Abstract
Tripartite motif–containing protein 21 (TRIM21) is a cytosolic antibody receptor that targets the internalized virus–antibody complex to the proteasome for degradation. However, the precise mechanism regulating TRIM21 activity is unknown. Here we show that TRIM21 is a substrate of histone deacetylase 6 (HDAC6) and that its function is regulated by acetylation. HDAC6 interacts with TRIM21 through its PRYSPRY motif and deacetylates TRIM21 at lysine 385 and lysine 387, thus promoting its homodimerization. Inhibiting HDAC6 activity increases TRIM21 acetylation, and hyperacetylation blocks TRIM21 dimerization and ubiquitination, preventing its binding to the virus–antibody complex and its degradation via the ubiquitin–proteasome pathway. HDAC6 depletion or inhibition increases virus accumulation in cells, indicative of an impaired capacity for antibody-dependent intracellular neutralization of viruses, whereas TRIM21 acetylation-deficient K385/387R mutant rescues HDAC6 depletion–caused ADIN impairment. These findings provide evidence for HDAC6 as a novel regulator of TRIM21-mediated intracellular innate immunity.
Keywords: HDAC6, TRIM21, virus, antibody-dependent intracellular neutralization, acetylation, histone deacetylase 6, HDAC6, infection
Tripartite motif-containing protein 21 (TRIM21) belongs to the TRIM family of proteins that share a common multidomain architecture comprising an N-terminal RING domain with E3 ubiquitin ligase activity, a B-box domain, and a helical coiled-coil domain. The C-terminal PRYSPRY motif of TRIM21 determines its binding specificity to the Fc region of IgG antibody (1). TRIM21 was first reported to interact with the Fc domain of human IgG1 in a yeast two-hybrid screen (2). The significance of this interaction was revealed by a model for antibody-dependent intracellular neutralization (ADIN) involving the nonenveloped DNA virus adenovirus (AdV) type 5 (3). During ADIN, TRIM21 functions as both a sensor and an effector (3, 4). When antibody-bound AdV enters the target cell, TRIM21 homodimerizes through its coiled-coil domain and binds with high affinity to the Fc domain of the antibody via its PRYSPRY motif (1, 3–5). This interaction activates the E3 ligase activity of TRIM21 and triggers its monoubiquitination and lysine-63 polyubiquitination, which targets the AdV–antibody complex to the proteasome for degradation (6, 7).
TRIM32 and TRIM25, two important members of TRIM family, play a critical role in antiviral infections (8, 9). The catalytic activity of the RING domains of these two proteins is governed by their oligomerization through coiled-coil domains (10, 11). TRIM21 also dimerizes to bind IgG (12, 13). However, the precise mechanism regulating TRIM21 activity is unknown. Posttranslational modifications regulate protein localization, activity, and interaction with other cellular molecules; acetylation and ubiquitination modify lysine residues of a protein, and their interaction controls the stability and function of the target protein. Given that TRIM21 ubiquitination is a critical step in ADIN (3, 6, 7, 14), we speculated that TRIM21 is acetylated and that this influences the process of ADIN. In our previous work, we identified TRIM21 as a putative substrate of histone deacetylase 6 (HDAC6) and determined by MS that TRIM21 acetylation in the liver was 5.77-fold higher in HDAC6 KO mice compared with their WT counterparts (15), suggesting that TRIM21 is deacetylated by HDAC6.
Based on the subcellular localization and observed acetylation of TRIM21, in this study we investigated whether HDAC6 regulates TRIM21-mediated ADIN. We found that HDAC6 deacetylated TRIM21; inhibition of HDAC6 activity resulted in TRIM21 hyperacetylation, which prevented its dimerization and polyubiquitination. Moreover, HDAC6 inhibition or depletion impaired virus removal through ADIN. These results demonstrate that the regulation of TRIM21 by HDAC6 plays an essential role in the cellular response to viral infection.
Results
HDAC6 colocalizes and interacts with TRIM21
In our previous work, we identified the cytosolic protein TRIM21 as a putative substrate of HDAC6 (15). To confirm this possibility, we first examined the subcellular distribution of TRIM21 and HDAC6 by immunofluorescence microscopy. Endogenous TRIM21 colocalized with HDAC6 (Fig. 1a), as did exogenous TRIM21 that was diffusely distributed in the cytoplasm (Fig. 1b, upper panel). However, exogenous TRIM21 also appeared to form rodlike shapes that was only weakly colocalized with HDAC6 (Fig. 1b, lower panel).
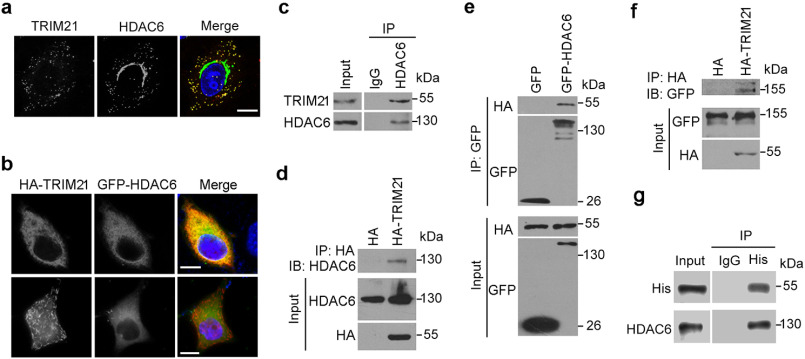
Figure 1.
HDAC6 interacts with TRIM21. a, representative image of HeLa cells labeled with antibodies against TRIM21 (red) and HDAC6 (green) and stained with DAPI (blue). b, representative image of HeLa cells cotransfected with HA-TRIM21 and GFP-HDAC6 followed by staining with antibodies against HA (red) and GFP (green) and DAPI (blue). Scale bars in panels a and b, 10 μm. c, HEK293T cell lysates were immunoprecipitated with control rabbit IgG or anti-HDAC6 antibody and probed with the indicated antibodies. d, HEK293T cells were transfected with HA-TRIM21 or HA constructs, and cell lysates were subjected to immunoprecipitation with an anti-HA antibody and then probed with the indicated antibodies. e, HEK293T cells were cotransfected with HA-TRIM21 and GFP or GFP-HDAC6. Anti-GFP immunoprecipitates were probed with the indicated antibodies. f, HEK293T cells were cotransfected with GFP-HDAC6 and HA or HA-TRIM21 constructs, and anti-HA immunoprecipitates were probed with the indicated antibodies. g, His-tagged TRIM21 and Myc/DDK tagged HDAC6 were incubated with IgG or anti-His antibody together with protein A beads, and the pulldown precipitates were probed with an anti-His antibody.
To determine whether HDAC6 interacts with TRIM21 in cells, we immunoprecipitated endogenous HDAC6 from HEK293T cell lysates. Endogenous TRIM21 was precipitated with an HDAC6 antibody, but not with the control IgG (Fig. 1c). Conversely, endogenous HDAC6 was precipitated with an anti-HA antibody, indicating an interaction between HDAC6 and HA-tagged TRIM21 (Fig. 1d). Similarly, exogenous HA-tagged TRIM21 was precipitated with exogenously expressed GFP-tagged HDAC6 (Fig. 1e), and vice versa (Fig. 1f). In vitro pulldown experiments using recombinant His-tagged TRIM21 and Myc/DDK-tagged HDAC6 showed that HDAC6 directly interacted with TRIM21 in the absence of any other cellular proteins (Fig. 1g).
HDAC6 deacetylates TRIM21 through interaction with the PRYSPRY motif
We next constructed GFP-tagged truncated HDAC6 (Fig. 2a) and FLAG-tagged truncated TRIM21 (Fig. 2b) to analyze the interaction between HDAC6 and TRIM21. The Δ(460–840) deletion mutant of HDAC6 lacking the DD2 deacetylase domain lost its ability to bind TRIM21 (Fig. 2a), suggesting that these two proteins interact through deacetylation. Conversely, the TRIM21 fragment lacking the PRYSPRY motif failed to precipitate with GFP-HDAC6 (Fig. 2b), indicating that HDAC6 binds to TRIM21 through this motif.
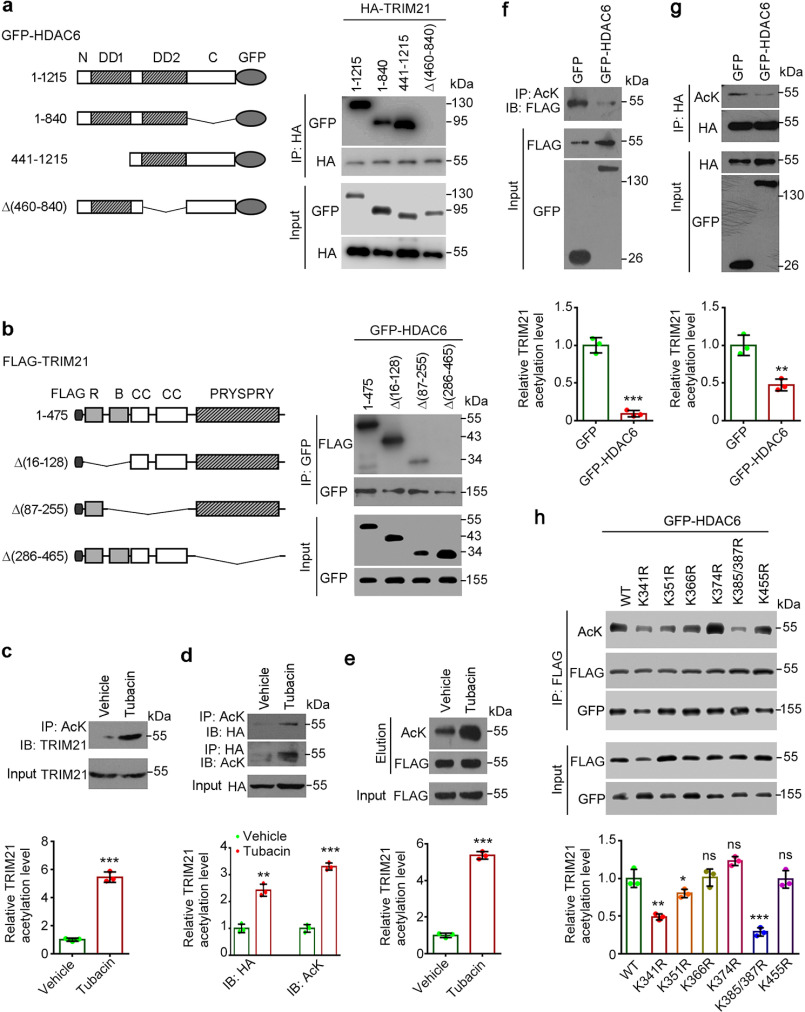
Figure 2.
HDAC6 deacetylates TRIM21. a, HEK293T cells were cotransfected with HA-TRIM21 and the indicated GFP-tagged HDAC6 constructs. Cell lysates were subjected to immunoprecipitation with an anti-HA antibody and probed with the indicated antibodies. b, HEK293T cells were cotransfected with GFP-HDAC6 and the indicated FLAG-tagged TRIM21 constructs; cell lysates were subjected to immunoprecipitation with an anti-GFP antibody and probed with the indicated antibodies. c, HEK293T cells were treated with tubacin or vehicle for 8 h; anti-AcK immunoprecipitates were probed with an anti-TRIM21 antibody. The relative TRIM21 acetylation level was determined by normalizing to the corresponding input. d and e, HEK293T cells were transfected with HA-TRIM21 (d) or FLAG-TRIM21 (e) and treated with tubacin or vehicle for 8 h. Anti-AcK and -HA immunoprecipitates were probed with the indicated antibodies (d), or anti-FLAG immunoprecipitates were subjected to competitive elution with FLAG peptide, and the elution was probed with the indicated antibodies (e). The relative TRIM21 acetylation level was determined by normalizing to the corresponding input (d) or the corresponding elution probed with anti-FLAG antibody (e). f, HEK293T cells were cotransfected with FLAG-TRIM21 and GFP or GFP-HDAC6; anti-AcK immunoprecipitates were probed with an anti-FLAG antibody. The relative TRIM21 acetylation level was determined by normalizing to the corresponding input probed with anti-FLAG antibody. g, HEK293T cells were cotransfected with HA-TRIM21 and GFP or GFP-HDAC6; anti-HA immunoprecipitates were probed with the indicated antibodies. The relative TRIM21 acetylation level was determined by normalizing to the corresponding immunoprecipitate probed with anti-HA antibody. h, HEK293T cells were transfected with GFP-HDAC6 and various FLAG-TRIM21 mutants, anti-FLAG immunoprecipitates were probed with the indicated antibodies. The relative TRIM21 acetylation level was determined by normalizing to the corresponding immunoprecipitate probed with anti-FLAG antibody.*, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
The association between HDAC6 and TRIM21 suggests that TRIM21 is a target of HDAC6. To test this possibility, we treated HEK293T cells with the HDAC6 inhibitor tubacin (16) and performed immunoprecipitation with an antibody against acetyl-lysine (AcK) or the TRIM21 epitope tag. Endogenous TRIM21 showed increased acetylation upon tubacin treatment (Fig. 2c). HDAC6 inhibition also led to increased acetylation of overexpressed TRIM21 (Fig. 2, d and e). To confirm the regulatory relationship between the two proteins, we coexpressed GFP-tagged HDAC6 with FLAG-tagged TRIM21 or HA-tagged TRIM21; anti-AcK or anti-HA immunoprecipitate was analyzed by Western blotting. HDAC6 overexpression markedly reduced the levels of acetylated TRIM21 (Fig. 2, f and g). To identify HDAC6-specific deacetylation sites in TRIM21, we constructed acetylation-deficient mutants of TRIM21, in which the lysines within the PRYSPRY motif were replaced with arginines (K314R, K351R, K366R, K374R, K385/387R, K455R). We found that HDAC6-mediated deacetylation of TRIM21 was remarkably impaired by K385/387R mutant and mildly impaired by K341R or K351R mutants (Fig. 2h), suggesting Lys-385 and Lys-387 as the major sites of TRIM21 that are deacetylated by HDAC6. These data suggest that HDAC6 deacetylates TRIM21.
Hyperacetylation of TRIM21 blocks its dimerization and ubiquitination
TRIM21 forms homodimers through its coiled-coil domain (17), which is required for its binding with AdV–antibody complexes (3). We investigated whether HDAC6-mediated deacetylation of TRIM21 affects its dimerization by immunoprecipitation. Inhibiting HDAC6 activity by tubacin treatment impaired the ability of TRIM21 to form dimers (Fig. 3, a and b). Conversely, overexpression of WT HDAC6, but not the catalytically defective mutant, enhanced TRIM21 dimerization (Fig. 3c). Given that dimerization is critical for the formation of rodlike shapes by TRIM21 (17, 18), we examined the effect of HDAC6 inhibition on the subcellular localization of TRIM21. Tubacin treatment increased the fraction of cells harboring rodlike TRIM21 (Fig. 3, d and e) while disrupting its dimerization (Fig. 3, a and b).
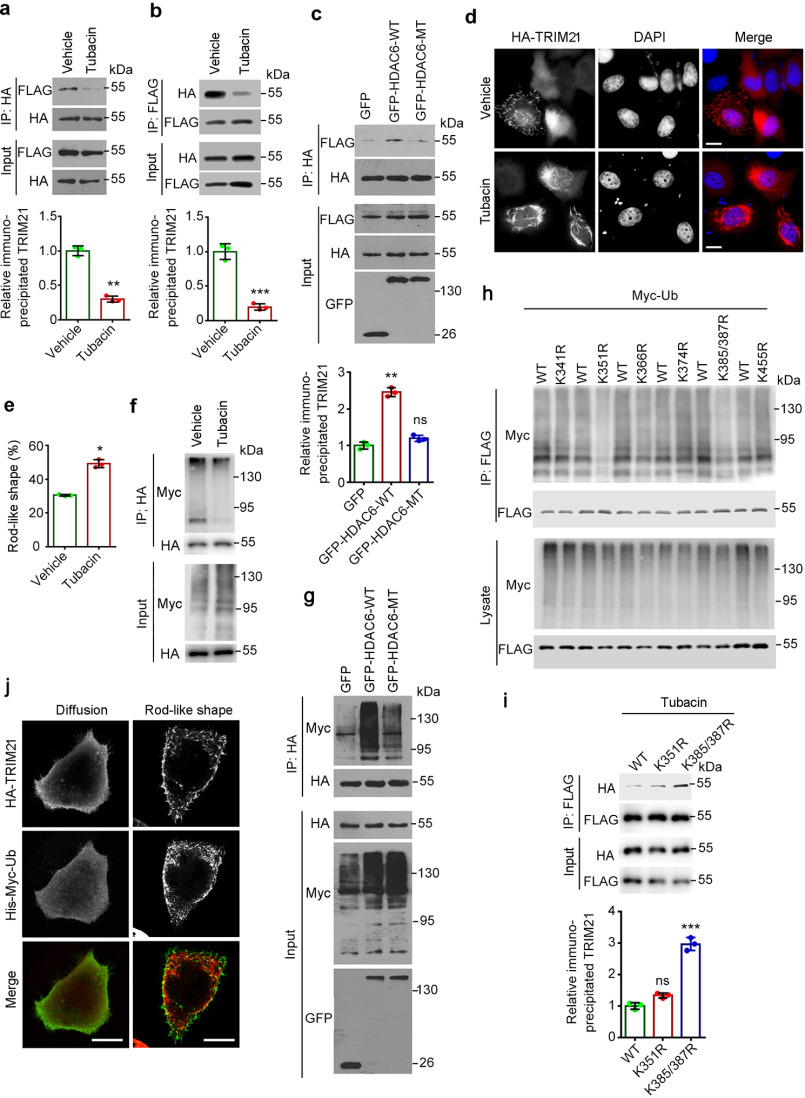
Figure 3.
Hyperacetylation of TRIM21 blocks its dimerization and ubiquitination. a and b, HEK293T cells were cotransfected with FLAG-TRIM21 and HA-TRIM21 and treated with tubacin or vehicle for 8 h; anti-HA immunoprecipitates (a) and anti-FLAG immunoprecipitates (b) were probed with an anti-FLAG or -HA antibody. The relative TRIM21 level was determined by normalizing to the corresponding immunoprecipitate probed with an anti-HA (a) or -FLAG (b) antibody. c, HEK293T cells were cotransfected with FLAG-TRIM21 and HA-TRIM21 and either GFP-HDAC6-WT, the inactive mutant (MT), or the GFP-only construct. Anti-HA immunoprecipitates were probed with the indicated antibodies. The relative TRIM21 level was determined by normalizing to the corresponding immunoprecipitate probed with anti-HA antibody. d, HeLa cells were transfected with HA-TRIM21, treated with tubacin or vehicle, and labeled with an anti-HA antibody (red) followed by staining with DAPI (blue). e, experiments were performed as in (d), and the percentage of cells with rodlike TRIM21 was quantified (n = 200 for each dataset). f, HEK293T cells were cotransfected with HA-TRIM21 and His-Myc-ubiquitin and treated with the proteasome inhibitor MG132 and tubacin or vehicle for 8 h. Anti-HA immunoprecipitates were probed with the indicated antibodies. g, HEK293T cells were cotransfected with HA-TRIM21 and His-Myc-ubiquitin along with GFP-HDAC6-WT, GFP-HDAC6-MT, or the GFP-only construct and treated with MG132. Anti-HA immunoprecipitates were probed with the indicated antibodies. h, HEK293T cells were cotransfected with His-Myc-ubiquitin and various FLAG-TRIM21 mutants and treated with the proteasome inhibitor MG132 for 8 h. Anti-FLAG immunoprecipitates were probed with the indicated antibodies. i, HEK293T cells were cotransfected with the indicated FLAG-TRIM21 and the indicated HA-TRIM21 plasmids and treated with tubacin for 8 h; anti-FLAG immunoprecipitates were probed with an anti-FLAG or -HA antibody. j, HeLa cells were cotransfected with HA-TRIM21 and His-Myc-ubiquitin (Ub) and labeled with anti-HA (green) and anti-Myc (red) antibodies. The relative TRIM21 level was determined by normalizing to the corresponding immunoprecipitate probed with an anti-FLAG antibody. Scale bars in panels c and g, 10 μm. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
TRIM21 is known to undergo ubiquitination (3, 7, 19), which influences its activity as a regulator of intracellular viral neutralization. We therefore examined the effect of inhibiting HDAC6 on TRIM21 ubiquitination. Treatment with tubacin significantly reduced TRIM21 ubiquitination (Fig. 3f), whereas overexpression of HDAC6-WT but not the catalytically defective mutant HDAC6-MT led to an increase in TRIM21 ubiquitination (Fig. 3g). We then sought to investigate the association of acetylation with ubiquitination by using constructed acetylation-deficient mutants of TRIM21. Analysis of TRIM21 ubiquitination revealed that K351R and K385/K387R mutants resulted in a significant decrease in ubiquitination (Fig. 3h). In addition, our further immunoprecipitation revealed that K385/K387R mutant but not K351R mutant increased the dimerization of TRIM21 (Fig. 3i), indicating Lys-385 and Lys-387 acetylation are critical for the regulation of TRIM21 dimerization. Additionally, in agreement with previous findings (20), the rodlike TRIM21 did not colocalize with ubiquitin (Fig. 3j), indicating that this form of TRIM21 is not ubiquitinated. Thus, the decreased ubiquitination of TRIM21 in response to tubacin treatment is associated with its assembly into rodlike shapes (Fig. 3, d and e). These results indicate that hyperacetylation of TRIM21 caused by HDAC6 inhibition blocks its dimerization and ubiquitination.
HDAC6 depletion or inhibition impedes TRIM21-mediated ADIN
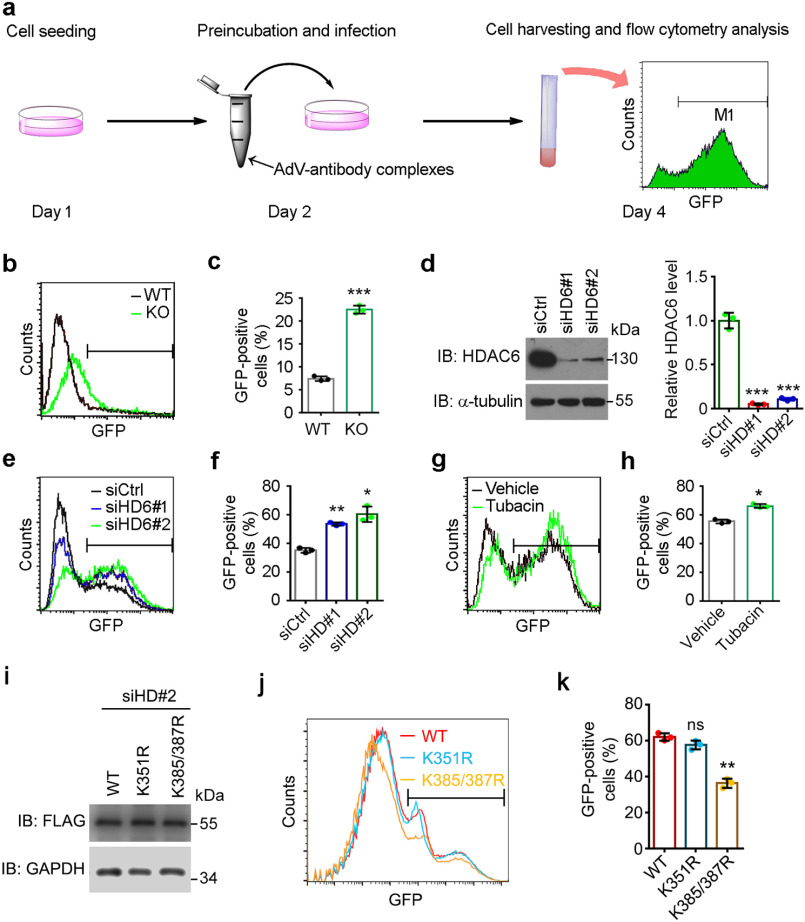
TRIM21 functions as a mediator of ADIN. We therefore investigated whether HDAC6 is an upstream regulator of this process by means of the viral neutralization assay. Briefly, AdVs carrying a GFP gene were preincubated with an antibody against hexon, the capsid protein of AdVs, to form AdV-antibody complexes, which were then added to cells. After culturing for 48 h, the percentage of GFP-positive cells was analyzed by flow cytometry (Fig. 4a). We first performed the experiments using WT and HDAC6 KO mouse embryonic fibroblasts (MEFs) and observed that the percentage of GFP-positive cells was higher in the latter than in the former (Fig. 4, b and c), suggesting that cells lacking HDAC6 were more susceptible to intracellular entry of antibody-coated AdVs. We used siRNAs targeting HDAC6 to further evaluate the protective effect of HDAC6 in this process. HDAC6 expression levels were reduced in HeLa cells transfected with siRNAs siHD6#1 and siHD6#2 compared with those transfected with the scrambled control siRNA (Fig. 4d). The percentage of GFP-positive cells was higher in cells depleted of HDAC6 (Fig. 4, e and f), consistent with the results obtained using MEFs. To determine whether the protective effect of HDAC6 depends on its deacetylase activity, we treated HeLa cells with tubacin for 8 h before infection. This increased the infection rate (Fig. 4, g and h), indicating that HDAC6 activity protects cells against viral infection. To investigate the involvement of HDAC6-mediated deacetylation in ADIN, we overexpressed FLAG-tagged TRIM21-WT, TRIM21-K351R, or TRIM21-K385/387R mutants in HDAC6-depleted cells (Fig. 4i). Consistent with the critical role of Lys-385 and Lys-387 acetylation in TRIM21 dimerization regulation (Fig. 3i), K385/387R mutant rescued HDAC6 depletion-induced ADIN impairment (Fig. 4, j and k). These findings demonstrate that HDAC6 deacetylates TRIM21 to promote ADIN.
Figure 4.
Depletion of HDAC6 or inhibition of its activity impairs viral clearance. a, protocol for cellular infection with antibody-coated AdVs. Cells were seeded at the indicated density on day 1. On day 2, AdVs carrying the GFP gene were preincubated with an antibody against hexon. Cells were incubated with AdV-antibody complexes for 48 h. On day 4, the cells were harvested and infection rate was analyzed by flow cytometry. b and c, WT and HDAC6 KO MEFs were incubated with AdV-antibody complexes. d, Western blot analysis of the knockdown efficiency of siRNAs used in (e) and (f). Ctrl, control; HD6, HDAC6. The relative HDAC6 level was determined by normalizing to the corresponding α-tubulin. e–h, HeLa cells transfected with control or HDAC6-specific siRNA without (e and f) or with (g and h) tubacin or vehicle pretreatment were incubated with AdV-antibody complexes. i, Western blot analysis of the expression efficiency of the indicated FLAG-TRIM21 plasmids used in (j and k). j and k, HeLa cells transfected with HDAC6-specific siRNA and the indicated plasmids were incubated with AdV-antibody complexes. Representative plots of GFP-positive cells detected by flow cytometry (b, e, g, and j) and quantification of GFP-positive cells (c, f, h, and k) are shown (n = 6 for each dataset). Data represent the mean ± S.D. of three independent experiments. ns, not significant; *, p < 0.05, **, p < 0.01; ***, p < 0.001.
Discussion
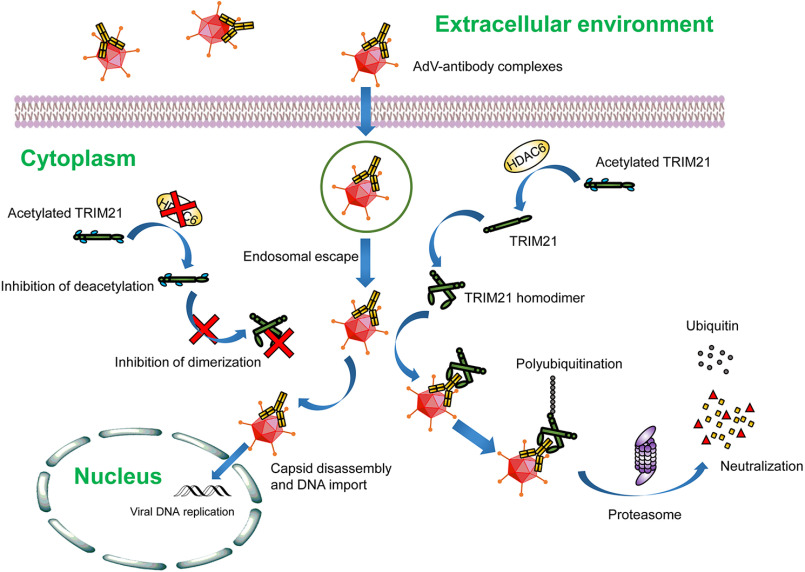
Numerous groups, including our laboratory, have identified HDAC6 as a versatile regulator in a diversity of cellular activities, including cell motility (21–24), primary ciliogenesis (25–27), and immunity (28, 29), mainly through its action in deacetylating tubulin and cortactin. In addition, emerging evidence reveals the critical roles of HDAC6 in regulation of the infection of various types of viruses through both deacetylase-dependent and -independent manners (30–33). Here we report a new mechanism by which HDAC6 protects host cells against viruses. Based on our observations, we propose a model in which antibody-coated AdVs that enter cells are detected by TRIM21, which recruits HDAC6. Their interaction in the cytoplasm leads to the deacetylation of TRIM21 and its homodimerization through the coiled-coil domain. The homodimers bind AdV-antibody complexes via the Fc domain of the antibody and target the complexes to the proteasome for degradation through automonoubiquitination and Lys-63–linked polyubiquitination. In the absence of HDAC6, TRIM21 remains hyperacetylated, which prevents the formation of TRIM21 homodimers. Consequently, AdV–antibody complexes are not eliminated via TRIM21-mediated ADIN, resulting in viral replication, transcription of the viral genome, and expression of viral proteins (Fig. 5).
Figure 5.
Model of the role of HDAC6 in TRIM21-mediated ADIN. TRIM21 senses the entry of antibody-bound AdVs and recruits HDAC6, which deacetylates and promotes the dimerization of TRIM21 via the coiled-coil domain. TRIM21 homodimers bind AdV-antibody complexes via the Fc domain of the antibody and target the complexes to the proteasome for degradation through automonoubiquitination and subsequent polyubiquitination. In the absence of HDAC6, TRIM21 is hyperacetylated, which inhibits the formation of TRIM21 homodimers; antibody-bound AdVs fail to be removed through TRIM21-mediated ADIN, resulting in viral replication and expression of viral proteins.
The antiviral effects of HDAC6 in vivo have been reported previously (34–36). HDAC6 KO mice are more susceptible to Sendai virus infection than their WT counterparts (34) and are highly sensitive to infection by vesicular stomatitis virus Indiana strain, a lethal RNA virus (35). Meanwhile, HDAC6 transgenic mice exhibit enhanced resistance to H5N1 influenza A virus infection (36). Deacetylation of β-catenin or retinoic acid–inducible gene I in target cells stimulates the innate immune response (34, 35). Before entering cells, circulating viruses are recognized by specific antibodies and neutralized. Antibody-bound viruses that fail to be cleared enter cells are sensed by the cytosolic Fc receptor TRIM21 and removed by TRIM21-mediated ADIN (3, 37). In this study, we demonstrated that HDAC6 promotes the neutralization of intracellular antibody-bound AdVs through deacetylation of TRIM21, providing evidence for the antiviral effects of HDAC6.
Our data revealed that Lys-385 and Lys-387 were critical for the regulation of TRIM21 acetylation, and ubiquitination occurred at Lys-351, Lys-385, and Lys-387 sites, suggesting a competition between acetylation and ubiquitination may exist at Lys-385 and Lys-387 sites. Interestingly, K385/387R mutant was able to rescue HDAC6 depletion-induced ADIN impairment. Given that both the TRIM21 dimerization and ubiquitination are required for ADIN, we postulate that HDAC6-mediated Lys-385 and Lys-387 deacetylation promotes TRIM21 dimerization, which undergoes ubiquitination at Lys-351 site, thus resulting in ADIN. The recognition of antibody-bound viruses by TRIM21 depends not on pathogen-associated molecular patterns, but on the presence of intracellular antibodies, which function as danger signals. Thus, it is possible that TRIM21-mediated ADIN acts as an intracellular defense mechanism to remove virus that is coated by the antibody before internalization. Our finding that TRIM21-mediated ADIN depends on HDAC6 activity can serve as a basis for the development of therapeutic strategies for viral infection.
Materials and Methods
Materials
Primary antibodies used in this study included the following: TRIM21, HDAC6 (Santa Cruz Biotechnology, Dallas, TX, USA), acetylated-lysine, Myc, GFP, HA (Sigma-Aldrich), FLAG, and α-tubulin (Abcam, Cambridge, MA, USA). Protein A agarose beads, anti-FLAG M2 magnetic beads (Sigma-Aldrich), anti-HA M2 magnetic beads (Santa Cruz Biotechnology), and Pan Anti-Acetyllysine Antibody Conjugated Agarose Beads (PTM Biolabs, Hangzhou, China) were purchased from the indicated sources. Secondary antibodies included Alexa Fluor 488 or Alexa Fluor 568 conjugated anti-mouse or anti-rabbit antibodies (Invitrogen) for immunofluorescence microscopy and horseradish peroxidase–conjugated goat anti-mouse or anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) for immunoblotting. 4′,6-diamidino-2-phenylindole (DAPI), MG132, and tubacin were purchased from Sigma-Aldrich. AdVs carrying the GFP gene were purchased from Thermo Fisher Scientific.
Plasmids and proteins
Mammalian expression plasmids for GFP-HDAC6-WT (1–1215) and catalytically defective mutant GFP-HDAC6-MT (H216/611A) were generated by insertion of the cDNAs into the pEGFPN1 vector as described previously (38). pEGFPN1-HDAC6 deletion mutants were constructed by in-fusion cloning (Takara Bio, Shiga, Japan), and the primers were as follows: GFP-HDAC6 (1–840): (forward: 5′-GATCTCGAGCTCAAGATGAGTGGAGCGAACCGCGGC-3′, reverse: 5′-CCGCGGTACCGTCGAGGACTTGGATGGTCTCAGTG-3′); GFP-HDAC6 (441–1215): (forward: 5′-GATCTCGAGCTCAAGCTGGAAGCCCTTGAGCCCTTC-3′, reverse: 5′-CCGCGGTACCGTCGAGTGTGGGTGGGGCATATCCTC-3′); and GFP-HDAC6-Δ(460–840): (forward 1: 5′-GATCTCGAGCTCAAGATGAGTGGAGCGAACCGCGGC-3′, reverse 1: 5′-GGACTTGGATGGTCTTCTCCACGGTCTCAGTTGATC-3′; forward 2: 5′-CTGAGACCGTG GAGACATCGCAGATACTGGCGCAGC-3′, reverse 2: 5′-CCGCGGTACCGTCGAGTGTGGGTGGGGCATATCCTC-3′). Mammalian expression plasmids for His-Myc-ubiquitin, HA-TRIM21, and FLAG-TRIM21 were generated by insertion of the cDNAs into the pcDNA3, pcDNA3-HA, and pcDNA3-FLAG vectors, respectively. The deletion mutants of FLAG-TRIM21 were directly synthesized and inserted into pcDNA3-HA vector by GenScript (Piscataway, NJ, USA), and the K/R point mutations were generated by PCR and site-directed mutagenesis with pcDNA3-FLAG-TRIM21 as a template. His-TRIM21 purified from insect cells was purchased from Sino Biological (Beijing, China), and c-Myc/DDK-tagged HDAC6 purified from HEK293T cells was purchased from OriGene Technologies Inc. (Rockville, MD, USA).
Isolation of primary MEFs
HDAC6 heterozygous mice were a kind gift from Tso-Pang Yao (Duke University) and intercrossed to generate WT and HDAC6 knockout littermates. All mouse experiments were carried out in accordance with the relevant regulatory standards and were approved by the Animal Care and Use Committee of Shandong Normal University. Primary MEFs were isolated from E14.5 WT and HDAC6 knockout embryos as described previously (39). Briefly, the pregnant mice with E14.5 embryos were sacrificed by cervical dislocation. The embryos were isolated from the uterus and heads and visceral organs were removed. The embryos were minced and digested with 0.5% trypsin-EDTA (w/v) at 37°C for 30 min. Cells were cultured in MEF medium in a 5% CO2 incubator until confluent.
Cell culture and transfection
HeLa and HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (v/v), and maintained at 37°C in a humidified incubator with 5% CO2. Plasmids were transfected into cells with polyethylenimine (Sigma-Aldrich). siRNAs targeting HDAC6 (siHD6#1: 5′-GCAGUUAAAUGAAUUCCAU-3′ and siHD6#2: 5′-GGAGUUAACUGGCAGGCAU-3′) or the scrambled control siRNA (siCtrl: 5′-CGUACGCGGAAUACUUCGA-3′) (all from RiboBio, Guangzhou, China) were transfected into cells with Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific). After 48 h, gene knockdown efficiency was evaluated by Western blotting.
Immunofluorescence analysis
HeLa cells grown on coverslips were fixed with 4% paraformaldehyde (w/v) for 30 min, permeabilized with 0.5% Triton X-100 (v/v) in PBS for 20 min, and blocked with 2% BSA in PBS for 30 min at room temperature. Immunolabeling was performed using mouse anti-TRIM21, mouse anti-HA, rabbit anti-HDAC6, and rabbit anti-Myc antibodies diluted 1:500 in 2% BSA (w/v). Fluorophore-conjugated secondary antibodies were used at 1:1000 dilution in 2% BSA to detect the primary antibodies. The cells were stained with 4′,6-diamidino-2-phenylindole (Sigma-Aldrich), and the coverslips were mounted with mounting medium (Thermo Fisher Scientific). Images were acquired with an Axio Observer A1 fluorescence microscope (Carl Zeiss, Oberkochen, Germany) or a TCS SP8 confocal microscope (Leica, Wetzlar, Germany).
Western blotting and co-immunoprecipitation
Cells were harvested and lysed, and proteins were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA) that was blocked with 5% fat-free milk in TBS containing 0.1% Tween 20 (v/v) for 45 min, then probed with the indicated primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Protein bands were visualized using enhanced chemiluminescence detection reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. For immunoprecipitation, the proteins were incubated overnight at 4°C with antibody-conjugated agarose beads, which were then washed extensively and subjected to Western blotting. The experiments were repeated independently at least three times, and the intensity of immunoblots was determined by densitometry with the ImageJ software (National Institutes of Health). The relative protein levels were normalized to the corresponding immunoprecipitated or input proteins and the ratio was then determined by dividing with the control group (the value of the control group was set to be 1).
Cell infection with antibody-coated AdVs
Cells were seeded at 1 × 105 cells per well in 6-well plates and allowed to adhere overnight. AdVs carrying a GFP gene (at a multiplicity of infection of 10 for HeLa cells and 40 for MEFs) were pre-incubated with goat anti-hexon antibody (Millipore) at 200 ng/ml for 30 min at room temperature; the AdV-antibody complexes were added to the cells. After 48 h, cells were harvested and GFP-positive cells were examined by flow cytometry (BD Biosciences).
Statistical analysis
Differences between groups were evaluated with the Student's t test. p < 0.05 was considered statistically significant.
Data availability
All the data are either presented in the manuscript or available from the corresponding author (Songbo Xie, Shandong Normal University, xiesongbo@sdnu.edu.cn) upon request.
Author contributions—S. X., L. Z., J. Z., D. L., and M. L. conceptualization; S. X., L. Z., W. X., D. L., and M. L. software; S. X., J. Z., D. L., and M. L. supervision; S. X., C. F., and W. X. funding acquisition; S. X., L. Z., D. D., R. G., Q. H., C. F., and W. X. investigation; S. X., L. Z., W. X., D. L., and M. L. methodology; S. X., L. Z., and D. L. writing-original draft; S. X., D. L., and M. L. project administration; S. X., J. Z., D. L., and M. L. writing-review and editing; D. D., R. G., Q. H., and C. F. validation; Q. H. and C. F. visualization; C. F., W. X., D. L., and M. L. resources; D. L. data curation.
Funding and additional information—This work was supported by National Natural Science Foundation of China Grants 31701216 (to S. X.), 31701209 (to W. X.), and 31900560 (to C. F.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ADIN
- antibody-dependent intracellular neutralization
- AdV
- adenovirus
- AcK
- acetyl-lysine
- MEF
- mouse embryonic fibroblast
- DAPI
- 4′,6-diamidino-2-phenylindole.
References
- 1. James L. C., Keeble A. H., Khan Z., Rhodes D. A., and Trowsdale J. (2007) Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. U. S. A. 104, 6200–6205 10.1073/pnas.0609174104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang Y., Eversole T., Lee D. J., Sontheimer R. D., and Capra J. D. (1999) Protein-protein interactions between native Ro52 and immunoglobulin G heavy chain. Scand. J. Immunol. 49, 620–628 10.1046/j.1365-3083.1999.00547.x [DOI] [PubMed] [Google Scholar]
- 3. Mallery D. L., McEwan W. A., Bidgood S. R., Towers G. J., Johnson C. M., and James L. C. (2010) Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21). Proc. Natl. Acad. Sci. U. S. A. 107, 19985–19990 10.1073/pnas.1014074107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McEwan W. A., Tam J. C., Watkinson R. E., Bidgood S. R., Mallery D. L., and James L. C. (2013) Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat. Immunol. 14, 327–336 10.1038/ni.2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keeble A. H., Khan Z., Forster A., and James L. C. (2008) TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc. Natl. Acad. Sci. U. S. A. 105, 6045–6050 10.1073/pnas.0800159105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hauler F., Mallery D. L., McEwan W. A., Bidgood S. R., and James L. C. (2012) AAA ATPase p97/VCP is essential for TRIM21-mediated virus neutralization. Proc. Natl. Acad. Sci. U. S. A. 109, 19733–19738 10.1073/pnas.1210659109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fletcher A. J., Mallery D. L., Watkinson R. E., Dickson C. F., and James L. C. (2015) Sequential ubiquitination and deubiquitination enzymes synchronize the dual sensor and effector functions of TRIM21. Proc. Natl. Acad. Sci. U. S. A. 112, 10014–10019 10.1073/pnas.1507534112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu B., Wang L., Ding H., Schwamborn J. C., Li S., and Dorf M. E. (2015) TRIM32 senses and restricts influenza A virus by ubiquitination of PB1 polymerase. PLoS Pathog. 11, e1004960 10.1371/journal.ppat.1004960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sanchez J. G., Sparrer K. M. J., Chiang C., Reis R. A., Chiang J. J., Zurenski M. A., Wan Y., Gack M. U., and Pornillos O. (2018) TRIM25 binds RNA to modulate cellular anti-viral defense. J. Mol. Biol. 430, 5280–5293 10.1016/j.jmb.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koliopoulos M. G., Esposito D., Christodoulou E., Taylor I. A., and Rittinger K. (2016) Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 35, 1204–1218 10.15252/embj.201593741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Streich F. C. Jr., Ronchi V. P., Connick J. P., and Haas A. L. (2013) Tripartite motif ligases catalyze polyubiquitin chain formation through a cooperative allosteric mechanism. J. Biol. Chem. 288, 8209–8221 10.1074/jbc.M113.451567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bottermann M., Foss S., van Tienen L. M., Vaysburd M., Cruickshank J., O'Connell K., Clark J., Mayes K., Higginson K., Hirst J. C., McAdam M. B., Slodkowicz G., Hutchinson E., Kozik P., Andersen J. T., et al. (2018) TRIM21 mediates antibody inhibition of adenovirus-based gene delivery and vaccination. Proc. Natl. Acad. Sci. U. S. A. 115, 10440–10445 10.1073/pnas.1806314115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dickson C., Fletcher A. J., Vaysburd M., Yang J. C., Mallery D. L., Zeng J., Johnson C. M., McLaughlin S. H., Skehel M., Maslen S., Cruickshank J., Huguenin-Dezot N., Chin J. W., Neuhaus D., and James L. C. (2018) Intracellular antibody signalling is regulated by phosphorylation of the Fc receptor TRIM21. Elife 7, e32660 10.7554/eLife.32660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xue B., Li H., Guo M., Wang J., Xu Y., Zou X., Deng R., Li G., and Zhu H. (2018) TRIM21 promotes innate immune response to RNA viral infection through Lys27-linked polyubiquitination of MAVS. J. Virol. 92, e00321–18 10.1128/JVI.00321-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang L., Liu S., Liu N., Zhang Y., Liu M., Li D., Seto E., Yao T. P., Shui W., and Zhou J. (2015) Proteomic identification and functional characterization of MYH9, Hsc70, and DNAJA1 as novel substrates of HDAC6 deacetylase activity. Protein Cell 6, 42–54 10.1007/s13238-014-0102-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haggarty S. J., Koeller K. M., Wong J. C., Grozinger C. M., and Schreiber S. L. (2003) Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. U. S. A. 100, 4389–4394 10.1073/pnas.0430973100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang D., Buyon J. P., Yang Z., Di Donato F., Miranda-Carus M. E., and Chan E. K. (2001) Leucine zipper domain of 52 kDa SS-A/Ro promotes protein dimer formation and inhibits in vitro transcription activity. Biochim. Biophys. Acta 1568, 155–161 10.1016/S0304-4165(01)00212-4 [DOI] [PubMed] [Google Scholar]
- 18. Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P. G., and Ballabio A. (2001) The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukuda-Kamitani T., and Kamitani T. (2002) Ubiquitination of Ro52 autoantigen. Biochem. Biophys. Res. Commun. 295, 774–778 10.1016/S0006-291X(02)00750-7 [DOI] [PubMed] [Google Scholar]
- 20. Wada K., Tanji K., and Kamitani T. (2006) Oncogenic protein UnpEL/Usp4 deubiquitinates Ro52 by its isopeptidase activity. Biochem. Biophys. Res. Commun. 339, 731–736 10.1016/j.bbrc.2005.11.076 [DOI] [PubMed] [Google Scholar]
- 21. Li D., Xie S., Ren Y., Huo L., Gao J., Cui D., Liu M., and Zhou J. (2011) Microtubule-associated deacetylase HDAC6 promotes angiogenesis by regulating cell migration in an EB1-dependent manner. Protein Cell 2, 150–160 10.1007/s13238-011-1015-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li D., Sun X., Zhang L., Yan B., Xie S., Liu R., Liu M., and Zhou J. (2014) Histone deacetylase 6 and cytoplasmic linker protein 170 function together to regulate the motility of pancreatic cancer cells. Protein Cell 5, 214–223 10.1007/s13238-013-0010-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang L., Liu N., Xie S., He X., Zhou J., Liu M., and Li D. (2014) HDAC6 regulates neuroblastoma cell migration and may play a role in the invasion process. Cancer Biol. Ther. 15, 1561–1570 10.4161/15384047.2014.956632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., and Yao T. P. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- 25. Ran J., Yang Y., Li D., Liu M., and Zhou J. (2015) Deacetylation of α-tubulin and cortactin is required for HDAC6 to trigger ciliary disassembly. Sci. Rep. 5, 12917 10.1038/srep12917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang Y., Ran J., Liu M., Li D., Li Y., Shi X., Meng D., Pan J., Ou G., Aneja R., Sun S. C., and Zhou J. (2014) CYLD mediates ciliogenesis in multiple organs by deubiquitinating Cep70 and inactivating HDAC6. Cell Res. 24, 1342–1353 10.1038/cr.2014.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith Q., Macklin B., Chan X. Y., Jones H., Trempel M., Yoder M. C., and Gerecht S. (2018) Differential HDAC6 activity modulates ciliogenesis and subsequent mechanosensing of endothelial cells derived from pluripotent stem cells. Cell Rep. 24, 1930 10.1016/j.celrep.2018.08.005 [DOI] [PubMed] [Google Scholar]
- 28. Yan B., Xie S., Liu Y., Liu W., Li D., Liu M., Luo H. R., and Zhou J. (2018) Histone deacetylase 6 modulates macrophage infiltration during inflammation. Theranostics 8, 2927–2938 10.7150/thno.25317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan B., Liu Y., Bai H., Chen M., Xie S., Li D., Liu M., and Zhou J. (2017) HDAC6 regulates IL-17 expression in T lymphocytes: implications for HDAC6-targeted therapies. Theranostics 7, 1002–1009 10.7150/thno.17615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen H., Qian Y., Chen X., Ruan Z., Ye Y., Chen H., Babiuk L. A., Jung Y. S., and Dai J. (2019) HDAC6 restricts influenza A virus by deacetylation of the RNA polymerase PA subunit. J. Virol. 93, e01896–18 10.1128/JVI.01896-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jo H., Jang H. Y., Youn G. S., Kim D., Lee C. Y., Jang J. H., Choi S. Y., Jun J. G., and Park J. (2018) Hindsiipropane B alleviates HIV-1 Tat-induced inflammatory responses by suppressing HDAC6-NADPH oxidase-ROS axis in astrocytes. BMB Rep. 51, 394–399 10.5483/bmbrep.2018.51.8.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zanin M., DeBeauchamp J., Vangala G., Webby R. J., and Husain M. (2020) Histone deacetylase 6 knockout mice exhibit higher susceptibility to influenza A virus infection. Viruses 12, 728 10.3390/v12070728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wenzel E. D., Speidell A., Flowers S. A., Wu C., Avdoshina V., and Mocchetti I. (2019) Histone deacetylase 6 inhibition rescues axonal transport impairments and prevents the neurotoxicity of HIV-1 envelope protein gp120. Cell Death Dis. 10, 674 10.1038/s41419-019-1920-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chattopadhyay S., Fensterl V., Zhang Y., Veleeparambil M., Wetzel J. L., and Sen G. C. (2013) Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators. MBio 4, e00636–12 10.1128/mBio.00636-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choi S. J., Lee H. C., Kim J. H., Park S. Y., Kim T. H., Lee W. K., Jang D. J., Yoon J. E., Choi Y. I., Kim S., Ma J., Kim C. J., Yao T. P., Jung J. U., Lee J. Y., et al. (2016) HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. EMBO J. 35, 429–442 10.15252/embj.201592586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang D., Meng Q., Huo L., Yang M., Wang L., Chen X., Wang J., Li Z., Ye X., Liu N., Li Q., Dai Z., Ouyang H., Li N., Zhou J., et al. (2015) Overexpression of Hdac6 enhances resistance to virus infection in embryonic stem cells and in mice. Protein Cell 6, 152–156 10.1007/s13238-014-0120-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rakebrandt N., Lentes S., Neumann H., James L. C., and Neumann-Staubitz P. (2014) Antibody- and TRIM21-dependent intracellular restriction of Salmonella enterica. Pathog. Dis. 72, 131–137 10.1111/2049-632X.12192 [DOI] [PubMed] [Google Scholar]
- 38. Ran J., Liu M., Feng J., Li H., Ma H., Song T., Cao Y., Zhou P., Wu Y., Yang Y., Yang Y., Yu F., Guo H., Zhang L., Xie S., et al. (2020) ASK1-mediated phosphorylation blocks HDAC6 ubiquitination and degradation to drive the disassembly of photoreceptor connecting cilia. Dev. Cell 53, 287–299 10.1016/j.devcel.2020.03.010 [DOI] [PubMed] [Google Scholar]
- 39. Gao Y. S., Hubbert C. C., Lu J., Lee Y. S., Lee J. Y., and Yao T. P. (2007) Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol. Cell Biol. 27, 8637–8647 10.1128/MCB.00393-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data are either presented in the manuscript or available from the corresponding author (Songbo Xie, Shandong Normal University, xiesongbo@sdnu.edu.cn) upon request.