Abstract
Polysaccharides from morels possess many characteristics beneficial to health, such as anti-tumor and immunomodulatory activities. The gut microbiota plays a critical role in the modulation of immune function. However, the impact of morel polysaccharides on the gut microbiota has not yet been explored. In this study, a high-throughput pyrosequencing technique was used to investigate the effects of MP, a new heteropolysaccharide extracted from wild morels, on the diversity and composition of microbiota along the intestine in mice, as well as the production of short-chain fatty acids (SCFAs). The results showed that MP treatment increased the number of operational taxonomic unit (OTUs) and diversity along the intestine, especially in the small intestine. MP treatment induced a significant decrease in the number of Firmicutes and a significant increase in the number of Bacteroidetes in the small intestine microbiota. It was also observed that the relative abundance of SCFA-producing bacteria, especially Lachnospiraceae, was increased in both the cecum and colon of MP-treated mice. Moreover, MP promoted the production of SCFAs in mice. These results provide a foundation for further understanding the health benefits conferred by morel polysaccharides.
Keywords: polysaccharide, wild morels, gut microbiota, short-chain fatty acids
INTRODUCTION
Morels are edible mushrooms appreciated worldwide for their high nutritional value and health-related benefits [1]. They contain several bioactive compounds, including polysaccharides, proteins, minerals, trace elements, dietary fiber, and vitamins. Several studies have recently shown that morels have nephroprotective [2], anti-hepatotoxic, hepatoprotective [3], anti-inflammatory [4, 5], and anti-tumor activities [4].
Many of the health benefits and bioactivities of morels are ascribed to their polysaccharides, and several studies have described these properties and health benefits. For example, the polysaccharide MEP-II isolated from the fermentation broth of Morchella esculenta was found to inhibit the proliferation of a human hepatoma cell line (HepG2) through an apoptotic pathway [6]. Furthermore, a M. esculenta polysaccharide (MEP) extracted by pulsed electric field (PEF) in submerged fermentation was found to have anti-proliferation and anti-tumor activities, and apoptosis tests proved that it could inhibit the proliferation and growth of human colon cancer HT-29 cells in a time- and dose-dependent manner [7]. A high-molecular-weight galactomannan, isolated from a polar extract of M. esculenta, exhibited immunostimulatory activity and resulted in the increased expression of NF-kB [8]. Two polysaccharides from M. esculenta, MEP-I and MEP-II, were found to have significant immune-modulating activity [9].
Polysaccharides can alter the composition of the gut microbiota and maintain health by promoting expansion of several health-promoting species—e.g., short-chain fatty acid (SCFA)-producing bacteria—and suppressing some potential pathogens [10]. For example, polysaccharides from purple sweet potatoes (PSPPs) were found to modulate intestinal bacteria content in mice by promoting the proliferation of SCFA-producing Lachnospiraceae and Oscillospira, as well as by reducing the abundance of Alcaligenaceae and Sutterella [11]. Jin et al. demonstrated that polysaccharides from the mycelia of Ganoderma lucidum (GLP) modulate the function of the intestinal biological barrier by both increasing microbiota richness and decreasing the Firmicutes-to-Bacteroidetes ratio (F/B) [12]. In addition, GLP was found to induce changes in certain intestinal bacteria, such as S24-7, SMB53, Rikenellaceae, Allobaculum, Rc4-4, and Ruminococcaceae [12].
Numerous studies have shown that gut microbes play a critical role in the development of the immune system and the modulation of immune function, and dysbiosis of the gut microbiota has been shown to be closely related to human health and disease [13, 14]. Therefore, it is important to understand the composition and characteristics of the gut microbiota after the administration of polysaccharides. Although the immunomodulatory, immunostimulatory, anti-proliferating, and anti-tumor activities of polysaccharides derived from morels have been intensively investigated [6,7,8,9], it is not yet clear how morel polysaccharides impact the characteristics and distribution of the gut microbiota.
Previously, we described a new heteropolysaccharide, MP, isolated from the fruiting body of wild morels [15]. Chemical characterization indicated that MP consists of D-mannose, D-glucose, D-galactose, and L-rhamnose, with a mass ratio of 43.15:19.56:20.25:1, and the average molecular weight of MP was estimated to be 3.974 × 103 kDa. The goal of this study was to investigate the effects of MP on microbiota diversity and composition in the intestine of mice using a high-throughput sequencing technique. The results will provide a foundation for understanding the mechanism behind the health benefits of MP, as well as any potential side effects.
MATERIALS AND METHODS
Materials and chemicals
Wild morels collected from the Qinling Mountains in Shaanxi province (China) were washed, dried in hot air at 60−70°C, crushed, and then passed through 100-mesh sieves to obtain morel powder. Polysaccharides were extracted from the dried morel powder as previously described [16]. Organic acids used as standards for gas chromatography, including acetic acid, propionic acid, isobutyric acid, n-butyric acid, isovaleric acid, n-valeric acid, hexanoic acid, and 2-ethylbutyric acid, were of chromatographic grade and purchased from Sigma-Aldrich (St. Louis, MO, USA). Other chemical reagents, including alcohol, phenol, and sulfuric acid, were of analytical grade and purchased from Shanghai Aladdin Biochemical Technology Co., Ltd (Shanghai, China). DEAE Sepharose Fast Flow resin was purchased from GE Healthcare (Chalfont St. Giles, UK).
Animals and experimental design
Male Kunming mice (KM; average weight of 20 ± 2.0 g) were obtained from the Animal Experimental Center at Xi’an Jiaotong University (certificate SCXK (Shaan) 2014-001). All animals were handled in accordance with the Guide for Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996). The mice were kept at a temperature of 22°C and a relative humidity of 50% ± 5% in a 12 hr light/dark cycle environment. After one week of adaptation, mice were divided into two groups for 8 consecutive days: (1) the P group (n=8), the members of which received gavage administration of MP (dissolved in sterile physiological saline, 1.5 mg/mL) 75 mg/kg body weight and were reared in the same cage, and (2) the CT group (n=8), the members of which received gavage administration with the same volume of sterile physiological saline and were reared in the same cage. On the last day, after intraperitoneal injection of chloral hydrate (0.1 mL), mice were sacrificed by cervical dislocation. The contents of the small intestine, cecum, and colon, as well as fecal samples, were collected separately under sterile conditions and immediately stored at −80°C until further analysis.
DNA extraction and gut microbe 16S rRNA sequencing
Four intestinal content samples were randomly selected from each group for 16S rRNA gene sequencing. Total microbial genomic DNA from the small intestine, cecum, and colon contents was extracted using a QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
PCR reactions targeting the V3–V4 region of bacterial 16S rRNA were performed using the forward primer 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR was carried out in a total reaction volume of 20 µL consisting of 13.25 µL H2O, 2.0 µL 10 × PCR Ex Taq Buffer, 0.5 µL DNA template (100 ng mL−1), 1 µL forward and reverse primers (10 mM), 2.0 µL dNTP, and 0.25 µL Ex Taq (5 U mL−1). After initial denaturation at 98°C for 2 min, amplification was performed using 30 cycles of incubations at 98°C for 30 sec, 50°C for 30 sec, and 72°C for 60 sec, followed by a final extension at 72°C for 5 min. The amplified products were purified and recovered using a 1.0% agarose gel electrophoresis method. Finally, the libraries were constructed, and the barcoded V3−V4 PCR amplicons were sequenced using an Illumina HiSeq platform (Illumina, San Diego, CA, USA). The extraction of genomic DNA, the amplification of the 16S rRNA V3–V4 region, the library construction, and the sequencing were completed by Beijing Biomarker Technologies Co., Ltd. (Beijing, China).
Bioinformatic analysis
The bioinformatic analysis done in this study was completed using the Biomarker biocloud platform (www.biocloud.net). To obtain the raw tags, the raw paired-end reads from the original DNA fragments were merged using FLASH (version 1.2.11) [17]. Clean tags were then obtained after filtering for quality and length. Tags with an average quality score greater than 20 in a 50 bp sliding window were truncated using Trimmomatic (version 0.33), and tags shorter than 300 bp were removed [18]. Finally, effective tags for further bioinformatic analysis were obtained after removing possible chimeras, which were identified using UCHIME (version 8.0) [19], a tool included in Mothur [20].
Effective tags were clustered using USEARCH [21, 22], and tags with similarity greater than or equal to 97% were considered an operational taxonomic unit (OTU). Taxonomy was assigned to all OTUs by searching against the SILVA databases (Release132, http://www.arb-silva.de) [23] using the RDP classifier within QIIME [24]. Alpha diversity (ACE, Chao1, and Shannon) analysis, Venn diagrams, principal component analysis (PCA), and linear discriminant analysis (LDA) effect size (LEfSe) were further processed with a BMKCloud online bioinformatics tool (Biomarker Technologies Corporation, Beijing, China).
Determination of the levels of SCFAs
Four fecal samples were randomly selected from each group for the determination of SCFA levels by gas chromatography (GC) according to the method detailed by Zhao et al. [25]. Fecal samples (0.1 g) were added to 0.5 mL of deionized water and shaken for about 3 min to yield a 17% (w/w) fecal suspension. The pH of the suspension was adjusted to 2–3 by adding HCl, and the suspension was then centrifuged at 12,000 × g for 20 min to yield a clear supernatant. The internal standard, 2-ethylbutyric acid solution, was spiked into the supernatant to a final concentration of 1 mM, and the supernatant was injected into the GC for analysis. Acetic acid, propionic acid, isobutyric acid, n-butyric acid, isovaleric acid, n-valeric acid, and hexanoic acid served as standards.
Chromatographic analysis was carried out using a Thermo Trace 1300 GC system (Thermo Fisher Scientific Inc., Waltham, MA, USA) equipped with a flame ionization detector (FID). A fused-silica capillary column with a free fatty acid phase (DB-FFAP, J&W Scientific, Agilent Technologies Inc., Palo Alto, CA, USA) of 30 m × 0.25 mm i.d. was used. Helium was supplied as the carrier gas at a flow rate of 1.0 mL min−1. The initial oven temperature of 50°C was maintained for 1.0 min, raised to 120°C at a rate of 15°C min−1, increased to 170°C at a rate of 5°C min−1, increased to 240°C at a rate of 15°C min−1, and then finally held at 240°C for 3 min. The temperature of the injection port was 250°C, and the split ratio was 25:1. The temperature of the FID detector was 260°C, and the flow rates of hydrogen, air, and nitrogen as makeup gases were 30, 300, and 20 mL min−1, respectively. The injected sample volume for GC analysis was 1 µL, and the run time for each analysis was 23.5 min. Data handling was carried out using the Xcalibur software.
Statistical analysis
Differences between the CT and P groups were analyzed by t-test using the GraphPad Prism 5.0 software. Differences were considered to be statistically significant if the p-values were less than 0.05.
RESULTS
Diversity of the gut microbiota along the gastrointestinal tract in non-treated and MP-treated mice
Multiplex pyrosequencing targeting the V3–V4 region of bacterial 16S rRNA gene was employed to characterize the diversity of gut microbiota along the gastrointestinal tract of mice. After merging, filtering, and removing chimeras, 1421571 and 1763418 clean bacterial 16S rRNA gene reads were obtained from the CT and P groups, respectively, and used in the final analysis (Supplementary Table 1). OTUs in each group were defined based on sequence identity greater than 97%. On average, 293 and 350 OTUs were defined for the CT and P groups, respectively (Supplementary Table 2).
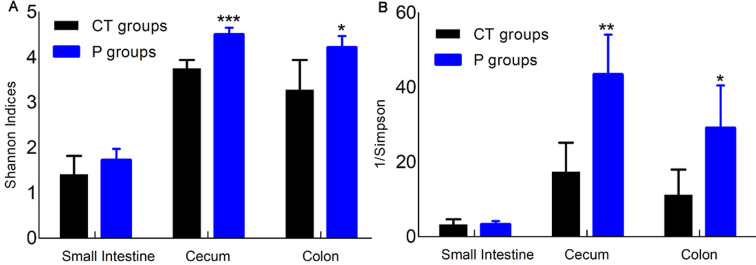
Table 1 shows that the cecum and colon samples had a higher diversity of gut microbiota than the small intestine samples in both the CT and P groups (p<0.01 or p<0.001). There were no significant differences in the ACE indices of the small intestine, cecum, or colon microbiota between the CT and P groups. However, MP treatment significantly increased the numbers of OTUs in the small intestine, cecum, and colon microbiota (p<0.05 or p<0.01) compared with the CT group. Moreover, there were significant increases in the Chao1 indices of the small intestine after administration of MP. Figure 1 shows that the Shannon and Simpson indices were similar in the small intestines of mice in both the CT and P groups. However, MP treatment increased the Shannon indices and decreased the Simpson indices of gut microbiota in the cecum and colon compared with the CT group.
Table 1. Diversity of gut microbiota in non-treated mice (CT) and MP-treated mice (P).
| Sample type | Groups | OTUs | ACE | Chao1 |
|---|---|---|---|---|
| Small intestine | CT | 131.80 ± 33.78 | 233.20 ± 49.79 | 191.50 ± 44.82 |
| P | 226.00 ± 64.80* | 309.20 ± 54.15 | 289.90 ± 60.84* | |
| Cecum | CT | 381.30 ± 26.02 | 405.50 ± 21.77 | 412.00 ± 29.37 |
| P | 424.80 ± 14.86* | 433.30 ± 13.36 | 433.20 ± 14.74 | |
| Colon | CT | 365.30 ± 17.48 | 402.00 ± 17.45 | 403.90 ± 20.39 |
| P | 399.50 ± 5.26** | 415.10 ± 4.77 | 419.30 ± 6.68 |
T-tests were used to evaluate significant differences in the diversity of gut microbiota. Each value is presented as the mean ± SD. *p<0.05 vs. CT; **p<0.01 vs. CT. OTU: operational taxonomic unit.
Fig. 1.
Shannon (A) and Simpson (B) indices of gut microbiota in non-treated (CT) and MP-treated (P) mice. T-tests were used to evaluate significant differences for the Shannon and Simpson indices of gut microbiota. *p<0.05, **p<0.01, ***p<0.001.
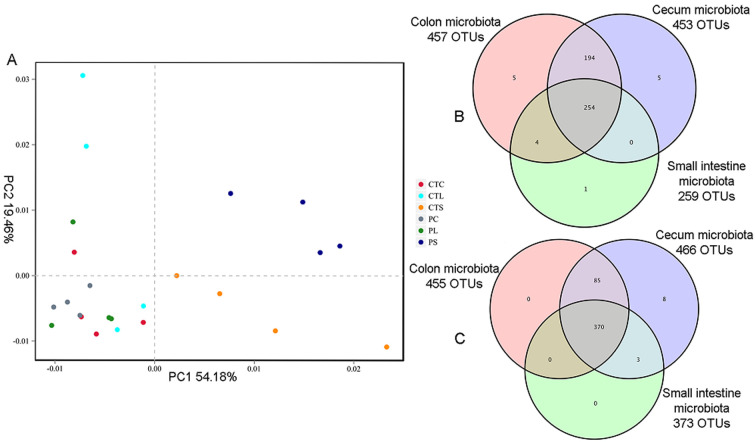
Principal component analysis (PCA) was employed to assess the phylogenetic differences and similarities within the gut microflora (Fig. 2A). The results revealed that each mouse was distinct from all of the others and that there was a high degree of variation between individuals (Fig. 2). The first principal coordinate (PC1), which accounted for 54.18% of the variance in the data, could completely separate the microbiota of the CT and P groups in the small intestine from those of the CT and P groups in the cecum and colon, indicating that there were significant differences in the diversity of microbiota between the small intestine and cecum or the small intestine and colon. The second principal coordinate (PC2), which accounted for 19.46% of the variance in the data, could completely separate the microbiota of the CT group from those of the P group in the small intestine. The diversity of bacterial populations along the intestinal tract was also compared using OTU overlaps (Fig. 2B and 2C). The total numbers of OTUs in the small intestine, cecum, and colon microbiota were 259, 453, and 457 for the CT group and 373, 466, and 455 for the P group, and the two groups had 254 and 370 OTUs in common (Fig. 2B and 2C).
Fig. 2.
PCA (A) analysis of gut microbiota and operational taxonomic unit (OTU) overlaps of non-treated mice (B) and MP-treated mice (C) along the intestinal tract.
MP treatment shifts the composition of microbiota in the small intestine, cecum, and colon
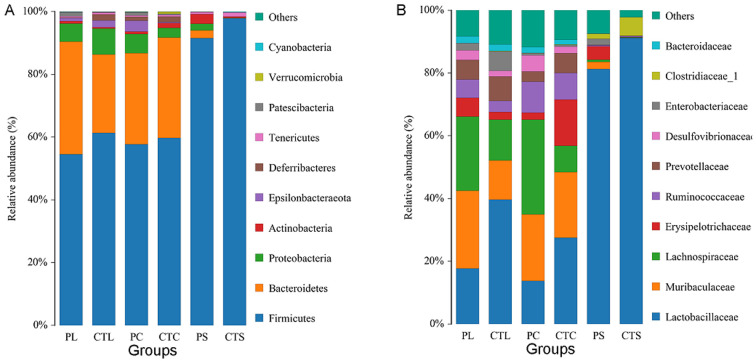
To assess specific changes in the gut microbiota, several predominant taxa from each group were selected for analysis (Fig. 3, Tables 2, 3, 4 ). At the phylum level, Firmicutes was the most predominant phyla, accounting for 97.98% of total bacteria in the small intestine in the CT group (Fig. 3A, Table 2). However, MP treatment decreased the abundance of Firmicutes and significantly increased the proportion of Bacteroidetes in the small intestine compared with the CT group (Table 2, p<0.05). This indicates that MP treatment decreased the F/B ratio in the small intestine. Moreover, MP treatment significantly increased the amount of Epsilonbacteraeota in the cecum compared with the CT group, while there was no obvious difference in the composition of colon microbiota between the CT and P groups (Table 2).
Fig. 3.
Comparison of small intestine (CTS, PS), cecum (CTC, PC) and colon (CTL, PL) microbiota at the level of the phylum (A) and family (B). Each taxonomic phylum and family accounted for the proportion of identified classification. CTS (PS), CTC (PC), and CTL (PL) correspond to small intestine, cecum, and colon of mice of the CT (P) group, respectively.
Table 2. Relative abundance of predominant taxa at the phylum level.
| Sample type | Small intestine | Cecum | Colon | |||
|---|---|---|---|---|---|---|
| Groups | CT | P | CT | P | CT | P |
| Firmicutes (%) | 97.98 ± 2.04 | 91.57 ± 4.21* | 59.24 ± 5.45 | 57.30 ± 8.36 | 361.33 ± 6.53 | 53.92 ± 5.42 |
| Bacteroidetes (%) | 0.13 ± 0.07 | 2.49 ± 1.63* | 32.29 ± 6.70 | 29.37 ± 9.51 | 25.01 ± 8.84 | 36.43 ± 7.08 |
| Proteobacteria (%) | 0.10 ± 0.13 | 2.08 ± 3.93 | 3.02 ± 1.83 | 6.29 ± 2.99 | 8.23 ± 8.99 | 5.93 ± 3.86 |
| Actinobacteria (%) | 0.34 ± 0.39 | 3.04 ± 4.76 | 1.66 ± 1.11 | 0.77 ± 0.44 | 0.42 ± 0.21 | 0.77 ± 0.56 |
| Epsilonbacteraeota (%) | 0.00 ± 0.00 | 0.02 ± 0.01 | 0.35 ± 0.19 | 3.36 ± 1.68* | 2.17 ± 2.56 | 0.93 ± 1.28 |
T-tests were used to evaluate the significant differences in each bacterial population at the phylum level. Each value is presented as the mean ± SD. *p<0.05 vs. CT. CT: non-treated mice, P: MP-treated mice..
Table 3. Relative abundance of predominant taxa at the family level.
| Sample type | Small intestine | Cecum | Colon | |||
|---|---|---|---|---|---|---|
| Groups | CT | P | CT | P | CT | P |
| Lactobacillaceae (%) | 91.72 ± 6.88 | 81.23 ± 8.05 | 27.32 ± 7.59 | 13.39 ± 7.03* | 39.68 ± 9.84 | 16.59 ± 11.25* |
| Muribaculaceae (%) | 0.11 ± 0.07 | 2.34 ± 1.67* | 21.10 ± 10.27 | 21.47 ± 10.59 | 12.46 ± 8.52 | 25.08 ± 8.88 |
| Lachnospiraceae (%) | 0.15 ± 0.12 | 0.67 ± 0.74 | 8.18 ± 5.74 | 30.01 ± 10.13** | 13.04 ± 4.95 | 23.76 ± 4.28* |
| Erysipelotrichaceae (%) | 0.05 ± 0.04 | 4.25 ± 5.61 | 15.07 ± 10.30 | 2.34 ± 2.24 | 2.38 ± 2.97 | 6.17 ± 6.87 |
| Ruminococcaceae (%) | 0.31 ± 0.53 | 0.33 ± 0.16 | 8.00 ± 3.80 | 9.88 ± 3.71 | 3.59 ± 3.57 | 6.15 ± 2.42 |
| SCFA-Producing Bacteria (%) | 0.51 ± 0.68 | 5.24 ± 5.17 | 31.26 ± 4.69 | 42.22 ± 5.30* | 19.00 ± 6.77 | 36.08 ± 7.40* |
T-tests were used to evaluate the significant differences in each bacterial population at the family level. Each value is presented as the mean ± SD. *p<0.05 vs. CT; **p<0.01 vs. CT. CT: non-treated mice, P: MP-treated mice, SCFA: short-chain fatty acid.
Table 4. Relative abundance of predominant taxa at the genus level.
| Sample type | Small intestine | Cecum | Colon | |||
|---|---|---|---|---|---|---|
| Groups | CT | P | CT | P | CT | P |
| Lachnospiraceae_NK4A136_group (%) | 0.05 ± 0.01 | 0.29 ± 0.32 | 0.97 ± 0.62 | 7.31 ± 3.69* | 2.41 ± 1.49 | 10.07 ± 5.05* |
| Dubosiella (%) | 0.03 ± 0.03 | 3.08 ± 3.76 | 10.03 ± 11.14 | 1.53 ± 1.77 | 0.84 ± 1.29 | 3.92 ± 4.42 |
| Prevotellaceae_UCG-001 (%) | 0.00 ± 0.00 | 0.05 ± 0.07 | 4.98 ± 2.44 | 2.09 ± 1.04 | 5.88 ± 5.71 | 3.13 ± 2.20 |
| Escherichia-Shigella (%) | 0.01 ± 0.01 | 1.99 ± 3.95 | 0.62 ± 0.59 | 0.58 ± 0.86 | 6.15 ± 10.26 | 2.13 ± 4.08 |
| Roseburia (%) | 0.02 ± 0.04 | 0.05 ± 0.07 | 2.90 ± 5.06 | 3.12 ± 1.32 | 3.26 ± 4.22 | 1.72 ± 0.58 |
T-tests were used to evaluate the significant differences in each bacterial population at the genus level. Each value is presented as the mean ± SD. *p<0.05 vs. CT. CT: non-treated mice, P: MP-treated mice.
Obvious changes were observed in the gut microbiota at the family level (Fig. 3, Table 3). Compared with the CT group, mice in the P group exhibited a decrease in the relative abundance of Lactobacillaceae and a significant increase in the relative abundance of Lachnospiraceae in both the cecum and colon (p<0.05 or p<0.01), while no significant changes were observed in the composition of the microbiota of the small intestine between the CT and P groups. Some species in the predominant families Lachnospiraceae, Ruminococcaceae, and Erysipelotrichaceae are capable of producing SCFAs, especially butyrate and propionate [26]. The total levels of Lachnospiraceae, Ruminococcaceae, and Erysipelotrichaceae (SCFA-producing bacteria; Table 3) were significantly increased after administration of MP in both the cecum and colon in the P group compared with the CT group.
To validate the differences in the gut microbiota between the CT and P groups, several predominant microorganisms at the genus level were analyzed and compared (Table 4). The MP-treated group exhibited distinct increases in the relative abundances of Lachnospiraceae_NK4A136_group (%) in the cecum and colon microbiota compared with the CT group. No significant change was observed in the small intestine microbiota between the CT and P groups.
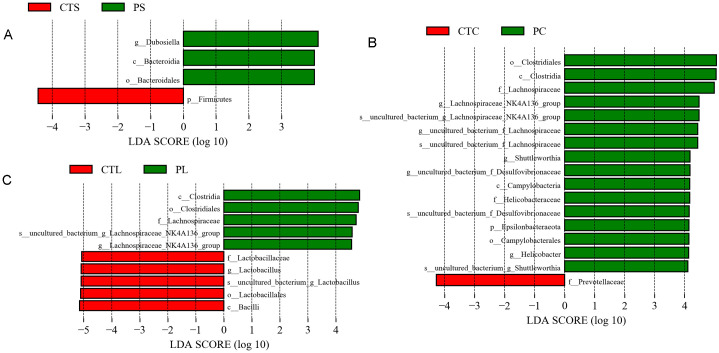
LEfSe was employed to identify specific taxa (from phylum to species levels) that were statistically different between the CT and P groups. A pairwise comparison between the CT and P groups indicated that there were 4 significantly different taxa in the small intestine, 17 significantly different taxa in the cecum, and 10 significantly different taxa in the colon microbiota (Fig. 4). For both the cecum and colon microbiota, the family Lachnospiraceae was higher in the P group compared with the CT group (LDA score >4, Fig. 4), which was consistent with the results shown in Table 3.
Fig. 4.
Comparisons of small intestine (A), cecum (B), and colon (C) bacteria using linear discriminant analysis effect size (LEfSe). The histogram shows the linear discriminant analysis (LDA) scores computed for features from phylum to species level.
MP treatment shifts the production of SCFAs
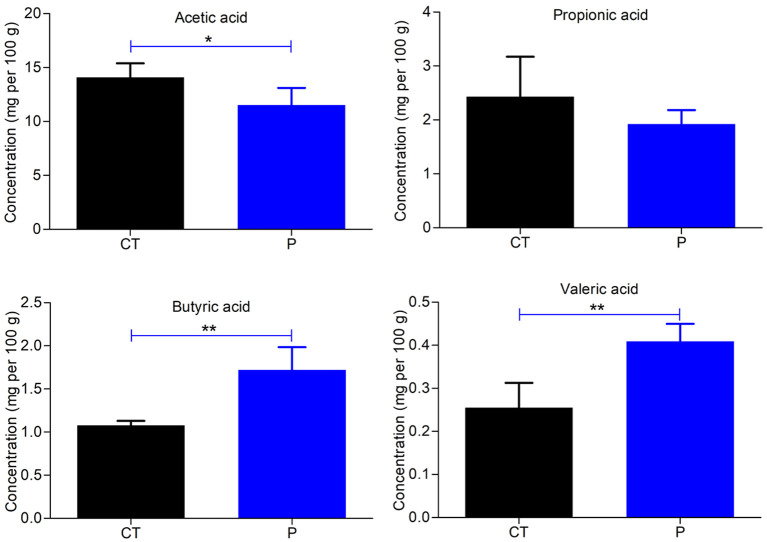
Acetic acid, propionic acid, butyric acid, and valeric acid are the four main SCFAs found in the intestine, and all are beneficial for human health. To assess the effects of MP on the production of SCFAs in mice, gas chromatography was used to determine the level of SCFAs in fecal samples. As shown in Fig. 5, the concentration of acetic acid was notably lower in the P group compared with the CT group (p<0.05 or p<0.01). Moreover, MP administration resulted in an increase in the production of butyric acid and valeric acid in mice compared with the CT group (p<0.01, Fig. 5).
Fig. 5.
Effects of the polysaccharide extracted from morels on short-chain fatty acid (SCFA) production (mg per 100 g) in mice. T tests were used to evaluate significant differences for SCFA production. Each value was presented as the mean ± SD. *p<0.05, **p<0.01.
DISCUSSION
Gut microbiota composition has been associated with human health and disease. However, feces do not fully reflect the microbial ecology of the intestine [27, 28]. Therefore, it is essential to investigate the characteristics and structure of microbiota along the mouse gastrointestinal tract [29]. Some polysaccharides are nondigestible carbohydrates and are not fully digested in the upper gut. It has become clear that nondigestible carbohydrates are healthy, and their health benefits are attributed to their contribution to the gut microbiota [10, 11]. It has been shown that nondigestible carbohydrates have a large impact on the characteristics and distribution of gut microbiota [30, 31]. Several studies have recently indicated that nondigestible carbohydrates affect the profiles of gut microbiota in a manner dependent on the structure of polysaccharides, including their glycosidic linkages and monosaccharide compositions [30,31,32,33]. As newly identified prebiotic resources, mushroom polysaccharides have garnered much attention [34, 35]. Several studies over the years have demonstrated that mushroom polysaccharides could alter the composition of the gut microbiota. Xu et al. reported that L. edodes-derived polysaccharide L2 could reverse age-related changes in the composition of the gut microbiota, such as the reduction in the ratio of Firmicutes to Bacteroidetes, the increase in the levels of Bacteroidia and Bacteroidaceae, and the decrease in the levels of Bacilli, Betaproteobacteria, Lactobacillaceae, and Alcaligenaceae [36]. The study by Chang et al. showed that Ganoderma lucidum could reduce obesity in mice by modulating the composition of the gut microbiota, indicating that G. lucidum and its high-molecular-weight polysaccharides may be used as prebiotic agents to prevent gut dysbiosis and obesity-related metabolic disorders [37]. Polysaccharides extracted from morels have also been intensively investigated for their potential medical applications and bioactivities [6,7,8,9]. However, the effect of polysaccharides from morels on gut microbiota has rarely been reported, and this needs to be studied in greater detail.
Previously, a new heteropolysaccharide, MP, isolated from the fruiting body of wild morels was described [15]. The total carbohydrate and protein contents of MP were 88.01% and 3.54%, respectively, and the UV spectra of MP demonstrated that the sample was free of nucleic acids [15]. The results of HPLC showed that MP mainly consisted of D-mannose, D-glucose, D-galactose, and L-rhamnose, with the mass ratio being 43.15:19.56:20.25:1 [15]. It has been considered that the molecular weights of polysaccharides have a great relationship with their biological activity; most polysaccharides with medicinal properties are molecules with a molecular weight above 100 kDa [38]. The average molecular weight of MP was found to be 3.974 × 103 kDa, implying that MP is certain to have medicinal value [15]. In this study, the impact of a mushroom polysaccharide extracted from morels (MP) on microbiota diversity and composition along the mouse intestine was explored using a high-throughput pyrosequencing technique. We examined the effects of MP treatment on the diversity of gut microbiota along the gastrointestinal tract in mice and found that MP increased the number of OTUs and bacterial diversity. Recent studies have shown that low levels of microbiota species and diversity are associated with abnormal immune function [39]. Our results indicated that MP was beneficial for host health.
Comparisons of the predominant gut microbiota at the phylum level revealed that Firmicutes and Bacteroidetes were the two most prevalent phyla in the small intestine, cecum, and colon among the two groups. However, in the small intestine after administration of MP, the relative abundance of Bacteroidetes was notably increased, whereas that of Firmicutes was significantly decreased. Bacteroidetes can metabolize many complex carbohydrates using a series of membrane protein complexes, termed Sus-like systems [40]. For example, it was found that nearly all of the major plant and host glycans could be utilized by B. thetaiotaomicron and B. ovatus [41]. It was also found that the phylum Bacteroidetes was enriched in carbohydrate metabolic pathways, whereas the phylum Firmicutes possessed a smaller number of polysaccharide-degrading enzymes [42, 43]. Therefore, the enrichment of Bacteroidetes in MP-treated mice could be related to the degradation of MP in the mouse intestine.
At the family level, MP was found to notably increase the relative abundance of Lachnospiraceae in the cecum and colon. It is well known that plant polysaccharides are not digested by human enzymes but are instead processed by absorbable SCFAs secreted by gut bacteria [28]. It has been reported that Ruminococcaceae and Lachnospiraceae are responsible for the degradation of polysaccharides [44, 45]. Meanwhile, some species in the families of Lachnospiraceae, Ruminococcaceae, and Erysipelotrichaceae are capable of producing SCFAs, especially butyrate and propionate [26]. In our study, the total levels of the SCFA-producing bacteria Lachnospiraceae, Ruminococcaceae, and Erysipelotrichaceae along the intestine were compared between the CT and P groups, and the results showed that the relative abundance of SCFA-producing bacteria in the cecum and colon was significantly increased after the administration of MP. These results indicated that MP promoted the proliferation of SCFA-producing bacteria, especially Lachnospiraceae, resulting in an increase in the production of SCFAs.
To further verify these results, the levels of SCFAs in fecal samples from non-treated and MP-treated mice were measured using gas chromatography. These results showed that, following MP treatment, the concentration of acetic acid in the fecal samples notably decreased, while the concentrations of butyric acid and valeric acid both significantly increased. The decrease in acetic acid may be due to SCFA-producing bacteria consuming acetate to produce butyrate via butyryl-CoA:acetate CoA-transferase [26]. It is believed that increased levels of SCFAs produced by SCFA-producing bacteria can acidify the intestinal environment to protect it against pathogenic bacteria [46].
SCFAs have been reported to play important roles in the regulation of host immunity [47,48,49]. It has been demonstrated that SCFAs regulate gut immunity by inducing regulatory T cells (Treg cells) through the inhibition of histone deacetylase (HDAC) [50,51,52]. It has also been shown that butyrate can function as a ligand of the G-protein-coupled receptor, GPR109a, expressed on dendritic cells (DCs) and induce the production of retinoic acid and IL-10, leading to the expansion of Treg cells [53]. Considering these positive effects on host physiology and immunity, our results indicated that MP treatment could enhance the immunity of the host and was beneficial for host health. However, the extent of the effects of MP on host immunity is still not fully understood and needs further investigation.
In conclusion, MP treatment increased the number of OTUs and diversity along the gastrointestinal tract of mice, especially in the small intestine. It also altered the composition of the gut microbiota along the intestine. At the phylum level, the relative abundance of Bacteroidetes was increased, while the relative abundance of Firmicutes was decreased in the small intestine. At the family level, MP treatment increased the levels of Lachnospiraceae in both the cecum and colon. Moreover, It promoted the production of SCFAs, such as butyric acid and valeric acid, in mice.
Supplementary Material
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
REFERENCES
- 1.Tietel Z, Masaphy S. 2018. True morels (Morchella)-nutritional and phytochemical composition, health benefits and flavor: a review. Crit Rev Food Sci Nutr 58: 1888–1901. [DOI] [PubMed] [Google Scholar]
- 2.Nitha B, Janardhanan KK. 2008. Aqueous-ethanolic extract of morel mushroom mycelium Morchella esculenta, protects cisplatin and gentamicin induced nephrotoxicity in mice. Food Chem Toxicol 46: 3193–3199. [DOI] [PubMed] [Google Scholar]
- 3.Nitha B, Fijesh PV, Janardhanan KK. 2013. Hepatoprotective activity of cultured mycelium of Morel mushroom, Morchella esculenta. Exp Toxicol Pathol 65: 105–112. [DOI] [PubMed] [Google Scholar]
- 4.Nitha B, Meera C, Janardhanan KK. 2007. Anti-inflammatory and antitumour activities of cultured mycelium of morel mushroom, Morchella esculenta. Curr Sci 92: 235–239. [Google Scholar]
- 5.Kim JA, Lau E, Tay D, De Blanco EJ. 2011. Antioxidant and NF-κB inhibitory constituents isolated from Morchella esculenta. Nat Prod Res 25: 1412–1417. [DOI] [PubMed] [Google Scholar]
- 6.Hu M, Chen Y, Wang C, Cui H, Duan P, Zhai T, Yang Y, Li S. 2013. Induction of apoptosis in HepG2 cells by polysaccharide MEP-II from the fermentation broth of Morchella esculenta. Biotechnol Lett 35: 1–10. [DOI] [PubMed] [Google Scholar]
- 7.Liu C, Sun Y, Mao Q, Guo X, Li P, Liu Y, Xu N. 2016. Characteristics and antitumor activity of morchella esculenta polysaccharide extracted by pulsed electric field. Int J Mol Sci 17: 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan CJ, Pugh N, Pasco DS, Ross SA. 2002. Isolation of a galactomannan that enhances macrophage activation from the edible fungus Morchella esculenta. J Agric Food Chem 50: 5683–5685. [DOI] [PubMed] [Google Scholar]
- 9.Cui HL, Chen Y, Wang SS, Kai GQ, Fang YM. 2011. Isolation, partial characterisation and immunomodulatory activities of polysaccharide from Morchella esculenta. J Sci Food Agric 91: 2180–2185. [DOI] [PubMed] [Google Scholar]
- 10.Tang C, Ding R, Sun J, Liu J, Kan J, Jin C. 2019. The impacts of natural polysaccharides on intestinal microbiota and immune responses—a review. Food Funct 10: 2290–2312. [DOI] [PubMed] [Google Scholar]
- 11.Tang C, Sun J, Zhou B, Jin C, Liu J, Kan J, Qian C, Zhang N. 2018. Effects of polysaccharides from purple sweet potatoes on immune response and gut microbiota composition in normal and cyclophosphamide treated mice. Food Funct 9: 937–950. [DOI] [PubMed] [Google Scholar]
- 12.Jin M, Zhu Y, Shao D, Zhao K, Xu C, Li Q, Yang H, Huang Q, Shi J. 2017. Effects of polysaccharide from mycelia of Ganoderma lucidum on intestinal barrier functions of rats. Int J Biol Macromol 94Pt A: 1–9. [DOI] [PubMed] [Google Scholar]
- 13.Xu X, Xu P, Ma C, Tang J, Zhang X. 2013. Gut microbiota, host health, and polysaccharides. 31: 318-337. [DOI] [PubMed] [Google Scholar]
- 14.Sommer F, Bäckhed F. 2013. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 11: 227–238. [DOI] [PubMed] [Google Scholar]
- 15.Huo W, Feng Z, Hu S, Cui L, Qiao T, Dai L, Qi P, Zhang L, Liu Y, Li J. 2020. Effects of polysaccharides from wild morels on immune response and gut microbiota composition in non-treated and cyclophosphamide-treated mice. Food Funct 11: 4291–4303. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Yin TT, Zhang ST. 2014. Isolation, purification, and characterization of polysaccharides from wide Morchella esculenta (L.) Pers. Int J Food Prop 18: 1385–1390. [Google Scholar]
- 17.Tanja M, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 21: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- 22.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10: 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao G, Nyman M, Jönsson JA. 2006. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomed Chromatogr 20: 674–682. [DOI] [PubMed] [Google Scholar]
- 26.Louis P, Flint HJ. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19: 29–41. [DOI] [PubMed] [Google Scholar]
- 27.Lavelle A, Lennon G, Docherty N, Balfe A, Mulcahy HE, Doherty G, O Donoghue D, Hyland JM, Shanahan F, Sheahan K, Coffey JC, Winter DC, O Connell PR. 2013. Depth-dependent differences in community structure of the human colonic microbiota in health. PLoS One 8: e78835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, Zhang X. 2015. Lentinula edodes-derived polysaccharide alters the spatial structure of gut microbiota in mice. PLoS One 10: e0115037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu S, Chen D, Zhang JN, Lv X, Wang K, Duan LP, Nie Y, Wu XL. 2013. Bacterial community mapping of the mouse gastrointestinal tract. PLoS One 8: e74957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9: 577–589. [DOI] [PubMed] [Google Scholar]
- 31.Jeffery IB, O’Toole PW. 2013. Diet-microbiota interactions and their implications for healthy living. Nutrients 5: 234–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J Abell GC, Christophersen CT, McOrist AL, Clarke JM. 2011. Dietary resistant and butyrylated starches have different effects on the faecal bacterial flora of azoxymethane-treated rats. Br J Nutr 105: 1480–1485. [DOI] [PubMed] [Google Scholar]
- 33.Tachon S, Zhou J, Keenan M, Martin R, Marco ML. 2013. The intestinal microbiota in aged mice is modulated by dietary resistant starch and correlated with improvements in host responses. FEMS Microbiol Ecol 83: 299–309. [DOI] [PubMed] [Google Scholar]
- 34.Chou WT, Sheih IC, Fang TJ. 2013. The applications of polysaccharides from various mushroom wastes as prebiotics in different systems. J Food Sci 78: M1041–M1048. [DOI] [PubMed] [Google Scholar]
- 35.Aida FMNA, Shuhaimi M, Yazid M, Maaruf AG. 2009. Mushroom as a potential source of prebiotics: a review. Trends in Food Science & Technology 20: 0-575. [Google Scholar]
- 36.Xu X, Yang J, Ning Z, Zhang X. 2015. Lentinula edodes-derived polysaccharide rejuvenates mice in terms of immune responses and gut microbiota. Food Funct 6: 2653–2663. [DOI] [PubMed] [Google Scholar]
- 37.Chang CJ, Lin CS, Lu CC, Martel J, Ko YF, Ojcius DM, Tseng SF, Wu TR, Chen YY, Young JD, Lai HC. 2015. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat Commun 6: 7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su CA, Xu XY, Liu DY, Wu M, Zeng FQ, Zeng MY, Wei W, Jiang N, Luo X. 2013. Isolation and characterization of exopolysaccharide with immunomodulatory activity from fermentation broth of Morchella conica. Daru 21: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehman A, Lepage P, Nolte A, Hellmig S, Schreiber S, Ott SJ. 2010. Transcriptional activity of the dominant gut mucosal microbiota in chronic inflammatory bowel disease patients. J Med Microbiol 59: 1114–1122. [DOI] [PubMed] [Google Scholar]
- 40.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284: 24673–24677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9: e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1: 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolam DN, Sonnenburg JL. 2011. Mechanistic insight into polysaccharide use within the intestinal microbiota. Gut Microbes 2: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shang Q, Shan X, Cai C, Hao J, Li G, Yu G. 2018. Correction: Dietary fucoidan modulates the gut microbiota in mice by increasing the abundance of Lactobacillus and Ruminococcaceae. Food Funct 9: 655. [DOI] [PubMed] [Google Scholar]
- 45.Zheng CJ, Liu R, Xue B, Luo J, Gao L, Wang Y, Ou S, Li S, Peng X. 2017. Impact and consequences of polyphenols and fructooligosaccharide interplay on gut microbiota in rats. Food Funct 8: 1925–1932. [DOI] [PubMed] [Google Scholar]
- 46.Kim HJ, White PJ. 2009. In vitro fermentation of oat flours from typical and high beta-glucan oat lines. J Agric Food Chem 57: 7529–7536. [DOI] [PubMed] [Google Scholar]
- 47.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. 2014. The role of short-chain fatty acids in health and disease. Adv Immunol 121: 91–119. [DOI] [PubMed] [Google Scholar]
- 48.Chen T, Noto D, Hoshino Y, Mizuno M, Miyake S. 2019. Butyrate suppresses demyelination and enhances remyelination. J Neuroinflammation 16: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canfora EE, Jocken JW, Blaak EE. 2015. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol 11: 577–591. [DOI] [PubMed] [Google Scholar]
- 50.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504: 446–450. [DOI] [PubMed] [Google Scholar]
- 53.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. 2014. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.