Abstract
The automatic identification and segmentation of edemas associated with diabetic macular edema (DME) constitutes a crucial ophthalmological issue as they provide useful information for the evaluation of the disease severity. According to clinical knowledge, the DME disorder can be categorized into three main pathological types: serous retinal detachment (SRD), cystoid macular edema (CME), and diffuse retinal thickening (DRT). The implementation of computational systems for their automatic extraction and characterization may help the clinicians in their daily clinical practice, adjusting the diagnosis and therapies and consequently the life quality of the patients. In this context, this paper proposes a fully automatic system for the identification, segmentation and characterization of the three ME types using optical coherence tomography (OCT) images. In the case of SRD and CME edemas, different approaches were implemented adapting graph cuts and active contours for their identification and precise delimitation. In the case of the DRT edemas, given their fuzzy regional appearance that requires a complex extraction process, an exhaustive analysis using a learning strategy was designed, exploiting intensity, texture, and clinical-based information. The different steps of this methodology were validated with a heterogeneous set of 262 OCT images, using the manual labeling provided by an expert clinician. In general terms, the system provided satisfactory results, reaching Dice coefficient scores of 0.8768, 0.7475, and 0.8913 for the segmentation of SRD, CME, and DRT edemas, respectively.
Keywords: Computer-aided diagnosis, Retinal imaging, Optical coherence tomography, Diabetic macular edema, Fluid segmentation
Introduction
The presence of macular edemas (ME) constitutes a retinal disease that produces, as main symptoms, a blurred or wavy central vision that could also induce a change in the color perception of the visual acuity [24]. Different medical studies reported that this eye disorder is caused by damage in the retinal microvascularity that consequently carries a problematic leak of blood within the retinal tissues. The presence of this intraretinal fluid leads to a significant deterioration of the morphology and architecture of the retinal tissues (especially regarding its thickness), reducing consequently the visual acuity of the patient. This eye fundus pathology is prevalent in advanced stages of significant retinal diseases including diabetic macular edema (DME) or age-related macular degeneration (AMD). DME is considered one of the leading causes of blindness and visual impairment among working-aged adults in industrially developed countries [5]. Although few clinical methods, such as anti-vascular endothelial growth factor (AntiVEGF) therapy, are successfully applied with patients with DME, the availability of robust and sensitive imaging biomarkers may benefit the early diagnosis, allowing consequently the prescription of more adjusted treatments [4].
Computer-aided diagnosis (CAD) systems have gained popularity over the recent years as auxiliary tools to support the clinician’s diagnosis and evaluation of many significative diseases. Many of them include, as source of information, the input of different medical imaging modalities. Actually, in ophthalmology, optical coherence tomography (OCT) has became nowadays a valuable image modality for the analysis and diagnosis of many retinal disorders, specially those that affects the retinal layers, as the presence of intraretinal MEs. This imaging technique offers an easy and direct visualization of the in vivo histopathology of the retinal tissues in a contactless and non-invasive capturing process. Given that, nowadays, it is extensively used in the clinical practice of many ophthalmological services for the diagnosis and follow-up of patients with different eye disorders.
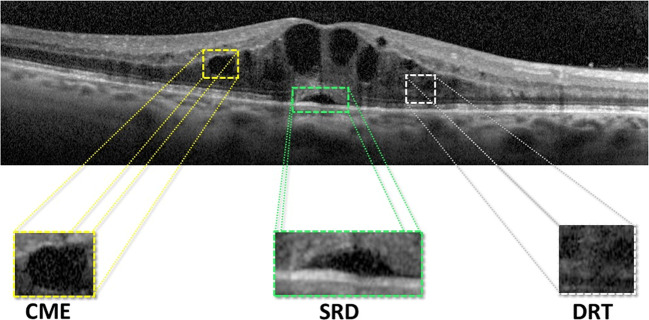
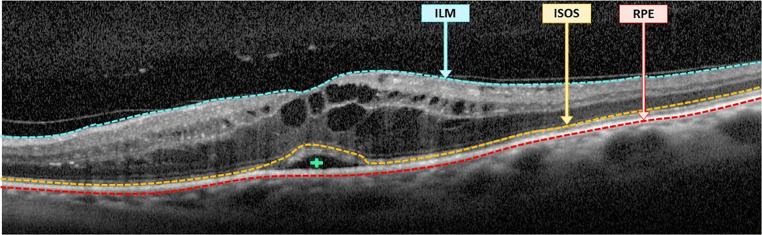
Using the OCT image modality as reference, Otani et al. [29] established a novel clinical classification that characterizes the abnormal intraretinal fluid accumulations. In particular, this clinical classification enables the identification and categorization of the MEs associated with DME into 3 main pathological types: serous retinal detachment (SRD), cystoid macular edema (CME), and diffuse retinal thickening (DRT). In the OCT images, SRD edemas are typically recognized as a “dome-shape” hyporeflective region that presents a significant contrast over the nearby tissue (the photoreceptor layer). In the case of CME edemas, they are clinically defined as a hyporeflective cystoid space surrounded by highly reflective membranes that represent the “cystoid-cavities.” Finally, DRT edemas typically proliferate in the outer retina layer with a “sponge-like” appearance. This pattern results as consequence of the absence of limiting membranes that constrain the fluid in this specific retinal region. Posteriorly, this classification was extended by Panozzo et al. [30], characterizing these 3 ME types using additional criteria in a more precise classification. In this study, the authors defined 5 clinical criteria: retinal thickness, diffusion, volume, morphology, and presence of vitreous traction [2]. Figure 1 includes the simultaneous presence of the 3 ME types in a particular OCT image, illustrating the complexity and heterogeneity of the DME retinal disorder.
Fig. 1.
OCT image with the localization of the 3 defined types of DME: SRD, CME, and DRT
Given the relevance of the disease, different works were presented over the recent years related with this issue. Part of them faced globally the identification of pathological scenarios by the general presence of fluid accumulations (addressing the cases that are typically more visible and omitting any type of characterization). As reference, in the work of Sidibé et al. [40], the authors proposed a strategy that firstly models the appearance of normal OCT images using Gaussian mixture models (GMM) to, then, detect cases with the presence of intraretinal fluid as outliers. Roy et al. [34] applied a convolutional architecture based on the RelayNet network to simultaneously segment the retinal layers as well as the fluid regions that may be present in the OCT images. Montuoro et al. [25] proposed an automatic method based on graph theory that simultaneously allows the segmentation of the retinal layers and the existing fluid regions. In particular, a probability map was used to perform the initial surface segmentation to posteriorly extract several context-based features.
Other proposals faced partially the ME disease, extracting particular pathological cases, mainly the most directly identifiable, as the case of CME edemas. In that line, Girish et al. [14] designed a methodology based on a marker-controlled watershed method to segment existing CME regions. In the works of Lee et al. [19] and Schlegl et al. [39], similar strategies were proposed to segment CME edemas using adapted CNNs. Venhuizen et al. [44] developed a deep learning method for the automatic segmentation and quantification of CME edemas using the FCNN architecture. Rashno et al. [32] focused their study on the automatic segmentation of CME regions using a neutrosophic transformation and a graph-based shortest path method. Sun et al. [43] presented a framework for the SRD edema segmentation that combined AdaBoost classification and a shape-constrained graph cut. Similarly, Lee et al. [20] proposed a CNN method for the detection and quantification of SRD cases. Novosel et al. [28] proposed a strategy based on loosely coupled level set (LCLS) to simultaneously segment the retinal layers and the pathological structures that are present in the OCT images, as drusen or SRD edemas. Ding et al. [11] used a graph cut approach with a split Bregman-based segmentation method to delimit the fluid regions. Then, a learning strategy was implemented, using as reference the previously identified dark regions to classify each candidate as a case of SRD or CME. A similar strategy was proposed by Zheng et al. [47], where the output of a split Bregman-based segmentation method is compared and validated with the manual delimitation of an expert.
Despite the existence of general or partial proposals that faced the analysis of the DME disease, at present, only the work of Samagaio et al. [35] addressed the automatic localization of the 3 clinically defined edema types of DME using OCT scans. To date, no other work faced the entire process of simultaneous localization, characterization, and also the segmentation of all the existing cases of the 3 defined DME types in OCT images.
Summarizing, the main contributions of the paper include (i) a new methodology for the simultaneous identification, characterization, and segmentation of the 3 defined types of edemas (SRD, CME, and DRT). In the case of SRD and CME edemas, different approaches were implemented adapting graph cuts and active contours for their identification and precise delimitation. In the case of the DRT edemas, given their fuzzy regional appearance that requires a complex extraction process, an exhaustive analysis using a learning strategy was designed, exploiting intensity, texture, and clinical-based information; (ii) to date, this work represents the only proposal that accurately identifies, characterizes, and segments all the cases of the ME types that are associated with DME, a significant disease of great impact at the moment; (iii) this fully automatic system provides crucial information that can facilitate the early diagnosis of a relevant disease as is DME, among the main causes of vision loss and blindness in the developed countries.
The manuscript is organized as follows: “Methodology” presents a detailed explanation about the steps that were followed for the identification and subsequent segmentation of the 3 ME types. “Results” includes the results and validation of the main designed stages of the methodology. “Discussion” exposes the discussion of the obtained results and the main challenges that were faced in this work. Finally, “Conclusions” depicts the general conclusions of the study and the possible future lines of work.
Methodology
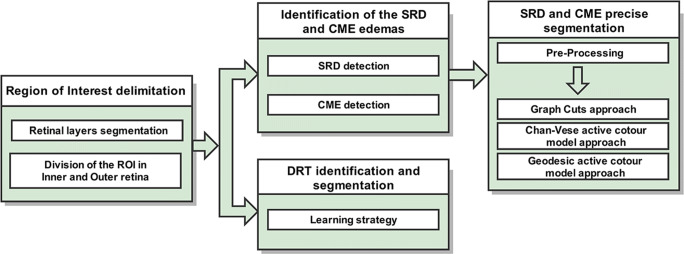
As illustrated in Fig. 2, the designed pipeline of the method is composed of 3 main stages. Firstly, the system identifies 4 representative retinal layers that delimit the ROI where the intraretinal fluid may appear. Posteriorly, this area is sub-divided into 2 main regions: the inner and the outer retinas. This ROI division facilitates the identification of each ME type given their characteristic relative positions within the retina. Regarding the identification stage, we follow a similar strategy for the detection of the SRD and CME edemas as in [35]. Then, the system exploits an adjusted combination of pre-processing filter and robust segmentation algorithms for their precise extraction. In the case of the DRT edemas, a learning strategy was implemented for the identification and subsequent segmentation of this ME type. Each one of these stages is going to be discussed next.
Fig. 2.
Main stages of the proposed methodology for the identification and segmentation of the DME types: SRD, CME, and DRT
Delimitation of the Region of Interest
MEs are accumulated in typical relative positions within the main retinal layers [29]. In particular, CMEs normally start to manifest symptoms in the inner retina. In contrast, SRD and DRT edemas typically appear in the outer retina. In more severe pathological stages, CMEs can also proliferate from the inner to the outer retina and merge with the DRT edemas. Based on those premises, two retinal regions were particularly segmented, facilitating the posterior identification and analysis of the pathological regions that are associated with the DME disease, with restricted and limited regions of interest for each target case.
For this purpose, initially, the system detects 4 retinal layers that delimit the entire region of analysis. Next, 2 sub-retinal regions are identified to facilitate the extraction of each ME type. This complete process is described with more detail in the next subsections.
Retinal Layer Segmentation
The 4 retinal layers that provide the delimitation of the entire search space are the following: the inner limiting membrane (ILM), the outer plexiform layer (OPL), the junction of inner and outer segments (ISOS), and the retinal pigment epithelium (RPE). The identification of these retinal layers initially follows the work of González-López et al. [15]. To do that, firstly, we employed a noise filtering strategy based on the Fourier Butterworth filter to not only efficiently reduce the speckle noise but also preserve the clinical information contained in the images [36]. Then, an active contour-based model is used to segment and extract the main retinal layers. Finally, different refinement processes are implemented to correct existing segmentation errors, improving the accuracy of the obtained results. In this context, different anatomical knowledge was used, such as the horizontal placement of the retinal layers and their relative positions over the analyzed OCT images. Using this strategy, we obtain the ILM, ISOS, and RPE layers.
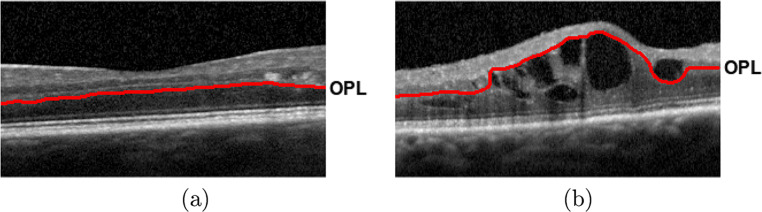
The proliferation of pathological structures, as the MEs in our case, carries a significant deterioration of the retinal architecture and morphology [10]. Specifically, these alterations impact drastically the intermediate intraretinal layers, as the aimed OPL layer, hardening its identification process. Figure 3 shows 2 representative examples of OCT images of patients with non-pathological and pathological conditions, illustrating a significative deterioration of the OPL layer with the presence of the DME disease. Given that the previous strategy does not offer satisfactory results with this layer in pathological scenarios, the previous approach is complemented with a specific strategy for the OPL layer. In this case, the method uses the previous detected ISOS layer as baseline for the application of a region growing approach [48]. Over this layer, N initial points were randomly generated and used as seeds for the region growing approach, as shown in Fig. 4a. The number of generated seeds is proportional to the image dimensionality, representing an amount of 10% of the input OCT image width. This way, we obtain the entire region over the ISOS layer by intensity similarity. Hence, as illustrated in Fig. 4b, the upper limits of the obtained region corresponds to the target identification of the OPL layer.
Fig. 3.
Representative examples of OCT images. a Normal OCT image without the presence of DME. b OCT image with the pathological presence of DME
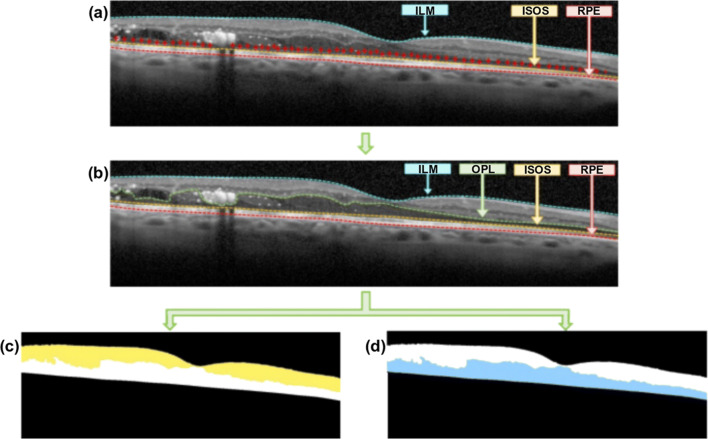
Fig. 4.
Example of OCT image with the identification of the inner and outer retinal regions. a Identification of the ILM, ISOS, and RPE layers. b Additional delimitation of the OPL layer. c Identified inner retina, between the ILM and the OPL layers. d Identified outer retina, between the OPL and the RPE layers
Division of the ROI in Two Subregions: Inner and Outer Retinas
As said, CME edemas begin to manifest in the inner layers of the retina. However, in more advanced pathological scenarios, these edemas proliferate to the outer retina, merging with the DRT edemas. In opposition, the SRD and DRT edemas present a characteristic relative position in the outer retinal layers, without spreading to the upper retinal region [29]. The identification of these retinal regions simplifies the identification of each type of intraretinal fluid accumulation, reducing significantly the search space for each ME type.
Based on these clinical premises, 2 main subregions were defined to facilitate the identification process: the inner and the outer retinas. As illustrated in Fig. 4, the inner retina (Fig. 4c) is delimited by the ILM and the OPL layers as superior an inferior boundaries, whereas the outer retina (Fig. 4d) presents the OPL and the RPE layers as limits, respectively.
Identification of the SRD and CME Edemas
In this stage, 2 specific and individual strategies were designed for the identification of the SRD and CME edemas (see Fig. 2) as in [35]. In particular, the SRD edema is searched in the outer retina, whereas the CME edemas are verified in both retinal regions, with adapted approaches to the image characteristics of each region.
SRD Detection
The system automatically identifies the SRD edemas by the combination of clinical knowledge and image processing techniques. The method firstly applies an adaptive thresholding to segment the areas with identical intensity profiles. Then, to decrease the false positive rates, a list of clinical restrictions was implemented, using criteria such as relative position, area, morphology, thickness of the photoreceptor layer, or the intensity profile [35]. Figure 5 presents an illustrative example of an OCT image with the identification of the existing SRD edema.
Fig. 5.
OCT image with the identification of the SRD edema (+) as well as the ILM, ISOS, and RPE layers. ILM/RPE delimits the entire ROI. ISOS/RPE indicates the photoreceptor layer
CME Detection
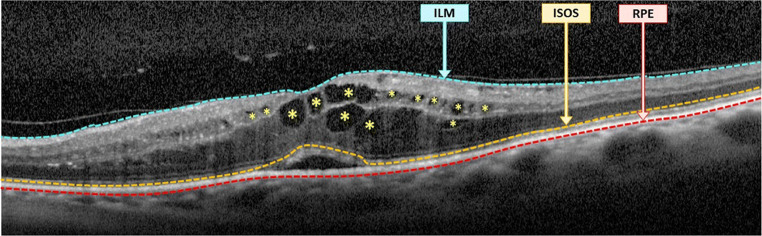
CME edemas are typically characterized as cystoid spaces with a significant low-intensity profile in comparison with the surrounding retinal tissues. These edemas present a significant variability in terms of dimensions, shape, or morphology, appearing from a cystoid to a petaloid-like appearance. In the early stages of the disease, they typically emerge in the inner retina (ILM/OPL), where the contrast with the surrounding retinal tissue is significative. However, in more severe clinical stages, they can also proliferate in the outer retina (OPL/RPE). This region, with a low-intensity profile, presents a reduced contrast with the edemas, hardening significantly their identification. To detect the CME edemas, we apply a similar strategy used for the identification of the SRD edemas [35]. Firstly, an adaptive thresholding is applied to identify all the CME candidates, separately in the inner and the outer retinas. Then, a post-processing stage based on a flooding process over the image gradient was applied to merge partial identifications. Finally, a list of clinical criteria was applied to filter any existing false CME candidate: area size, major and minor axes of the candidates, retinal thickness, and eccentricity were used to preserve the true detections. Figure 6 includes a representative example of an OCT image with the identification of the visible CME edemas.
Fig. 6.
OCT image with the identification of existing CME edemas (identified with asterisks)
SRD and CME Precise Segmentation
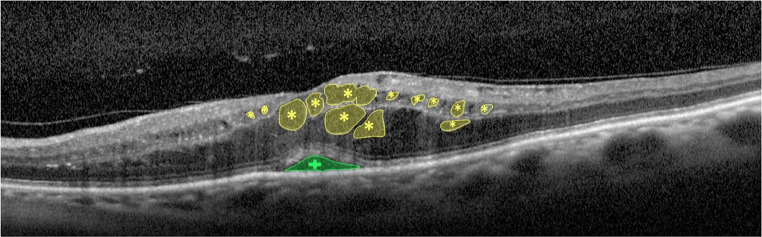
Regarding the SRD and CME edemas, the proposed system exploits an adjusted combination of pre-processing filters and robust segmentation algorithms for the precise extraction of these pathological fluid regions. Figure 7 exposes a representative example of OCT image including SRD and CME edemas, illustrating their initial localizations and their target regions.
Fig. 7.
OCT image with the SRD (plus sign) and CME (asterisk) edemas
Image Pre-Procesing for the Segmentation Process
Using as reference the preliminary identifications of the SRD and CME edemas, we continue with the precise segmentation of each case. The OCT images are often affected by motion artifacts, speckle noise, or poor soft tissues that makes the analysis of different pathological structures difficult [37]. To eliminate the noise and small artifacts while preserving and enhancing the retinal structures and in order to facilitate the segmentation process, we implemented the optimized anisotropic diffusion filtering proposed by Kroon et al. [18]. This filter consists of an iterative method that enhances the contrast between the pathological and non-pathological retinal structures. In this way, the filter describes the local image structure using the “second-moment matrix,” also designed as a structure tensor. This descriptor is transformed into the diffusion tensor D, which is commonly determined in an iterative forward difference approximation, as follows:
| 1 |
Where u indicates the image and t the diffusion time. In this filter, the parameters magnitude of diffusion and direction are estimated and correlated.
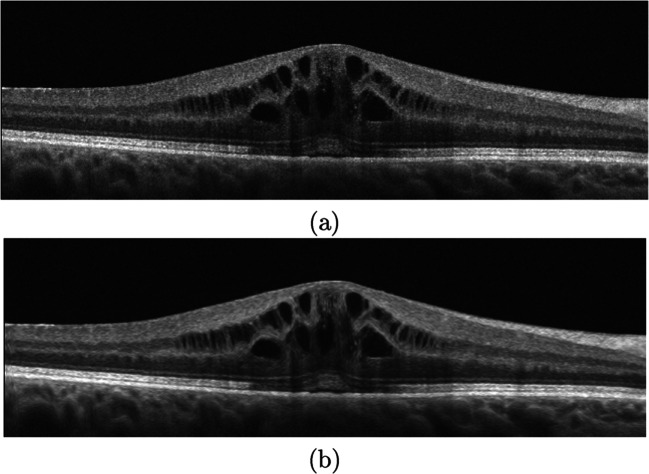
Figure 8 illustrates a representative example of application of the optimized anisotropic filter in a pathological OCT image. As shown, this filter significantly reduces the noise while preserving and simultaneously highlighting the limiting boundaries of the SRD and CME edemas, therefore facilitating the target segmentation process.
Fig. 8.
Pre-processing stage. a Input OCT image. b Pre-processed OCT image after applying the anisotropic diffusion filter
For the segmentation of the SRD and CME regions, 3 robust alternative strategies were adapted and analyzed in this work, using as seed the initial extractions of the localization process: (a) an approach based on graph cuts [33]; (b) another based on a Chan-Vese active contour model [7]; (c) a last one adapting a geodesic active contour model [6].
Graph Cut Approach
We based our approach in the GrabCut proposal of Rother et al. [33], given its suitability and adequate results for similar segmentation problems [8, 13]. The main goal of this segmentation strategy is to classify the pixels in the OCT image into 2 categories, the foreground (the pathological fluid regions) and the background. To do that, as said, the algorithm, as the other studied approaches, uses the initial extractions of the localization process as a first approximation of the foreground region, as shown in Fig. 9. Gaussian mixture models (GMMs) are created for the initial foreground and background regions. Then, each pixel of the OCT image is assigned to the most likely Gaussian component in the background or the foreground GMM. Finally, an interactive min-cut algorithm is applied to achieve the target segmentation, precisely separating both regions.
Fig. 9.
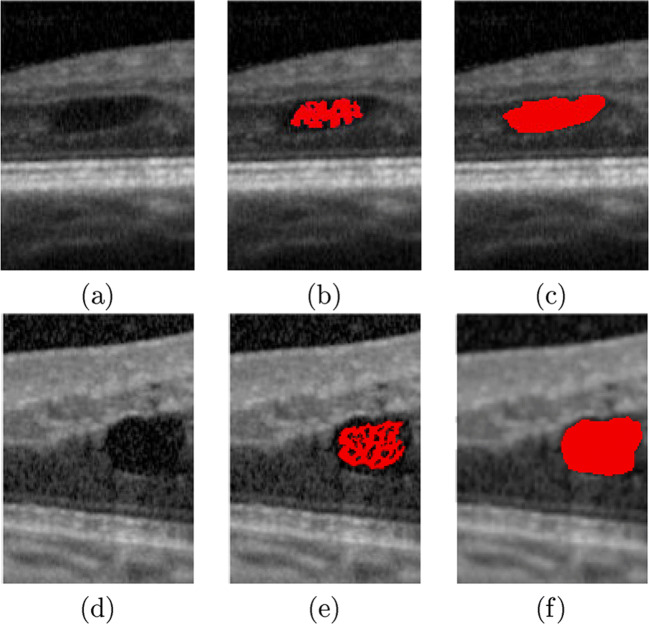
Segmentation stage. a, d OCT images with the presence of CME edemas. b, e Seed regions that were obtained in the CME localization stage. c, f Final CME segmentations
Chan-Vese Active Contour Model Approach
This active contour model proposed by Chan et al. [7] is also considered as a powerful and flexible method, which is able to perform segmentation tasks in a large variability of image types. This method, based on the Mumford-Shad function [26] and the level set approach [23], offered satisfactory results in similar complex medical imaging segmentation issues [9, 46]. In this line, the Chan-Vese model assumes that the fluid regions and the background present a Gaussian distribution with the same standard deviation. Using this information as reference, the model seeks a precise segmentation by minimizing the energy function based on the curve that represents the limits of the fluid regions. In particular, the Chan-Vese model is defined by:
| 2 |
where I represents the OCT image, Ω1 is the fluid region, and is the background region. In addition, c1 and c2 are the mean intensity values for the regions Ω1 and , respectively. Moreover, represents the smoothness regularization term, where α is a weighting coefficient and indicates the length of the curve . This level set formulation facilitates the identification of regions with similar image properties, as the case of the target edemas.
Geodesic Active Contour Model Approach
Caselles et al. [6] proposed a segmentation method based on the relationship between active contours and the computation of geodesics or minimal distance curves, the geodesic model. This model was also applied in similar strategies for the segmentation of pathological regions in many fields of medical imaging [1, 16]. In particular, the geodesic model is defined along a curve C(t) and minimized by evolving the curve in the normal direction, as following:
| 3 |
where I represents the OCT image, t is a time parameter, and 𝜃(t,x,y) = 0 represents the zero level set of the function 𝜃. The level set function 𝜃 is selected to be a signed distance function, being negative in the interior (the pathological fluid regions) and positive in the exterior (background) of the zero level set.
DRT Identification and Segmentation
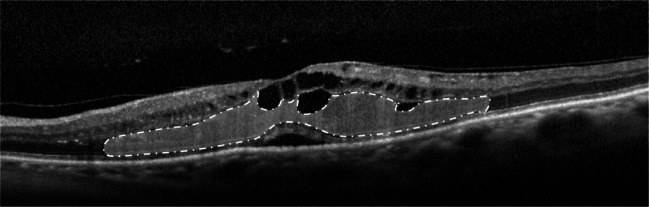
Finally, we identify the presence of the DRT edemas and segment its constituent region without any pre-processing stage. DRT edemas are characterized by a “sponge-like” appearance due to swelling of the surrounding retinal tissues. This swelling is normally produced by the fluid accumulations in the OPL/ISOS region, as illustrated in Fig. 10.
Fig. 10.
OCT image with the presence of the DRT edema (dash-dot-dash)
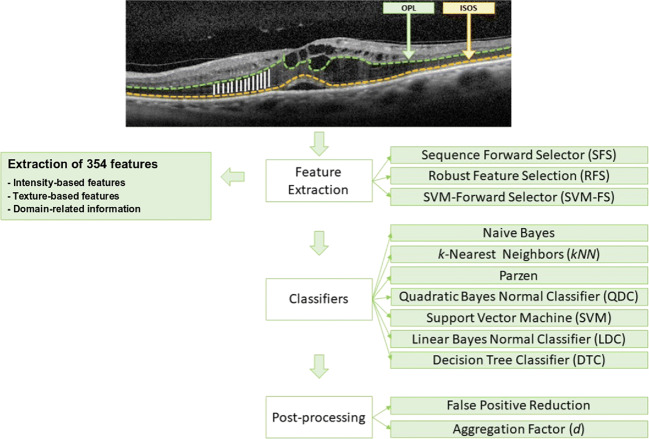
Figure 11 illustrates a schematic representation of the designed strategy for the detection of the DRT edemas, analyzing its presence by columns in a process that is composed of 3 main steps: feature extraction, use of classifiers to identify the DRT presence, and post-processing stages to complete and refine the final segmented region.
Fig. 11.
Schematic representation of the designed strategy for the identification and segmentation of the DRT edemas
Firstly, to characterize each analyzed region as DRT or non-DRT, we exploit the information contained in each column within the search space (the OPL/ISOS region), where this DME type usually appears. To do that, we use a window of a defined size that is centered in each column under analysis, as illustrated in the input image of Fig. 11. Besides, considering that the thickness of the OPL/ISOS region is not constant, the height of the window varies according to the analyzed column in that region, being adjusted to the OPL/ISOS height. In this way, we extract the set of features that better discriminate the presence of the DRT edemas. Given that, a suitable combination of features based on intensity, texture, and clinical-based information was analyzed. In fact, a total of 354 features were extracted from the analyzed regions (listed and described in Table 1).
Table 1.
List of the defined set of 354 features to identify the DRT presence
| No. of features | Category | Features |
|---|---|---|
| [1 − 15] | Global intensity-based features (GIBS) | Maximum, minimum, mean, median, std, variance, 25th percentile, 75th percentile, skewness, and maximum likelihood estimates for normal distribution |
| [16 − 20] | Gray-level intensity histogram (GLIH) | Obliquity, kurtosis, energy, and entropy |
| [21 − 36] | Gray-level co-occurrence matrix (GLCM) | Contrast, energy, correlation, and homogeneity |
| [37 − 117] | Histogram of oriented gradients (HOG) | 9 windows per bound box and 9 histogram bins |
| [118 − 245] | Gabor | Mean and std. orientations = 8 and scale = 8 |
| [246 − 309] | Local binary pattern (LBP) | Mean and std. number of neighbors = (4, 8, 12, 16) and filter radius: 1–8 |
| [310 − 337] | LAWS | A collection of convolutional kernels that search for characteristic texture patterns |
| [338 − 340] | Fractal dimension (FD) | Mean, std, and lacunarity |
| [341 − 347] | Gray-level run length image (GLRLI) | SRE, LRE, GLN, RP, RLN, LGRE, and HGRE |
| [348 − 354] | Retinal thickness analysis | The maximum height of the OPL/ISOS, ILM/ISOS, ILM/RPE, and the ratio between these regions |
SRE short run emphasis, LRE long run emphasis, GLN gray-level non-uniformity, RP run percentage, RLN run length non-uniformity, LGRE low gray-level run emphasis, HGRE high gray-level emphasis
Given the high dimensionality of the feature set, we applied different feature selectors to avoid irrelevant and redundant features as well as measure their potential of discrimination. In particular, we used 3 selection strategies: sequential forward selection (SFS) [41], robust feature selection (RFS) [27], and SVM-forward selector (SVM-FS) [3]. Generally, the SFS selector adds each feature to the selected subset by an incremental importance feature order. In the case of the RFS selector, it identifies the features by the use of an emphasizing joint ℓ2,1-norm regularization, selecting those with joint sparsity. Finally, the SVM-FS selector uses a linear ℓ1-norm SVM to select the features and a non-linear ℓ1-norm SVM to predict the best subset.
Using the selected feature sets as input, 7 representative classifiers were trained and tested: the naive Bayes classifier [12], k-nearest neighbors (k NN) [45], Parzen [21], the quadratic Bayes normal classifier (QDC) [42], the support vector machine (SVM) [17], the linear Bayes normal classifier (LDC) [22], and the decision tree classifier (DTC) [31]. In the case of the k NN classifier, 3 configurations were tested, using values of k = [3, 5, 7]. For the refinement of the DRT segmentation, we use as reference the detected DRT columns and the thickness of the OPL/ISOS region.
Furthermore, the isolated classifications of all the columns produce irregularities and discontinuities over the DRT extraction. Given the consistent and regional appearance of the DRT edemas, we introduced 2 independent post-processing approaches [38] with 2 specific targets of removing spurious detections and joining partial DRT identifications. This way, we exploit these regional characteristics, improving the performance of the proposed method:
The first post-processing approach aims at reducing the false positive (FP) rates, typically as false detections that are introduced by the presence of other retinal structures. In this way, the system analyzes the minimum width of each DRT region and the distance to the next closest DRT region, removing small and isolated DRT detections.
The second post-processing approach analyzes the influence of the misclassified regions, with the aim of reducing the false negative (FN) rates. To do that, we implemented an aggregation factor (d) that connects 2 consecutive significative detected DRT regions if the distance between them is smaller than the defined value of the aggregation factor, considering that those significantly nearby regions belong to the same global DRT region.
Results
The proposed methodology was validated using a dataset composed of 262 OCT images. These images were captured with a Spectralis®; OCT capture device from Heidelberg Engineering. The OCT scans were obtained using a 7 Line Rater scan configuration with a 30∘× 5∘ of angle of capture and with a space of 240μ m. These images are centered on the macula and present a variability of resolutions ranging from 361 × 1285 to 480 × 1023 pixels. The images were captured from both left and right eyes of different individuals, presenting a varying degree of the analyzed DME types in a single scan. This study was approved by the local ethics committee. To ensure the anonymity of the patients that participated in the study, the corresponding images were renamed by the specialists before being provided for the validation of the proposed system.
In order to test the performance of the proposed methodology, the OCT images were labeled by an expert clinician, identifying, and categorizing the presence of pathological regions of the 3 edema types of DME. The performance of this system was validated using statistical metrics that are commonly used in the state-of-the-art to measure the performance of similar computational proposals. In particular, precision, recall, and Dice coefficients were measured for the quantitative validation of the results. Mathematically, these metrics are formulated as indicated in Eqs. 4, 5, and 6, where TP, TN, FP, and FN indicate true positives, true negatives, false positives, and false negatives, respectively.
| 4 |
| 5 |
| 6 |
SRD edemas are considered a particular and less frequent ME type that commonly affects a reduced group of patients [29]. Additionally, in each OCT image, it is only possible to verify the existence, if present, of a single SRD edema. Given that context, in the analyzed dataset, 22 SRD edemas were present, being correctly identified in all the cases by the system. Regarding the segmentation performance, Table 2 shows the achieved results of the proposed methodology for the segmentation of the SRD edemas with and without the explained pre-processing stage that employs the anisotropic diffusion filter. Using a best configuration of pre-processing filter and Chan-Vese approach, the proposed strategy reached an accurate value of 0.8768 for the Dice coefficient as well as values of 91.16% and 87.48% for precision and recall, respectively.
Table 2.
Results of the SRD segmentation stage using Dice, precision, and recall metrics
| Method | Dice | Precision | Recall |
|---|---|---|---|
| Chan-Vese | 0.8705 | 90.63 | 86.62 |
| Geodesic | 0.7157 | 78.10% | 76.11% |
| Grabcut | 0.6761 | 80.35% | 68.63% |
| Pre-processing and Chan-Vese | 0.8768 | 91.16% | 87.48% |
| Pre-processing and Geodesic | 0.7316 | 78.40% | 78.29% |
| Pre-processing and Grabcut | 0.6703 | 79.52% | 68.83% |
Regarding the CME edemas, a total of 829 cases were identified by the specialist in the entire image dataset. In this experiments, once again, we analyzed the best combination of the 3 segmentation approaches with and without the use of the anisotropic diffusion filter. As shown in Table 3, the best results were provided by the combination of pre-processing and Chan-Vese approach, returning values of 0.7475, 85.22%, and 73.46% for the Dice coefficient, precision, and recall, respectively. Despite that the Geodesic approach presents better results than the Chan-Vese approach without the pre-processing stage, the combination of pre-processing and Chan-Vese approach significantly increased the performance of the proposed system. This provided by the characteristics of the pre-processing filter, as it generates images with a higher homogeneity in terms of intensities over the analyzed retinal layers. This scenario benefits the Chan-Vese approach that often achieves better segmentation results in images with intensity homogeneity [7].
Table 3.
Results of the CME segmentation stage using Dice, precision, and recall metrics
| Method | Dice | Precision | Recall |
|---|---|---|---|
| Chan-Vese | 0.6604 | 90.13% | 58.97% |
| Geodesic | 0.6975 | 74.06% | 76.45% |
| Grabcut | 0.6178 | 91.38% | 53.77% |
| Pre-processing and Chan-Vese | 0.7475 | 85.22% | 73.46% |
| Pre-processing and geodesic | 0.7264 | 71,16% | 82,37% |
| Pre-processing and Grabcut | 0.6290 | 91.03% | 55.40% |
Regarding the DRT edemas, we analyzed the performance of the proposed methodology in 3 main steps: firstly, we tested the detection of the columns containing DRT edemas; then, we determined the best combination between feature selectors and classifiers for the DRT segmentation. Finally, we analyzed the post-processing approaches to determine their influence and impact in the reduction of the FP and FN rates, using the best-trained configuration.
To do that, using the labeled regions that were identified by the specialist as DRT columns, we constructed a set of features by the extraction of the defined 354 features from 2576 samples with non-DRT and DRT edema presence. The constructed set was randomly divided into 2 subsets with the same size, one for training and other for testing. Then, to ensure the accuracy of the global performance, we trained the classifiers using a 10-fold cross-validation with a total of 50 repetitions, being calculated the mean error/accuracy to measure the final performance of the method. Then, using the best configuration of each classifier, we validated the performance of the proposed system with 92,571 columns containing DRT and 52,845 non-DRT columns that were obtained from 131 OCT images.
Regarding the DRT detection, we analyzed the performance of the proposed method using different feature selectors and classifiers, as shown in Table 4. Firstly, to determine the subset of features that better discriminate the presence of this ME type, as said, 3 feature selectors were applied: RFS, SVM-FS, and SFS. In this analysis, generally, a majority of the selected features were taken from Gabor, LBP, HOG, and LAWS as they present the highest capacity of differentiation between the DRT and non-DRT patterns supporting the discrimination on the characteristic gradients and textures of this pathological presence. As we can observe in Table 4, the best configuration was achieved by the LDC classifier combined with the characteristics that were indicated by the SFS feature selector. In particular, a total of 76 features were selected in this best combination, achieving an accuracy of 90.31%. In opposition, the naive Bayes classifier with the RFS feature selector obtained the lowest rates, with an accuracy of 82.97%.
Table 4.
Accuracy results that were obtained with the tested classifiers for the DRT detection using different feature set sizes
| Classifiers | Properties | SFS | RFS | SVM-FS |
|---|---|---|---|---|
| DTC | No. of features | 19 | 51 | 23 |
| Accuracy | 0.8538 | 0.8446 | 0.8461 | |
| 3-k NN | No. of features | 74 | 139 | 31 |
| Accuracy | 0.8867 | 0.8920 | 0.8570 | |
| 5-k NN | No. of features | 69 | 112 | 80 |
| Accuracy | 0.8858 | 0.8904 | 0.8635 | |
| 7-k NN | No. of features | 64 | 112 | 80 |
| Accuracy | 0.8835 | 0.8904 | 0.8635 | |
| LDC | No. of features | 76 | 140 | 83 |
| Accuracy | 0.9031 | 0.8802 | 0.8710 | |
| Naive Bayes | No. of features | 8 | 141 | 6 |
| Accuracy | 0.8339 | 0.8297 | 0.8349 | |
| Parzen | No. of features | 18 | 92 | 69 |
| Accuracy | 0.8936 | 0.8954 | 0.8712 | |
| QDC | No. of features | 26 | 124 | 65 |
| Accuracy | 0.8870 | 0.8612 | 0.8609 | |
| SVM | No. of features | 75 | 142 | 53 |
| Accuracy | 0.8989 | 0.8973 | 0.8801 |
Regarding the DRT segmentation refinement, we analyzed the best configuration (LDC classifier and SFS feature selector) that was obtained for the DRT detection using the thickness of the OPL/ISOS region. Table 5 lists the obtained results for the DRT segmentation before any post-processing refinement using again Dice coefficient, precision, and recall. The proposed strategy achieved a Dice coefficient of 0.8369 as well as values of 95.69% and 74.20% for precision and recall, respectively.
Table 5.
Results of the DRT segmentation process before the post-processing stage
| No post-processing | |||
|---|---|---|---|
| Classifier | Dice | Precision | Recall |
| LDC | 0.8369 | 0.9569 | 0.7420 |
Using these results as baseline, next, we tested the capabilities of the designed post-processing approaches. Thus, Table 6 presents a comparative analysis for the application of both designed strategies. As we can observe, the first post-processing approach achieved an improvement in the Dice coefficient of 1%. On the other hand, the best increment in the accuracy results was obtained using the second post-processing approach with the aggregation factor (d). In this case, the best performance was obtained with a value of d = 47, reaching a Dice coefficient of 0.8913. This strategy leads to an improvement over 5% of the Dice coefficient. With disparity in their relevance, we can conclude that the application of the post-processing approaches refines and improves the segmentation performance of the extracted DRT regions.
Table 6.
Results of the DRT segmentation process after the first and second post-processing approaches
| First post-processing | Second post-processing | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Classifier | Wmin | dmin | Dice | Precision | Recall | d | Dice | Precision | Recall |
| LDC | 33 | 1 | 0.8441 | 0.9538 | 0.7570 | 47 | 0.8913 | 0.9080 | 0.8784 |
Discussion
Over the recent years, the OCT image modality was established as an accurate source of information for a precise retinal cross-sectional visualization and analysis of different eye anatomical structures, facilitating the diagnosis of many diseases, specially those that proliferate among the retinal layers, as represents the case of the DME disease. Globally, DME is a leading cause of vision impairment that constitutes a serious health problem, affecting approximately 10% of the people with diabetes. This pathology is caused by the fluid accumulations that proliferate in the macular region. In this context, these fluid accumulations typically present a great variability and heterogeneity within the retinal tissues, characterizing different ME types. Despite this complex and challenging scenario, the proposed system is able to efficiently identify and segment the hypothetical presence of each ME type, even when they appear simultaneously.
As said, SRD edemas are less frequent than the other 2 DME types, given that this case affects a reduced group of patients. Moreover, when present, only one SRD edema appears in each individual. The proposed system for the segmentation of this ME type achieved satisfactory identification and segmentation results, providing accurate extractions as the case that is illustrated in Fig. 12. In particular, the combination of the anisotropic diffusion filter with the Chan-Vese approach offered the best results, presenting a precise segmentation of the SRD regions.
Fig. 12.
Examples of results of the SRD segmentation stage. a, d Original OCT images. b, e Segmented SRD regions. c, b OCT images with the segmented SRD regions
Regarding the SRD and CME edemas, we want to highlight that we use as a reference their respective preliminary identifications to make the precise segmentation. In this line, the segmentation performance was evaluated even when the preliminary identifications were not correctly obtained (as happens in some CME cases), being therefore the general performance of the proposed method penalized. If the performance analysis would have been measured only over the correct localizations, the performance metrics would offer even better results.
Regarding the methodological novelty and potential relevance of the proposed method with respect to other similar approaches, we would like to point out that in the literature, to date, there is no existing methodology specifically designed for the automatic segmentation and characterization of all the existing pathological cases of DME, following the clinical classification of reference in the ophthalmological field [29].
In terms of a comparative analysis, we have to consider that, to date, there is no other entire dataset that was labeled simultaneously with the 3 types of ME. In particular, in the case of SRD and CME edemas, the existing state-of-the-art method was validated with different datasets, without any specifications about the selected cases that were used in the study and under different settings (pixel-level resolution, quality, OCT device, image size, signal averaging, image acquisition protocol, labelling of different clinical experts, enhanced depth, etc.).
Table 7 exposes a comparative analysis between the few existing proposals of the literature that faced the automatic segmentation of the SRD edemas and our proposed approach. As said, we have to consider that the presented methods were tested using different private datasets. Considering this, in any case, our method shows a competitive performance compared with the rest of the proposals.
Table 7.
SRD segmentation performance comparison of the state-of-the-art methods and the proposed approach
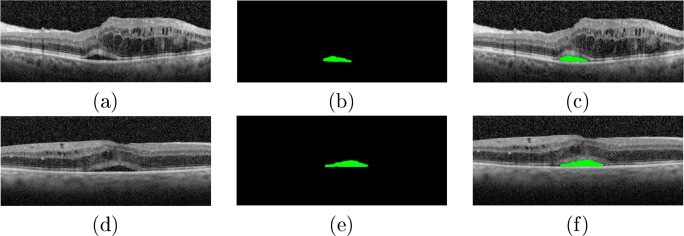
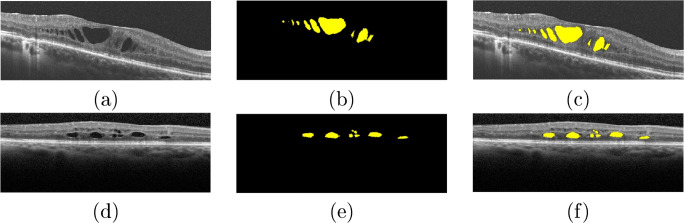
In the case of the CME edemas, the proposed system was capable of satisfactorily delimiting the area occupied by these edemas, as illustrated in the examples of Fig. 13. Despite their high variability in terms of shape, size, and low contrast with the surrounding tissue of many cases, as said, the proposed system achieved satisfactory results.
Fig. 13.
Examples of results of the CME segmentation stage. b, d Original OCT images. b, e Segmented CME regions. c, f OCT images with the segmented CME regions
Complementarily, Table 8 lists a comparative analysis between different representative works of the literature that faced the specific segmentation of CME edemas and our proposed system. In the same way as for the SRD cases, this comparison presents some limitations given that each work performed their study using a different dataset, without any specifications about the selected cases that were used in the study. Additionally, we consider that our dataset fits the real conditions that are commonly faced by the expert clinicians, including the simultaneous presence of different types of ME. In this context, the proposed system shows a competitive performance compared with the current state of the art.
Table 8.
CME segmentation performance comparison of the state-of-the-art methods and the proposed approach
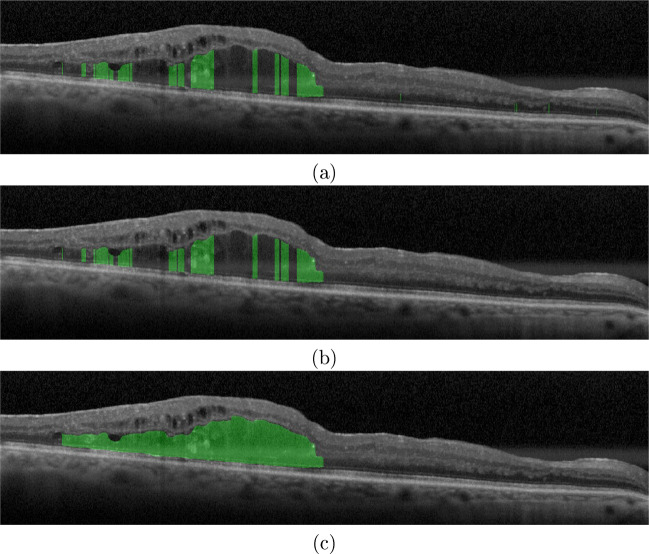
In the case of the DRT edemas, to date, no scientific study proposed a strategy that automatically segments the region occupied by this ME type. This challenge is related to the absence of limiting membranes, as well as to the significant difficulties of pattern variability for the correct delimitation of the pathological tissues that are affected by this ME type. Furthermore, to improve the performance of the proposed method, two independent post-processing approaches were included and analyzed. Figure 14b shows an example of OCT image after the application of the first post-processing approach. As we can see, the presence of artifacts in the outer retina may produce wrong detections. In this context, the first post-processing aims at the reduction of the FP rates. On the other hand, as illustrated in Fig. 14c, the application of the second post-processing approach further provided the unification of non-DRT regions, reducing the FN rates. The proposed method achieved the best results with a Dice coefficient of 0.8913 after applying the second post-processing approach with an aggregation factor of d = 47 using the LDC classifier.
Fig. 14.
Example of results of the DRT segmentation process. a DRT segmentation without the application of any post-processing approach. b DRT segmentation after the application of the first post-processing approach. c DRT segmentation after the application of the second post-processing approach
In general, as we can observe in the literature, most of the presented methods only aimed at the partial analysis of CME or SRD regions and, therefore, addressed only the pathological scenarios of DME that are typically more visible and structurally well-defined. Contrary to the other types, DRT edemas were barely faced. In addition, to date, our contribution represents the only one that accurately identifies, characterizes, and segments all the cases of the ME types that are associated with DME, even when they appear simultaneously.
Conclusions
In this work, we propose a complete methodology for the identification, characterization, and segmentation of the 3 defined types of edemas (SRD, CME, and DRT) in OCT images that are associated with the DME disease by the clinical classification of reference in the ophthalmological field. This fully automatic system provides crucial information that facilitates the diagnostic process and monitoring of the patients with DME.
For this purpose, firstly, we restricted the search space of these edemas using, as reference, 4 principal retinal layers that are initially segmented: ILM, OPL, ISOS, and RPE layers. These layers enable the delimitation of the ROI and the posterior sub-division in the inner and the outer retinas. Regarding the SRD and CME edemas, different specific approaches were designed and analyzed for the localization and the subsequent segmentation of the area that is affected by these 2 types of MEs. In the case of the DRT edemas, given their fuzzy presence and not clearly defined boundaries, a learning strategy was implemented and applied only in the OPL/ISOS region where this edema typically appears. The experimental results showed that the proposed system achieved satisfactory results, even with the simultaneous appearance of several edemas of the different analyzed DME types.
Summarizing, in the segmentation of the SRD and CME edemas, the system achieved Dice coefficient values of 0.8768 and 0.7475, respectively. Regarding the DRT edemas, the system was capable to successfully localize and segment this DME type, reaching a value of 0.8913 also for the Dice coefficient. In this way, despite the high variability and heterogeneity of the edemas in terms of size, morphology, contrast, and localization of each type, the proposed system satisfactorily identifies and segments the pathological areas of each considered ME disorder. Therefore, the proposed methodology offers an auxiliary tool for the ophthalmologists, facilitating the visualization, diagnosis, and monitoring of the DME disease, adjusting the treatments options and consequently improving significantly the life quality of the patients.
As future work, we plan to extend the dataset in a meaningful way for the application of convolutional neural networks (CNNs). Given the potential existence of other structures, we also plan to extend the proposed methodology for the automatic segmentation of other relevant eye diseases, such as age-related macular degeneration, central serous retinopathy, and peripheral retinal holes, among others. In this way, it will be possible to detect more precisely the disease condition, specially in early stages.
Funding Information
This work is supported by the Instituto de Salud Carlos III, Government of Spain, and FEDER funds through the DTS18/00136 research project and by Ministerio de Ciencia, Innovación y Universidades, Government of Spain through the DPI2015-69948-R and RTI2018-095894-B-I00 research projects. Also, this work has received financial support from the European Union (European Regional Development Fund - ERDF) and the Xunta de Galicia, Centro de Investigación del Sistema Universitário de Galicia, Ref. ED431G 2019/01; and Grupos de Referencia Competitiva, Ref. ED431C 2016-047.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Joaquim de Moura, Email: joaquim.demoura@udc.es.
Gabriela Samagaio, Email: gabrielasamagaio1995@gmail.com.
Jorge Novo, Email: jnovo@udc.es.
Pablo Almuina, Email: pablo.almuina@gmail.com.
María Isabel Fernández, Email: maribelfernandez@institutogomez-ulla.es.
Marcos Ortega, Email: mortega@udc.es.
References
- 1.Alemán-Flores M, Álvarez L, Caselles V. Texture-oriented anisotropic filtering and geodesic active contours in breast tumor ultrasound segmentation. J Math Imaging Vis. 2007;28(1):81–97. [Google Scholar]
- 2.Baamonde S, de Moura J, Novo J, Ortega M: Automatic detection of epiretinal membrane in OCT images by means of local luminosity patterns. In: International Work-Conference on Artificial Neural Networks, 2017, pp 222–235
- 3.Bi J, Bennett K, Embrechts M, Breneman C, Song M. Dimensionality reduction via sparse support vector machines. J Mach Learn Res. 2003;3(Mar):1229–1243. [Google Scholar]
- 4.Blinder KJ, Dugel PU, Chen S, Jumper JM, Walt JG, Hollander DA, Scott LC. Anti-VEGF treatment of Diabetic Macular Edema in clinical practice: effectiveness and patterns of use (ECHO Study Report 1) Clin Ophthalmol (Auckland, NZ) 2017;11:393. doi: 10.2147/OPTH.S128509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourne RR, Stevens GA, White RA, Smith JL, Flaxman SR, Price H, Jonas JB, Keeffe J, Leasher J, Naidoo K, et al. Causes of vision loss worldwide, 1990–2010: a systematic analysis. Lancet Glob Health. 2013;1(6):e339–e349. doi: 10.1016/S2214-109X(13)70113-X. [DOI] [PubMed] [Google Scholar]
- 6.Caselles V, Kimmel R, Sapiro G. Geodesic active contours. Int J Comput Vis. 1997;22(1):61–79. [Google Scholar]
- 7.Chan TF, Vese LA. Active contours without edges. IEEE Trans Image Process. 2001;10(2):266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Udupa JK, Bagci U, Zhuge Y, Yao J. Medical image segmentation by combining graph cuts and oriented active appearance models. IEEE Trans Image Process. 2012;21(4):2035–2046. doi: 10.1109/TIP.2012.2186306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Wang Z, Zhao W: Liver segmentation in CT images using Chan-Vese model. In: 2009 First International Conference on Information Science and Engineering. IEEE, 2009, pp 3669–3672
- 10.Das R, Spence G, Hogg RE, Stevenson M, Chakravarthy U. Disorganization of inner retina and outer retinal morphology in diabetic macular edema. JAMA Ophthalmol. 2018;136(2):202–208. doi: 10.1001/jamaophthalmol.2017.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding W, Young M, Bourgault S, Lee S, Albiani DA, Kirker AW, Forooghian F, Sarunic M, Merkur AB, Beg MF: Automatic detection of subretinal fluid and sub-retinal pigment epithelium fluid in Optical Coherence Tomography images. In: 35th annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, 2013, pp 7388–7391 [DOI] [PubMed]
- 12.Friedman N, Geiger D, Goldszmidt M. Bayesian network classifiers. Mach Learn. 1997;29(2-3):131–163. [Google Scholar]
- 13.Funka-Lea G, Boykov Y, Florin C, Jolly MP, Moreau-Gobard R, Ramaraj R, Rinck D: Automatic heart isolation for CT coronary visualization using graph-cuts. In: 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro, 2006. IEEE, 2006, pp 614–617
- 14.Girish G, Thakur B, Chowdhury SR, Kothari AR, Rajan J. Segmentation of intra-retinal cysts from Optical Coherence Tomography images using a fully convolutional neural network model. IEEE J Biomed Health Inform. 2019;23(1):296–304. doi: 10.1109/JBHI.2018.2810379. [DOI] [PubMed] [Google Scholar]
- 15.González-López A, de Moura J, Novo J, Ortega M, Penedo M. Robust segmentation of retinal layers in optical coherence tomography images based on a multistage active contour model. Heliyon. 2019;5(2):1–34. doi: 10.1016/j.heliyon.2019.e01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez M, Frangi AF. Non-parametric geodesic active regions: method and evaluation for cerebral aneurysms segmentation in 3DRA and CTA. Med Image Anal. 2007;11(3):224–241. doi: 10.1016/j.media.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Keerthi SS, Shevade SK, Bhattacharyya C, Murthy KRK. Improvements to platt’s SMO algorithm for SVM classifier design. Neural Comput. 2001;13(3):637–649. [Google Scholar]
- 18.Kroon DJ, Slump CH, Maal TJ: Optimized anisotropic rotational invariant diffusion scheme on cone-beam CT. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2010, pp 221–228 [DOI] [PubMed]
- 19.Lee CS, Tyring AJ, Deruyter NP, Wu Y, Rokem A, Lee AY. Deep-learning based, automated segmentation of Macular Edema in Optical Coherence Tomography. Biomed Opt Express. 2017;8(7):3440–3448. doi: 10.1364/BOE.8.003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H, Kang KE, Chung H, Kim HC. Automated segmentation of lesions including subretinal hyperreflective material in neovascular age-related macular degeneration. Am J Ophthalmol. 2018;191:64–75. doi: 10.1016/j.ajo.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Lissack T, Fu KS. Error estimation in pattern recognition via lα-distance between posterior density functions. IEEE Trans Inform Theory. 1976;22(1):34–45. [Google Scholar]
- 22.Liu C, Wechsler H. Robust coding schemes for indexing and retrieval from large face databases. IEEE Trans Image Process. 2000;9(1):132–137. doi: 10.1109/83.817604. [DOI] [PubMed] [Google Scholar]
- 23.Malladi R, Sethian JA, Vemuri BC. Shape modeling with front propagation: a level set approach. IEEE Trans Pattern Anal Mach Intell. 1995;17(2):158–175. [Google Scholar]
- 24.Marmor MF: Mechanisms of fluid accumulation in retinal edema. In: Macular Edema. Springer, 2000, pp 35–45
- 25.Montuoro A, Waldstein S, Gerendas B, Schmidt-Erfurth U, Bogunović H. Joint retinal layer and fluid segmentation in OCT scans of eyes with severe macular edema using unsupervised representation and auto-context. Biomed Opt Express. 2017;8(3):1874–1888. doi: 10.1364/BOE.8.001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mumford D, Shah J. Optimal approximations by piecewise smooth functions and associated variational problems. Commun Pure Appl Math. 1989;42(5):577–685. [Google Scholar]
- 27.Nie F, Huang H, Cai X, Ding C: Efficient and robust feature selection via joint ℓ2, 1-norms minimization. In: Advances in Neural Information Processing Systems, 2010, pp 1813–1821
- 28.Novosel J, Vermeer KA, de Jong JH, Wang Z, van Vliet LJ. Joint segmentation of retinal layers and focal lesions in 3-D OCT data of topologically disrupted retinas. IEEE Trans Med Imaging. 2017;36(6):1276–1286. doi: 10.1109/TMI.2017.2666045. [DOI] [PubMed] [Google Scholar]
- 29.Otani T, Kishi S, Maruyama Y. Patterns of diabetic macular edema with optical coherence tomography. Am J Ophthalmol. 1999;127(6):688–693. doi: 10.1016/s0002-9394(99)00033-1. [DOI] [PubMed] [Google Scholar]
- 30.Panozzo G, Parolini B, Gusson E, Mercanti A, Pinackatt S, Bertoldo G, Pignatto S: Diabetic macular edema: an OCT-based classification. In: Seminars in Ophthalmology, vol 19, 2004, pp 13–20 [DOI] [PubMed]
- 31.Quinlan JR. Improved use of continuous attributes in c4.5. J Artif Intell Res. 1996;4:77–90. [Google Scholar]
- 32.Rashno A, Koozekanani DD, Drayna PM, Nazari B, Sadri S, Rabbani H, Parhi KK. Fully automated segmentation of fluid/cyst regions in Optical Coherence Tomography images with diabetic macular edema Using neutrosophic sets and graph algorithms. IEEE Trans Biomed Eng. 2018;65(5):989–1001. doi: 10.1109/TBME.2017.2734058. [DOI] [PubMed] [Google Scholar]
- 33.Rother C, Kolmogorov V, Blake A: Grabcut: Interactive foreground extraction using iterated graph cuts. In: ACM Transactions on graphics (TOG), vol 23. ACM, 2004, pp 309–314
- 34.Roy A, Conjeti S, Phani Karri S, Sheet D, Katouzian A, Wachinger C, Navab N. Relaynet: retinal layer and fluid segmentation of macular Optical Coherence Tomography using fully convolutional network. Biomed Optics Express. 2017;8(8):3627–3642. doi: 10.1364/BOE.8.003627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samagaio G, Estévez A, de Moura J, Novo J, Fernandez MI, Ortega M. Automatic macular edema identification and characterization using OCT images. Comput Methods Programs Biomed. 2018;163:47–63. doi: 10.1016/j.cmpb.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 36.Samagaio G, de Moura J, Novo J, Ortega M: Optical coherence tomography denoising by means of a fourier butterworth Filter-Based approach. In: International Conference on Image Analysis and Processing. Springer, 2017, pp 422–432
- 37.Samagaio G, de Moura J, Novo J, Ortega M: Optical coherence tomography denoising by means of a fourier butterworth Filter-Based approach. In: International Conference on Image Analysis and Processing. Springer, 2017, pp 422–432
- 38.Samagaio G, de Moura J, Novo J, Ortega M. Automatic segmentation of diffuse retinal thickening edemas using Optical Coherence Tomography images. Procedia Comput Sci. 2018;126:472–481. [Google Scholar]
- 39.Schlegl T, Waldstein S, Bogunovic H, Endstraßer F, Sadeghipour A, Philip AM, Podkowinski D, Gerendas BS, Langs G, Schmidt-Erfurth U. Fully automated detection and quantification of macular fluid in OCT using deep learning. Ophthalmology. 2018;125(4):549–558. doi: 10.1016/j.ophtha.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 40.Sidibé D, Sankar S, Lemaître G, Rastgoo M, Massich J, Cheung C, Tan G, Milea D, et al. An anomaly detection approach for the identification of DME patients using spectral domain Optical Coherence Tomography Images. Computer Methods and Programs in Biomedicine. 2017;139:109–117. doi: 10.1016/j.cmpb.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Siedlecki W, Sklansky J. On automatic feature selection. Int J Pattern Recognit Artif Intell. 1988;2(02):197–220. [Google Scholar]
- 42.Srivastava S, Gupta MR, Frigyik BA. Bayesian quadratic discriminant analysis. J Mach Learn Res. 2007;8(Jun):1277–1305. [Google Scholar]
- 43.Sun Z, Chen H, Shi F, Wang L, Zhu W, Xiang D, Yan C, Li L, Chen X. An automated framework for 3D serous pigment epithelium detachment segmentation in SD-OCT images. Sci Rep. 2016;6(21):739. doi: 10.1038/srep21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venhuizen FG, van Ginneken B, Liefers B, van Asten F, Schreur V, Fauser S, Hoyng C, Theelen T. Sánchez, C.I.: Deep learning approach for the detection and quantification of intraretinal cystoid fluid in multivendor Optical Coherence Tomography. Biomed Opt Express. 2018;9(4):1545–1569. doi: 10.1364/BOE.9.001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y: Expert network: Effective and efficient learning from human decisions in text categorization and retrieval. In: Proceedings of the 17th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval. Springer-Verlag New York Inc, 1994, pp 13–22
- 46.Yazdanpanah A, Hamarneh G, Smith B, Sarunic M: Intra-retinal layer segmentation in Optical Coherence Tomography using an active contour approach. In: International conference on medical image computing and computer-assisted intervention. Springer, 2009, pp 649–656 [DOI] [PubMed]
- 47.Zheng Y, Sahni J, Campa C, Stangos AN, Raj A. Harding, S.P.: Computerized assessment of intraretinal and subretinal fluid regions in spectral-domain Optical Coherence Tomography images of the retina. Am J Ophthalmol. 2013;155(2):277–286. doi: 10.1016/j.ajo.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 48.Zhu S, Yuille A. Region competition: Unifying Snakes, Region Growing, and bayes/MDL for Multiband Image Segmentation. IEEE Trans Pattern Anal Mach Intell. 1996;18(9):884–900. [Google Scholar]