Abstract
In this work, we assess how pre-training strategy affects deep learning performance for the task of distinguishing false-recall from malignancy and normal (benign) findings in digital mammography images. A cohort of 1303 breast cancer screening patients (4935 digital mammogram images in total) was retrospectively analyzed as the target dataset for this study. We assessed six different convolutional neural network model structures utilizing four different imaging datasets (total > 1.4 million images (including ImageNet); medical images different in terms of scale, modality, organ, and source) for pre-training on six classification tasks to assess how the performance of CNN models varies based on training strategy. Representative pre-training strategies included transfer learning with medical and non-medical datasets, layer freezing, varied network structure, and multi-view input for both binary and triple-class classification of mammogram images. The area under the receiver operating characteristic curve (AUC) was used as the model performance metric. The best performing model out of all experimental settings was an AlexNet model incrementally pre-trained on ImageNet and a large Breast Density dataset. The AUC for the six classification tasks using this model ranged from 0.68 to 0.77. In the case of distinguishing recalled-benign mammograms from others, four out of five pre-training strategies tested produced significant performance differences from the baseline model. This study suggests that pre-training strategy influences significant performance differences, especially in the case of distinguishing recalled- benign from malignant and benign screening patients.
Keywords: Breast cancer, Digital mammography, Deep learning, Transfer learning, Training strategy
Background
Digital mammography is the primary clinical imaging exam for early-stage breast cancer screening in the general population [1]. The effectiveness of digital mammography screening for early detection and mortality reduction is well known, but as with any form of medical imaging, it is an imperfect modality. Serious challenges still exist in the distinction of benign from malignant lesions and in the reduction of false-recall [2]. False-recall refers to the situation when a woman is recommended for additional imaging or biopsy but the lesion is subsequently proven benign. The false-recall rate for screening mammography is alarmingly high. Over a 10-year period of annual screening, more than half of women will receive at least one false-positive recall [3]. Such high false-recall rates result in a large number of unnecessary and possibly invasive follow-up tests, inflicting psychological harm on the patient [4]. Therefore, improving the distinction among malignant, benign, and false-recall digital mammograms is a clinically significant task. So significant, in fact, that the breast cancer surveillance consortium has called for efforts to develop advanced technology to reduce false-positive rates in screening mammography [5].
Deep learning convolutional neural network (CNN)-based models have recently shown encouraging results on a number of breast imaging studies [6–8]. Specifically for the task of distinguishing false-recall and malignancy from benign findings in digital mammograms, a recent study has shown promising performance using deep learning [9]. The study indicates potential for pre-training, but the scope concerning pre-training strategy was limited. To further investigate the utility of such a CNN model for clinical applications, it is imperative to assess how performance metrics react to varying model conditions. When developing a deep learning model, there are a multitude of decisions to be made before the network is even trained [10], including the following:
Network structure: There are a number of popular structures, like AlexNet or residual networks. Different structures provide characteristics more suitable to some classification tasks than others.
Transfer learning datasets: Transfer learning involves using pre-trained model parameters as a starting point for natural image computer vision tasks. Previous work has shown the ability of transfer learning to boost performance on medical image tasks [11].
Layer freezing: Layers of a network can be “frozen” during fine-tuning so that weights are not updated. This method may preserve some knowledge from early layers of the network that learn more generic features.
Incremental training: Incremental training begins by initializing a network via a pre-trained model’s weights and sequentially training the network using one or more datasets before fine-tuning to the target task.
Multi-view input: Medical images, like mammograms, often have multi-view representation. The complementary nature of multi-view data can be harnessed by combining them into a single input representation.
In this study, we investigated the effects of training strategy on CNN-based models for distinguishing malignancy and false-recall from normal (benign) findings. This study represents an in-depth analysis of deep learning for the clinical task of classifying mammography images. Specifically, we assessed six different model structures utilizing four different pre-training datasets on six classification tasks to determine how the performance of deep learning models varies based on training strategy.
Methods
Study Cohorts and Datasets
This was an Institutional Review Board (IRB)-approved retrospective study and informed consent from patients was waived due to the retrospective nature. Our main study cohort included 1303 breast cancer screening patients who underwent standard mammography screening from 2007 to 2014; a total of 4935 digital mammogram images of this cohort constituted the “target dataset” for assessing model classification effects. Our study also included four “pre-training datasets” for pre-training deep learning models. The four datasets are comprised of 1.3 million natural images, 9648 film mammogram images of 2412 patients, 108,948 X-ray images from more than 30,000 patients, and 22,000 digital mammogram images from 1427 patients. Our main cohort was previously reported [9, 12], and the four pre-training datasets were publicly available or available from the literature [13–16]. Different from a clinical study, the focus of this study is a technical evaluation of deep learning, where employing previously exposed datasets is common.
More specifically, the main study cohort consisted of three sub-cohorts: 552 patients were evaluated as negative (including benign findings), 376 patients were recalled and eventually determined to be benign based on pathology (referred to as recalled-benign in our experiments), and 375 patients were biopsy-proven positive for breast cancer malignancy. A patient case typically contains a single patient exam with the standard four screening mammography views including left and right breast with craniocaudal (CC) and mediolateral oblique (MLO) views. Images were acquired from Hologic Lorad Selenia machines with a bit depth of 12. Three categories of images were assembled corresponding to the three sub-cohorts (total 4935 images). Malignant images were taken from patients that were determined to have breast cancer based on pathology. Only cancer-affected breast images were used. Benign images were taken from patients who were determined to be cancer free after at least a 1-year follow-up period. Recalled-benign images were taken from patients who were recalled based on the screening mammography exam but were eventually determined benign by pathology. A breakdown of the patient and image numbers associated with the main study cohort is shown in Table 1.
Table 1.
Number of patients and mammogram images per category in the target dataset
| Category | Number of patients | Number of images |
|---|---|---|
| Benign | 552 | 2391 |
| Malignant | 375 | 917 |
| Recalled-benign | 376 | 1627 |
| Total | 1303 | 4935 |
Pre-training Datasets
The four datasets used for pre-training are described in Fig. 1. These datasets are ranked from least-to-most related to the target dataset. Following are pre-training datasets descriptions:
Fig. 1.
Pre-training datasets ranked from least-to-most related to the target FFDM dataset in terms of organ imaged, image modality, and classification task
ImageNet
The ImageNet dataset is a natural-image dataset consisting of 1.3 million labeled images of 1000 natural objects and animal categories [13].
ChestX-Ray8 Dataset
The ChestX-Ray8 dataset consists of 108,948 frontal-view X-ray images from more than 30,000 patients with eight disease labels [14]. All images were used in our experiments by formulating a binary classification of images positive for infiltration versus all others (infiltration-negative), as this task provides the most balanced classes for training.
Digital Database of Screening Mammography Dataset
The digital database of screening mammography (DDSM) dataset contains 9648 digitized images of film mammograms from 2412 patient cases [15] with normal (benign), malignant, and benign (recalled but determined benign by biopsy) labels. Images were segmented to outline the whole-breast region as input for the models. Note that while film mammography is no longer clinically useful, this dataset has been shown useful for pre-training of FFDM-related deep learning models [9].
Breast Density Dataset
The Breast Density dataset was created to assess four qualitative breast imaging and reporting data system (BI-RADS) breast density categories. This dataset consists of 22,000 negative full-field digital mammograms from 1427 patients [16].
Classification Tasks and Pre-training Strategies
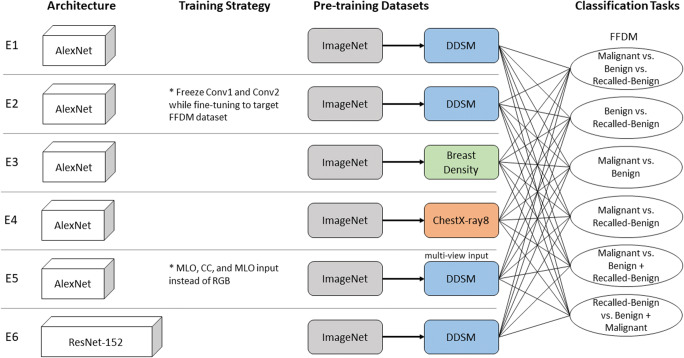
We designed six experiments to assess pre-training for each of the following classification tasks: each possible binary combination, malignant vs. benign plus recalled-benign, and recalled-benign vs. benign plus malignant.
Experiment 1
As a baseline for pre-training strategy comparison, Experiment 1 models were AlexNet models [17] incrementally trained (pre-trained in sequence) first on ImageNet and subsequently on the DDSM dataset with a classification task of benign vs. malignant before being fine tuned to the target dataset. This experimental setup was chosen as a baseline since a previous study [9] has already shown that this pre-training strategy outperformed the setting of training from scratch.
Experiment 2
Experiment 2 models, also AlexNet models, were trained in the same fashion as experiment 1 except layers “Conv1” and “Conv2” were frozen while fine tuning to the target dataset.
Experiment 3
Experiment 3 AlexNet models tested a different dataset for incremental pre-training by incrementally training first on ImageNet and subsequently on the breast density dataset with a classification task of scattered density vs. heterogeneously dense before being fine tuned to the target dataset.
Experiment 4
Experiment 4 models were trained in the same fashion as experiment 3 models with the exception of the ChestX-Ray8 dataset used for incremental training instead of the breast density dataset.
Experiment 5
Experiment 5 models replaced RGB color channel input with MLO, CC, and MLO (for consistency) views. The models used an AlexNet structure incrementally trained on ImageNet and the multi-view DDSM dataset before being fine-tuned to the multi-view target dataset.
Experiment 6
Experiment 6 models tested a different network structure, ResNet-152 [18]. The models were incrementally trained on ImageNet and the DDSM dataset with a classification task of benign vs. malignant before being fine tuned to the target dataset.
The structure of all experiments is illustrated in Fig. 2.
Fig. 2.
Experimental settings of the six experiments (denoted by E1 to E6) in terms of network architecture, training strategy, pre-training datasets, and classification tasks
The following preprocessing algorithm was applied to the target dataset before being input into the CNN:
The DICOM images were converted to grayscale jpg images (intensity range 0–255) and the intensity distribution of all jpg images were adjusted via histogram equalization using Open CV [19]. The whole-breast regions were segmented by LIBRA [20].
Images were resized to 227 × 227 to comply with the input size of the pre-trained AlexNet model (i.e., bvlc_alexnet [21]); the input size was 224 × 224 for ResNet-152.
To ensure each feature pixel has zero mean, the mean training data was generated and subtracted from each input.
CNN Modeling and Model Evaluation
Models in all experiments followed the same experimental setup. A total of five runs were performed. For each run, a randomly selected 10% of the overall target dataset was selected via stratified sampling for testing. The remaining 90% of the target dataset was used for training and validation with a ratio of nine images to one, respectively. Testing images do not overlap in any run. Data was stratified to ensure that the testing and training splits for each run had the same class distribution as the overall dataset. The validation set was used to monitor the model during the training phase for each run. Training was stopped when the model ceased to increase performance on the validation set. The model with the best performance on the validation set was selected as the final model to be evaluated on the testing set. The number of images used for testing and training for each classification task is shown in Table 2.
Table 2.
Number of testing and training images for each classification task
| Classification task | Total images per category | Testing images per category |
|---|---|---|
| Malignant vs. benign vs. recalled-benign | ||
| Malignant | 917 | 91 |
| Benign | 2391 | 239 |
| Recalled-benign | 1627 | 162 |
| Total | 4935 | 492 |
| Benign vs. recalled-benign | ||
| Benign | 2391 | 239 |
| Recalled-benign | 1627 | 162 |
| Total | 4018 | 401 |
| Malignant vs. benign | ||
| Malignant | 917 | 91 |
| Benign | 2391 | 239 |
| Total | 3308 | 330 |
| Malignant vs. recalled-benign | ||
| Malignant | 917 | 91 |
| Recalled-benign | 1627 | 162 |
| Total | 2544 | 253 |
| Malignant vs. benign + recalled-benign | ||
| Malignant | 917 | 91 |
| Benign + recalled-benign | 4018 | 401 |
| Total | 4935 | 492 |
| Recalled-benign vs. benign + malignant | ||
| Recalled-benign | 1627 | 162 |
| Benign + malignant | 3308 | 330 |
| Total | 4935 | 492 |
CNN model parameters were fixed for all experiments: batch size of 50 for stochastic gradient descent, weight decay of 0.001, and momentum of 0.9. The learning rate started at 0.001 and dropped by a factor of ten every 2500 iterations. Rectified linear units were used as the activation function. Cross entropy loss was used for all experiments. Neither data augmentation nor batch normalization was used in order to best compare to previous work [9].
The receiver operating characteristic (ROC) curve was generated and the area under the curve (AUC) was calculated as a metric of classification performance. For triple classification, we generated a ROC curve for each binary class combination and reported the average of the AUCs as is commonplace in the literature [22]. Ninety-five percent confidence intervals were computed via Delong’s method [23]. P values were calculated via bootstrap method [23] for each experiment compared to the experiment 1 (serving as a baseline).
Networks were implemented using the caffe platform running on a machine with the following specifications: Intel® Core™ i7-2670QM CPU@2.20GHZ with a Titan X Geforce GTX Graphics Processing Unit.
Results
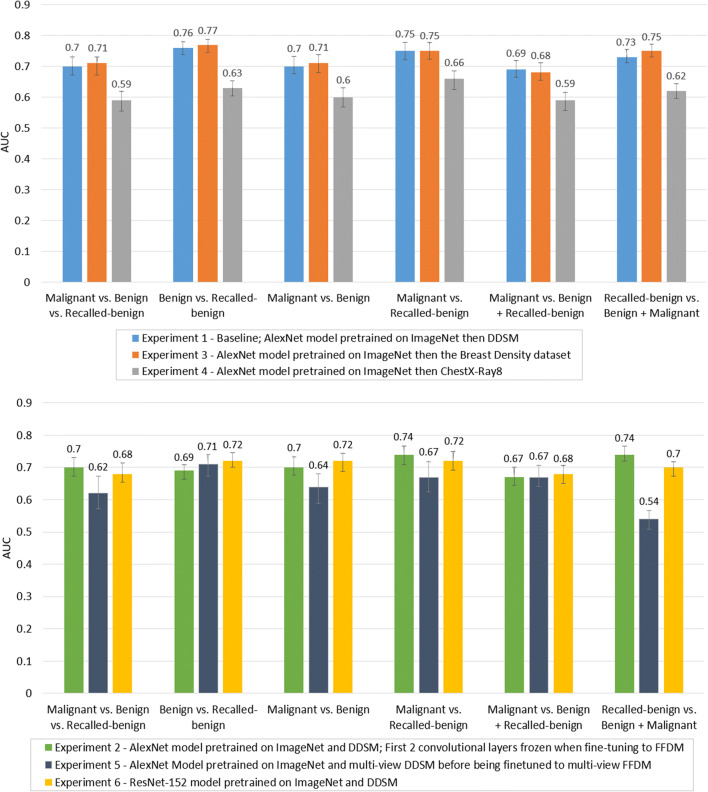
Figure 3 compares the model performance of all experiments on each classification task. The results are grouped by classification task for comparison of the performance of each experimental model. As illustrated in Fig. 3, the average AUC of all experiments for all tasks were within the range of 0.54–0.77. In general, models trained to distinguish false-recall from malignancy or false-recall from benign images appear to perform better than other tasks.
Fig. 3.
Average CNN model performance comparison across experiment 1, experiment 3, and experiment 4 (top plot) and across experiment 2, experiment 5, and experiment 6 (bottom plot), grouped by classification task. Error bars depict 95% confidence intervals
Table 3 shows the significance of the AUC difference between each experimental model and the baseline model, experiment 1. The model that consistently performed better than the baseline was experiment 3, though only two task differences were statistically significantly (malignant vs. recalled-benign and recalled-benign vs. benign + malignant). Further, experiment 3 models had the best performance out of all models on all tasks aside from the malignant vs. benign classification task. Experiment 4 was significantly worse than the baseline for all tasks. Experiment 5 models consistently performed worse than the baseline on all tasks, with all differences statistically significant aside from the malignant vs. benign + recalled-benign task. Experiment 2 models were only significantly worse than the baseline for two tasks: malignant vs benign + recalled-benign, and benign vs recalled-benign. The model that had the worst performance on every task was experiment 4. Experiment 6 models had mixed results as two tasks were not significantly different than the baseline, two tasks were significantly worse than the baseline, and two tasks were significantly better than the baseline.
Table 3.
Statistical significance of difference in AUC values compared to the baseline model, experiment 1
| Malignant vs benign vs recalled-benign | Benign vs recalled-benign | Malignant vs benign | Malignant vs recalled-benign | Malignant vs benign + recalled-benign | Recalled-benign vs. benign + malignant | |
|---|---|---|---|---|---|---|
| Experiment 1 | – | – | – | – | – | – |
| Experiment 2 | 0.32 | < 0.01 | 0.96 | 0.09 | 0.02 | 0.53 |
| Experiment 3 | 0.29 | 0.34 | 0.65 | < 0.01 | 0.49 | 0.03 |
| Experiment 4 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | < 0.01 |
| Experiment 5 | < 0.01 | 0.01 | 0.01 | 0.01 | 0.43 | < 0.01 |
| Experiment 6 | < 0.01 | 0.35 | < 0.01 | < 0.01 | 0.48 | < 0.01 |
Statistically significant results (p < 0.05) are displayed in bold
The models were also compared by variability of performance on each of the five runs. The AUC of each run for each experiment on each classification task plus the standard deviation is shown in Fig. 4.
Fig. 4.
Individual AUC for each run (plus standard deviation from the mean) of each experiment on each classification task to show the performance variability of the models
Although not directly comparable, the performance of the best performing model (AUC range 0.69–0.77 in experiment 3) in this study is in line with related studies. Among the best reported results in a previous work using the same target dataset [9], the AUC performance of the AlexNet model incrementally trained on ImageNet then the DDSM dataset is in the range 0.68–0.83. In a recent deep learning-based study on breast cancer risk prediction using negative digital mammograms [24], the image-only model achieves an AUC of 0.68. In terms of human performance, not all classification tasks tested in this study have performance statistics in the literature. Specifically, for binary-class diagnosis task (malignant vs. benign), reported performance varies (see [5, 9] for more details) but in one multicenter trial [25], the radiologists’ average AUC was reported at 0.82. Note that while these numbers may not directly comparable due to differences in specified classification tasks, data sets, and experimental settings, we have put these numbers in context to inspire insights and future research.
Discussion
We performed an evaluation study to assess CNN performance with respect to pre-training strategy on a given classification task. The application of deep learning to a specific classification task requires many initialization choices including model structure and pre-training data. A comprehensive evaluation of the effect of these factors on CNN performance is critical to gain insight into how robustly deep learning addresses a classification task. Despite this need, researchers have been using a trial-and-error approach to optimize parameters in the process of building better performing models due to lack of interpretability of deep learning. In this study, we leveraged a large breast cancer screening cohort and several pre-training datasets to assess behaviors of our CNN models for a specific breast imaging classification.
We tested six classification tasks and the degree to which pre-training strategy made a significant impact on performance seems to be dependent on the classification task. Our study focus is on the identification of recalled-benign mammograms. In the case of distinguishing these images from malignant and benign mammograms, four out of five pre-training strategies produced significant performance differences compared to the baseline model. We consider these tasks to be sensitive to pre-training strategy. Tasks that were moderately sensitive to pre-training strategy include the triple task, benign vs recalled-benign, and malignant vs benign, each having three out of five experiments with significant performance changes. The classic diagnosis model, malignant vs. recalled-benign + benign, has only two out of five experiments producing statistically significant differences compared to the baseline.
The only experimental model that outperformed the baseline model for all tasks (except one) was experiment 3, models trained first on ImageNet and then on the Breast Density dataset before being fine-tuned to the target dataset. This small performance increase over the baseline seems intuitive based on how the pre-training datasets were ranked from least-to-most related to the target dataset. As the breast density dataset is the same imaging modality (digital mammography) and concerns the same organ (breast) as the target dataset, it is “most similar” and thus may have pre-learned features “closer to” ones useful for classification of the target dataset. Conversely, it is observed in Experiment 4 that training first on ImageNet and then on the large ChestX-Ray8 dataset significantly decreased the AUCs across all tasks. ChestX-Ray8 is of a different modality and images a different organ, which may explain the drop in performance. These comparisons imply that different modalities and organs of interest of medical image datasets may effect overall performance when used for pre-training.
In terms of different CNN network structure, we tested both AlexNet and ResNet-152. The ResNet-152 model (experiment 6) significantly outperformed the AlexNet model trained with the same training strategies and datasets (experiment 1), for the malignant vs benign and recalled-benign vs. benign + malignant tasks. The ResNet-152 model performed significantly worse than the AlexNet model for the triple task and the malignant vs recalled-benign task. The remaining two tasks had no significant differences in performance. For these reasons, we consider the AUCs for the two structures overall comparable. Although AlexNet has a simpler structure than ResNet-152, it may still be possible to achieve comparable performance to other popular CNNs, as made evident by our classification tasks and data. As this observation is also likely data and classification task dependent, the model structure should be tested on a case-by-case basis to determine which CNN model is optimal for a given scenario.
Our study has some limitations. First, our target dataset comes from a single institution. To be clinically useful, models must also be validated in the context of generalizability to external independent cohorts that cover several institutions, machines, and imaging protocols. Second, we were not realistically able to test all possible combinations of model structure and pre-training strategy. While our selected strategies are typical, we acknowledge that there are other interrelated factors meriting further assessment. Third, efforts on designing advanced networks for multi-view input are important to pursue. As shown in experiment 5, multi-view pre-training using the DDSM dataset significantly decreased the AUCs in comparison to single-view for all tasks (except one). This could be due to several reasons, including the fact that the training samples were reduced by half as compared to using the views independently. However, the format in which we combine the views may not optimally capture collaborative features.
Conclusion
In summary, we have performed an extensive technical analysis of deep learning on digital mammograms to distinguish false-recall from malignant and benign findings. We evaluated several pre-training strategies and found that the influence depended on the classification task. In particular, pre-training strategy influences significant performance differences in distinguishing recalled-benign images from others. Thus, advanced pre-training strategies are important to pursue for deep learning-based classification tasks.
Funding Information
This work was supported by the National Institutes of Health (NIH)/National Cancer Institute (NCI) grants (#1R01CA193603, #3R01CA193603-03S1, and #1R01CA218405), a Radiological Society of North America (RSNA) Research Scholar Grant (#RSCH1530), an Amazon AWS Machine Learning Research Award, and a University of Pittsburgh Physicians (UPP) Academic Foundation Award. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation grant number ACI-1548562. Specifically, it used the Bridges system, which is supported by NSF award number ACI-1445606, at the Pittsburgh Supercomputing Center (PSC).
Compliance with Ethical Standards
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siu AL, on behalf of the U.S. Preventive Services Task Force: Screening for Breast Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 164:279–296. 10.7326/M15-2886 [DOI] [PubMed]
- 2.Nelson HD, Tyne K, Naik A, Bougatsos C, Chan BK, Humphrey L: Screening for breast cancer: an update for the U.S. Preventive Services Task Force. Ann Intern Med. 151:727–737. 10.7326/0003-4819-151-10-200911170-00009 [DOI] [PMC free article] [PubMed]
- 3.Hubbard RA, Kerlikowske K, Flowers CI, Yankaskas BC, Zhu W, Miglioretti DL: Cumulative probability of false-positive recall or biopsy recommendation after 10 years of screening mammography: a cohort study. Ann Intern Med. 155:481–492. 10.7326/0003-4819-155-8-201110180-00004 [DOI] [PMC free article] [PubMed]
- 4.Brewer NT, Salz T, Lillie SE: Systematic review: the long-term effects of false-positive mammograms. Ann Intern Med. 146:502–510. 10.7326/0003-4819-146-7-200704030-00006 [DOI] [PubMed]
- 5.Lehman D, Arao RF, Sprague BL, et al. National performance benchmarks for modern screening digital mammography: update from the breast cancer surveillance consortium constance. Radiology. 2017;283(1):49–58. doi: 10.1148/radiol.2016161174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litjens G, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Samala RK, et al. Multi-task transfer learning deep convolutional neurals network: application to computer-aided diagnosis of breast cancer on mammograms. Phys Med Biol. 2017;62(23):8894. doi: 10.1088/1361-6560/aa93d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen L: End-to-end training for whole image breast cancer diagnosis using an all convolutional design. arXiv preprint arXiv:1708.09427, 2017
- 9.Aboutalib SS, et al: Deep learning to distinguish recalled but benign mammography images in breast cancer screening. Clinical Cancer Research, 2018 [DOI] [PMC free article] [PubMed]
- 10.Hoo-Chang S, et al: Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35: 5 1285, 2016 [DOI] [PMC free article] [PubMed]
- 11.Tajbakhsh N, et al: Convolutional neural networks for medical image analysis: Full training or fine tuning?. IEEE Trans Med Imaging 35: 5 1299-1312, 2016 [DOI] [PubMed]
- 12.Clancy K, et al: Deep learning for identifying breast cancer malignancy and false recalls: a robustness study on training strategy. Medical Imaging 2019: Computer-Aided Diagnosis. Vol. 10950. International Society for Optics and Photonics, 2019
- 13.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, and Fei-Fei L: (* = equal contribution) ImageNet Large Scale Visual Recognition
- 14.Wang X, et al: Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. Computer Vision and Pattern Recognition (CVPR), 2017 IEEE Conference on. IEEE, 2017
- 15.Heath M, et al: The digital database for screening mammography. Proceedings of the 5th international workshop on digital mammography. Medical Physics Publishing, 2000
- 16.Mohamed AA, et al. A deep learning method for classifying mammographic breast density categories. Med Phys. 2018;45(1):314–321. doi: 10.1002/mp.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krizhevsky A, Sutskever I, Hinton GE: Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012.
- 18.He K, et al: Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016
- 19.Bradski G: The OpenCV Library. Dr. Dobb’s Journal of Software Tools. 2000
- 20.Keller BM, et al. Estimation of breast percent density in raw and processed full field digital mammography images via adaptive fuzzy c-means clustering and support vector machine segmentation. Med Phys. 2012;39(8):4903–4917. doi: 10.1118/1.4736530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia Y, et al: Caffe: convolutional architecture for fast feature embedding. Proceedings of the 22nd ACM international conference on Multimedia. ACM, 2014
- 22.Landgrebe TCW, Duin RPW. Approximating the multiclass ROC by pairwise analysis. Pattern Recog Lett. 2007;28(13):1747–1758. doi: 10.1016/j.patrec.2007.05.001. [DOI] [Google Scholar]
- 23.Robin X, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yala A, et al: A deep learning mammography-based model for improved breast cancer risk prediction. Radiology. 182716, 2019 [DOI] [PubMed]
- 25.Rafferty EA, et al. Assessing radiologist performance using combined digital mammography and breast tomosynthesis compared with digital mammography alone: results of a multicenter, multireader trial. Radiology. 2013;266(1):104–113. doi: 10.1148/radiol.12120674. [DOI] [PMC free article] [PubMed] [Google Scholar]