | 106/324 | 81/319 | 82/344 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 1.30 (0.92-1.85) | 0.90 (0.61-1.34) | 0.87 (0.55-1.37) | 0.28 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 1.32 (0.93-1.88) | 0.95 (0.64-1.42) | 0.94 (0.60-1.50) | 0.50 |
| BRAF-mutated vs BRAF-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 71/526 | 72/521 | 70/522 | 74/536 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.85 (0.59-1.23) | 0.73 (0.49-1.10) | 0.70 (0.45-1.11) | 0.11 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.88 (0.61-1.27) | 0.80 (0.53-1.21) | 0.80 (0.50-1.28) | 0.33 |
| KRAS-mutated vs KRAS-wildtype | |||||
| Mutated (n)/Wildtype cases (n) | 175/326 | 169/344 | 183/328 | 174/331 | |
| Minimally adjusted OR (95%CI)a | 1.00 (Reference) | 0.93 (0.71-1.21) | 1.08 (0.81-1.45) | 1.03 (0.74-1.44) | 0.66 |
| Multivariate adjusted OR (95%CI)b | 1.00 (Reference) | 0.92 (0.70-1.21) | 1.07 (0.79-1.43) | 1.00 (0.71-1.41) | 0.79 |
Minimally adjusted OR was adjusted for age, sex, study, and total energy (continuous).
In addition to minimally adjusted OR, multivariate adjusted OR was further adjusted for tobacco smoking (never-, past and current smoker <25, 25-<50, 50-<75, ≥75 pack-years), red meat intake (study- and sex-specific quartiles as continuous), and processed meat intake (study- and sex-specific quartiles as continuous).
Abbreviations: CI: confidence interval; OR: odds ratio.
When we combined markers to define subtypes in polytomous logistic regression analyses (Figure 1, Supplementary Table S7), we observed a linear trend for increased fiber intake in relation to CRC risk among Type 4 tumors, MSS/MSI-low, CIMP-negative, BRAF-wildtype, and KRAS-wildtype [multivariate adjusted OR = 0.94 (95% CI = 0.88-0.99), Ptrend = 0.03)] compared with controls, although the difference between Type 4 and each of the other Types was not statistically significant. This finding was consistent with results in the single marker analyses (Table 3), where the same trend was seen for MSS/MSI-low tumors (Ptrend = 3.4e-03), CIMP-negative tumors (Ptrend =9.0e-03), BRAF-wildtype tumors (Ptrend = 3.5e-03), and KRAS-wildtype tumors (Ptrend = 0.03) compared with controls.
Figure 1. Forest plot of the association between fiber intake and colorectal cancer risk by combined molecular subtypes using polytomous logistic regression analysis.
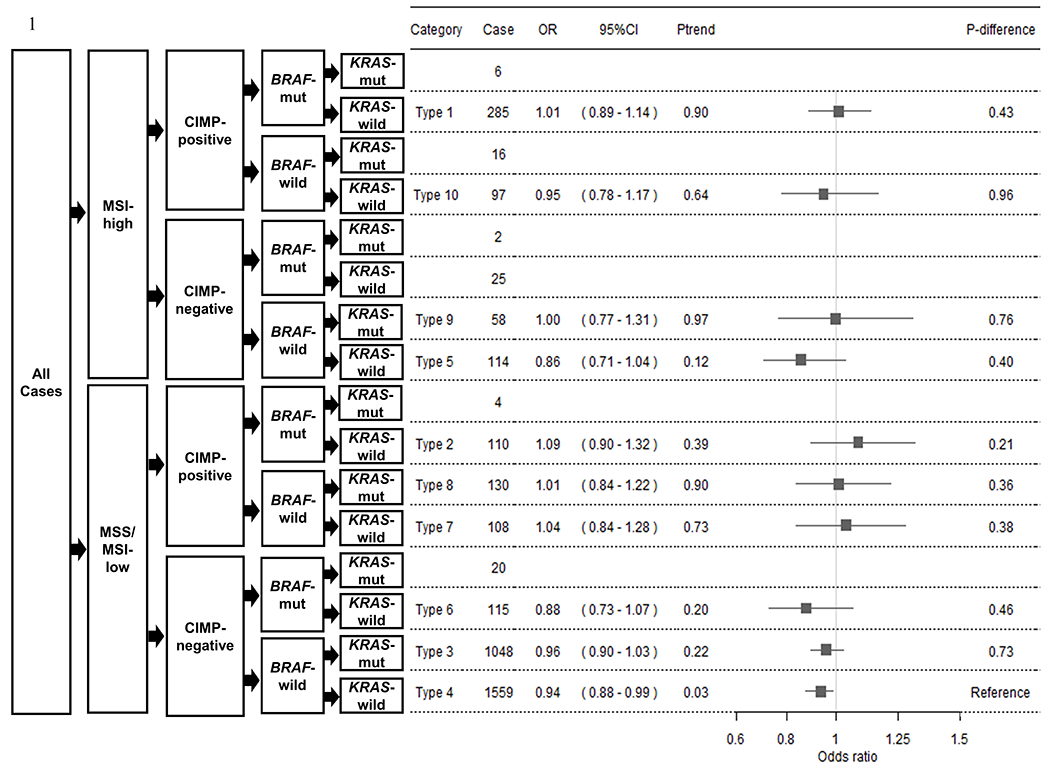
Odds ratios (ORs) and 95% confidence intervals (95% CIs) for multivariate adjusted models are presented for each increasing quartile of fiber intake and each combined molecular subtypes of colorectal cancer (CRC) risk. A total of 3,697 cases and 6,485 controls were included. Gray boxes are centered at multivariate adjusted ORs, and lines depict their 95% CIs. Subtype classifications with less than 50 cases were excluded from analyses. The number of cases per molecular subtype are listed under Cases. Ptrend was calculated by assigning ordinal values for quartile categories of fiber intake and modeling that variable continuously. P-difference is the degree of difference in P-value of multivariate adjusted OR between Type 4 and each of the other Types.
DISCUSSION
This is the largest analysis using individual level of dietary data to investigate their association with well-described molecular subtypes for CRC. While higher fruit intake was not associated with overall CRC risk, we observed that fruit intake was statistically significantly associated with a decreased risk for BRAF-mutated tumors but not BRAF-wildtype tumors. This finding was observed only among case-control studies. Additionally, we observed in single-marker polytomous logistic regression analyses that higher fiber intake was associated with a decreased risk for MSS/MSI-low, CIMP-negative, BRAF-wildtype, and KRAS-wildtype molecular subtypes compared with controls. The association in the combined-marker analysis also showed a trend towards a negative association. These negative associations were stronger than they were for MSI-high, CIMP-positive, BRAF-mutated or KRAS-mutated tumors, but the differences were not statistically significant. Increased fiber intake was differentially associated with a decreased risk of CIMP-negative compared with CIMP-positive tumors in cohort studies, but not in case-control studies. We did not identify any differences in vegetable intake with CRC risk in subtype analyses examining MSI, CIMP, and KRAS and BRAF mutations separately or in combination.
CRC development is caused by different etiological pathways underlying different genetic and epigenetic aberrations, which have been defined by specific molecular subtypes associated with distinct development trajectories. If dietary risk factors for CRC, such as fruit, vegetable, and fiber intake, impact specific etiologic pathways, then we can expect that the specific dietary factors are differentially associated with these molecular subtypes. For specific molecular subtypes, the mutation in the KRAS oncogene has been widely known as an acting driver of CRC development (28). In addition, mutation of the BRAF oncogene induces proliferation and inhibits normal apoptosis of colonic epithelial cells (29). Both KRAS and BRAF are key players of the mitogen-activated protein kinase (MAPK) pathway (24). Another driving force of CRC development is CIMP. Widespread methylation of numerous promoter CpG island loci are responsible for inactivation of tumor suppressor genes and other tumor-related genes (28). CIMP is also strongly related to the serrated pathway. Silencing of tumor suppressor genes such as p16INK4a and IGFBP7 via the synergistic effects of mutation of BRAF and CIMP resulting from hypermethylation could facilitate progression to serrated colorectal polyps (24). MSI is recognized by high-frequency of genetic alterations in repeated microsatellite sequences of DNA resulting from a DNA mismatch repair deficiency (30).
Fruit intake was associated in BRAF-mutated tumors, which is of interest given that BRAF-mutated tumors have particularly poor survival (31). Differences in findings for fruit intake by study design may be due to differences in sample size, given that approximately two-thirds of cases were from case-control studies. To evaluate study-specific differences among case-control studies, we explored consistency of the case-control study findings. The meta-analysis OR from case-controls studies was similar to the pooled estimate from all studies, and we did not observe substantial heterogeneity among the case-control studies [ORcase-control meta-analysis = 0.85 (95% CI = 0.78-0.93), P-heterogeneity = 0.52]. However, selection and recall biases, which are more likely to occur in case-control studies, may also explain observed differences due to study design. Selection bias could occur when CRC cases and/or controls are not representative of the target population. While it could be possible that controls with a healthy diet are more willing to participate in studies, this is unlikely to explain the observed differential effect of fruit intake specifically on BRAF-mutated vs BRAF-wildtype tumors in the case-only analysis. For this case-only finding to be explained by selection bias, the participation of BRAF-mutated vs BRAF-wildtype CRC cases would need to differ by fruit intake. This may be possible if survival after CRC diagnosis impacts participation and differs by BRAF mutational status and by fruit intake. However, we recently showed in a pooled analysis that BRAF-mutational status is not associated with survival after CRC diagnosis (32) and previous studies do not show strong evidence for the impact of fruit intake on survival (33,34). For recall bias to explain the case-only finding, cases with and without BRAF mutation would need to recall their fruit intake differently. If treatment with BRAF-inhibitors could have an impact is unknown; however, the majority of CRC cases included in this study were diagnosed before treatment with BRAF-inhibitors were introduced (35). As BRAF mutation are measured by standard methods, it is probably also less likely that measurement error could explain the observed difference in the case-only analysis for BRAF-mutated vs wildtype tumors.
To our knowledge, only one study has published findings on fruit intake and CRC risk by BRAF mutation status (15). This cohort study of 186 cases found no clear differences in risk. Several meta-analyses of studies not accounting for tumor markers have been published. A meta-analysis published in 2003 reported statistically significant inverse associations between fruit intake and CRC risk in case-control studies, but the relative risk was null when restricted to cohort studies (36). Additionally, a previous pooled analysis of 14 cohort studies published in 2007 also showed no overall association (37). However, a more recent meta-analysis of 19 cohort studies published in 2018 reported statistically significant inverse associations between fruit intake and CRC risk, and reported nonlinear relationships between fruit intake and CRC risk (2). It is possible as sample sizes of meta-analyses increase that the power becomes sufficient to capture a significant association in a subset of the cancers (in this case BRAF-mutated tumors). Additional studies ideally conducting in cohorts are needed to replicate our finding.
Fruit intake may plausibly play a role in inhibiting events of BRAF-mutated tumors. Fruits are rich in many nutrients and biologically active compounds, such as vitamins, carotenoids, and folic acid, which may be cancer-preventive (38). Accordingly, a previous laboratory study showed that high levels of vitamin C specifically kill BRAF-mutated CRC cells, but not BRAF wildtype cells (39). This effect is due to uptake of dehydroascorbate (DHA), which is the oxidized form of vitamin C. Increased intracellular DHA uptake accumulates cellular reactive oxygen species and inactivates glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Inhibition of GAPDH in BRAF-mutated CRC cells leads to an energetic crisis and cell death through inhibiting glycolysis and depleting ATP (39). However, two randomized, double-blind, placebo-controlled trials with a mean follow-up period ranging from 8.0 to 9.4 years did not find that 500mg of vitamin C supplementation daily was associated with a decreased CRC incidence in both men and women (40,41). Fruit intake may also impact BRAF-mutated CRC development through impacting the MAPK-pathway. It has been previously reported that Lycium barbarum fruit, also known as Chinese Wolfberry, inhibits cancer cell growth by cell cycle arrest and apoptosis through regulating the activation of MAPK-signaling pathway (42). Furthermore, previous studies have shown that fisetin, a dietary flavonoid found in apples, strawberries, kiwi and other fruits inhibits cell invasion by targeting MAPK signaling pathway (43) and reduces the anti-invasive and anti-metastatic effects of BRAF-mutated cells (44). Accordingly, fruit intake may reduce BRAF-mutated CRC through multiple nutrients and biologically active compounds.
With regard to colorectal carcinogenic pathways, we investigated three different pathways: a) a serrated pathway [Types 1 and 2], b) an alternate pathway [Type 3], and c) a traditional pathway [Types 4 and 5] (23,24). Because this colorectal carcinogenic hypothesis only shows the predominant pathways, there is overlap between them (24). We observed that higher intake of fiber was associated with a decreased risk of combined CRC subtypes that were MSS/MSI-low, CIMP-negative, BRAF-wildtype and KRAS-wildtype [Types 4 and 5: traditional pathway]. Moreover, higher fiber intake was associated with a decreased risk for CIMP-negative tumors, but not for CIMP-positive tumors in cohort studies. Higher dietary fiber intake has “probable strong evidence” for decreasing the risk of CRC as defined by WCRF/AICR (4), which is in line with our findings given that the traditional pathway is the predominate pathway for CRC development. A previous meta-analysis of 7 observational studies investigating how lifestyle exposures may increase or decrease the risk of serrated colorectal polyps presented a suggestive trend toward an inverse association between fiber intake and risk of serrated colorectal polyps, but findings were not statistically significant (45), which is not consistent with our finding. Currently functional data are missing that could explain why the association with fiber intake is restricted to a subset of molecularly-defined CRC tumors.
A strength of our pooled analysis from 9 observational studies with up to 9,592 CRC cases and 7,869 controls is that we were well powered to detect associations of fruit, vegetables, and fiber intake with risk of major CRC molecular subtypes. Due to this sizable dataset, we could further assess the association of CRC molecular subtype markers in combination. Furthermore, we used a consistent approach to harmonize all dietary variables across the studies to enable a pooled analysis of individual level data. Additionally, we have robust data on other factors including age at diagnosis, total energy consumption, and sex as covariates.
Our study has some limitations. First, it is difficult to exclude potential selection bias. While molecular subtypes are ideally available for all cases, there may be tissue retrieval biases with tissue availability potentially associated with tumor size and stage (46). However, some of the included studies previously have shown that there were no differences in age, diet, or other lifestyle characteristics between cases with and without available tumor tissue (47,48). Second, fruit, vegetables, and fiber intake are likely measured with error because they were assessed via in-person interviews or self-administered questionnaires resulting in exposure misclassification. To best account for differences between dietary assessment methods we harmonized data as study- and sex-specific quartiles rather than the continuous intake value, as it is commonly done for pooled dietary analyses (37). As exposure misclassification would likely be nondifferential, it would result in an underestimation of the association of these dietary variables with CRC risk. Third, we analyzed the CRC risk only using the lifestyle information measured at a single point in time, while lifestyle habits of study participants might change during the relevant etiologic time window. However, as such changes are not differential between cases and controls, this would have led to an attenuation of the associations. Fourth, some studies had a sizable number of missing covariate data; however, the missing covariate data were imputed by sex-specific mean for each study to ensure no samples were missed in the analysis due to missing confounders.
In summary, we found in our large pooled analysis for well-defined molecular subtypes that higher fruit intake was associated with a decreased risk of BRAF-mutated CRC but not with BRAF-wildtype CRC. Additionally, higher fiber intake may be associated with a stronger decreased risk of CRC developing by the traditional adenoma-CRC pathway that is MSS/MSI-low, CIMP-negative, BRAF-wildtype and KRAS-wildtype tumors. These results potentially explain in part the inconsistent findings between fruit or fiber intake and overall CRC risk that have previously been reported.
Supplementary Material
Significance:
These analyses by colorectal cancer molecular subtypes potentially explain the inconsistent findings between dietary fruit or fiber intake and overall colorectal cancer risk that have previously been reported.
Acknowledgements:
CCFR: The Colon CFR graciously thanks the generous contributions of their 42,505 study participants, dedication of study staff, and the financial support from the U.S. National Cancer Institute, without which this important registry would not exist. The content of this manuscript does not necessarily reflect the views or policies of the NIH or any of the collaborating centers in the CCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government, any cancer registry, or the CCFR.
CPS-II: The authors thank the CPS-II participants and Study Management Group for their invaluable contributions to this research. The authors would also like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, and cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program.
Harvard cohorts (HPFS, NHS, PHS): The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We would like to thank the participants and staff of the HPFS, NHS and PHS for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
NSHDS investigators thank the Biobank Research Unit at Umeå University, the Västerbotten Intervention Programme, the Northern Sweden MONICA study and Region Västerbotten for providing data and samples and acknowledge the contribution from Biobank Sweden, supported by the Swedish Research Council (VR 2017-00650).
Financial support:
This work was supported by the Uehara Memorial Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO): National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA164930, U01 CA137088, R01 CA059045, U01 CA164930, R21 CA191312, R01 CA201407).
Genotyping/Sequencing services were provided by the Center for Inherited Disease Research (CIDR) (X01-HG008596 and X-01-HG007585). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268201200008I. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA015704.
The Colon Cancer Family Registry (CCFR, www.coloncfr.org) was supported in part by funding from the National Cancer Institute (NCI), National Institutes of Health (NIH) (award U01 CA167551) and through U01/U24 cooperative agreements from NCI with the following CCFR centers: Australasian (CA074778 and CA097735) , Ontario (OFCCR) (CA074783), Seattle (SFCCR) (CA074794 and R01 CA076366 to PAN), and the Mayo Clinic (CA074800). Support for case ascertainment was provided in part from the Surveillance, Epidemiology, and End Results (SEER) Program, the Minnesota Cancer Surveillance System (MCSS); the Victoria Cancer Registry (Australia) and the Ontario Cancer Registry (Canada).The content of this manuscript does not necessarily reflect the views or policies of the NIH or any of the collaborating centers in the CCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government, any cancer registry, or the CCFR.
CPS-II: The American Cancer Society funds the creation, maintenance, and updating of the Cancer Prevention Study-II (CPS-II) cohort. This study was conducted with Institutional Review Board approval.
DALS: National Institutes of Health (R01 CA48998 to M. L. Slattery).
EPIC: The coordination of EPIC is financially supported by the European Commission (DGSANCO) and the International Agency for Research on Cancer. The national cohorts are supported by Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF), Deutsche Krebshilfe, Deutsches Krebsforschungszentrum and Federal Ministry of Education and Research (Germany); the Hellenic Health Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRCItaly and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); ERC-2009-AdG 232997 and Nordforsk, Nordic Centre of Excellence programme on Food, Nutrition and Health (Norway); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden).
Harvard cohorts (HPFS, NHS: HPFS is supported by the National Institutes of Health (P01 CA055075, UM1 CA167552, U01 CA167552, R01 CA137178, R01 CA151993, R35 CA197735, K07 CA190673, and P50 CA127003), NHS by the National Institutes of Health (R01 CA137178, P01 CA087969, UM1 CA186107, R01 CA151993, R35 CA197735, K07 CA190673, and P50 CA127003).
MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 509348, 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database.
NFCCR: This work was supported by an Interdisciplinary Health Research Team award from the Canadian Institutes of Health Research (CRT 43821); the National Institutes of Health, U.S. Department of Health and Human Serivces (U01 CA74783); and National Cancer Institute of Canada grants (18223 and 18226). The authors wish to acknowledge the contribution of Alexandre Belisle and the genotyping team of the McGill University and Génome Québec Innovation Centre, Montréal, Canada, for genotyping the Sequenom panel in the NFCCR samples. Funding was provided to Michael O. Woods by the Canadian Cancer Society Research Institute.
NSHDS: Swedish Research Council; Swedish Cancer Society; Cutting-Edge Research Grant and other grants from Region Västerbotten; Knut and Alice Wallenberg Foundation; Lion’s Cancer Research Foundation at Umeå University; the Cancer Research Foundation in Northern Sweden; and the Faculty of Medicine, Umeå University, Umeå, Sweden.
Conflict of Interest Statement: M.G. receives research funding from Bristol-Myers Squibb and Merck. These Funds had no role in the design, data collection, analysis, interpretation, or manuscript drafting or in the decision to submit the manuscript for publication. The remaining authors declared no conflicts of interest related to the study.
Abbreviations:
- CCFR
Colon Cancer Family Registry
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- CRC
colorectal cancer
- DHA
dehydroascorbate
- FFQ
food frequency questionnaire
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GECCO
Genetics and Epidemiology of Colorectal Cancer Consortium
- IGFBP7
insulin-like growth factor binding protein 7
- MAPK
mitogen-activated protein kinase
- MSI
microsatellite instability
- OR
odds ratio
- SD
standard deviation
- WCRF/AICR
World Cancer Research Fund and the American Institute for Cancer Research
References
- 1.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. [DOI] [PubMed] [Google Scholar]
- 2.Schwingshackl L, Schwedhelm C, Hoffmann G, Knüppel S, Laure Preterre A, Iqbal K, et al. Food groups and risk of colorectal cancer. Int J Cancer. 2018;142:1748–1758. [DOI] [PubMed] [Google Scholar]
- 3.Aune D, Chan DSM, Lau R, Vieira R, Greenwood DC, Kampman E, et al. Dietary fibre, whole grains, and risk of colorectal cancer: systematic review and dose-response meta-analysis of prospective studies. BMJ. 2011;343:d6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Cancer Research Fund/American Institute for Cancer Research. Continuous Update Project Expert Report 2018. Diet, nutrition, physical activity and colorectal cancer. [Google Scholar]
- 5.Fung TT, Hu FB, Wu K, Chiuve SE, Fuchs CS, Giovannucci E. The Mediterranean and Dietary Approaches to Stop Hypertension (DASH) diets and colorectal cancer. Am J Clin Nutr. 2010;92:1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabat GC, Shikany JM, Beresford SAA, Caan B, Neuhouser ML, Tinker LF, et al. Dietary carbohydrate, glycemic index, and glycemic load in relation to colorectal cancer risk in the Women’s Health Initiative. Cancer Causes Control. 2008;19:1291–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leenders M, Siersema PD, Overvad K, Tjønneland A, Olsen A, Boutron-Ruault M- C, et al. Subtypes of fruit and vegetables, variety in consumption and risk of colon and rectal cancer in the European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 2015;137:2705–2714. [DOI] [PubMed] [Google Scholar]
- 8.Kocarnik JM, Shiovitz S, Phipps AI. Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterol Rep (Oxf). 2015;3:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes LAE, Simons CCJM, van den Brandt PA, van Engeland M, Weijenberg MP. Lifestyle, diet, and colorectal cancer risk according to (epi)genetic instability: current evidence and future directions of molecular pathological epidemiology. Curr Colorectal Cancer Rep. 2017;13:455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amitay EL, Carr PR, Jansen L, Roth W, Alwers E, Herpel E, et al. Smoking, alcohol consumption and colorectal cancer risk by molecular pathological subtypes and pathways. Br J Cancer. 2020;122:1604–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limsui D, Vierkant RA, Tillmans LS, Wang AH, Weisenberger DJ, Laird PW, et al. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. J Natl Cancer Inst. 2010;102:1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carr PR, Alwers E, Bienert S, Weberpals J, Kloor M, Brenner H, et al. Lifestyle factors and risk of sporadic colorectal cancer by microsatellite instability status: a systematic review and meta-analyses. Ann Oncol. 2018;29:825–834. [DOI] [PubMed] [Google Scholar]
- 13.Diergaarde B, Braam H, van Muijen GNP, Ligtenberg MJL, Kok FJ, Kampman E. Dietary factors and microsatellite instability in sporadic colon carcinomas. Cancer Epidemiol Biomarkers Prev. 2003;12:1130–1136. [PubMed] [Google Scholar]
- 14.Gay LJ, Arends MJ, Mitrou PN, Bowman R, Ibrahim AE, Happerfield L, et al. MLH1 promoter methylation, diet, and lifestyle factors in mismatch repair deficient colorectal cancer patients from EPIC-Norfolk. Nutr Cancer. 2011;63:1000–1010. [DOI] [PubMed] [Google Scholar]
- 15.Naguib A, Mitrou PN, Gay LJ, Cooke JC, Luben RN, Ball RY, et al. Dietary, lifestyle and clinicopathological factors associated with BRAF and K-ras mutations arising in distinct subsets of colorectal cancers in the EPIC Norfolk study. BMC Cancer. 2010;10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satia JA, Keku T, Galanko JA, Martin C, Doctolero RT, Tajima A, et al. Diet, lifestyle, and genomic instability in the North Carolina Colon Cancer Study. Cancer Epidemiol Biomarkers Prev. 2005;14:429–436. [DOI] [PubMed] [Google Scholar]
- 17.Slattery ML, Anderson K, Curtin K, Ma KN, Schaffer D, Samowitz W. Dietary intake and microsatellite instability in colon tumors. Int J Cancer. 2001;93:601–607. [DOI] [PubMed] [Google Scholar]
- 18.Slattery ML, Curtin K, Anderson K, Ma KN, Edwards S, Leppert M, et al. Associations between dietary intake and Ki-ras mutations in colon tumors: a population-based study. Cancer Res. 2000;60:6935–6941. [PubMed] [Google Scholar]
- 19.Slattery ML, Curtin K, Wolff RK, Herrick JS, Caan BJ, Samowitz W. Diet, physical activity, and body size associations with rectal tumor mutations and epigenetic changes. Cancer Causes Control. 2010;21:1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slattery ML, Curtin K, Sweeney C, Levin TR, Potter J, Wolff RK, et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120:656–663. [DOI] [PubMed] [Google Scholar]
- 21.Bautista D, Obrador A, Moreno V, Cabeza E, Canet R, Benito E, et al. Ki-ras mutation modifies the protective effect of dietary monounsaturated fat and calcium on sporadic colorectal cancer. Cancer Epidemiol Biomarkers Prev. 1997;6:57–61. [PubMed] [Google Scholar]
- 22.He X, Wu K, Zhang X, Nishihara R, Cao Y, Fuchs CS, et al. Dietary intake of fiber, whole grains and risk of colorectal cancer: An updated analysis according to food sources, tumor location and molecular subtypes in two large US cohorts. Int J Cancer. 2019;145:3040–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. [DOI] [PubMed] [Google Scholar]
- 24.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. [DOI] [PubMed] [Google Scholar]
- 25.Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148:77–87.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutter CM, Chang-Claude J, Slattery ML, Pflugeisen BM, Lin Y, Duggan D, et al. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012;72:2036–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skelly AC, Dettori JR, Brodt ED. Assessing bias: the importance of considering confounding. Evid Based Spine Care J. 2012;3:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bae JM, Kim JH, Kang GH. Molecular subtypes of colorectal cancer and their clinicopathologic features, with an emphasis on the serrated neoplasia pathway. Arch Pathol Lab Med. 2016;140:406–412. [DOI] [PubMed] [Google Scholar]
- 29.Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol. 2011;42:1–10. [DOI] [PubMed] [Google Scholar]
- 30.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. [DOI] [PubMed] [Google Scholar]
- 31.Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014;53:852–864. [DOI] [PubMed] [Google Scholar]
- 32.Phipps AI, Alwers E, Harrison T, Banbury B, Brenner H, Campbell PT, et al. Association between molecular subtypes of colorectal tumors and patient survival, based on pooled analysis of 7 international studies. Gastroenterology. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dray X, Boutron-Ruault MC, Bertrais S, Sapinho D, Benhamiche-Bouvier AM, Faivre J. Influence of dietary factors on colorectal cancer survival. Gut. 2003;52:868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fung TT, Kashambwa R, Sato K, Chiuve SE, Fuchs CS, Wu K, et al. Post diagnosis diet quality and colorectal cancer survival in women. PLoS One. 2014;9:e115377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N Engl J Med. 2019;381:1632–1643. [DOI] [PubMed] [Google Scholar]
- 36.Riboli E, Norat T. Epidemiologic evidence of the protective effect of fruit and vegetables on cancer risk. Am J Clin Nutr. 2003;78:559S–569S. [DOI] [PubMed] [Google Scholar]
- 37.Koushik A, Hunter DJ, Spiegelman D, Beeson WL, van den Brandt PA, Buring JE, et al. Fruits, vegetables, and colon cancer risk in a pooled analysis of 14 cohort studies. J Natl Cancer Inst. 2007;99:1471–1483. [DOI] [PubMed] [Google Scholar]
- 38.Lampe JW. Health effects of vegetables and fruit: assessing mechanisms of action in human experimental studies. Am J Clin Nutr. 1999;70:475S–490S. [DOI] [PubMed] [Google Scholar]
- 39.Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaziano JM, Glynn RJ, Christen WG, Kurth T, Belanger C, MacFadyen J, et al. Vitamins E and C in the prevention of prostate and total cancer in men: the Physicians’ Health Study II randomized controlled trial. JAMA. 2009;301:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin J, Cook NR, Albert C, Zaharris E, Gaziano JM, Van Denburgh M, et al. Vitamins C and E and beta carotene supplementation and cancer risk: a randomized controlled trial. J Natl Cancer Inst. 2009;101:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong G, Liu Q, Deng Y, Dang T, Dai W, Liu T, et al. Arabinogalactan derived from Lycium barbarum fruit inhibits cancer cell growth via cell cycle arrest and apoptosis. Int J Biol Macromol. 2020;149:639–650. [DOI] [PubMed] [Google Scholar]
- 43.Pal HC, Sharma S, Strickland LR, Katiyar SK, Ballestas ME, Athar M, et al. Fisetin inhibits human melanoma cell invasion through promotion of mesenchymal to epithelial transition and by targeting MAPK and NFκB signaling pathways. PLoS One. 2014;9:e86338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pal HC, Diamond AC, Strickland LR, Kappes JC, Katiyar SK, Elmets CA, et al. Fisetin, a dietary flavonoid, augments the anti-invasive and anti-metastatic potential of sorafenib in melanoma. Oncotarget. 2016;7:1227–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailie L, Loughrey MB, Coleman HG. Lifestyle Risk Factors for Serrated Colorectal Polyps: A Systematic Review and Meta-analysis. Gastroenterology. 2017;152:92–104. [DOI] [PubMed] [Google Scholar]
- 46.Hoppin JA, Tolbert PE, Taylor JA, Schroeder JC, Holly EA. Potential for selection bias with tumor tissue retrieval in molecular epidemiology studies. Ann Epidemiol. 2002;12:1–6. [DOI] [PubMed] [Google Scholar]
- 47.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–2142. [DOI] [PubMed] [Google Scholar]
- 48.Slattery ML, Edwards SL, Palmer L, Curtin K, Morse J, Anderson K, et al. Use of archival tissue in epidemiologic studies: collection procedures and assessment of potential sources of bias. Mutat Res. 2000;432:7–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


