Abstract
Hemoglobin High-performance liquid chromatography (Hb HPLC) is a standard first-line technique for diagnosis of thalassemia and hemoglobinopathies. We compared two HPLC systems for detection and quantification of normal and abnormal Hb fractions. EDTA samples from 100 normal healthy subjects and 107 subjects affected with hemoglobinopathies or carriers were analysed using HPLC systems Tosoh HLC-723G11 and Bio-Rad Variant-II. Retention time (RT) and area of peaks for HbA2, HbF and other structural variants were compared. In discrepant cases samples were run on Sebia Capillary zone electrophoresis (CZE) for confirmation of results (39 out of 107 cases with HbE, HbD Iran, Hb Lepore and HbQ). Measurement of HbA2 and HbF in normal samples and HbF in those with variant Hbs showed good correlation by both analyzers (R2 = 0.83, 0.9 and 0.99 respectively). HbE co-elutes with HbA2 in Bio-Rad. Correlation done using the apparent HbA2 concentration from Bio-Rad with (HbE + HbA2) from Tosoh G11 showed good correlation (R2 = 0.97). Correlation of HbS (Eluting at S-window at RT 3.11 min in Tosoh G11 and 4.33 min in Bio-Rad) as well as HbD Punjab (Eluting at D-window at RT 2.82 min in Tosoh G11 and 4.06 min in Bio-Rad) by both instruments was good. HbD Iran (Eluting at E-window at RT 2.69 min in Tosoh G11 and with HbA2 at 3.53 min in Bio-Rad); HbQ (Eluting at C-window at RT 3.78 min in Tosoh G11 and unknown window at 4.7 min in Bio-Rad), HbH (Eluting at P00 window at RT 0.13 min in Tosoh G11 and giving pre-integration peak in Bio-Rad), Hb Lepore (Eluting at P08 window at RT 2.67 min in Tosoh G11 and with HbA2 at 3.46 min in Bio-Rad) gave comparable results. Correlation with findings of CZE was done in few cases when needed. Two automated HPLC instruments demonstrated similar usefulness for screening patients for hemoglobinopathies. However, complex elution patterns as well as co-elution of variants like HbA2, HbE, Hb Lepore, HbD Iran (in Bio-Rad); HbD Iran and HbE (Tosoh G11) pose difficulty in interpretation. A complementary second method like CZE may be required.
Electronic supplementary material
The online version of this article (10.1007/s12288-020-01298-5) contains supplementary material, which is available to authorized users.
Keywords: Hemoglobinopathy, HPLC, Chromatogram
Introduction
Disorders of hemoglobin (Hb) synthesis include both quantitative (thalassemia syndrome) as well as qualitative defects (Hb variants).
The separation and estimation of the different Hb fractions is essential in screening for carriers as well as the diagnosis of hemoglobinopathies. Clinical and family history, red cell indices, complete blood counts, HbA2, HbF estimation by hemoglobin electrophoresis, sickling test aid in their diagnosis. However the utility of Hb electrophoresis is limited due to overlap in electrophoretic motilities of HbS/HbD/HbG/HbQ in acidic pH and HbC/HbE/HbO/HbA2 in alkaline pH which makes their diagnosis as well as of compound heterozygous states difficult.
High-performance liquid chromatography (HPLC) as a screening technique for hemoglobinopathies and thalassemias has taken over hemoglobin electrophoresis. The advantage of the HPLC is its high resolution and reproducibility resulting in accurate diagnosis.
Sometimes however, this technique results in patterns that are relatively complex and difficult to interpret. Capillary zone electrophoresis (CZE) separates the hemoglobin fractions according to their electrophoretic mobility with an alkaline buffer. It provides high throughput analysis providing separation of various hemoglobin variants with high resolution. Clinical studies comparing HPLC and CZE have found that they are complementary methods, and can be used in tandem for accurate and precise hemoglobin variant quantification.
In the present study we have analyzed two HPLC systems Tosoh HLC-723G11 and Bio-Rad Variant-II for the detection of common (HbA, A2, S and E) as well as few uncommon Hb variants (D-Punjab, D-Iran, Q-India, H and Lepore). To resolve any discrepancies that were noted, samples were run on CZE for further interpretation. We also compared HbF and A2 in samples from cases with normal Hb.
In the Bio-Rad Variant-II (Bio-Rad, Hercules, CA) HPLC analysis is done using β-thalassemia Short Program which separates hemoglobin variants by cation exchange chromatography using a salt gradient. HbA2 and HbF single point calibrators were performed daily to adjust and ensure proper retention times and accurate quantification. Bio-Rad level 1 (HbA, F, and A2) and level 2 (HbA, F, A2, and S) QC materials (Hercules, CA) were also run daily before patient samples were analyzed.
Tosoh HLC-723 G11(Tosoh Bioscience) is an automated HPLC system that can separate and quantify HbA2 and HbF in a 5 min run in the β-thalassemia mode. It also allows the presumptive identification of the most frequent Hb variants by assigned retention time windows for HbF, HbA0, HbA2, HbE, HbD, HbS and HbC. Hemoglobin separation is performed by utilizing differences in ionic interactions between the cation exchange group on the column resin surface and the hemoglobin components in a step gradient elution containing three citric acid buffers with different pH and salt concentrations.
Capillary zone electrophoresis (CZE) was performed using the Capillary Hemoglobin (E) kit in the Sebia Capillary Flex system (Sebia, Norcross, GA). The instrument is equipped to re-suspend, lyse and separate hemoglobin variants and analyze them using EDTA whole blood. The lysed red cells are electrophoresed in alkaline buffer (pH 9.4) allowing separation to be directed by pH and endosmosis. CZE does not require daily calibration. To ensure proper charge and function of the capillaries, normal HbA and HbA2 migration controls are analyzed through each capillary daily before additional QC or patient samples are run.
Methods
This observational study was carried out at Dr. Lal PathLabs Ltd., National Reference Lab, Delhi, India in April and May 2018.
EDTA samples from 100 hematologically normal and healthy subjects and 107 subjects who were carriers or affected with hemoglobinopathies were analysed using HPLC systems Tosoh HLC-723G11 and Bio-Rad Variant-II. Their results were compared. Wherever discrepancies were noted, samples were run on Sebia Capillary Electrophoresis for confirmation of results. The specimens tested were residual samples referred to our laboratory for routine laboratory tests. They were not subjected to any pre- treatment and directly processed from their primary tubes. Calibrators and controls provided by the manufacturer were used with every batch. Both levels of control were run five times every day in order to check for the precision (Table 1).
Table 1.
Determination of analytical precision for the HbA2 and Hb F percentage (%) using the Bio-Rad Variant-II
| Within run | Within run | Between runs | Between runs | |||||
|---|---|---|---|---|---|---|---|---|
| HbF mean | CV(%) | HbA2 mean | CV(%) | HbF mean | CV(%) | HbA2 mean | CV(%) | |
| Level 1 | 1.55 | 2.29 | 3.36 | 2.23 | 1.54 | 2.11 | 3.36 | 1.73 |
| Level 2 | 4.7 | 0.91 | 5.73 | 1.34 | 4.71 | 0.72 | 5.79 | 1.36 |
Comparison between the HbF and HbA2 values was done in case of normal samples using regression analysis. Also for the samples from subjects who were carriers or affected by hemoglobinopathies were compared for levels of HbF, HbA2, and other abnormal hemoglobin along with their interpretation.
Interpretation of Reports: Chromatograms generated were studied. Interpretation was done by observing HbA2 and HbF concentration for beta thalassemia as well as retention time and area percentage of various peaks and windows for other structural variants. Each chromatogram from Bio-Rad Variant II shows peaks of HbA0, A2, and Hb F along with C window, D window, S window, and two minor peaks, P2 and P3. Tosoh G11, apart from F, A0, A2 D, S, C windows has E window. The minor/insignificant hemoglobin variants are given separately in P00–P10 windows. Several hemoglobin variants may elute in the same window. They were provisionally given a diagnosis by retention time and area percentage.
Other relevant tests were done wherever required, for example, sickling test as supporting evidence of HbS. Family study, clinical and correlation with CBC parameters was carried out whenever needed and possible. Also correlation with findings of Hb CZE result was done in 39 out of 107 cases with HbE, HbD Iran, Hb Lepore and HbQ. Eight cases with borderline HbA2 value of 3.6–3.9 were excluded from the study.
Results
The evaluation of analytical precision for the measurement of HbA2 and HbF are shown in Table 1. Between-run CVs and within-run CVs for both normal and raised HbA2 levels were found always below 1.73 and 2.23% respectively. Between-run CVs and within-run CVs for both normal and raised HbF levels were found always below 2.11 and 2.29% respectively.
HbA2 and HbF in Samples Without Variant Hemoglobin
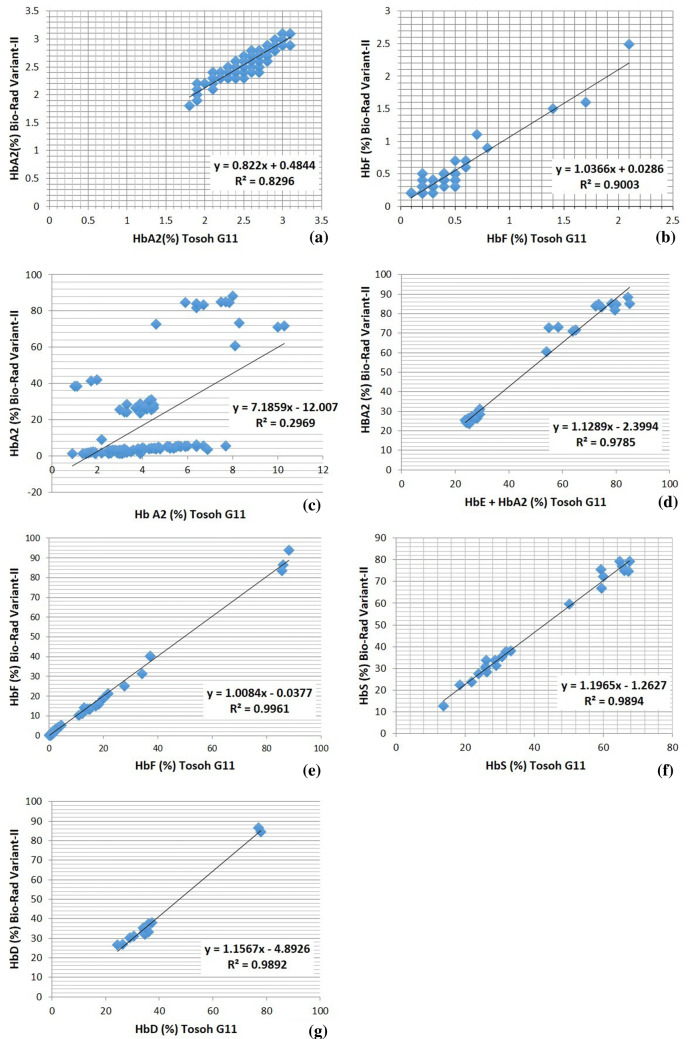
Comparison of HbA2 and HbF values measured by Tosoh G11 with Bio-Rad Variant-II HPLC system was done in case of normal samples (number = 100). The linear regression graphs are as given in Fig. 1a, b. Levels of HbA2 and HbF correlated well by both the analyzers in normal samples with a R2 values of 0.83 and 0.9 respectively.
Fig. 1.
Regression analysis of results from Tosoh G11 versus Bio-Rad Variant II for a HbA2 in samples without Hb variants, b HbF in samples without Hb variants, c HbA2 in samples with variants, d HbE + HbA2 of Tosoh vs. HbA2 of Bio-Rad Variant II, e HbF in samples with variants, f HBS, and g HbD Punjab
HbF in Samples with Hemoglobin Variants
On comparison of HbF values between both analyzers in samples with hemoglobin variants (n = 107), a good correlation was seen with R2 value of 0.99 (Fig. 1e).
HbA2 in Samples with Hb Variants
HbA2 cannot be accurately quantified in the presence of HbE using HPLC by Bio-Rad Variant-II. This is because HbE co-elutes with HbA2. HPLC by Tosoh G11 however gives two separate peaks for HbA2 and HbE. With CZE HbA2 and HbE are separated in different zones. Comparing HbA2 concentrations across both HPLC instruments, with all variants included, unacceptable correlation was obtained with R2 value of 0.29 as shown in Fig. 1c.
Analysis of HbE, HbS, HbD
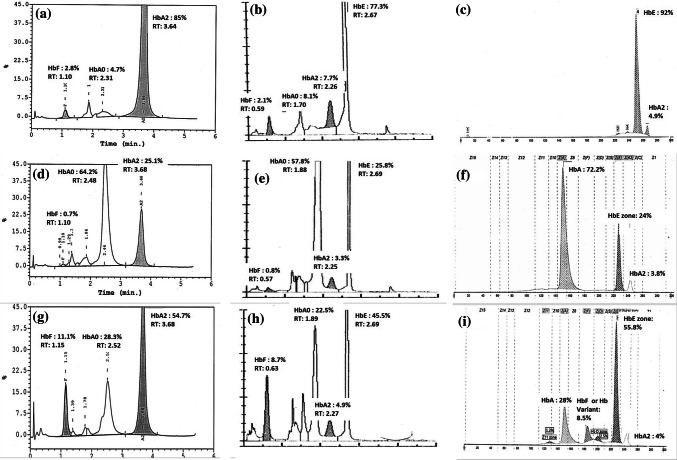
HbE was not directly detected in any of the patients using HPLC by Bio-Rad Variant-II. But was detected in 20 HbE heterozygous, 8 HbE homozygous and 5 HbE β thalassemia cases using Tosoh G11. These were confirmed by CZE. Since HbE co-elutes with HbA2 using Bio-Rad Variant II, correlation was attempted using the apparent HbA2 concentration from Bio-Rad with the (HbE + HbA2) concentration acquired using Tosoh G11 which showed a good correlation as shown in Fig. 1d. Figure 2 shows a comparison of HbE homozygous (a–c), HbE heterozygous (d–f) and E-beta (compound heterozygous) (g–i) chromatograms on all three systems.
Fig. 2.
Chromatograms of HbE homozygous (a–c), HbE heterozygous (d–f) and E-beta (compound heterozygous) (g–i) on Bio-Rad Variant II, Tosoh G11 and Sebia CZE
HbS was detected in 12 HbS heterozygous, 8 HbS homozygous, 1 HbSβ thalassemia and 2 HbDS Compound heterozygous cases. HbD-Punjab was detected in 11 heterozygous and 2 homozygous cases. Correlation of HbS (Eluting at S window at RT 3.11 min in Tosoh G11 and 4.33 min in Bio-Rad Variant II) as well as HbD Punjab (Eluting at D-window at RT 2.82 min in Tosoh G11 and 4.06 min in Bio-Rad Variant II) by both instruments was good as shown in Fig. 1f, g.
Other rare structural variants identified:
Among other structural variants, 4 HbD Iran, 2 HbQ India, 1 HbQ India with α, 1 HbH, 1 Hb Lepore and 1 sample with unknown α variant were isolated.
HbD Iran co-elutes with HbA2 at 3.53 min in Bio-Rad Variant II, and with HbE in Tosoh G11 RT 2.69 min.
HbQ India gives an unknown peak in Bio-Rad at RT 4.67 min and elutes in C window in Tosoh G11 at RT 3.78 min.
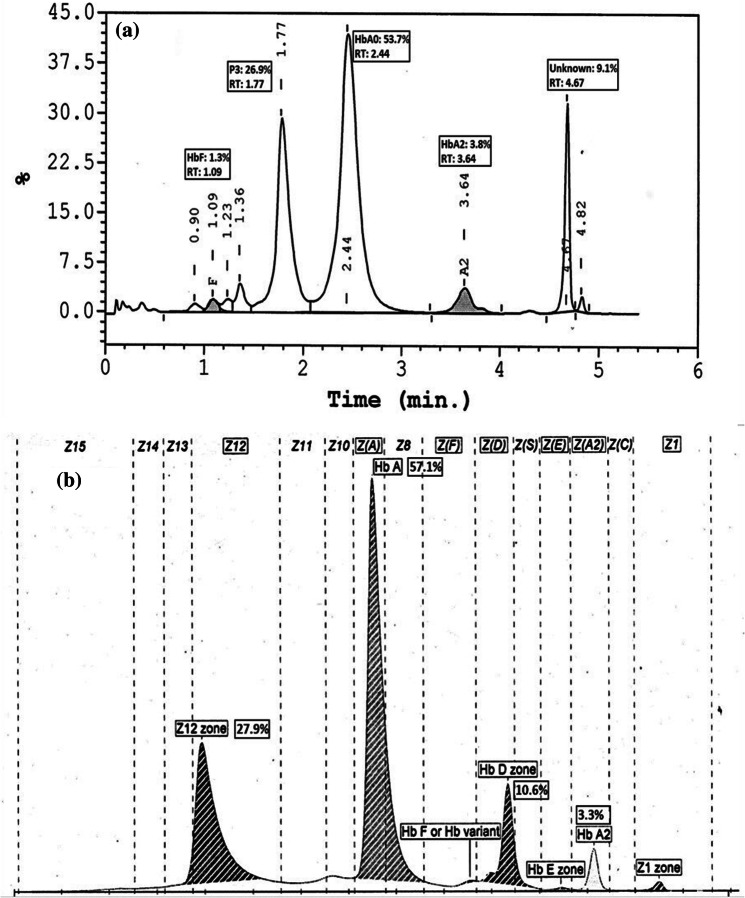
Another unusual case was HbQ India with unknown alpha variant showing two unknown peaks P07 and P012 in Tosoh G11, Peak for HbQ at RT of around 4.7 min and another unknown at RT 1.7 min for alpha variant in Bio-Rad Variant II. CZE also showed two peaks, one for HbQ that elutes in HbD zone and another in zone 12 for alpha variant. Figure 3 shows HPLC by Bio-Rad Variant II and CZE chromatogram showing the two peaks although in different zones but almost the same area/percentage that is quantitatively. We lost Tosoh G11 graph for the same due to fading of chromatogram on thermal paper.
Fig. 3.
HPLC by Bio-Rad Variant II and Sebia CZE chromatogram showing unusual case of HbQ India with unknown alpha variant
HbH elutes rapidly giving a pre-integration peak with RT < 1 min in Bio-Rad as well as P00 in Tosoh G11 with RT 0.13 min.
Hb Lepore co-elutes with HbA2 in Bio-Rad with RT 3.46 min and gives a separate peak around RT 2.67 min in Tosoh G11. It eluted in D zone in CZE.
One sample of unknown alpha variant was identified by HbA2 at RT 3.64 min of 1.9% in Bio-Rad along with a separate peak P3 of 23.7% at RT 1.72 min. In Tosoh G11 this alpha variant gave HbA2 1.9% along with P04 15.7% at RT 1.27 min.
Distribution of hemoglobin variants identified in both instruments and CZE done in a few cases is as shown in Table 2.
Table 2.
Distribution of hemoglobin variants and their RT
| S. no. | Hb variant | No. | HbF (%) TOSOH G11 | HbA2 (%) TOSOH G11 | Other 1 (%) | Other 2 (%) | HbF (%) Bio-Rad (RT 1.08–1.17 min) |
|---|---|---|---|---|---|---|---|
| 1 | β thalassemia homozygous | 3 | 86.5 ± 1.3 | 4.09 ± 0.68 | 87.43 ± 5.27 | ||
| 2 | β thalassemia trait | 25 | 0.9 ± 0.7 | 5.2 ± 0.74 | 1.09 ± 0.9 | ||
| 3 | HbE β thalassemia | 5 | 19.1 ± 8.5 | 8.2 ± 2.2 | 51.0 ± 3.4 HbE (RT 2.67 min) | 17.34 ± 7.8 | |
| 4 | HbE heterozygous | 20 | 0.8 ± 0.3 | 3.9 ± 0.4 | 22.4 ± 1.5 HbE (RT 2.67 min) | 0.83 ± 0.5 | |
| 5 | HbE homozygous | 8 | 2.7 ± 1.2 | 7 ± 0.79 | 71.4 ± 4.1 HbE (RT 2.67 min) | 3.08 ± 1.49 | |
| 6 | HbD Punjab Heterozygous | 11 | 0.7 ± 0.8 | 2.7 ± 0.49 | 32.15 ± 4.3 HbD (RT 2.82 –2.84 min) | 0.918 ± 1.01 | |
| 7 | HbD Punjab homozygous | 2 | 1.3 ± 1.4 | 5.4 ± 2.12 | 77.4 ± 0.6 HbD (RT 2.82 –2.84 min) | 1.85 ± 1.9 | |
| 8 | HbDS Compound heterozygous | 2 | 37.1 ± 0 | 1.6 ± 0 | 33.9 ± 0 HbD (RT 2.82 –2.84 min) | 13.7 HbS (RT 3.11–3.16 min) | 40.2 |
| 9 | HbS heterozygous | 12 | 1.02 ± 1.002 | 2.93 ± 0.67 | 26.8 ± 4.18 HbS (RT 3.11–3.16 min) | 1.208 ± 1.18 | |
| 10 | HbS homozygous | 8 | 18.48 ± 5.12 | 3.3 ± 1.4 | 61.6 ± 5.6 HbS (RT 3.11–3.16 min) | 17.3 ± 4.6 | |
| 11 | HbSβ thalassemia | 1 | 10.9 | 7.7 | 67.2 HbS (RT 3.11–3.16 min) | 10.2 | |
| 12 | HbQ | 2 | 0.37 ± 0.4 | 1.43 ± 0.09 | 12.0 ± 0.9 HbC window (RT 3.78 min) | 0.3 ± 0.14 | |
| 13 | HbQ India with α | 1 | 1.7 | 3.8 | 22.3 P07 | 12.7 P012 | 1.3 |
| 14 | HbD Iran Heterozygous | 4 | 0.67 ± 0.15 | 1.45 ± 0.48 | 35.35 ± 1.8 HbE (2.69 min) | 0.425 ± 0.26 | |
| 15 | HbH | 1 | 1.2 | 0.9 | 7.1 P00 (RT 0.13 min) | 0.8 | |
| 16 | Unknown α variant | 1 | 1.1 | 1.9 | 15.7 P04 (RT 1.27 min) | 0.8 | |
| 17 | Hb Lepore trait | 1 | 1.4 | 2.2 | 7.6 P08 (RT 2.67 min) | 2.1 | |
| 18 | Normal | 100 | 0.41 ± 0.56 | 2.53 ± 0.3 | 0.45 ± 0.59 |
| S. no. | HbA2 (%) Bio-Rad (RT 3.55–3.66 min) | Other 1 (%) | Other 2 (%) | HbA2 (%) CZE | HbF (%) CZE | Others (%) CZE |
|---|---|---|---|---|---|---|
| 1 | 3.7 ± 0.7 | |||||
| 2 | 4.8 ± 0.6 | |||||
| 3 | 69.8 ± 5.25 | 5.1 ± 1.6 | 21.4 ± 18.2 | 57.3 ± 2.19 HbE zone | ||
| 4 | 26.7 ± 1.9 | 3.62 ± 0.37 | 0.9 ± 0.2 | 24.1 ± 1.14 HbE zone | ||
| 5 | 84.6 ± 1.8 | 4.45 ± 0.8 | 13.7 ± 1.2 | 89.3 ± 3.16 HbE zone | ||
| 6 | 1.6 ± 0.3 | 32.4 ± 3.7 HbD (RT 4.02–4.06 min) | ||||
| 7 | 2.3 ± 1.6 | 85.4 ± 1.4 HbD (RT 4.02–4.06 min) | ||||
| 8 | 1.6 | 35.3 ± 0 HbD (RT 4.02–4.06 min) | 12.5 HbS (RT 4.33–4.37 min) | |||
| 9 | 3.02 ± 0.73 | 30.98 ± 5.01 HbS (RT 4.33–4.37 min) | ||||
| 10 | 2.9 ± 1.04 | 73.07 ± 6.8 HbS (RT 4.33–4.37 min) | ||||
| 11 | 5.5 | 74.8 HbS (RT 4.33–4.37 min) | ||||
| 12 | 1.15 ± 0.07 | 15 Unknown (RT 4.7 min) | 1.55 ± 0.5 | 17.05 ± 0.4 HbD zone | ||
| 13 | 3.8 | 26.9 P03 (RT 1.77 min) | 9.1 Unknown (RT 4.67 min) | 3.3 | 0.2, 27.9 of Unknown 3 in Zone 12 | 0.2 HbE zone; 10.6 HbD zone |
| 14 | 40.1 ± 1.87 (RT 3.53 min) | 2.15 | 43.2 ± 0.04 HbD zone | |||
| 15 | 1.2 | RT < 1 min | ||||
| 16 | 1.9 | 23.7 P03 (RT 3.64 min) | ||||
| 17 | 8.9 (RT 3.46 min) | 2 | 7.6 HbD zone | |||
| 18 | 2.65 ± 0.27 |
Supplementary figures show comparative chromatograms of Bio-Rad Variant II and Tosoh G11 in cases of thalassemia trait and thalassemia homozygous (Supplementary Figure 1a–d); HbS trait, homozygous and sickle beta compound heterozygous (Supplementary Figure 2a–f), HbD Punjab heterozygous, homozygous and HbH (Supplementary Figure 3a–f). Chromatograms of Hb Lepore, HbD Iran, and HbQ India on both HPLC instruments and Sebia CZE are shown in Supplementary Figure 4a–i. For comparison chromatograms on all these three instrument in a normal control is shown in Supplementary Figure 5a–c.
Discussion
The comparison between two automated, HPLC instruments demonstrated similar usefulness of both of them in the routine evaluation of patients for the presence of hemoglobinopathies. There was co-elution of various Hb in both instruments, like HbA2, HbE, Hb Lepore, HbD Iran (in Bio-Rad); HbD Iran and HbE; HbQ and HbC (Tosoh G11). In these cases a second complementary method becomes essential for better interpretation.
Glycated HbS has been known to co-elute with HbA2 by HPLC falsely altering HbA2 values [1]. HbH that was identified giving a pre-integration peak that can be identified by HPLC only when present at a higher concentration of more than 5% that can be easily visualized. However its identification by CZE is possible even in concentration as low as 1% [2]. We had 8 samples of borderline HbA2 values that we excluded from the study because they were borderline by both HPLC systems requiring parental or molecular studies for confirmation. A recent study on HbQ India showed additional findings of split A2, tiny peak in the S window and a tiny peak just after Q India peak on cation exchange HPLC [3]. Out of the two cases of HbQ India in our study, post Q peak was seen in both cases, split A2 and tiny peak in S window (0.6%) only in one of the two cases. No findings similar to these were seen in Tosoh G11 in these two cases.
In our study we used CZE to draw further conclusions wherever any discrepancies in the results were noted. We used CZE when percentage of HbA2 > 10% or P3 > 16% or any unknown peak of > 5%. HPLC produces ambiguous peaks even in subjects with no hemoglobinopathy due to post-translational modification of HbA including HbA1c. CZE does not separate these modified variants until their concentration is greater than the normal threshold [4]. However in case when HbA is absent, the laboratory must perform a second analysis to obtain the appropriate zones by mixing normal sample with patient sample for verifying the identity of the hemoglobin in CZE. This additional step is not required in case of HPLC.
One important advantage of Tosoh G11 over Bio-Rad Variant-II was separation of HbA2 from HbE giving comparable results with CZE. Even other routine screening methods like alkaline and acid gel electrophoresis are not able to do so [5–7]. The studies comparing HPLC and CZE conclude that both are complementary in providing adequate detection and quantitation of Hb variants [2, 4, 8]. Previous studies have compared the Capillary Electrophoresis with HPLC. None of them have compared these two HPLC instruments. One previous study comparing Tosoh HLC-723 G8 analyzer was found to have a high degree of correlation to the Bio-Rad Variant-II and Sebia Capillary systems for the determination of HbA2, HbF and HbS levels [4]. The results obtained after Hb analysis lead to the same interpretation as other HPLC systems. However they did not compare other rare Hb variants as done in our study.
Good knowledge of their elution patterns is a must. Sometimes erroneous results may occur due to sampling error, or unstable hemoglobin that may break down and form other hemoglobin degradation peaks or poorly stored samples can accelerate the break down process & form many minor degradation peaks. HPLC gives only a provisional identification of variant hemoglobin, and confirmation by an alternative method based on a different principle is needed. CZE used along with HPLC helps in resolving some of these variants, however the confirmation by molecular tests remains necessary. Currently we use Bio-Rad Variant-II HPLC system as the screening method and CZE to confirm other relatively uncommon variants, such as HbS, HbC, HbE, HbD, HbQ and Hb Lepore.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1: Chromatograms of Bio-Rad Variant II and Tosoh G11in cases of thalassemia trait (a, b) and thalassemia homozygous (c, d) (JPEG 164 kb)
Supplementary Fig. 2: Chromatograms of Bio-Rad Variant II and Tosoh G11in cases of HbS trait (a, b), homozygous (c, d) and sickle beta compound heterozygous (e, f) (JPEG 274 kb)
Supplementary Fig. 3: Chromatograms of Bio-Rad Variant II and Tosoh G11in cases of HD Punjab heterozygous (a, b), homozygous (c, d) and HbH (e, f) (JPEG 342 kb)
Supplementary Fig. 4: Chromatograms of Bio-Rad Variant II, Tosoh G11 and Sebia CZE in cases of Hb Lepore (a–c), HbD Iran (d–f) and HbQ India (g–i) (JPEG 468 kb)
Supplementary Fig. 5: Chromatograms of Bio-Rad Variant II, Tosoh G11 and Sebia CZE in normal control without any hemoglobin variants (a–c) (JPEG 130 kb)
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Parul Chopra, Email: parul6588@yahoo.co.in, Email: parul.chopra@lalpathlabs.com.
Sunanda Bhardwaj, Email: sunanda.lpl@lalpathlabs.com.
Pushkar Negi, Email: pushkar.negi@lalpathlabs.com.
Anil Arora, Email: anil.arora@lalpathlabs.com.
References
- 1.Keren DF, Hedstrom D, Gulbranson R, Ou C-N, Bak R. Comparison of Sebia Capillarys capillary electrophoresis with the Primus high-pressure liquid chromatography in the evaluation of hemoglobinopathies. Am J Clin Pathol. 2008;130(5):824–831. doi: 10.1309/AJCPQY80HZWHHGZF. [DOI] [PubMed] [Google Scholar]
- 2.Greene DN, Pyle AL, Chang JS, Hoke C, Lorey T. Comparison of Sebia Capillarys Flex capillary electrophoresis with the BioRad Variant II high pressure liquid chromatography in the evaluation of hemoglobinopathies. Clin Chim Acta Int J Clin Chem. 2012;413(15–16):1232–1238. doi: 10.1016/j.cca.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 3.Sharma S, Sharma P, Das R, Chhabra S, Hira JK. HbQIndia (HBA1:c.193G>C): hematological profiles and unique CEHPLC findings of potential diagnostic utility in 65 cases. Ann Hematol. 2017;96:1227–1229. doi: 10.1007/s00277-017-3020-z. [DOI] [PubMed] [Google Scholar]
- 4.Merono F, Agouti I, Bonello-Palot N, Paolasso C, Levy N, Badens C. Analytical evaluation of the Tosoh HLC-723 G8 automated HPLC analyzer for hemoglobin analysis in beta-thalassemia mode. Clin Biochem. 2011;44(5–6):441–443. doi: 10.1016/j.clinbiochem.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Ou C-N, Rognerud CL. Diagnosis of hemoglobinopathies: electrophoresis vs HPLC. Clin Chim Acta. 2001;313(1):187–194. doi: 10.1016/S0009-8981(01)00672-6. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder WA, Shelton JB, Shelton JR, Huynh V. The estimation of Hb A2 in the presence of Hb C or Hb E by reverse phase high performance liquid chromatography. Hemoglobin. 1986;10(3):253–257. doi: 10.3109/03630268609042846. [DOI] [PubMed] [Google Scholar]
- 7.Joutovsky A. HPLC retention time as a diagnostic tool for hemoglobin variants and hemoglobinopathies: a study of 60000 samples in a clinical diagnostic laboratory. Clin Chem. 2004;50(10):1736–1747. doi: 10.1373/clinchem.2004.034991. [DOI] [PubMed] [Google Scholar]
- 8.Multicenter validation of fully automated capillary electrophoresis method for diagnosis of thalassemias and hemoglobinopathies in Thailand (2019). https://www.ncbi.nlm.nih.gov/pubmed/22299449 [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1: Chromatograms of Bio-Rad Variant II and Tosoh G11in cases of thalassemia trait (a, b) and thalassemia homozygous (c, d) (JPEG 164 kb)
Supplementary Fig. 2: Chromatograms of Bio-Rad Variant II and Tosoh G11in cases of HbS trait (a, b), homozygous (c, d) and sickle beta compound heterozygous (e, f) (JPEG 274 kb)
Supplementary Fig. 3: Chromatograms of Bio-Rad Variant II and Tosoh G11in cases of HD Punjab heterozygous (a, b), homozygous (c, d) and HbH (e, f) (JPEG 342 kb)
Supplementary Fig. 4: Chromatograms of Bio-Rad Variant II, Tosoh G11 and Sebia CZE in cases of Hb Lepore (a–c), HbD Iran (d–f) and HbQ India (g–i) (JPEG 468 kb)
Supplementary Fig. 5: Chromatograms of Bio-Rad Variant II, Tosoh G11 and Sebia CZE in normal control without any hemoglobin variants (a–c) (JPEG 130 kb)