Abstract
Fungi inhabit extraordinarily diverse ecological niches, including the human body. Invasive fungal infections have a devastating impact on human health worldwide, killing ∼1.5 million individuals annually. The majority of these deaths are attributable to species of Candida, Cryptococcus, and Aspergillus. Treating fungal infections is challenging, in part due to the emergence of resistance to our limited arsenal of antifungal agents, necessitating the development of novel therapeutic options. Whereas conventional antifungal strategies target proteins or cellular components essential for fungal growth, an attractive alternative strategy involves targeting proteins that regulate fungal virulence or antifungal drug resistance, such as regulators of fungal stress responses. Stress response networks enable fungi to adapt, grow, and cause disease in humans and include regulators that are highly conserved across eukaryotes as well as those that are fungal-specific. This review highlights recent developments in elucidating crystal structures of fungal stress response regulators and emphasizes how this knowledge can guide the design of fungal-selective inhibitors. We focus on the progress that has been made with highly conserved regulators, including the molecular chaperone Hsp90, the protein phosphatase calcineurin, and the small GTPase Ras1, as well as with divergent stress response regulators, including the cell wall kinase Yck2 and trehalose synthases. Exploring structures of these important fungal stress regulators will accelerate the design of selective antifungals that can be deployed to combat life-threatening fungal diseases.
Keywords: fungi, structural biology, antifungal, stress response, heat shock protein 90 (Hsp90), Ras, calcineurin, drug design, antibiotics, molecular chaperone, microbiology, microbial pathogenesis, antifungal drug development, structure-guided drug design
There are millions of fungal species, which inhabit remarkably diverse environments (1, 2). Fungi include symbionts with roles in decomposing organic matter and recycling nutrients as well as parasites that cause detrimental effects to plant and animal species (3). Although relatively few fungal species cause disease in people (4), the burden on human health is substantial, as fungi infect over one billion individuals (5). These range from superficial skin or mucosal infections to invasive infections, killing ∼1.5 million people annually (6). The major agents of fatal fungal infections are species of Candida, Aspergillus, and Cryptococcus, which mainly infect immunocompromised populations. Invasive fungal infections are a growing concern as the number of individuals with immunosuppression due to HIV/AIDS, organ transplants, cancer treatments, and other immunosuppressive therapies increases (7).
Systemic fungal infections have unacceptably high mortality rates, due at least in part to the limited therapeutic options (8). The development of clinically useful antifungals is challenging because fungi are eukaryotes with many core cellular regulators that are conserved in the human host. This limits the number of unique druggable targets in the pathogen that can be disrupted without causing toxicity due to inhibition of the host counterpart. Currently, only three main classes of antifungal drugs are used in the clinic to treat systemic fungal infections. Polyenes and azoles impair integrity of the fungal cell membrane. Specifically, polyenes extract the essential cell membrane sterol, ergosterol, whereas azoles inhibit the ergosterol biosynthetic enzyme 14-α sterol demethylase, encoded by ERG11 (Candida albicans and Cryptococcus neoformans) or cyp51A and cyp51B (Aspergillus fumigatus). Echinocandins target fungal cell wall synthesis via inhibition of the (1,3)-β-D-glucan synthase, Fks1 (9). The utility of these drugs is hampered by challenges including host toxicity, fungistatic activity, and restricted spectrum of activity (9). Additionally, the increased emergence and prevalence of resistant fungal species threatens current treatment regimens (10). Most recent advances in clinical antifungal drug treatments have been improvements to the most commonly used drugs, which seldom address the problem of drug resistance (11). This has led to a dire need for new therapeutic targets and treatments to combat invasive fungal infections.
Antivirulence treatments, which seek to hinder microbial virulence factors that enable disease by damaging the host or evading the host immune system, have been investigated with considerable interest in the last 15 years (12, 13). Antivirulence strategies provide significant advantages, such as a reduced selection pressure for the development of resistance and a minimal impact on the host microbiota (13), yet only a limited number of antivirulence drugs are used in the clinic, all to treat bacterial toxin-mediated diseases, which consist of neutralizing immunoglobulins or monoclonal antibodies (12). Over the last few decades, extensive research has provided important insights into understanding fungal virulence factors, including elucidating the mechanisms of fungal stress responses (14). Targeting fungal stress responses is a promising strategy to cripple fungal pathogens as they regulate fungal virulence, are critical for fungal survival and growth in different ecological niches (15, 16), and enable the development of antifungal drug resistance (15, 17, 18). The human body is a hostile environment for pathogenic fungi, which must endure stresses such as high temperatures, nutrient limitation, and reactive oxygen species. Additionally, antifungal drug treatment induces fungal cell membrane or cell wall stress (19). Fungi respond and adapt to such stresses with cellular response networks that consist of sensors, signal transducers, and specific adaptive responses (15). As one example, heat shock triggers one of the most highly conserved stress responses in which hyperphosphorylation of the heat shock transcription factor Hsf1 induces increased transcription of genes encoding heat shock proteins, including Hsp90, which are critical for maintaining protein homeostasis during thermal stress (20, 21). Other pathways are crucial for mounting responses to osmotic, oxidative, alkaline, membrane, and cell wall stresses (15), providing a myriad of potential antifungal targets. Targeting these fungal stress responses in combination with current antifungals has the potential to disarm fungal virulence, reduce antifungal drug resistance, and sensitize drug-resistant pathogens.
Although some stress response pathways are conserved across eukaryotes, differences in human and fungal regulators can be exploited for pathogen-specific therapeutic strategies to minimize harmful effects to the host. Toxicity can be further curtailed through combination therapy, whereby the synergy between stress response inhibitors and current antifungals can allow for reduced dosage of both drugs and can abrogate drug resistance (22). For stress response regulators of therapeutic interest as antifungal drug targets, structural analysis can be instrumental for the rational design and optimization of potent and selective inhibitors. For highly conserved proteins, inhibitors developed against human proteins can serve as probes to explore the biology of orthologous fungal proteins (23–25). Importantly, such inhibitors can also be leveraged as scaffolds for the development of new inhibitors with different specificities (26). Structure-based approaches can identify regions of similarity between the fungal and human counterparts as well as differences that could be exploited to specifically target the pathogen. Useful dissimilarities include structurally diverse regions and distinct ligand-induced conformational changes. These insights can aid in optimizing scaffolds to improve fungal selectivity, reducing the time and cost required to develop novel antifungals (27). Proteins absent from humans are privileged targets for antifungal discovery. Despite the advantage of reduced concern over host toxicity, there can be challenges resulting from a lack of known chemical matter to specifically probe enzyme active sites, requiring a more in-depth structural and biochemical understanding of enzyme-substrate interactions. In silico approaches to guide optimization of lead inhibitors based on structures of protein targets can improve timelines and reduce costs in the discovery and development of novel antifungal drugs (28). Structure-guided insights can accelerate the design, development, and optimization of novel first-in-class inhibitors of fungal stress response that are both potent and specific, with minimal off-target toxicities.
Several fungal stress response regulators have emerged as promising antifungal drug targets. In this review, we highlight insights from structural analyses of fungal enzymes crucial for responses to stress induced within the host or upon drug exposure, along with the most recent advances in structure-guided development of novel antifungals that exploit vulnerabilities of the major fungal pathogens that cause devastating human infections.
Hsp90
Hsp90 is a highly conserved and essential molecular chaperone that regulates the folding and maturation of many diverse client proteins. This chaperone has been dubbed a hub of protein homeostasis, interacting with ∼10% of the proteome of the yeast Saccharomyces cerevisiae (29, 30). Hsp90 clients are enriched in regulators of cellular signaling cascades, such as kinases and transcription factors, allowing the chaperone to orchestrate numerous stress response pathways (31). Hsp90 is an ATP-dependent dimeric chaperone, recognized for its conformational flexibility. Each monomer consists of an N-terminal domain containing an unusual nucleotide-binding pocket within the Bergerat fold, followed by a middle domain important for recognition and binding of client proteins, and ending with a C-terminal domain crucial for dimerization (32). The chaperoning activity of Hsp90 is modulated by interactions with co-chaperones, as well as by a number of posttranslational modifications, including phosphorylation, acetylation, and S-nitrosylation (31). With central roles in proteostasis and a druggable nucleotide-binding pocket, Hsp90 has been investigated as a therapeutic target for numerous diseases, including cancer and neurodegeneration (33, 34). The initiation of multiple Hsp90 anticancer drug development programs has provided a diverse array of natural products and synthetic small molecules that specifically inhibit Hsp90 by competitively blocking nucleotide binding and hydrolysis, leading to degradation of oncogenic client proteins (35, 36).
As an essential regulator of cellular stress responses, Hsp90 has also emerged as an ideal target to cripple fungal pathogens. In diverse fungi, Hsp90 regulates responses to stress induced by antifungal drugs and enables the evolution of drug resistance by stabilizing core components of stress responses, including the protein phosphatase calcineurin and multiple kinases in the Pkc1 mitogen-activated protein kinase cell wall integrity pathway (17, 23, 37, 38). Inhibiting Hsp90 in C. albicans not only impedes the emergence of azole resistance, but also reverses azole resistance acquired in the laboratory or the human host (39). Even at concentrations that are well-tolerated in humans, clinical Hsp90 inhibitors substantially increase in vitro azole efficacy against C. albicans (40). The synergy between Hsp90 inhibitors and azole or echinocandin antifungals has been documented in invertebrate models of invasive infection with C. albicans, A. fumigatus, and C. neoformans (40, 41). Moreover, beyond regulating antifungal drug resistance, Hsp90 affects the virulence and pathogenicity of diverse fungal pathogens. In C. albicans, the ability to transition between round yeast and elongated filamentous forms is a major virulence trait, and Hsp90 is a key regulator of temperature-dependent morphogenesis as well as biofilm formation and dispersal (42–44). In A. fumigatus, repression of HSP90 results in a myriad of phenotypic defects associated with attenuated virulence, including reduced formation of asexual conidia spores, germination, and hyphal elongation (45, 46). More recently, Hsp90 has also been implicated in the pathogenicity of C. neoformans. Hsp90 is critical for C. neoformans thermotolerance, which is required for the environmental pathogen to infect humans and for the induction and maintenance of its polysaccharide capsule, a key virulence trait of this fungus (41, 47).
Thus far, the therapeutic potential of targeting fungal Hsp90 in a mammalian model has been most promising in the context of a localized infection, where pharmacological inhibition of Hsp90 in combination with an azole eradicated azole-resistant biofilms in a rat venous catheter infection model (43). In a murine model of systemic infection, genetic depletion of C. albicans HSP90 resulted in attenuated virulence, increased antifungal efficacy, and improved fungal clearance; however, pharmacological inhibition of Hsp90 with molecules lacking fungal selectivity was not well-tolerated due to host toxicity (40). Similarly, genetic repression of fungal HSP90 rescued mice from lethal invasive aspergillosis infections (46), whereas the use of current Hsp90 inhibitors resulted in detrimental effects to the host (48). Thus, fungal-selective Hsp90 inhibitors must be developed for systemic use to abrogate Hsp90-dependent fungal stress responses, drug resistance, and pathogenicity, while circumventing host toxicities associated with inhibiting the host chaperone.
The high sequence conservation of Hsp90 between fungi and humans presents a challenge in the design of fungal-selective Hsp90 inhibitors, but recent crystal structures of Hsp90 from fungal pathogens are facilitating these endeavors. The nucleotide-binding domain (NBD) of human Hsp90 shares 72, 76, and 78% sequence identity to the domains of C. albicans, A. fumigatus, and C. neoformans, respectively. Moreover, the residues that line the surfaces of the nucleotide-binding pockets of these four proteins are nearly identical. Biochemical studies investigating the dynamic conformational changes of human, yeast, and bacterial Hsp90 have revealed high species specificity at the level of conformational equilibrium and intrinsic ATPase activity (49). Comparing C. albicans and human Hsp90 isoforms has also revealed similar disparities in ATPase activity (50). An additional layer of conformational regulation is provided by co-chaperones and accessory proteins, which also vary in composition across species (51). The crystal structure of the C. albicans Hsp90 N-terminal domain, which includes the ATP-binding domain, has recently enabled the rational design of the first fungal-selective inhibitor targeting Hsp90 in a fungal pathogen (50). Whereas apo (unliganded) structures were highly similar between human and C. albicans Hsp90, with a main-chain atom root mean square deviation of 1.0 Å, co-crystallization with multiple Hsp90 inhibitors revealed considerable ligand-induced flexibility in the NBD that was not observed in the human complex structure (50). C. albicans co-crystal structures of Hsp90 with distinct inhibitors revealed regions of the fungal NBD that were rigid and those that were prone to ligand-induced structural changes. In particular, the binding of the Hsp90 inhibitor AUY922, which is in preclinical development for oncology, to the C. albicans NBD revealed larger structural differences from the apo structure relative to the human complex, suggesting a greater degree of conformational flexibility in the fungal Hsp90 NBD compared with the human protein (50). This potential for ligand-induced flexibility in C. albicans Hsp90 has been exploited to design fungal-selective inhibitors.
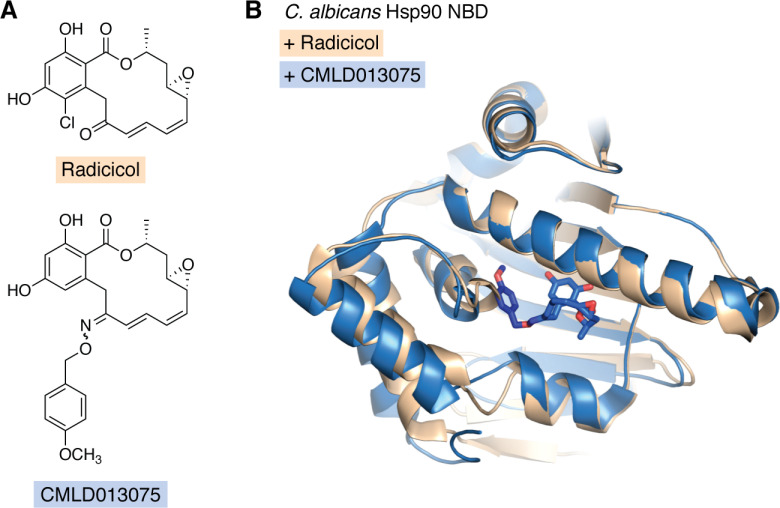
The natural product radicicol is among the most bioactive inhibitors of fungal Hsp90 (50), while also inhibiting the human chaperone. Building on the scaffold of the resorcyclic acid lactone, radicicol, and the closely related monocillin I, a small series of semisynthetic oxime analogs were developed using the structural insights to increase fungal selectivity (50). One of these compounds, CMLD013075 (Fig. 1A), displayed >25-fold selectivity for C. albicans Hsp90 compared with the human ortholog. Beyond potent target binding, CMLD013075 limited proliferation of a drug-resistant C. albicans clinical isolate, had synergistic activity with the azole fluconazole, and was less toxic to human cells compared with radicicol (50). The co-crystal structure of the C. albicans Hsp90 NBD with CMLD013075 displayed substantial backbone rearrangements relative to the radicicol complex with a Cα root mean square deviation of 1.9 Å. This included a remodeling of the ATP-binding site, N terminus, and lid region of the fungal chaperone (Fig. 1B). Key amino acid differences in the nucleotide-binding pockets of C. albicans and human Hsp90, including Leu-130 and Phe-131 in C. albicans instead of human residues alanine and tyrosine at equivalent positions, contribute to the C. albicans selectivity of CMLD013075. However, these residues are not conserved in other prominent fungal pathogens (50), suggesting that the selectivity of CMLD013075 for C. albicans Hsp90 may not extend to A. fumigatus and C. neoformans. In fact, these three fungal sequences have pairwise sequence identities ranging from 73 to 76% in their NBDs, which is comparable with the sequence identities comparing the fungal NBDs to humans. Although the pharmacokinetic properties of CMLD013075 require optimization for in vivo testing, such as to address a P450 metabolic liability, this proof-of-concept molecule has demonstrated the feasibility of selectively targeting fungal Hsp90 as a novel antifungal strategy.
Figure 1.
The co-crystal structure of the C. albicans Hsp90 NBD with CMLD013075 displays unique structural rearrangements. A, chemical structures of radicicol (top) and fungal-selective semisynthetic compound CMLD013075 (bottom). B, superposition of the structures of C. albicans Hsp90 NBD in complex with radicicol (tan; PDB entry 6CJL) and CMLD013075 (blue; PDB entry 6CJP). The CMLD013075 ligand is shown in a stick representation, with the carbon atoms of the methoxybenzyl ring in darker blue. The ligand in the radicicol complex occupies a similar site as the lactone ring in the CMLD013075 complex but is not included here for clarity.
As a complementary approach to advance Hsp90 inhibitors as antifungals, biochemical structure-activity relationship analyses were performed to probe selective target engagement of molecules with C. albicans and C. neoformans Hsp90 (52). Many points of diversity were investigated in a series of over 100 fully synthetic, nonmacrocyclic, resorcylate analogs, which identified novel compounds with enhanced potency and selectivity for fungal Hsp90 isoforms as compared with their human counterparts. As fungal selectivity of the inhibitors compared with human Hsp90 increased, advanced analogs diverged in their species selectivity between C. albicans and C. neoformans (52). In line with the findings with CMLD013075, this suggests challenges for the development of fungal-selective Hsp90 inhibitors with broad-spectrum activity against diverse pathogenic fungi. Ongoing efforts toward obtaining co-crystal structures with novel synthetic fungal-selective compounds and the Hsp90 NBD from diverse fungal pathogens will provide an important resource to understand species selectivity and to guide structure-based efforts in the development of Hsp90 inhibitors with therapeutically useful fungal selectivity. An ideal fungal-selective inhibitor would have high selectivity at the level of target engagement, while also having suitable druglike properties, such as solubility, fungal cell permeability, and metabolic stability.
Fungal Hsp90 inhibitors have the potential to impede the emergence of antifungal drug resistance and resensitize drug-resistant clinical isolates to antifungals currently deployed in the clinic (39, 40). Thus, although Hsp90 inhibitors have single-agent antifungal activity, the greatest promise for antifungal therapy targeting Hsp90 lies in combinatorial treatment. Targeting other more divergent elements of the Hsp90 chaperone network may provide an alternate antifungal strategy. Proteins of particular interest are those that regulate Hsp90 function. One compelling strategy is to target enzymes that posttranslationally modify Hsp90, including modifications such as acetylation, thereby regulating Hsp90 function and interactions with clients and co-chaperones. Pharmacological inhibition of lysine deacetylases (KDACs) broadly impedes the emergence and maintenance of Hsp90-dependent antifungal drug resistance and reduces virulence in C. albicans, A. fumigatus, and C. neoformans (53–57). KDACs represent an interesting target in the Hsp90 fungal stress response network, given that their divergence between fungal pathogens and humans is far greater than for the chaperone Hsp90 (53, 58). Structural analysis of KDACs will be crucial to advance the development of potent and selective inhibitors. Other targets of interest in the Hsp90 network include proteins that depend on Hsp90, such as calcineurin, to mediate downstream effects on fungal stress responses, drug resistance, morphogenesis, and virulence (44).
Calcineurin
As with Hsp90, the serine/threonine protein phosphatase calcineurin is a highly conserved stress response regulator in eukaryotes. In both yeast and humans, calcineurin is a key member of calcium-activated signal transduction pathways. Calcineurin functions as a heterodimer of the catalytic subunit, CnA, and regulatory subunit, CnB (59). In response to stress, intracellular Ca2+ concentrations increase, causing Ca2+ to bind to calmodulin, which binds to the calcineurin catalytic subunit, CnA, displacing an autoinhibitory domain and thereby activating calcineurin. This activation of CnA leads to the transcriptional regulation of downstream targets (59). In humans, calcineurin plays a key role in many cellular signaling processes, including T-cell activation (60), and is the target of immunosuppressive drugs used in the clinic (61). Calcineurin is also being explored as an antifungal target as it plays important roles in regulating fungal development, stress responses, and virulence. However, the immunosuppressive nature of current calcineurin inhibitors complicates their use in the treatment of invasive fungal infections. Recent structural analyses of calcineurin in fungal pathogens have revealed insights that will help guide the development of fungal-selective calcineurin inhibitors.
Although not essential under standard laboratory growth conditions, calcineurin is required to respond to the stresses encountered in a human host and hence is required for virulence of the three major fungal pathogens, C. albicans, C. neoformans, and A. fumigatus (7, 62). In fungal pathogens, calcineurin is activated by stress and dephosphorylates the downstream transcription factor Crz1 (CrzA in A. fumigatus), which then translocates into the nucleus, where it binds to calcineurin-dependent response elements in target gene promoters (63). The genes regulated by fungal Crz1 include those with roles in cell signaling, cell wall integrity, and ion homeostasis, with distinct target genes in different species (64–67). In C. albicans, calcineurin has important roles in governing cell wall integrity and growth in the presence of serum (68–71). Consequently, C. albicans calcineurin mutants exhibit decreased virulence in a murine model of disseminated candidiasis (71). In some non-albicans Candida species, calcineurin regulates another important virulence trait, the ability to undergo filamentous growth in response to various inducing cues (72). Calcineurin is also required for C. neoformans virulence, as it plays crucial roles in thermotolerance, such that deletion of genes encoding calcineurin subunits impairs growth at 37 °C (73–75). Finally, in A. fumigatus, mutants lacking the catalytic subunit of calcineurin are defective in hyphal and conidial morphology, with reduced pathogenicity in murine models of invasive aspergillosis (76–78). As calcineurin is required for hyphal growth of A. fumigatus, pharmacological inhibition of calcineurin impairs growth of this filamentous fungus in vitro (79).
In addition to regulating environmental stress responses and virulence, calcineurin enables crucial responses to the stress imposed by antifungal agents, such as echinocandins and azoles. As a consequence, pharmacological inhibition or genetic depletion of calcineurin sensitizes C. albicans and A. fumigatus to echinocandins (68, 70, 79–81). Interestingly, whereas C. neoformans is intrinsically resistant to echinocandins, deletion of genes encoding calcineurin subunits sensitizes cells to the echinocandin caspofungin (82, 83). Calcineurin also regulates the response to cell membrane stress imposed by the azole antifungals. Inhibition of calcineurin potentiates azole activity against C. albicans and renders the drug combination fungicidal (39, 84–86) and enhances the activity of azoles against C. albicans biofilms (87). Fungal calcineurin is also an Hsp90 client protein, requiring Hsp90 for stability and functioning as a key mediator of Hsp90-dependent drug resistance (68, 88). In azole-resistant strains of A. fumigatus, the combination of inhibitors of Hsp90 and calcineurin is fungicidal (89). Thus, calcineurin is an attractive target to impair fungal virulence and drug resistance.
Calcineurin inhibitors are central to procedures such as organ transplantation and the treatment of immune-mediated diseases (90). These calcineurin inhibitors include the natural products cyclosporine A and the more potent FK506, which inhibit calcineurin function through binding to the immunophilins cyclophilin A and FK506-binding protein 12 (FKBP12), respectively (91, 92). FKBPs, including FKBP12, are peptidyl-prolyl cis/trans-isomerases that aid in the folding of proteins containing proline and have a key role in protein homeostasis (93). In humans, FKBP12 is also involved in receptor signaling and regulation of cellular partners, such as transforming growth factor-β (93). In fungi, the FKBPs are not essential for fungal growth, and their physiological roles are not well-characterized (94–96). However, FKBP12 plays a conserved role in FK506-mediated inhibition of calcineurin in pathogenic fungi (95, 96).
Given the immunosuppressive nature of current calcineurin inhibitors, there is a pressing need to identify fungal-selective inhibitors that reduce the immunosuppressive effects. One promising study characterized four FK506 analogs that had reduced immunosuppressive activity and were produced by strains of Streptomyces with deletion of specific FK506 biosynthetic genes (97, 98). These compounds displayed synergistic activity with fluconazole as well as single-agent activity against diverse pathogenic fungi under conditions requiring calcineurin function (99). A recent follow-up study used combinations of mutations in FK506-producing Streptomyces species to produce additional analogs with reduced immunosuppressive activity despite robust antifungal activity and synergistic activity with fluconazole (100). These findings highlight the potential for the development of fungal-selective calcineurin inhibitors. An alternate strategy to enhance the antifungal potential of FK506 that has been explored involves a combination of FK506 and FK506 antagonists that compete for binding to FKBP12, the latter of which permeates human but not fungal cells; this strategy exerts antifungal activity against A. fumigatus with a reduction in immunosuppressive effects on the host (101).
Structural analyses are also providing insights to guide the development of antifungal therapies targeting fungal calcineurin. Because FKBP12 mediates the interaction between calcineurin and inhibitors like FK506 (95, 96), structural differences in the fungal and human FKBP12 can be leveraged in the design of fungal-selective calcineurin inhibitors. The first structural elucidation of FKBP12 from pathogenic fungi reported crystallized apo structures of C. albicans, Candida glabrata, and A. fumigatus FKBP12 (102). Overall, these FKBP12 structures had a similar fold to those from humans and the model yeast S. cerevisiae, with a 5–6-stranded β sheet around an α helix with three extended loops (the 40s loop, 50s loop, and 80s loop) surrounding the active site (102). Although the fungal FKBP12s share 40–50% sequence identity to the human protein, striking differences between fungal and human FKBP12 have been identified and exploited in the search for fungal-specific inhibitors (26, 102).
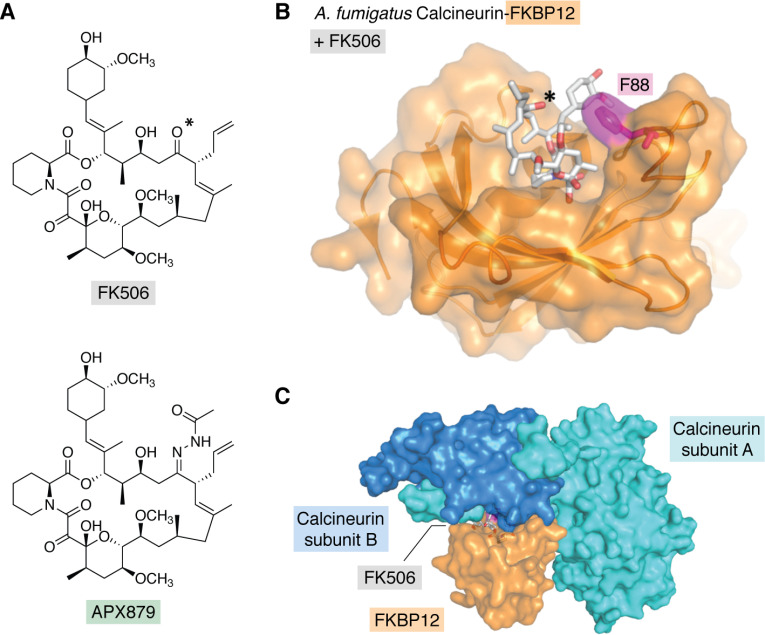
One such structural difference has led to the development of a promising proof-of-concept fungal-selective FK506 analog. A recent study elucidated the crystal structures of calcineurin catalytic and regulatory subunits in complex with both FKBP12 and the inhibitor FK506 for four diverse fungal pathogens (26). Structure-guided mutational analysis of the calcineurin-FK506-FKBP12 complex in A. fumigatus identified a residue, Phe-88, in the FKBP12 80s loop critical for binding and inhibiting fungal calcineurin (Fig. 2, B and C). This residue is found at the interface of the calcineurin interaction with FKBP12 and FK506, and substitution of this residue conferred resistance to FK506 by decreasing binding affinity to calcineurin. Importantly, the human protein has a histidine at the equivalent position, which is one of the few differences in the FK506 pocket that can be exploited to build in fungal specificity. Identification of this residue enabled the development of an acetohydrazine-substituted variant of FK506, APX879, which interacts more favorably with the fungal complex than the human complex (Fig. 2A). This analog had reduced immunosuppressive effects but broad-spectrum antifungal activity. However, compared with FK506, APX879 showed reduced in vitro single-agent and combinatorial antifungal activity against various pathogenic fungi. Nevertheless, APX879 had therapeutic benefits in a murine model of invasive cryptococcosis, in sharp contrast to the acute toxicity caused by FK506 during infection (26). The therapeutic efficacy of APX879 was further improved by combination with fluconazole. Thus, although the antifungal activity of APX879 was not as potent as that of FK506, the reduced immunosuppressive activity highlights the feasibility of designing fungal-selective calcineurin inhibitors guided by structural differences between human and fungal proteins.
Figure 2.
The crystal structure of the A. fumigatus calcineurin-FK506-FKBP12 complex revealed a residue, Phe-88, in FKBP12 critical for ligand binding and calcineurin inhibition, which is divergent in human FKBP12. A, chemical structures of FK506 (top) and the fungal-selective acetohydrazine-substituted analog APX879 (bottom). B, surface representation of A. fumigatus calcineurin-FK506-FKBP12 complex structure (PDB entry 6TZ7). FKBP12 is shown in orange with the fungal-specific residue Phe-88 (F88) in magenta. FK506 is in stick representation. The carbonyl oxygen at position 22 is indicated with an asterisk in A and B. Here, the calcineurin subunits are not shown for clarity. C, the complex is shown with the inclusion of the calcineurin A and B subunits.
Another intriguing structural feature of FKBP12 that can be exploited for antifungal therapies is its propensity to form dimers, although the physiological role of this dimerization remains elusive. The formation of FKBP12 dimers has been studied in mammals and S. cerevisiae, and key residues in the 40s loop that are important for dimerization have been defined (103, 104). In contrast, C. albicans and A. fumigatus FKBP12 dimers contain a central proline in the 80s loop that inserts into the active site of an adjacent FKBP12 molecule and is not conserved in the human sequence (102). Interestingly, this FKBP12 self-interaction site overlaps with the binding site of FK506, and substitution of the proline or the adjacent residue in A. fumigatus confers FK506 resistance, indicating a potential loss of FKBP12 affinity for calcineurin (102). It is possible that the formation of this dimer might enable self-catalysis, although this remains to be explored. Substitution of additional residues in the 40s-50s FKBP12 loop involved in FKBP12 dimerization resulted in even higher FK506 resistance than mutation of the proline residue, as well as a 2-fold increase in dimer strength and impaired calcineurin binding (105). Together these studies establish a connection between FKBP12 dimerization, FK506 binding, and calcineurin inhibition in pathogenic fungi (102, 105). Further investigation into the functional role of FKBP12 dimerization and the specific residues involved in pathogenic fungi compared with humans will aid in determining how to best exploit structural differences for antifungal drug design.
In addition to targeting fungal FKBP12 proteins, additional therapeutic strategies could be explored. Regulators upstream of calcineurin that are important for its function could be investigated. Members of the RCAN (regulator of calcineurin) family of proteins have been characterized in fungal pathogens (106–109), and further biochemical and structural characterization of these proteins may reveal an opportunity for the design of fungal-selective inhibitors. Further, additional characterization of structural differences between calcineurin proteins of fungi and humans could provide another strategy by which to design fungal-selective molecules. For example, calcineurin of A. fumigatus and other filamentous fungi harbors a unique serine-proline-rich linker domain in its catalytic subunit that could be exploited to increase the selectivity of antifungal molecules (110). Mutations in this region result in virulence defects, highlighting a fungal-specific region required for calcineurin function (110). Finally, effectors downstream of calcineurin could provide promising targets for the design of antifungals, as with Crz1 and additional effectors that have been implicated in fungi (111–115). Defining the effectors that are crucial to calcineurin-mediated stress responses in pathogenic fungi could provide novel targets for antifungal drug development.
Ras
Structure-guided approaches are also enabling efforts to target Ras proteins, highly conserved environmental stress regulators, as an antifungal strategy. Ras proteins are a family of small monomeric proteins with GTPase activity. They localize to the plasma membrane and function primarily as signal transducers (116). Activated by various extracellular cues, Ras proteins play key roles in regulating growth and response to stress via downstream signaling cascades (117). In humans, Ras proteins have been explored as potential targets for cancer therapies, as nearly 30% of human tumors have Ras substitutions causing constitutive activation of downstream pathways (118). In pathogenic fungi, nonessential Ras GTPases play critical roles in virulence (119). In C. albicans, Ras1 mediates signals to the cAMP-protein kinase A pathway, which is crucial to enabling morphogenesis in response to most environmental conditions (120). C. albicans mutants lacking Ras1 are defective in filamentation, biofilm formation, and virulence (119, 120). In C. neoformans, Ras1 is required for growth at 37 °C and, as a consequence, for pathogenicity in a mammalian host (121). Finally, A. fumigatus Ras proteins modulate conidiation, hyphal growth, and virulence (122). The crucial role that Ras proteins play in regulating the virulence of fungal pathogens illuminates their potential as targets for antivirulence therapies to disarm fungal pathogens.
Based on recent investigations of Ras GTPases in oncology, two strategies are being explored to target fungal Ras proteins: direct inhibition of Ras activity or disruption of Ras posttranslational modifications (PTMs) (123). Directly targeting Ras has not advanced substantially as an antifungal strategy due to the high conservation of the GTPase-binding domain across eukaryotes and the lack of extensive structural analysis. Nevertheless, the identification of an N-terminal domain extension in fungal Ras that is required for Ras signaling in A. fumigatus but is absent in homologs of higher eukaryotes suggests structural divergences that could potentially be exploited for the development of a fungal-specific inhibitor (124). More promising is the prospect of targeting the enzymes required for Ras PTMs, as they share <45% sequence identity with human orthologs (123). Localization of Ras proteins to the plasma membrane is essential for their biological activity (125, 126) and is regulated by sequential PTMs, including prenylation (farnesylation or geranylgeranylation), proteolysis, methylation, and palmitoylation (123). Genetic disruption of prenylation or palmitoylation recapitulates the phenotypic defects of a rasA deletion mutant in A. fumigatus (126, 127) and impedes filamentation in C. albicans (128). Similarly, in C. neoformans, pharmacological inhibition of the farnesyltransferase mimics the temperature-sensitive phenotype of the ras1 deletion mutant (129). Together, this demonstrates the allure of targeting Ras modifications to cripple fungal pathogens.
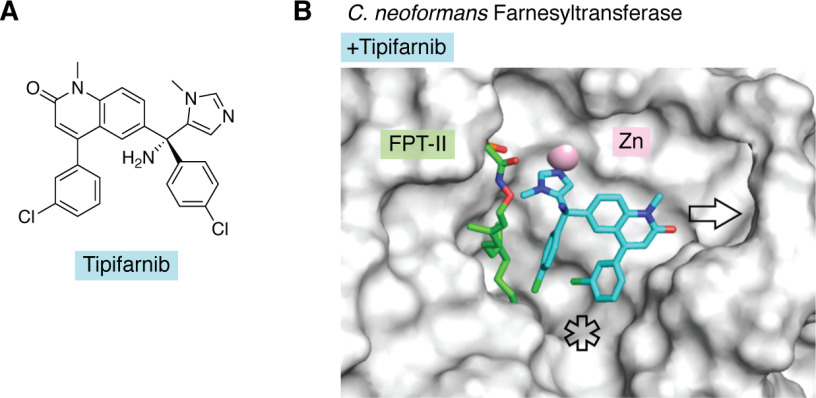
Farnesyltransferase (FTase) inhibitors developed as anticancer agents have demonstrated antifungal activity against C. neoformans at 37 °C (129). Remarkably, the natural product manumycin A disrupts C. neoformans Ras1 membrane localization and has growth-inhibitory activity against C. neoformans comparable with that of the clinical polyene antifungal drug amphotericin B (129). To guide optimization of the fungal selectivity of FTase inhibitors, the crystal structures of C. neoformans and A. fumigatus FTases were solved (129, 130). Structural analysis of the C. neoformans FTase demonstrated an overall similar architecture to the human ortholog, including the locations of the substrate-binding site and putative exit groove for the displaced prenylated product. However, in contrast to the situations with Hsp90 and FKBP12, the human and fungal FTases are more divergent, with pairwise sequence identities ranging from 30 to 40%. Whereas the human enzyme remains rigid throughout the prenyltransferase reaction cycle and forces substantial changes in the substrate conformation, a conformational change was observed in the cryptococcal enzyme upon peptide binding. A single-residue difference in the substrate-binding pocket of the C. neoformans FTase β subunit, Asn-413, in the place of the equivalent human Tyr-365 results in a deeper cavity in the active site relative to the human protein. An even larger active site was observed with the A. fumigatus FTase enzyme, calculated to be double the volume of that of the human FTase (130). Last, substantial structural variation was observed in the C. neoformans and A. fumigatus FTase product exit groove adjacent to the active site, resulting in a notable widening compared with the human isoform (129, 130). Co-crystal structures of fungal FTases with distinct inhibitor scaffolds, including the clinical anticancer agent tipifarnib, suggest that specific substitutions could be explored to exploit the structural differences between human and fungal FTases and optimize fungal selectivity (Fig. 3) (129, 130).
Figure 3.
The co-crystal structure of the C. neoformans FTase with tipifarnib suggests that inhibitor derivatization may lead to increased affinity and fungal selectivity. A, chemical structure of tipifarnib. B, surface representation of the structure of C. neoformans FTase in complex with farnesyl pyrophosphate analog, FPT-II, and tipifarnib (PDB entry 3SFX). The ligands are represented as sticks and color-coded according to heteroatom composition. Tipifarnib coordinates with the catalytic Zn2+ ion (pink) and could be derivatized to increase interactions with the product exit groove (arrow) and peptide-binding site (asterisk).
Trehalose biosynthesis
The development of inhibitors targeting unique pathways involved in fungal stress response and virulence that are absent from humans is a particularly attractive antifungal strategy. As such, the biosynthetic pathway of the nonreducing sugar trehalose provides an intriguing target to explore for antifungal development. Trehalose plays key roles in fungal metabolism, cell wall homeostasis, stress responses, and virulence in diverse pathogenic fungi (131). In yeasts and molds, the canonical trehalose biosynthetic pathway is a two-step enzymatic process comprised of trehalose-6-phosphate synthase (TPS; Tps1 in C. albicans and C. neoformans, TpsA/B in A. fumigatus) and trehalose-6-phosphate phosphatase (TPP; Tps2 in C. albicans and C. neoformans, OrlA in A. fumigatus). This pathway has direct links to the glycolytic flux and is integral to the regulation of glycolysis (132). Trehalose is found in fungi, bacteria, plants, and invertebrates but is absent from mammalian cells (133). Thus, inhibiting trehalose biosynthesis provides an opportunity to cripple human fungal pathogens with minimal toxicity to the host.
Inhibition of the biosynthesis of trehalose through perturbation of either the TPS or TPP has dramatic effects on fungal biology and virulence. In C. albicans, loss of Tps1 results in defects in thermotolerance in glucose-containing medium, impaired filamentation, hypersensitivity to oxidative stress, increased killing by macrophages, and severely attenuated virulence in a murine model of invasive candidiasis (134–136). Whereas loss of C. albicans Tps2 does not lead to an obvious defect in morphogenesis, it does cause increased thermosensitivity and reduced pathogenicity in a mouse model of systemic infection (137). In C. neoformans, Tps1 and Tps2 are required for growth at 37 °C in glucose-containing medium. Loss of C. neoformans Tps1 causes hypersensitivity to oxidative stress and to the clinical antifungal caspofungin (138), raising the possibility for a combinatorial treatment with cell wall–perturbing agents. Interestingly, the toxic accumulation of trehalose 6-phosphate upon loss of C. neoformans Tps2 results in cell death at elevated temperatures, suggesting that a treatment targeting Tps2 has the potential to be fungicidal. In A. fumigatus, trehalose production is blocked by deletion of two synthase-encoding genes, tpsA and tpsB, causing developmental defects and increased susceptibility to oxidative stress. Surprisingly, the double mutant is hypervirulent in a murine model of invasive aspergillosis, which is speculated to be due to reduced association with and phagocytosis by macrophages as a result of cell wall alterations (139). In contrast, deletion of the A. fumigatus tps2 homolog orlA results in a variety of reproductive, developmental, and morphological defects, and the mutant is severely attenuated in virulence in murine models of aspergillosis (140). Together, these findings support the potential of targeting the late step of trehalose biosynthesis to disarm fungal virulence.
To enable structure-guided drug design, crystal structures for fungal substrate-bound Tps1 and Tps2 have been solved. C. albicans Tps1 structures enabled the identification of key residues for substrate recognition and catalysis (141). C. albicans amino acid substitutions generating a catalytically dead version of Tps1 conferred temperature-sensitive growth and defects in filamentation and biofilm formation (141), suggesting that disruption of Tps1 activity with a competitive inhibitor could attenuate pathogenicity. Tps2 is a more attractive broad-spectrum antifungal target than Tps1, as genetic disruption of the second step in trehalose biosynthesis results in severe temperature sensitivity and/or attenuated virulence in all three major human fungal pathogens. The structure of the C. albicans Tps2 C-terminal domain, which contains the catalytically active phosphatase domain (Tps2PD), provided critical insights (142). Analysis of a Tps2PD transition state structure with trehalose enabled a mechanistic understanding of the enzymatic phosphatase reaction as well as substrate binding and specificity (142). Key residues for substrate-binding and catalytic activity were identified, including the critical aspartate nucleophile for phosphatase activity (142). The C. neoformans substrate-bound Tps2PD structure revealed a conserved active site and an overall structure nearly identical to the C. albicans counterpart. Substitution of the key catalytic aspartate residue in the cryptococcal Tps2PD abolished phosphatase activity, and the resulting mutant strain had phenotypic defects similar to a C. neoformans tps2 deletion mutant, including hypersensitivity to high temperature (142). This finding suggests that specific pharmacological inhibition of Tps2 phosphatase activity could cripple the pathogenicity of C. neoformans. Finally, a substrate-free A. fumigatus Tps2 (OrlA) structure was solved, which further established an overall structural conservation across fungal TPP enzymes (142). Together, structural and biochemical evidence identify a highly specific Tps2 active site that is distinct from other mammalian phosphatases, with key catalytic residues conserved across major fungal pathogens, making it an excellent target for antifungal drug development (131, 142).
Yck2
Targeting fungal cell wall integrity is a promising avenue for the design of antifungal agents, as cell walls are crucial for fungal cells and absent from human cells. The newest class of antifungals in the clinic, the echinocandins, perturb fungal cell wall synthesis by inhibiting the synthesis of (1,3)-β-D-glucan in the cell wall, impeding viability of fungal species such as C. albicans with minimal toxicity in humans (7). Alterations in cell wall structure caused by echinocandin treatment induce severe cellular stress, which activates the cell wall integrity pathway to enable a protective response. Targeting kinases involved in cell wall integrity signaling and stress responses has been demonstrated to increase the efficacy of echinocandins and impair fungal virulence, making them great targets for single-agent and combinatorial antifungal treatments (23, 143). Recently, the nonessential casein kinase 1 family member Yck2 has emerged as an attractive new target governing cell wall integrity and virulence. Loss of Yck2 in C. albicans results in altered cellular morphology and biofilm formation, in addition to increased sensitivity to cell wall–perturbing agents (143, 144).
Inhibitors of Yck2 have emerged from recent screening initiatives. A screen to identify protein kinase inhibitors that potentiate the echinocandin caspofungin against a resistant C. albicans clinical isolate identified several compounds with a 2,3-aryl-pyrazolopyridine scaffold as caspofungin potentiators (24). Chemical genomic approaches revealed Yck2 as a putative target for the lead compound GW461484A (GW), which was confirmed by an in vitro kinase assay demonstrating potent, dose-dependent Yck2 inhibition by GW. To further explore this interaction, the crystal structures of the apo and GW-bound C. albicans Yck2 kinase domain were elucidated (24). GW bound in the ATP-binding pocket of Yck2, and although the residues compromising the fungal Yck2 ATP-binding site are highly conserved with those of human casein kinases, residues at the C-terminal end of the fungal kinase domain showed divergence with the human sequence. Modifying GW to increase access to these divergent residues proximal to the ATP-binding site may provide a promising avenue by which to optimize this scaffold to improve fungal selectivity. In addition, homology models of the closest Yck2 orthologs of various fungal pathogens revealed high conservation across the kinase active sites. Accordingly, GW showed strong caspofungin potentiation against the emerging fungal pathogen Candida auris, in addition to potent single-agent activity against C. neoformans. However, GW showed modest to no activity against A. fumigatus or C. glabrata, potentially due to altered intracellular GW accumulation or alternative cell wall maintenance roles for Yck2 in these fungal pathogens (24).
Proof-of-concept experiments have established the therapeutic potential of targeting C. albicans Yck2. The efficacy of GW was first examined in a co-culture system in which GW impaired C. albicans growth and increased caspofungin efficacy, rescuing human cell viability. Although metabolic liabilities of the GW pyrazolopyridine scaffold preclude testing in vivo efficacy, genetic depletion of C. albicans YCK2 severely reduced virulence in a mouse model of candidiasis, underscoring the therapeutic potential of inhibiting this important regulator of cell wall integrity (24). Further analysis of structure-activity relationships and structure-guided drug design will be leveraged to optimize potent and fungal-selective inhibitors of Yck2. Together, this highlights the potential of expanding the antifungal target space to include protein kinases as key regulators of fungal stress response, drug resistance, and virulence.
Conclusion and perspectives
There is a dire need to expand the current armamentarium of antifungal drugs to treat systemic fungal infections. Selective targeting of fungal stress responses has emerged as a promising strategy to address this challenge. With roles in governing growth, virulence, and resistance to antifungal drugs, regulators of stress responses are promising targets for both single-agent and combinatorial therapies. Structural and functional analyses of fungal stress response regulators have provided unprecedented opportunity for the rational design of potent, specific, and fungal-selective inhibitors of these targets (Fig. 4). To facilitate the advancement of novel antifungal strategies, these analyses should be expanded to leverage crystal structures of additional proteins that regulate fungal virulence. This includes proteins involved in the detoxification or response to host challenges, such as fluctuations in metal concentrations and oxidative stress, such as the key A. fumigatus siderophore biosynthetic enzyme ornithine-N5-monooxygenase (145), the A. fumigatus peroxiredoxin Asp f3 (146), and the C. albicans superoxide dismutase 5 (147).
Figure 4.
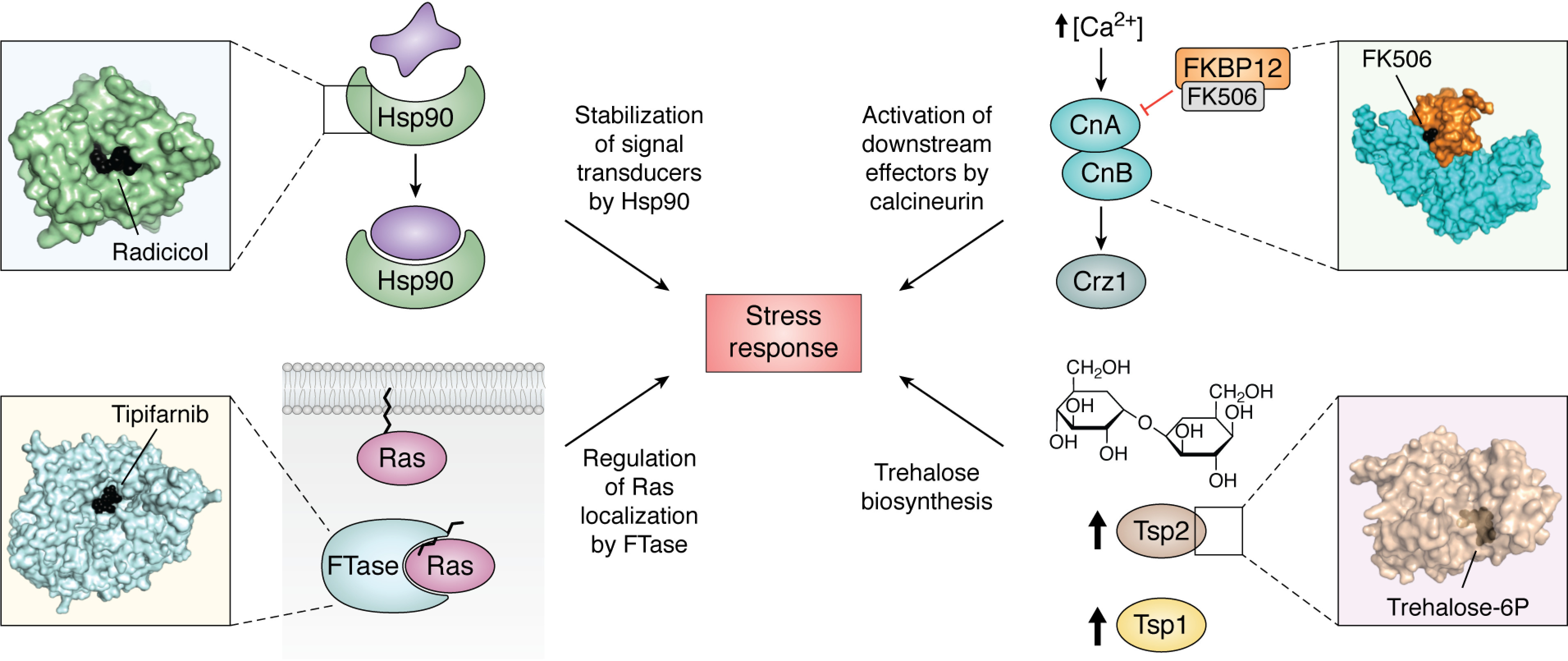
Structural analysis of fungal stress response proteins enables the unprecedented opportunity to develop inhibitors of these targets in pathogenic fungi. Top left, the molecular chaperone Hsp90 regulates stress responses via stabilization of multiple signal transducers; structural analysis of C. albicans Hsp90 NBD identified unique ligand-induced flexibility. Top right, calcineurin activates downstream effectors to mount stress responses in response to increased intracellular [Ca2+]; A. fumigatus calcineurin-FK506-FKBP12 complex structure identified a key fungal residue necessary for inhibitor interaction and target inhibition. Bottom left, Ras GTPases are important signal transducers that regulate the response to a variety of stresses and require prenylation by FTase for proper localization to the cell membrane; structural analysis of the C. neoformans FTase revealed structural features that can be exploited in the design of fungal-selective inhibitors. Bottom right, the two-step biosynthesis of trehalose is a stress response absent in mammalian cells; the structure of the C. neoformans Tps2 C-terminal domain provides a scaffold for inhibitor development.
A central challenge is to develop therapeutic agents that cripple the fungal pathogen without causing host toxicity. Although inhibitors of conserved targets such as calcineurin are used in the clinic, negative effects on the host preclude their use as antifungals to treat systemic infections. A number of high-throughput drug-screening approaches have been applied for the identification and development of specific fungal protein inhibitors, including scintillation proximity assays for kinase activity (148), fluorescence polarization–based assays (52, 149, 150), and surface plasmon resonance analyses (52, 151). In the absence of solved crystal structures, three-dimensional homology modeling is allowing for the virtual screening of chemical libraries (152). Alternatively, solving the structures of antifungal target enzymes enables insights into the architecture of relevant binding pockets, allowing for rational experimental or in silico inhibitor design (28). The new azole VT-1161 provides a powerful example of the success of structure-guided approaches in the design of improved antifungal agents. Following in silico predictions of binding affinity to prioritize azole scaffolds with improved fungal selectivity, homology model docking enabled rational stepwise improvements to potency and selectivity, resulting in a novel antifungal compound that has advanced into human clinical trials (153). Additionally, cutting-edge technological advances in cryo-EM are being harnessed to investigate previously intractable targets in drug discovery (154), providing an untapped potential for selective antifungal drug design.
The ongoing evolution of drug resistance in fungal pathogens and the emergence of novel fungal threats necessitate expanded efforts to discover and develop novel, resistance-evasive therapeutic strategies. Genomics has accelerated the identification of promising antifungal targets based on essentiality (155–158), requirement for virulence (159–163), or impact on drug resistance (38). Structural analyses of additional fungal stress response regulators and potential antifungal targets, including protein kinases in the cell wall integrity pathway and in the Hog1 osmotic response signaling cascade, are well-warranted to stimulate the design of much-needed antifungals (164). Proteins that are essential for fungal growth and virulence but do not have homologs in humans are privileged antifungal targets. Comparative genomics has identified 10 genes experimentally determined to be essential for growth in C. albicans and A. fumigatus that are conserved across eight fungal pathogens but are absent in the human genome (152). In particular, the protein Rim8, important for the response to alkaline pH, is highlighted as an attractive enzyme for pharmacological inhibition. In addition, an in vivo transposition strategy coupled to a machine-learning approach enabled a systematic inference of essentiality in a stable haploid derivative of C. albicans in an unbiased manner (156). This work facilitated the identification of 100 genes inferred to be essential in C. albicans that are conserved in A. fumigatus and C. neoformans but have no human homologs (156). Together, these findings provide a promising list of potential targets for antifungal drug development. The mobilization of interdisciplinary teams with fungal geneticists, structural biologists, and medicinal chemists has accelerated the optimization of new scaffolds and bolstered the antifungal drug development pipeline.
Acknowledgments
We thank all members of the Cowen laboratory for helpful discussions.
Funding and additional information—E. V. L. is supported by an Ontario Graduate Scholarship. L. E. C. is supported by Canadian Institutes of Health Research Foundation Grant FDN-154288 and National Institutes of Health Grant NIAID R01 R01AI120958-01A1; L. E. C. is a Canada Research Chair (Tier 1) in Microbial Genomics and Infectious Disease and co-Director of the CIFAR Fungal Kingdom: Threats and Opportunities program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—L. E. C. is a co-founder and shareholder in Bright Angel Therapeutics, a platform company for development of novel antifungal therapeutics. L. E. C. is a consultant for Boragen, a small-molecule development company focused on leveraging the unique chemical properties of boron chemistry for crop protection and animal health.
- NBD
- nucleotide-binding domain
- FKBP
- FK506-binding protein
- FTase
- farnesyltransferase
- GW
- GW461484A
- KDAC
- lysine deacetylase
- PTM
- posttranslational modification
- TPP
- trehalose-6-phosphate phosphatase
- TPS
- trehalose-6-phosphate synthase
- PDB
- Protein Data Bank.
References
- 1. Blackwell M. (2011) The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 98, 426–438 10.3732/ajb.1000298 [DOI] [PubMed] [Google Scholar]
- 2. Hawksworth D. L. (2012) Global species numbers of fungi: are tropical studies and molecular approaches contributing to a more robust estimate? Biodivers. Conserv. 21, 2425–2433 10.1007/s10531-012-0335-x [DOI] [Google Scholar]
- 3. Fisher M. C., Gurr S. J., Cuomo C. A., Blehert D. S., Jin H., Stukenbrock E. H., Stajich J. E., Kahmann R., Boone C., Denning D. W., Gow N. A. R., Klein B. S., Kronstad J. W., Sheppard D. C., Taylor J. W., et al. (2020) Threats posed by the fungal kingdom to humans, wildlife, and agriculture. MBio. 11, e00449–20 10.1128/mBio.00449-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Köhler J. R., Casadevall A., and Perfect J. (2014) The spectrum of fungi that infects humans. Cold Spring Harb. Perspect. Med. 5, a019273 10.1101/cshperspect.a019273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Havlickova B., Czaika V. A., and Friedrich M. (2008) Epidemiological trends in skin mycoses worldwide. Mycoses 51, 2–15 10.1111/j.1439-0507.2008.01606.x [DOI] [PubMed] [Google Scholar]
- 6. Brown G. D., Denning D. W., Gow N. A., Levitz S. M., Netea M. G., and White T. C. (2012) Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv13 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- 7. Perfect J. R. (2017) The antifungal pipeline: a reality check. Nat. Rev. Drug Discov. 16, 603–616 10.1038/nrd.2017.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roemer T., and Krysan D. J. (2014) Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb. Perspect. Med. 4, a019703 10.1101/cshperspect.a019703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robbins N., Wright G. D., and Cowen L. E. (2016) Antifungal drugs: the current armamentarium and development of nzew agents. Microbiol. Spectr. 4, 10.1128/microbiolspec.funk-0002-2016 [DOI] [PubMed] [Google Scholar]
- 10. Revie N. M., Iyer K. R., Robbins N., and Cowen L. E. (2018) Antifungal drug resistance: evolution, mechanisms and impact. Curr. Opin. Microbiol. 45, 70–76 10.1016/j.mib.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calderone R., Sun N., Gay-Andrieu F., Groutas W., Weerawarna P., Prasad S., Alex D., and Li D. (2014) Antifungal drug discovery: the process and outcomes. Future Microbiol. 9, 791–805 10.2217/fmb.14.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dickey S. W., Cheung G. Y. C., and Otto M. (2017) Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 16, 457–471 10.1038/nrd.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clatworthy A. E., Pierson E., and Hung D. T. (2007) Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548 10.1038/nchembio.2007.24 [DOI] [PubMed] [Google Scholar]
- 14. Vila T., Romo J. A., Pierce C. G., McHardy S. F., Saville S. P., and Lopez-Ribot J. L. (2017) Targeting Candida albicans filamentation for antifungal drug development. Virulence. 8, 150–158 10.1080/21505594.2016.1197444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown A. J. P., Cowen L. E., di Pietro A., and Quinn J. (2017) Stress adaptation. Microbiol. Spectr. 5, 10.1128/microbiolspec.funk-0048-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naranjo-Ortiz M. A., and Gabaldón T. (2019) Fungal evolution: major ecological adaptations and evolutionary transitions. Biol. Rev. 94, 1443–1476 10.1111/brv.12510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cowen L. E. (2013) The fungal Achilles' heel: targeting Hsp90 to cripple fungal pathogens. Curr. Opin. Microbiol. 16, 377–384 10.1016/j.mib.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 18. Juvvadi P. R., Lamoth F., and Steinbach W. J. (2014) Calcineurin as a multifunctional regulator: unraveling novel functions in fungal stress responses, hyphal growth, drug resistance, and pathogenesis. Fungal Biol. Rev. 28, 56–69 10.1016/j.fbr.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cowen L. E., and Steinbach W. J. (2008) Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot. Cell. 7, 747–764 10.1128/EC.00041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nicholls S., Leach M. D., Priest C. L., and Brown A. J. P. (2009) Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol. Microbiol. 74, 844–861 10.1111/j.1365-2958.2009.06883.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leach M. D., Klipp E., Cowen L. E., and Brown A. J. P. (2012) Fungal Hsp90: a biological transistor that tunes cellular outputs to thermal inputs. Nat. Rev. Microbiol. 10, 693–704 10.1038/nrmicro2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spitzer M., Robbins N., and Wright G. D. (2017) Combinatorial strategies for combating invasive fungal infections. Virulence 8, 169–185 10.1080/21505594.2016.1196300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. LaFayette S. L., Collins C., Zaas A. K., Schell W. A., Betancourt-Quiroz M., Gunatilaka A. A. L., Perfect J. R., and Cowen L. E. (2010) PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 6, e1001069 10.1371/journal.ppat.1001069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caplan T., Lorente-Macias A., Stogios P. J., Evdokimova E., Hyde S., Wellington M. A., Liston S., Iyer K. R., Puumala E., Shekhar-Guturja T., Robbins N., Savchenko A., Krysan D. J., Whitesell L., Zuercher W. J., et al. (2020) Overcoming fungal echinocandin resistance through inhibition of the non-essential stress kinase Yck2. Cell Chem Biol. 27, 269–282.e5 10.1016/j.chembiol.2019.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baxter B. K., DiDone L., Ogu D., Schor S., and Krysan D. J. (2011) Identification, in vitro activity and mode of action of phosphoinositide-dependent-1 kinase inhibitors as antifungal molecules. ACS Chem. Biol. 6, 502–510 10.1021/cb100399x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Juvvadi P. R., Fox D. 3rd, Bobay B. G., Hoy M. J., Gobeil S. M. C., Venters R. A., Chang Z., Lin J. J., Averette A. F., Cole D. C., Barrington B. C., Wheaton J. D., Ciofani M., Trzoss M., Li X., et al. (2019) Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents. Nat. Commun. 10, 4275 10.1038/s41467-019-12199-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miró-Canturri A., Ayerbe-Algaba R., and Smani Y. (2019) Drug repurposing for the treatment of bacterial and fungal infections. Front. Microbiol. 10, 41 10.3389/fmicb.2019.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Silva D. R., de Cássia Orlandi Sardi J., Freires I. A., Silva A. C. B., and Rosalen P. L. (2019) In silico approaches for screening molecular targets in Candida albicans: a proteomic insight into drug discovery and development. Eur. J. Pharmacol. 842, 64–69 10.1016/j.ejphar.2018.10.016 [DOI] [PubMed] [Google Scholar]
- 29. Zhao R., Davey M., Hsu Y.-C., Kaplanek P., Tong A., Parsons A. B., Krogan N., Cagney G., Mai D., Greenblatt J., Boone C., Emili A., and Houry W. A. (2005) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell 120, 715–727 10.1016/j.cell.2004.12.024 [DOI] [PubMed] [Google Scholar]
- 30. McClellan A. J., Xia Y., Deutschbauer A. M., Davis R. W., Gerstein M., and Frydman J. (2007) Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131, 121–135 10.1016/j.cell.2007.07.036 [DOI] [PubMed] [Google Scholar]
- 31. Taipale M., Jarosz D. F., and Lindquist S. (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 10.1038/nrm2918 [DOI] [PubMed] [Google Scholar]
- 32. Pearl L. H., and Prodromou C. (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75, 271–294 10.1146/annurev.biochem.75.103004.142738 [DOI] [PubMed] [Google Scholar]
- 33. Lackie R. E., Maciejewski A., Ostapchenko V. G., Marques-Lopes J., Choy W. Y., Duennwald M. L., Prado V. F., and Prado M. A. M. (2017) The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases. Front. Neurosci. 11, 254 10.3389/fnins.2017.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neckers L., and Workman P. (2012) Hsp90 molecular chaperone inhibitors: are we there yet? Clin. Cancer Res. 18, 64–76 10.1158/1078-0432.CCR-11-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taldone T., Patel H. J., Bolaender A., Patel M. R., and Chiosis G. (2014) Protein chaperones: a composition of matter review (2008–2013). Expert Opin. Ther. Pat. 24, 501–518 10.1517/13543776.2014.887681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zuehlke A. D., Moses M. A., and Neckers L. (2018) Heat shock protein 90: its inhibition and function. Philos. Trans. R. Soc. B Biol. Sci. 373, 20160527 10.1098/rstb.2016.0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cowen L. E., Carpenter A. E., Matangkasombut O., Fink G. R., and Lindquist S. (2006) Genetic architecture of Hsp90-dependent drug resistance. Eukaryot. Cell. 5, 2184–2188 10.1128/EC.00274-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caplan T., Polvi E. J., Xie J. L., Buckhalter S., Leach M. D., Robbins N., and Cowen L. E. (2018) Functional genomic screening reveals core modulators of echinocandin stress responses in Candida albicans. Cell Rep. 23, 2292–2298 10.1016/j.celrep.2018.04.084 [DOI] [PubMed] [Google Scholar]
- 39. Cowen L. E., and Lindquist S. (2005) Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309, 2185–2189 10.1126/science.1118370 [DOI] [PubMed] [Google Scholar]
- 40. Cowen L. E., Singh S. D., Köhler J. R., Collins C., Zaas A. K., Schell W. A., Aziz H., Mylonakis E., Perfect J. R., Whitesell L., and Lindquist S. (2009) Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc. Natl. Acad. Sci. U. S. A. 106, 2818–2823 10.1073/pnas.0813394106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cordeiro R. D A., Evangelista A. J. D J., Serpa R., Marques F. J. D F., Melo C. V. S. D., Oliveira J. S. D., Franco J. D S., Alencar L. P. D., Bandeira T. D J. P. G., Brilhante R. S. N., Sidrim J. J. C., and Rocha M. F. G. (2016) Inhibition of heat-shock protein 90 enhances the susceptibility to antifungals and reduces the virulence of Cryptococcus neoformans/Cryptococcus gattii species complex. Microbiology 162, 309–317 10.1099/mic.0.000222 [DOI] [PubMed] [Google Scholar]
- 42. Shapiro R. S., Uppuluri P., Zaas A. K., Collins C., Senn H., Perfect J. R., Heitman J., and Cowen L. E. (2009) Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 19, 621–629 10.1016/j.cub.2009.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robbins N., Uppuluri P., Nett J., Rajendran R., Ramage G., Lopez-Ribot J. L., Andes D., and Cowen L. E. (2011) Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 7, e1002257 10.1371/journal.ppat.1002257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Meara T. R., Robbins N., and Cowen L. E. (2017) The Hsp90 chaperone network modulates Candida virulence traits. Trends Microbiol. 25, 809–819 10.1016/j.tim.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lamoth F., Juvvadi P. R., Fortwendel J. R., and Steinbach W. J. (2012) Heat shock protein 90 is required for conidiation and cell wall integrity in Aspergillus fumigatus. Eukaryot. Cell 11, 1324–1332 10.1128/EC.00032-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lamoth F., Juvvadi P. R., Gehrke C., Asfaw Y. G., and Steinbach W. J. (2014) Transcriptional activation of heat shock protein 90 mediated via a proximal promoter region as trigger of caspofungin resistance in Aspergillus fumigatus. J. Infect. Dis. 209, 473–481 10.1093/infdis/jit530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chatterjee S., and Tatu U. (2017) Heat shock protein 90 localizes to the surface and augments virulence factors of Cryptococcus neoformans. PLoS Negl. Trop. Dis. 11, e0005836 10.1371/journal.pntd.0005836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blum G., Kainzner B., Grif K., Dietrich H., Zeiger B., Sonnweber T., and Lass-Flörl C. (2013) In vitro and in vivo role of heat shock protein 90 in amphotericin B resistance of Aspergillus terreus. Clin. Microbiol. Infect. 19, 50–55 10.1111/j.1469-0691.2012.03848.x [DOI] [PubMed] [Google Scholar]
- 49. Southworth D. R., and Agard D. A. (2008) Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol. Cell 32, 631–640 10.1016/j.molcel.2008.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Whitesell L., Robbins N., Huang D. S., McLellan C. A., Shekhar-Guturja T., LeBlanc E. V., Nation C. S., Hui R., Hutchinson A., Collins C., Chatterjee S., Trilles R., Xie J. L., Krysan D. J., Lindquist S., et al. (2019) Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus. Nat. Commun. 10, 402 10.1038/s41467-018-08248-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krukenberg K. A., Street T. O., Lavery L. A., and Agard D. A. (2011) Conformational dynamics of the molecular chaperone Hsp90. Q. Rev. Biophys. 44, 229–255 10.1017/S0033583510000314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang D. S., LeBlanc E. V., Shekhar-Guturja T., Robbins N., Krysan D. J., Pizarro J., Whitesell L., Cowen L. E., and Brown L. E. (2020) Design and synthesis of fungal-selective resorcylate aminopyrazole Hsp90 inhibitors. J. Med. Chem. 63, 2139–2180 10.1021/acs.jmedchem.9b00826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Robbins N., Leach M. D., and Cowen L. E. (2012) Lysine deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. Cell Rep. 2, 878–888 10.1016/j.celrep.2012.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garnaud C., Champleboux M., Maubon D., Cornet M., and Govin J. (2016) Histone deacetylases and their inhibition in Candida species. Front. Microbiol. 7, 1238 10.3389/fmicb.2016.01238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bauer I., Varadarajan D., Pidroni A., Gross S., Vergeiner S., Faber B., Hermann M., Tribus M., Brosch G., and Graessle S. (2016) A class 1 histone deacetylase with potential as an antifungal target. MBio 7, e00831–16 10.1128/mBio.00831-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pfaller M. A., Messer S. A., Georgopapadakou N., Martell L. A., Besterman J. M., and Diekema D. J. (2009) Activity of MGCD290, a Hos2 histone deacetylase inhibitor, in combination with azole antifungals against opportunistic fungal pathogens. J. Clin. Microbiol. 47, 3797–3804 10.1128/JCM.00618-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brandão F. A. S., Derengowski L. S., Albuquerque P., Nicola A. M., Silva-Pereira I., and Poças-Fonseca M. J. (2015) Histone deacetylases inhibitors effects on Cryptococcus neoformans major virulence phenotypes. Virulence. 6, 618–630 10.1080/21505594.2015.1038014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang X.-J., and Seto E. (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 10.1038/nrm2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stie J., and Fox D. (2008) Calcineurin regulation in fungi and beyond. Eukaryot. Cell 7, 177–186 10.1128/EC.00326-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Baksh S., and Burakoff S. J. (2000) The role of calcineurin in lymphocyte activation. Semin. Immunol. 12, 405–415 10.1006/smim.2000.0221 [DOI] [PubMed] [Google Scholar]
- 61. Azzi J. R., Sayegh M. H., and Mallat S. G. (2013) Calcineurin inhibitors: 40 years later, can't live without. J. Immunol. 191, 5785–5791 10.4049/jimmunol.1390055 [DOI] [PubMed] [Google Scholar]
- 62. Steinbach W. J., Reedy J. L., Cramer R. A., Perfect J. R., and Heitman J. (2007) Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat. Rev. Microbiol. 5, 418–430 10.1038/nrmicro1680 [DOI] [PubMed] [Google Scholar]
- 63. Thewes S. (2014) Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot Cell 13, 694–705 10.1128/EC.00038-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yoshimoto H., Saltsman K., Gasch A. P., Li H. X., Ogawa N., Botstein D., Brown P. O., and Cyert M. S. (2002) Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277, 31079–31088 10.1074/jbc.M202718200 [DOI] [PubMed] [Google Scholar]
- 65. Adler A., Park Y. D., Larsen P., Nagarajan V., Wollenberg K., Qiu J., Myers T. G., and Williamson P. R. (2011) A novel specificity protein 1 (SP1)-like gene regulating protein kinase C-1 (Pkc1)-dependent cell wall integrity and virulence factors in Cryptococcus neoformans. J. Biol. Chem. 286, 20977–20990 10.1074/jbc.M111.230268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Soriani F. M., Malavazi I., Savoldi M., Espeso E., Dinamarco T. M., Bernardes L. A., Ferreira M. E., Goldman M. H., and Goldman G. H. (2010) Identification of possible targets of the Aspergillus fumigatus CRZ1 homologue, CrzA. BMC Microbiol. 10, 12 10.1186/1471-2180-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Karababa M., Valentino E., Pardini G., Coste A. T., Bille J., and Sanglard D. (2006) CRZ1, a target of the calcineurin pathway in Candida albicans. Mol. Microbiol. 59, 1429–1451 10.1111/j.1365-2958.2005.05037.x [DOI] [PubMed] [Google Scholar]
- 68. Singh S. D., Robbins N., Zaas A. K., Schell W. A., Perfect J. R., and Cowen L. E. (2009) Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 5, e1000532 10.1371/journal.ppat.1000532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Munro C. A., Selvaggini S., de Bruijn I., Walker L., Lenardon M. D., Gerssen B., Milne S., Brown A. J., and Gow N. A. (2007) The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol Microbiol. 63, 1399–1413 10.1111/j.1365-2958.2007.05588.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wiederhold N. P., Kontoyiannis D. P., Prince R. A., and Lewis R. E. (2005) Attenuation of the activity of caspofungin at high concentrations against Candida albicans: possible role of cell wall integrity and calcineurin pathways. Antimicrob. Agents Chemother. 49, 5146–5148 10.1128/AAC.49.12.5146-5148.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Blankenship J. R., Wormley F. L., Boyce M. K., Schell W. A., Filler S. G., Perfect J. R., and Heitman J. (2003) Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell 2, 422–430 10.1128/ec.2.3.422-430.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yu S. J., Chang Y. L., and Chen Y. L. (2015) Calcineurin signaling: lessons from Candida species. FEMS Yeast Res. 15, fov016 10.1093/femsyr/fov016 [DOI] [PubMed] [Google Scholar]
- 73. Chen Y.-L., Lehman V. N., Lewit Y., Averette A. F., and Heitman J. (2013) Calcineurin governs thermotolerance and virulence of Cryptococcus gattii. G3 (Bethesda) 3, 527–539 10.1534/g3.112.004242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Odom A., Muir S., Lim E., Toffaletti D. L., Perfect J., and Heitman J. (1997) Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16, 2576–2589 10.1093/emboj/16.10.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fox D. S., Cruz M. C., Sia R. A., Ke H., Cox G. M., Cardenas M. E., and Heitman J. (2001) Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol. Microbiol. 39, 835–849 10.1046/j.1365-2958.2001.02295.x [DOI] [PubMed] [Google Scholar]
- 76. Steinbach W. J., Cramer R. A. Jr., Perfect B. Z., Asfaw Y. G., Sauer T. C., Najvar L. K., Kirkpatrick W. R., Patterson T. F., Benjamin D. K. Jr., Heitman J., and Perfect J. R. (2006) Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5, 1091–1103 10.1128/EC.00139-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Juvvadi P. R., Fortwendel J. R., Pinchai N., Perfect B. Z., Heitman J., and Steinbach W. J. (2008) Calcineurin localizes to the hyphal septum in Aspergillus fumigatus: implications for septum formation and conidiophore development. Eukaryot. Cell. 7, 1606–1610 10.1128/EC.00200-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Juvvadi P. R., Lamoth F., and Steinbach W. J. (2014) Calcineurin-mediated regulation of hyphal growth, septation, and virulence in Aspergillus fumigatus. Mycopathologia 178, 341–348 10.1007/s11046-014-9794-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Steinbach W. J., Cramer R. A. Jr., Perfect B. Z., Henn C., Nielsen K., Heitman J., and Perfect J. R. (2007) Calcineurin inhibition or mutation enhances cell wall inhibitors against Aspergillus fumigatus. Antimicrob Agents Chemother. 51, 2979–2981 10.1128/AAC.01394-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Steinbach W. J., Singh N., Miller J. L., Benjamin D. K. Jr., Schell W. A., Heitman J., and Perfect J. R. (2004) In vitro interactions between antifungals and immunosuppressants against Aspergillus fumigatus isolates from transplant and nontransplant patients. Antimicrob. Agents Chemother. 48, 4922–4925 10.1128/AAC.48.12.4922-4925.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kontoyiannis D. P., Lewis R. E., Osherov N., Albert N. D., and May G. S. (2003) Combination of caspofungin with inhibitors of the calcineurin pathway attenuates growth in vitro in Aspergillus species. J. Antimicrob. Chemother. 51, 313–316 10.1093/jac/dkg090 [DOI] [PubMed] [Google Scholar]
- 82. Del Poeta M., Cruz M. C., Cardenas M. E., Perfect J. R., and Heitman J. (2000) Synergistic antifungal activities of bafilomycin A1, fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob. Agents Chemother. 44, 739–746 10.1128/aac.44.3.739-746.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pianalto K. M., Billmyre R. B., Telzrow C. L., and Alspaugh J. A. (2019) Roles for stress response and cell wall biosynthesis pathways in caspofungin tolerance in Cryptococcus neoformans. Genetics 213, 213–227 10.1534/genetics.119.302290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Onyewu C., Blankenship J. R., Del Poeta M., and Heitman J. (2003) Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob. Agents Chemother. 47, 956–964 10.1128/aac.47.3.956-964.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Marchetti O., Moreillon P., Glauser M. P., Bille J., and Sanglard D. (2000) Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob. Agents Chemother. 44, 2373–2381 10.1128/AAC.44.9.2373-2381.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cruz M. C., Goldstein A. L., Blankenship J. R., Del Poeta M., Davis D., Cardenas M. E., Perfect J. R., McCusker J. H., and Heitman J. (2002) Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21, 546–559 10.1093/emboj/21.4.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Uppuluri P., Nett J., Heitman J., and Andes D. (2008) Synergistic effect of calcineurin inhibitors and fluconazole against Candida albicans biofilms. Antimicrob Agents Chemother. 52, 1127–1132 10.1128/AAC.01397-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Imai J., and Yahara I. (2000) Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol. Cell Biol. 20, 9262–9270 10.1128/mcb.20.24.9262-9270.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lamoth F., Juvvadi P. R., Gehrke C., and Steinbach W. J. (2013) In vitro activity of calcineurin and heat shock protein 90 Inhibitors against Aspergillus fumigatus azole- and echinocandin-resistant strains. Antimicrob. Agents Chemother. 57, 1035–1039 10.1128/AAC.01857-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hamawy M. M. (2003) Molecular actions of calcineurin inhibitors. Drug News Perspect. 16, 277–282 10.1358/dnp.2003.16.5.829315 [DOI] [PubMed] [Google Scholar]
- 91. Etzkorn F. A., Chang Z. Y., Stolz L. A., and Walsh C. T. (1994) Cyclophilin residues that affect noncompetitive inhibition of the protein serine phosphatase activity of calcineurin by the cyclophilin.cyclosporin A complex. Biochemistry 33, 2380–2388 10.1021/bi00175a005 [DOI] [PubMed] [Google Scholar]
- 92. Kissinger C. R., Parge H. E., Knighton D. R., Lewis C. T., Pelletier L. A., Tempczyk A., Kalish V. J., Tucker K. D., Showalter R. E., and Moomaw E. W. and (1995) Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature 378, 641–644 10.1038/378641a0 [DOI] [PubMed] [Google Scholar]
- 93. Kang C. B., Hong Y., Dhe-Paganon S., and Yoon H. S. (2008) FKBP family proteins: immunophilins with versatile biological functions. Neurosignals 16, 318–325 10.1159/000123041 [DOI] [PubMed] [Google Scholar]
- 94. Dolinski K., Muir S., Cardenas M., and Heitman J. (1997) All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 94, 13093–13098 10.1073/pnas.94.24.13093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cruz M. C., Cavallo L. M., Görlach J. M., Cox G., Perfect J. R., Cardenas M. E., and Heitman J. (1999) Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol. Cell Biol. 19, 4101–4112 10.1128/mcb.19.6.4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Falloon K., Juvvadi P. R., Richards A. D., Vargas-Muñiz J. M., Renshaw H., and Steinbach W. J. (2015) Characterization of the FKBP12-encoding genes in Aspergillus fumigatus. PLoS ONE 10, e0137869 10.1371/journal.pone.0137869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ban Y. H., Shinde P. B., Hwang J. Y., Song M. C., Kim D. H., Lim S. K., Sohng J. K., and Yoon Y. J. (2013) Characterization of FK506 biosynthetic intermediates involved in post-PKS elaboration. J. Nat. Prod. 76, 1091–1098 10.1021/np4001224 [DOI] [PubMed] [Google Scholar]
- 98. Shinde P., Ban Y., Hwang J., Cho Y., Chen Y.-A., Cheong E., Nam S.-J., Kwon H. J., and Yoon Y. (2014) A non-immunosuppressive FK506 analogue with neuroregenerative activity produced from a genetically engineered Streptomyces strain. RSC Adv. 10.1039/C4RA11907J [DOI] [Google Scholar]
- 99. Lee Y., Lee K. T., Lee S. J., Beom J. Y., Hwangbo A., Jung J. A., Song M. C., Yoo Y. J., Kang S. H., Averette A. F., Heitman J., Yoon Y. J., Cheong E., and Bahn Y. S. (2018) In vitro and in vivo assessment of FK506 analogs as novel antifungal drug candidates. Antimicrob. Agents Chemother. 62, e01627 10.1128/AAC.01627-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Beom J. Y., Jung J. A., Lee K. T., Hwangbo A., Song M. C., Lee Y., Lee S. J., Oh J. H., Ha S. J., Nam S. J., Cheong E., Bahn Y. S., and Yoon Y. J. (2019) Biosynthesis of nonimmunosuppressive FK506 analogues with antifungal activity. J. Nat. Prod. 82, 2078–2086 10.1021/acs.jnatprod.9b00144 [DOI] [PubMed] [Google Scholar]
- 101. Nambu M., Covel J. A., Kapoor M., Li X., Moloney M. K., Numa M. M., Soltow Q. A., Trzoss M., Webb P., Webb R. R., and Mutz M. (2017) A calcineurin antifungal strategy with analogs of FK506. Bioorganic Med. Chem. Lett. 27, 2465–2471 10.1016/j.bmcl.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 102. Tonthat N. K., Juvvadi P. R., Zhang H., Lee S. C., Venters R., Spicer L., Steinbach W. J., Heitman J., and Schumacher M. A. (2016) Structures of pathogenic fungal FKBP12s reveal possible self-catalysis function. MBio 7, e00492–16 10.1128/mBio.00492-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rollins C. T., Rivera V. M., Woolfson D. N., Keenan T., Hatada M., Adams S. E., Andrade L. J., Yaeger D., van Schravendijk M. R., Holt D. A., Gilman M., and Clackson T. (2000) A ligand-reversible dimerization system for controlling protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 97, 7096–7101 10.1073/pnas.100101997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Barrero J. J., Papanikou E., Casler J. C., Day K. J., and Glick B. S. (2016) An improved reversibly dimerizing mutant of the FK506-binding protein FKBP. Cell Logist. 6, e1204848 10.1080/21592799.2016.1204848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Juvvadi P. R., Bobay B. G., Gobeil S. M. C., Cole D. C., Venters R. A., Heitman J., Spicer L. D., and Steinbach W. J. (2020) FKBP12 dimerization mutations effect FK506 binding and differentially alter calcineurin inhibition in the human pathogen Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 526, 48–54 10.1016/j.bbrc.2020.03.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Reedy J. L., Filler S. G., and Heitman J. (2010) Elucidating the Candida albicans calcineurin signaling cascade controlling stress response and virulence. Fungal Genet. Biol. 47, 107–116 10.1016/j.fgb.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Fox D. S., and Heitman J. (2005) Calcineurin-binding protein Cbp1 directs the specificity of calcineurin-dependent hyphal elongation during mating in Cryptococcus neoformans. Eukaryot. Cell. 4, 1526–1538 10.1128/ec.4.9.1526-1538.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Görlach J., Fox D. S., Cutler N. S., Cox G. M., Perfect J. R., and Heitman J. (2000) Identification and characterization of a highly conserved calcineurin binding protein, CBP1/calcipressin, in Cryptococcus neoformans. EMBO J. 19, 3618–3629 10.1093/emboj/19.14.3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pinchai N., Perfect B. Z., Juvvadi P. R., Fortwendel J. R., Cramer R. A., Asfaw Y. G., Heitman J., Perfect J. R., and Steinbach W. J. (2009) Aspergillus fumigatus calcipressin CbpA is involved in hyphal growth and calcium homeostasis. Eukaryot. Cell 8, 511–519 10.1128/EC.00336-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Juvvadi P. R., Gehrke C., Fortwendel J. R., Lamoth F., Soderblom E. J., Cook E. C., Hast M. A., Asfaw Y. G., Moseley M. A., Creamer T. P., and Steinbach W. J. (2013) Phosphorylation of calcineurin at a novel serine-proline rich region orchestrates hyphal growth and virulence in Aspergillus fumigatus. PLoS Pathog. 9, e1003564 10.1371/journal.ppat.1003564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Stathopoulos A. M., and Cyert M. S. (1997) Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11, 3432–3444 10.1101/gad.11.24.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Miyazaki T., Yamauchi S., Inamine T., Nagayoshi Y., Saijo T., Izumikawa K., Seki M., Kakeya H., Yamamoto Y., Yanagihara K., Miyazaki Y., and Kohno S. (2010) Roles of calcineurin and Crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob. Agents Chemother. 54, 1639–1643 10.1128/AAC.01364-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Onyewu C., Wormley F. L. Jr., Perfect J. R., and Heitman J. (2004) The calcineurin target, Crz1, functions in azole tolerance but is not required for virulence of Candida albicans. Infect. Immun. 72, 7330–7333 10.1128/IAI.72.12.7330-7333.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Goldman A., Roy J., Bodenmiller B., Wanka S., Landry C. R., Aebersold R., and Cyert M. S. (2014) The calcineurin signaling network evolves via conserved kinase-phosphatase modules that transcend substrate identity. Mol. Cell. 55, 422–435 10.1016/j.molcel.2014.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chow E. W., Clancey S. A., Billmyre R. B., Averette A. F., Granek J. A., Mieczkowski P., Cardenas M. E., and Heitman J. (2017) Elucidation of the calcineurin-Crz1 stress response transcriptional network in the human fungal pathogen Cryptococcus neoformans. PLoS Genet. 13, e1006667 10.1371/journal.pgen.1006667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wennerberg K., Rossman K. L., and Der C. J. (2005) The Ras superfamily at a glance. J. Cell Sci. 118, 843–846 10.1242/jcs.01660 [DOI] [PubMed] [Google Scholar]
- 117. Weeks G., and Spiegelman G. B. (2003) Roles played by Ras subfamily proteins in the cell and developmental biology of microorganisms. Cell. Signal. 15, 901–909 10.1016/S0898-6568(03)00073-1 [DOI] [PubMed] [Google Scholar]
- 118. O'Bryan J. P. (2019) Pharmacological targeting of RAS: Recent success with direct inhibitors. Pharmacol. Res. 139, 503–511 10.1016/j.phrs.2018.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Pentland D. R., Piper-Brown E., Mühlschlegel F. A., and Gourlay C. W. (2017) Ras signalling in pathogenic yeasts. Microb. Cell 5, 63–73 10.15698/mic2018.02.612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Leberer E., Harcus D., Dignard D., Johnson L., Ushinsky S., Thomas D. Y., and Schröppel K. (2001) Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42, 673–687 10.1046/j.1365-2958.2001.02672.x [DOI] [PubMed] [Google Scholar]
- 121. Alspaugh J. A., Cavallo L. M., Perfect J. R., and Heitman J. (2000) RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36, 352–365 10.1046/j.1365-2958.2000.01852.x [DOI] [PubMed] [Google Scholar]
- 122. Fortwendel J. R., Zhao W., Bhabhra R., Park S., Perlin D. S., Askew D. S., and Rhodes J. C. (2005) A fungus-specific Ras homolog contributes to the hyphal growth and virulence of Aspergillus fumigatus. Eukaryot. Cell. 4, 1982–1989 10.1128/EC.4.12.1982-1989.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Abdallah Q. A., and Fortwendel J. R. (2015) Exploration of Aspergillus fumigatus Ras pathways for novel antifungal drug targets. Front. Microbiol. 6, 128 10.3389/fmicb.2015.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Al Abdallah Q., Norton T. S., Hill A. M., LeClaire L. L., and Fortwendel J. R. (2016) A fungus-specific protein domain is essential for RasA-mediated morphogenetic signaling in Aspergillus fumigatus. mSphere 1, e00234–16 10.1128/mSphere.00234-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Nichols C. B., Ferreyra J., Ballou E. R., and Alspaugh J. A. (2009) Subcellular localization directs signaling specificity of the Cryptococcus neoformans Ras1 protein. Eukaryot. Cell 8, 181–189 10.1128/EC.00351-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Fortwendel J. R., Juvvadi P. R., Rogg L. E., Asfaw Y. G., Burns K. A., Randell S. H., and Steinbach W. J. (2012) Plasma membrane localization is required for RasA-mediated polarized morphogenesis and virulence of Aspergillus fumigatus. Eukaryot. Cell 11, 966–977 10.1128/EC.00091-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Norton T. S., and Fortwendel J. R. (2014) Control of Ras-mediated signaling in Aspergillus fumigatus. Mycopathologia 178, 325–330 10.1007/s11046-014-9765-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Piispanen A. E., Bonnefoi O., Carden S., Deveau A., Bassilana M., and Hogan D. A. (2011) Roles of ras1 membrane localization during Candida albicans hyphal growth and farnesol response. Eukaryot. Cell 10, 1473–1484 10.1128/EC.05153-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hast M. A., Nichols C. B., Armstrong S. M., Kelly S. M., Hellinga H. W., Alspaugh J. A., and Beese L. S. (2011) Structures of Cryptococcus neoformans protein farnesyltransferase reveal strategies for developing inhibitors that target fungal pathogens. J. Biol. Chem. 286, 35149–35162 10.1074/jbc.M111.250506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mabanglo M. F., Hast M. A., Lubock N. B., Hellinga H. W., and Beese L. S. (2014) Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design. Protein Sci. 23, 289–301 10.1002/pro.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Thammahong A., Puttikamonkul S., Perfect J. R., Brennan R. G., and Cramer R. A. (2017) Central role of the trehalose biosynthesis pathway in the pathogenesis of human fungal infections: opportunities and challenges for therapeutic development. Microbiol. Mol. Biol. Rev. 81, e00053–16 10.1128/MMBR.00053-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Thevelein J. M., and Hohmann S. (1995) Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem. Sci. 20, 3–10 10.1016/S0968-0004(00)88938-0 [DOI] [PubMed] [Google Scholar]
- 133. Elbein A. D., Pan Y. T., Pastuszak I., and Carroll D. (2003) New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17R–27R 10.1093/glycob/cwg047 [DOI] [PubMed] [Google Scholar]
- 134. Zaragoza O., Blazquez M. A., and Gancedo C. (1998) Disruption of the Candida albicans TPS1 gene encoding trehalose-6-phosphate synthase impairs formation of hyphae and decreases infectivity. J. Bacteriol. 180, 3809–3815 10.1128/JB.180.15.3809-3815.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Alvarez-Peral F. J., Zaragoza O., Pedreno Y., and Argüelles J.-C. (2002) Protective role of trehalose during severe oxidative stress caused by hydrogen peroxide and the adaptive oxidative stress response in Candida albicans. Microbiology 148, 2599–2606 10.1099/00221287-148-8-2599 [DOI] [PubMed] [Google Scholar]
- 136. Martínez-Esparza M., Aguinaga A., González-Párraga P., García-Peñarrubia P., Jouault T., and Argüelles J. C. (2007) Role of trehalose in resistance to macrophage killing: study with a tps1/tps1 trehalose-deficient mutant of Candida albicans. Clin. Microbiol. Infect. 13, 384–394 10.1111/j.1469-0691.2007.01663.x [DOI] [PubMed] [Google Scholar]
- 137. Van Dijck P., De Rop L., Szlufcik K., Van Ael E., and Thevelein J. M. (2002) Disruption of the Candida albicans TPS2 gene encoding trehalose-6-phosphate phosphatase decreases infectivity without affecting hypha formation. Infect. Immun. 70, 1772–1782 10.1128/iai.70.4.1772-1782.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Petzold E. W., Himmelreich U., Mylonakis E., Rude T., Toffaletti D., Cox G. M., Miller J. L., and Perfect J. R. (2006) Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect. Immun. 74, 5877–5887 10.1128/IAI.00624-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Al-Bader N., Vanier G., Liu H., Gravelat F. N., Urb M., Hoareau C. M. Q., Campoli P., Chabot J., Filler S. G., and Sheppard D. C. (2010) Role of trehalose biosynthesis in Aspergillus fumigatus development, stress response, and virulence. Infect. Immun. 78, 3007–3018 10.1128/IAI.00813-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Puttikamonkul S., Willger S. D., Grahl N., Perfect J. R., Movahed N., Bothner B., Park S., Paderu P., Perlin D. S., and Cramer R. A. Jr. (2010) Trehalose 6-phosphate phosphatase is required for cell wall integrity and fungal virulence but not trehalose biosynthesis in the human fungal pathogen Aspergillus fumigatus. Mol. Microbiol. 77, 891–911 10.1111/j.1365-2958.2010.07254.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Miao Y., Tenor J. L., Toffaletti D. L., Maskarinec S. A., Liu J., Lee R. E., Perfect J. R., and Brennan R. G. (2017) Structural and in vivo studies on trehalose-6-phosphate synthase from pathogenic fungi provide insights into its catalytic mechanism, biological necessity, and potential for novel antifungal drug design. MBio 8, e00643–17 10.1128/mBio.00643-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Miao Y., Tenor J. L., Toffaletti D. L., Washington E. J., Liu J., Shadrick W. R., Schumacher M. A., Lee R. E., Perfect J. R., and Brennan R. G. (2016) Structures of trehalose-6-phosphate phosphatase from pathogenic fungi reveal the mechanisms of substrate recognition and catalysis. Proc. Natl. Acad. Sci. U. S. A. 113, 7148–7153 10.1073/pnas.1601774113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Blankenship J. R., Fanning S., Hamaker J. J., and Mitchell A. P. (2010) An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6, e1000752 10.1371/journal.ppat.1000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Jung S. I., Rodriguez N., Irrizary J., Liboro K., Bogarin T., Macias M., Eivers E., Porter E., Filler S. G., and Park H. (2017) Yeast casein kinase 2 governs morphology, biofilm formation, cell wall integrity, and host cell damage of Candida albicans. PLoS ONE 12, e0187721 10.1371/journal.pone.0187721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Franceschini S., Fedkenheuer M., Vogelaar N. J., Robinson H. H., Sobrado P., and Mattevi A. (2012) Structural insight into the mechanism of oxygen activation and substrate selectivity of flavin-dependent N-hydroxylating monooxygenases. Biochemistry 51, 7043–7045 10.1021/bi301072w [DOI] [PubMed] [Google Scholar]
- 146. Hillmann F., Bagramyan K., Straßburger M., Heinekamp T., Hong T. B., Bzymek K. P., Williams J. C., Brakhage A. A., and Kalkum M. (2016) The crystal structure of peroxiredoxin Asp f3 provides mechanistic insight into oxidative stress resistance and virulence of Aspergillus fumigatus. Sci. Rep. 6, 33396 10.1038/srep33396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Gleason J. E., Galaleldeen A., Peterson R. L., Taylor A. B., Holloway S. P., Waninger-Saroni J., Cormack B. P., Cabelli D. E., Hart P. J., and Culotta V. C. (2014) Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc. Natl. Acad. Sci. U. S. A. 111, 5866–5871 10.1073/pnas.1400137111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Sussman A., Huss K., Chio L.-C., Heidler S., Shaw M., Ma D., Zhu G., Campbell R. M., Park T.-S., Kulanthaivel P., Scott J. E., Carpenter J. W., Strege M. A., Belvo M. D., Swartling J. R., et al. (2004) Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Eukaryot. Cell 3, 932–943 10.1128/EC.3.4.932-943.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Qi J., Kizjakina K., Robinson R., Tolani K., and Sobrado P. (2012) A fluorescence polarization binding assay to identify inhibitors of flavin-dependent monooxygenases. Anal. Biochem. 425, 80–87 10.1016/j.ab.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Perfect J. R., Tenor J. L., Miao Y., and Brennan R. G. (2017) Trehalose pathway as an antifungal target. Virulence 8, 143–149 10.1080/21505594.2016.1195529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Pearson J. T., Hill J. J., Swank J., Isoherranen N., Kunze K. L., and Atkins W. M. (2006) Surface plasmon resonance analysis of antifungal azoles binding to CYP3A4 with kinetic resolution of multiple binding orientations. Biochemistry 45, 6341–6353 10.1021/bi0600042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Abadio A. K. R., Kioshima E. S., Teixeira M. M., Martins N. F., Maigret B., and Felipe M. S. S. (2011) Comparative genomics allowed the identification of drug targets against human fungal pathogens. BMC Genomics 12, 75 10.1186/1471-2164-12-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Hoekstra W. J., Garvey E. P., Moore W. R., Rafferty S. W., Yates C. M., and Schotzinger R. J. (2014) Design and optimization of highly-selective fungal CYP51 inhibitors. Bioorg. Med. Chem. Lett. 24, 3455–3458 10.1016/j.bmcl.2014.05.068 [DOI] [PubMed] [Google Scholar]
- 154. Renaud J.-P., Chari A., Ciferri C., Liu W., Rémigy H.-W., Stark H., and Wiesmann C. (2018) Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 17, 471–492 10.1038/nrd.2018.77 [DOI] [PubMed] [Google Scholar]
- 155. Roemer T., Jiang B., Davison J., Ketela T., Veillette K., Breton A., Tandia F., Linteau A., Sillaots S., Marta C., Martel N., Veronneau S., Lemieux S., Kauffman S., Becker J., et al. (2003) Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50, 167–181 10.1046/j.1365-2958.2003.03697.x [DOI] [PubMed] [Google Scholar]
- 156. Segal E. S., Gritsenko V., Levitan A., Yadav B., Dror N., Steenwyk J. L., Silberberg Y., Mielich K., Rokas A., Gow N. A. R., Kunze R., Sharan R., and Berman J. (2018) Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans. MBio 9, e02048–18 10.1128/mBio.02048-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Hu W., Sillaots S., Lemieux S., Davison J., Kauffman S., Breton A., Linteau A., Xin C., Bowman J., Becker J., Jiang B., and Roemer T. (2007) Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 3, e24 10.1371/journal.ppat.0030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Carr P. D., Tuckwell D., Hey P. M., Simon L., D'Enfert C., Birch M., Oliver J. D., and Bromley M. J. (2010) The transposon impala is activated by low temperatures: use of a controlled transposition system to identify genes critical for viability of Aspergillus fumigatus. Eukaryot. Cell 9, 438–448 10.1128/EC.00324-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Liu O. W., Chun C. D., Chow E. D., Chen C., Madhani H. D., and Noble S. M. (2008) Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135, 174–188 10.1016/j.cell.2008.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Noble S. M., French S., Kohn L. A., Chen V., and Johnson A. D. (2010) Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42, 590–598 10.1038/ng.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. O'Meara T. R., Veri A. O., Ketela T., Jiang B., Roemer T., and Cowen L. E. (2015) Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat. Commun. 6, 6741 10.1038/ncomms7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Peng Y., Zhang H., Xu M., and Tan M.-W. (2018) A Tet-Off gene expression system for validation of antifungal drug targets in a murine invasive pulmonary aspergillosis model. Sci. Rep. 8, 443 10.1038/s41598-017-18868-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lee K.-T., Hong J., Lee D.-G., Lee M., Cha S., Lim Y.-G., Jung K.-W., Hwangbo A., Lee Y., Yu S.-J., Chen Y.-L., Lee J.-S., Cheong E., and Bahn Y.-S. (2020) Fungal kinases and transcription factors regulating brain infection in Cryptococcus neoformans. Nat. Commun. 11, 1521 10.1038/s41467-020-15329-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Day A. M., and Quinn J. (2019) Stress-activated protein kinases in human fungal pathogens. Front. Cell. Infect. Microbiol. 9, 261 10.3389/fcimb.2019.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]