Abstract
Surface-exposed Toll-like receptors (TLRs) such as TLR2 and TLR4 survey the extracellular environment for pathogens. TLR activation initiates the production of various cytokines and chemokines, including type I interferons (IFN-I). Downstream of TLR4, IFNβ secretion is only vigorously triggered in macrophages when the receptor undergoes endocytosis and switches signaling adaptor; surface TLR4 engagement predominantly induces proinflammatory cytokines via the signaling adaptor MyD88. It is unclear whether this dichotomy is generally applicable to other TLRs, cell types, or differentiation states. Here, we report that diverse TLR2 ligands induce an IFN-I response in human monocyte-like cells, but not in differentiated macrophages. This TLR2-dependent IFN-I signaling originates from the cell surface and depends on MyD88; it involves combined activation of the transcription factors IRF3 and NF-κB, driven by the kinases TBK1 and TAK1-IKKβ, respectively. TLR2-stimulated monocytes produced modest IFNβ levels that caused productive downstream signaling, reflected by STAT1 phosphorylation and expression of numerous interferon-stimulated genes. Our findings reveal that the outcome of TLR2 signaling includes an IFN-I response in human monocytes, which is lost upon macrophage differentiation, and differs mechanistically from IFN-I-induction through TLR4. These findings point to molecular mechanisms tailored to the differentiation state of a cell and the nature of receptors activated to control and limit TLR-triggered IFN-I responses.
Keywords: innate immunity, Toll-like receptor (TLR), interferon, human, monocyte, cell surface, cell differentiation
A crucial step for activation of innate immunity is detection of invading pathogens. Cells sense microbes through a set of pattern recognition receptors (PRRs) that recognize conserved microbial structures, known as pathogen-associated molecular patterns (PAMPs) (1). TLRs constitute a family of transmembrane PRRs well-known for inducing innate defense against a plethora of microbes (2, 3). Upon ligand binding, TLRs initiate signaling events that result in the production of a large variety of cytokines, chemokines, and co-stimulatory molecules, which activate and polarize innate and adaptive immune cells (4, 5). A properly tailored innate immune response results in the clearance of pathogens, while minimizing inflammation-associated damage to the host. Whereas the molecular aspects of TLR sensing and signaling have been resolved to great detail in specific cell types, knowledge lags behind on how immune responses of appropriate quality (specificity, amplitude, and duration) are induced at the levels of cells, tissues, and organisms.
Among the TLR-induced cytokines, IFN-I, such as IFNα and IFNβ, mediate particularly potent antimicrobial effects. Secreted IFN-I can bind the IFN-α/β receptor (IFNAR) in an autocrine or paracrine manner to induce the expression of hundreds of ISGs (6). Many ISGs directly inhibit replication of intracellular microbes (7–9). In vivo studies illustrate the power of IFN-I in antiviral immunity, showing that administration of exogenous IFN-I protects mice from a lethal challenge of mouse-adapted human influenza virus (10) and that IFNAR1-deficient mice are extremely susceptible to numerous viruses, such as vesicular stomatitis virus, vaccinia virus, Semliki forest virus, and lymphocytic choriomeningitis virus (11). Similarly, IFN-I play an important role in the innate defense against various bacteria. For instance, IFNAR-deficient mice succumb when infected with group B streptococci, Escherichia coli, or Streptococcus pyogenes (12, 13), whereas WT mice survive these infections.
A potent effector function of IFN-I is to suppress cell proliferation and sensitize cells to apoptosis (14, 15). While providing an effective way to clear infected cells, this can also have detrimental effects. For instance, IFN-I–related apoptosis of macrophages and T cells increases host susceptibility to Listeria monocytogenes (9). Adverse effects of prolonged production of high levels of IFN-I also manifest in patients suffering from interferonopathies, a group of autoinflammatory disorders where uncontrolled IFN-I production causes severe tissue damage (16). Thus, IFN-I responses following TLR activation should be tightly regulated to induce optimal antimicrobial effects yet limit immunopathology.
IFN-I production following TLR activation differs vastly among cell types. For instance, subsets of dendritic cells (DCs) display extensive variation in TLR7- and TLR9-induced IFN-I levels. Intracellular TLR7 and TLR9 molecules survey endosomes for the presence of viral single-stranded RNA (17) and unmethylated DNA containing CpG motifs (18), respectively. In plasmacytoid DCs (pDCs), ligation of TLR7 or TLR9 results in the secretion of large quantities of IFN-I, whereas signaling via these receptors in conventional DCs (cDCs) induces much less IFN-I and, instead, mainly triggers the production of proinflammatory cytokines (19, 20). Although not completely resolved, the two DC subsets appear to trigger IFN-I production through distinct TLR7/9-induced molecular mechanisms (21). To understand the variation in the outcome of TLR ligation, it is, therefore, important to dissect the signaling cascades that culminate in the production of IFN-I for individual cell types.
Some surface-expressed TLRs yield proinflammatory as well as IFN-I responses within one cell type. The molecular basis for IFN-I synthesis has been most thoroughly dissected for TLR4 signaling in mouse macrophages, with many aspects being confirmed in human macrophages (22–24). Here, signaling events underpinning the synthesis of either IFN-I or the proinflammatory cytokines appear to be spatially separated (25). Engagement of TLR4 with bacterial lipopolysaccharides (LPS) at the cell surface induces receptor dimerization, leading to the recruitment of sorting and signaling adaptors TIR domain–containing adaptor protein (TIRAP, also termed Mal) and myeloid differentiation primary response 88 (MyD88) (26). Downstream of MyD88, signaling involves transforming growth factor-β–activated kinase-1 (TAK1), resulting in the nuclear translocation of the transcription factors nuclear factor-κB (NF-κB) and activator protein-1 (AP-1) (27). Jointly, NF-κB and AP-1 drive the expression of proinflammatory cytokines, such as interleukin-8 (IL8) and tumor necrosis factor-α (TNFα), but not of IFN-I. The switch to IFN-I production occurs following endocytosis of LPS-engaged TLR4 dimers. From endosomes, TLR4 engages a different set of sorting and signaling adaptor proteins, namely TRIF-related adaptor molecule (TRAM) and TIR domain–containing adaptor–inducing IFNβ (TRIF) (28, 29). These interactions promote signaling via TANK-binding kinase 1 (TBK1) and IκB kinase ε (IKKε), driving the phosphorylation of IFN regulatory factor 3 (IRF3) (30). The transcription factor IRF3 then dimerizes and translocates to the nucleus to induce the expression of vast amounts of IFNβ (31). Thus, macrophages appear to have evolved complex mechanisms to regulate the TLR4-dependent induction of IFN-I, with cellular trafficking events tailoring signaling outcome.
TLR2 is the second surface-exposed TLR that, although long thought to only induce proinflammatory cytokines, also leads to IFN-I production in mouse monocytes and macrophages (32, 33). TLR2 heterodimerizes with TLR1 or TLR6 to allow detection of a broad spectrum of PAMPs, including glycolipids, lipoproteins (34), and various virus structural proteins (35, 36). Mechanistic details for TLR2 have not been resolved to the same detail as for TLR4. IFN-I induction downstream of murine TLR2 differs from TLR4 in that all TLR2-signaling events require the adaptor MyD88 (32, 33), yet, analogous to TLR4, endocytosis of murine TLR2 molecules appears to drive activation of IRF transcription factors (37). It is even less clear how IFN-I is induced downstream of human TLR2 and in which cells.
In this study, we found that human TLR2 signaling outcome differs between monocytes and macrophages—by comparing these two differentiation states of THP1 myeloid cells—and elucidated how human TLR2 signals leading to IFN-I production are transduced. This work deepens our understanding of the strategies that cells employ to regulate their IFN-I output and what roles receptor usage and cellular differentiation play.
Results
Human monocytes, but not macrophages, support TLR2-dependent IFN-I signaling
To investigate TLR2 signaling in human myeloid cells, we employed the human acute monocytic leukemia-derived cell line THP1, which is highly manipulable and widely used for deciphering mechanistic aspects of (innate) immune signaling (38, 39). THP1 cells have a monocyte-like phenotype and can be differentiated toward macrophage-like cells using the phorbol ester PMA (40). For ease of reading, we here refer to the undifferentiated and differentiated THP1 cells as monocytes and macrophages, respectively. Both THP1 monocytes and macrophages endogenously express TLR2, TLR1, and TLR6 (Fig. S1), permitting the study of TLR2 signaling in either differentiation state.
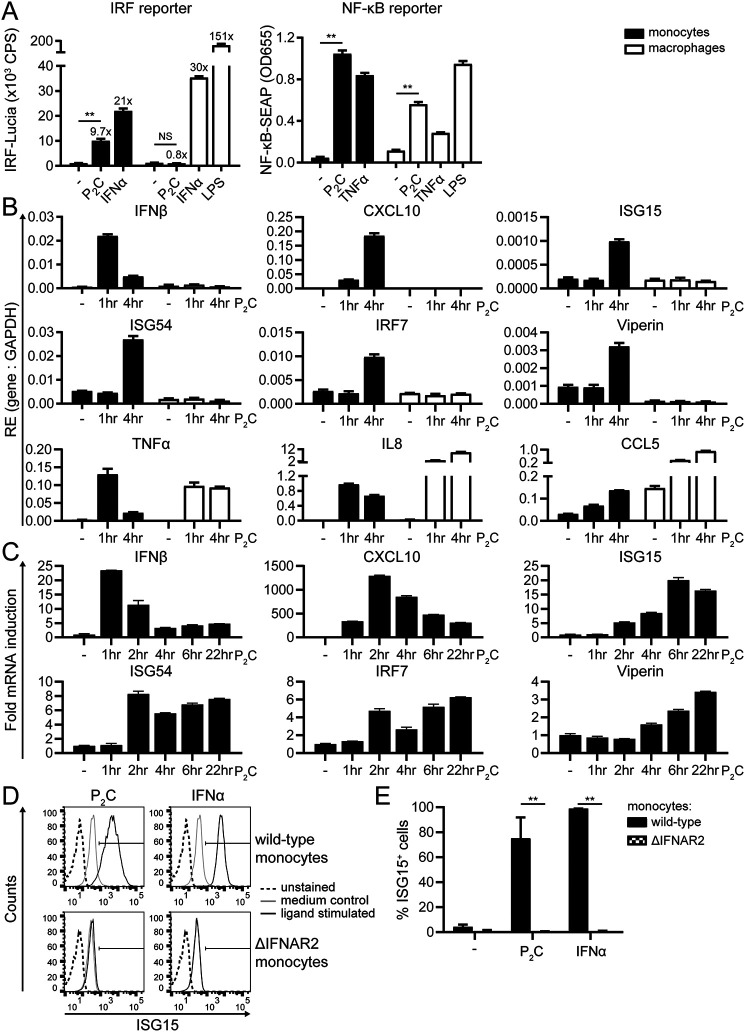
First, we assessed which signaling pathways are activated upon TLR2 ligation in monocytes and macrophages using THP1 Dual reporter cells. These cells secrete inducible reporter proteins reflecting activation of the IRF (Lucia luciferase) and NF-κB (secreted embryonic alkaline phosphatase (SEAP)) pathways. Monocytes and macrophages were incubated for 24 h with the TLR2 ligand Pam2CSK4 (P2C), IFNα as a positive control for inducing IRF activity, or TNFα as a positive control for inducing NF-κB activity. P2C-stimulated monocytes induced the IRF reporter 10-fold over unstimulated cells (Fig. 1A, left), indicating that IRF activity was induced by TLR2. To our surprise, this response was lost in differentiated cells. In contrast, high levels of SEAP were detected in the supernatants of both P2C-stimulated monocytes and macrophages (Fig. 1A, right), pointing to robust TLR2-induced NF-κB activation, irrespective of the differentiation state of the cells. For comparison, LPS stimulation showed that TLR4 engagement resulted in IRF signaling in addition to NF-κB signaling in macrophages (Fig. 1A), as reported by others (29, 41). Thus, human monocytes display TLR2-induced NF-κB and IRF activity, the latter of which is lost in differentiated cells.
Figure 1.
Human TLR2 activation triggers IFN-I and ISG production only in undifferentiated THP1 cells. A–E, undifferentiated (monocytes) or PMA-differentiated (macrophages) THP1 Dual cells were stimulated with the TLR2 ligand P2C (100 nm), TLR4 ligand LPS (1 μg/ml), TNFα (50 ng/ml), or IFNα (1000 units/ml) for 24 h (A, D, and E) or for the indicated times (B and C). A, to simultaneously compare activation of the IRF and NF-κB pathways in monocytes and macrophages, levels of IRF-Lucia and NF-κB-SEAP reporter proteins were determined in culture supernatants. Data are presented as means ± S.D. (error bars) from a representative of at least five independent experiments (one-way ANOVA with Dunnett's multiple-comparison test). -Fold inductions of the IRF reporter are indicated above the bars. B and C, to assess transcription levels of IFNβ and various ISGs (CXCL10, ISG15, ISG54, IRF7, and viperin) as well as NF-κB–regulated genes (TNFα, IL8, and CCL5) downstream of TLR2 activation, expression of the indicated gene products was assessed by qRT-PCR and normalized to GAPDH. B, for comparison of monocytes and macrophages, data are presented as RE levels (means ± S.D. (error bars)) from a representative of three independent experiments. C, to depict the kinetics of the IFN-I response in monocytes, relative expression levels were normalized to the unstimulated control (−, t = 0) and presented as -fold mRNA inductions (means ± S.E. (error bars)). D and E, to visualize the IFN-I response at the protein level, up-regulation of total ISG15 protein levels in permeabilized WT and ΔIFNAR2 THP1 monocytes was measured by flow cytometry. D, results are presented as histograms depicting ISG15 fluorescence intensities from a representative of three independent experiments. E, for easier comparison of the responses, percentages of ISG15-positive cells ± S.D. (error bars) were quantified from the experiments by gating, as shown in D (unpaired Student's t test). NS, not significant; **, p < 0.01.
Second, we analyzed whether TLR2 signaling in monocytes led to the production of both IFN-I and proinflammatory cytokines, as implicated by the reporter assay results above. qRT-PCR analysis revealed rapid and transient IFNβ transcription in P2C-stimulated monocytes, but not macrophages (Fig. 1B), with a peak 1 h after stimulation (Fig. 1C). The transient burst of IFNβ expression in monocytes was followed by sustained expression of various ISGs, including CXCL10, ISG15, ISG54, IRF7, and viperin (Fig. 1, B and C), which is indicative of productive IFN-I signaling. Note that all ISGs were expressed by monocytes 4 h after stimulation (Fig. 1B) but that each reached maximum expression at different times after TLR2 activation (Fig. 1C). In contrast to the monocyte-restricted expression of IFNβ and ISGs, both P2C-stimulated monocytes and macrophages up-regulated the NF-κB–inducible TNFα, IL8, and CCL5 mRNAs (Fig. 1B), confirming the NF-κB-SEAP reporter data (Fig. 1A). Secreted IFNβ protein levels in culture supernatants were below 10 IU/ml, the detection limit of the HEK Blue IFNαβ reporter assay (data not shown). As a proxy of IFN-I signaling at the protein level, we measured up-regulation of ISG15 in permeabilized cells by flow cytometry. ISG15 is an IFN-I–inducible ubiquitin-like protein, which serves as a post-translational modifier of proteins through covalent attachment. The large majority of P2C-stimulated monocytes became strongly ISG15-positive (Fig. 1, D and E), confirming the qRT-PCR results (Fig. 1, B and C). ISG15 can act as an IFN-I–independent direct response gene (42). In these experiments, however, TLR2-mediated ISG15 induction was entirely dependent on IFN-I signaling, as P2C no longer induced ISG15 in monocytes lacking the IFNAR2 receptor (Fig. 1, D and E); nor did two other TLR2 ligands, P3C and FSL-1 (Fig. S2A).
The combined results—from reporter assays, qRT-PCRs, and flow cytometry—thus demonstrate that the TLR2-induced IFNβ levels were sufficient to drive a productive IFN-I response in the monocytes. In conclusion, whereas both monocytes and macrophages induce NF-κB–dependent gene expression following TLR2 stimulation, the induction of IFNβ and subsequent IFN-I signaling is restricted to human monocytes.
Ligands for TLR1/2 and TLR2/6 heterodimers both induce dose-dependent IFN-I signaling in human monocytes
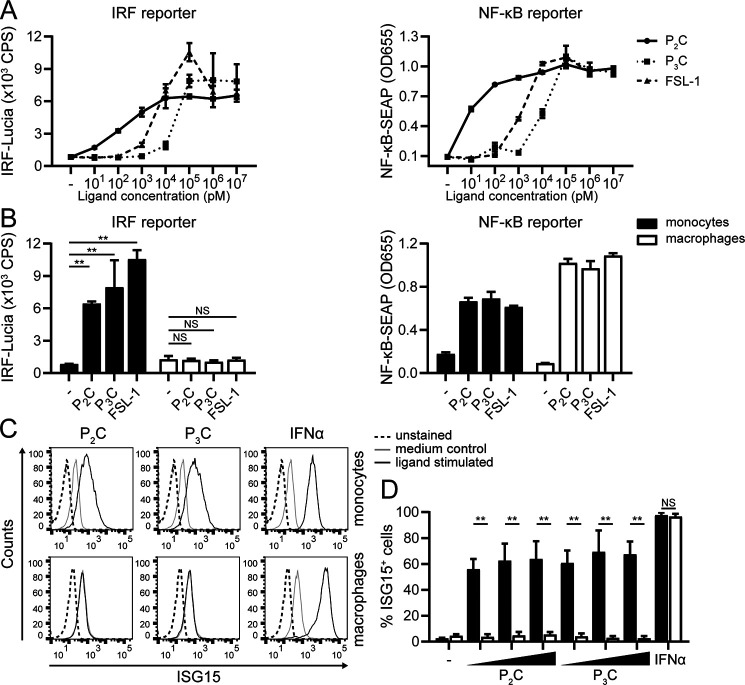
Heterodimerization of TLR2 with either TLR1 or TLR6 yields variable binding affinities for diverse ligands (43). For example, Pam3CSK4 (P3C) preferentially binds to TLR2/1 complexes (44), whereas FSL-1 is considered a TLR2/6 ligand (43). Both P3C and FSL-1 triggered IRF activity in monocytes in a dose-dependent fashion, like P2C did (Fig. 2A). Although we observed some variation among the ligands as well as between replicate experiments, there was a tendency that higher ligand doses might be required for activation of the IRF than of the NF-κB pathway (Fig. 2A and data not shown; roughly 2–5-fold more for the IRF reporter than for the NF-κB reporter). Macrophages did not yield an IRF response to any of these ligands, even at high doses (Fig. 2B). This differentiation state–dependent difference was confirmed at the protein level, as only monocytes up-regulated ISG15 in response to the diverse TLR2 ligands (Fig. 2 (C and D) (P2C, P3C) and Fig. S2B (FSL-1)), whereas both monocytes and macrophages were responsive to control treatment with IFNα (virtually 100% ISG15-positive cells). In summary, upon ligand engagement, both TLR2/1 and TLR2/6 receptor heterodimers appear to evoke an IFN-I response in human monocytes.
Figure 2.
Diverse TLR2 ligands dose-dependently induce IFN-I signaling in human monocytes. THP1 Dual monocytes (A and B) and macrophages (B) were stimulated with the TLR2 ligand P2C, TLR2/1 ligand P3C, or TLR2/6 ligand FSL-1 for 24 h. A, dose ranges of ligands were used to compare IRF and NF-κB reporter activation in monocytes. B, stimulations of monocytes and macrophages were compared at ligand concentrations of 100 nm P2C, 1 μm P3C, 100 nm FSL-1, or 1000 units/ml IFNα. Reporter data are presented as means ± S.D. (error bars) from a representative of five (A) or three (B) independent experiments (one-way ANOVA with Dunnett's multiple-comparison test). C and D, ISG15 levels of monocytes versus macrophages were assessed after stimulation with P2C (1, 10, or 100 nm), P3C (10, 100, or 1000 nm), or IFNα (1000 units/ml) for 24 h. Results are shown as representative fluorescence intensity histograms of cells stimulated at the highest dose (C) and as percentage of ISG15-positive cells ± S.D. (error bars). D, from three independent experiments (unpaired Student's t test). NS, not significant; **, p < 0.01.
MyD88 is essential for TLR2-induced IFN-I signaling in human monocytes, unlike for TLR4 in macrophages
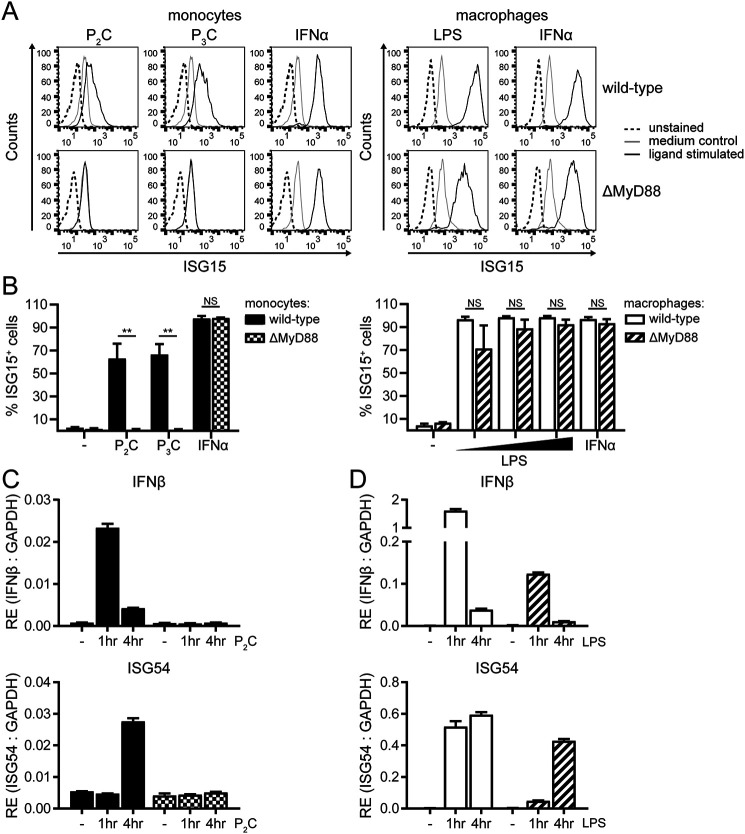
Next, we aimed to determine the requirements for the IFN-I response downstream of human TLR2. TLRs relay distinct signaling outcomes through the use of various signaling adaptors. TLR4 employs MyD88 to confer NF-κB activation, whereas IRF activity is induced via TRIF (28). However, experiments suggest that this dichotomy does not apply for mouse TLR2, as MyD88 expression was essential for the activation of NF-κB, but also for the induction of IFN-I (32, 33). To determine the role MyD88 plays in the TLR2-dependent IFN-I response in human monocytes, we examined THP1 cells devoid of MyD88 expression (ΔMyD88 cells).
In response to P2C, P3C, or FSL-1 stimulation, ΔMyD88 monocytes completely lost the ability to up-regulate intracellular ISG15 protein levels (Fig. 3 (A and B, left panels) and Fig. S2C) and did not produce IFNβ or ISG54 transcripts (Fig. 3C). From this, we conclude that MyD88 is an absolute requirement downstream of human TLR2 to confer the signaling events leading to IFN-I, reminiscent of observations in mouse cells. In contrast, ΔMyD88 macrophages retained induction of ISG15, IFNβ, and ISG54 when exposed to the TLR4 ligand LPS (Fig. 3, A and B (right panels) and D), although the response was less pronounced than in WT cells. These combined data reveal that, whereas MyD88-independent TLR4 signaling in macrophages still results in an IFN-I response, no IFN-I is induced upon TLR2 ligation when the adaptor MyD88 is absent from human monocytes.
Figure 3.
Downstream of human TLR2, MyD88 is indispensable for inducing IFN-I. WT or MyD88-knockout (ΔMyD88) THP1 monocytes or macrophages were stimulated with the TLR2 ligand P2C (100 nm), TLR2/1 ligand P3C (1 μm), TLR4 ligand LPS (0.01, 0.1, or 1 μg/ml), or IFNα (1000 units/ml). A and B, ISG15 levels after 24 h of stimulation were measured. Results are depicted as representative fluorescence intensity histograms of cells stimulated at the highest dose (A) and as percentage of ISG15-positive cells ± S.D. (error bars). B, from three independent experiments (unpaired Student's t test). NS, not significant; **, p < 0.01. C and D, at the indicated time points after stimulation, RE of IFNβ and ISG54 transcripts was determined by qRT-PCR and normalized to GAPDH. For monocytes stimulated with P2C (C), data are presented as means ± S.D. (error bars) from a representative of three independent experiments; for macrophages exposed to LPS (D), means ± S.D. (error bars) from a representative of two independent experiments are shown.
TLR2 induces IFN-I production in human monocytes via the signaling intermediates TBK1 and IRF3
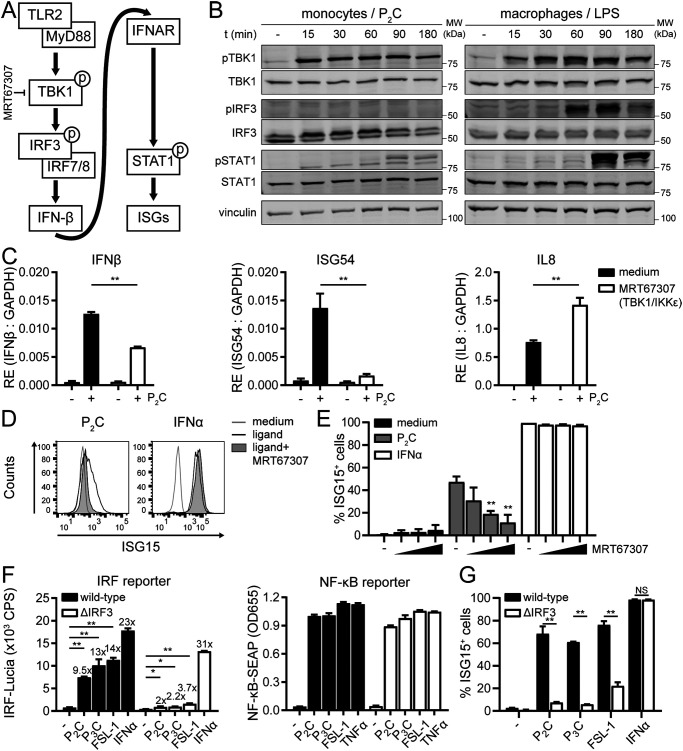
To obtain insight into how monocytes regulate IFNβ expression following TLR2 stimulation, we dissected the underlying MyD88-dependent signaling pathway. Various PRRs initiate expression of IFNβ by activating members of the IRF transcription factor family (45). In case of TLR4 signaling, a set of noncanonical IKKs, TBK1 and IKKε, drive the activation of IRF3 to induce IFNβ transcription (30). MyD88-dependent activation of TBK1 and IKKε occurs in murine bone marrow–derived macrophages stimulated with TLR2 ligands (46). Hence, we explored the role of TBK1 and IRFs in TLR2-mediated IFN-I induction and subsequent IFNAR signaling by human monocytes (Fig. 4A).
Figure 4.
Human TLR2 triggers IFN-I via both TBK1 and IRF3. A, schematic overview of the IFN-I signaling cascade investigated, with the level at which the inhibitor used (in C–E) acts. B, phosphorylation of TBK1, IRF3, and STAT1 was analyzed over time on total cell lysates of 100 nm P2C-treated THP1 monocytes (left) or 1 μg/ml LPS-treated macrophages (right) by Western blotting; vinculin served as a loading control. Of four independent experiments, representative Western blots of the monocyte TLR2 responses are shown; in this experiment, the control macrophage TLR4 response was included for comparison. C–E, to determine the role of TBK1/IKKε kinase activity in TLR2 signaling outcome, monocytes were (pre)treated with the inhibitor MRT67307 (2.5 μm in C and D; doses of 1.2, 2.5, or 5 μm in E). C, at 1 h (IFNβ) and 4 h (ISG54 and IL8) after stimulation with P2C (100 nm), RE of the indicated transcripts was assessed by qRT-PCR and normalized to GAPDH. Data are presented as means ± S.D. (error bars) from a representative of three independent experiments (unpaired Student's t test). D and E, cellular ISG15 levels 24 h after stimulation with P2C (100 nm) or IFNα (1000 units/ml) were measured. Data are depicted as fluorescence intensity histograms (D) and as percentage of ISG15-positive cells ± S.D. (error bars) (E) from three independent experiments (one-way ANOVA with Dunnett's multiple-comparison test). F and G, to determine the role of IRF3 in TLR2 signaling outcome, WT and ΔIRF3 monocytes were stimulated with P2C (100 nm), TLR2/1 ligand P3C (1 μm), TLR2/6 ligand FSL-1 (100 nm), TNFα (50 ng/ml), or IFNα (1000 units/ml) for 24 h. F, levels of IRF-Lucia and NF-κB-SEAP reporter proteins in culture supernatants are presented as means ± S.D. (error bars) from a representative of four independent experiments (one-way ANOVA with Dunnett's multiple-comparison test). -Fold inductions of the IRF reporter are indicated above the bars. G, percentages of ISG15-positive cells ± S.D. (error bars) are shown from three independent experiments (unpaired Student's t test). NS, not significant; *, p < 0.05; **, p < 0.01.
First, Western blotting analysis was employed to visualize phosphorylation of TBK1, IRF3, and signal transducer and activator of transcription-1 (STAT1) in monocytes that were stimulated for various times with the TLR2 ligand P2C. As a positive control, lysates of macrophages stimulated with the TLR4 ligand LPS were taken along. Rapid phosphorylation of TBK1 at Ser-172 was observed in P2C-stimulated monocytes and in LPS-stimulated macrophages (Fig. 4B), pointing to activation of this kinase through (auto)phosphorylation (47). This was followed by robust phosphorylation of IRF3 at Ser-396 in LPS-stimulated macrophages, yet—unexpectedly—not detectably in P2C-stimulated monocytes. Still, in both cases, the transcription factor STAT1 was subsequently activated, as phospho-STAT1 bands appeared at later time points (Fig. 4B), indicative of a secondary IFN-I signaling cascade downstream of the IFNAR (Fig. 4A). STAT1 phosphorylation levels (Fig. 4B) mirrored the observed IFNβ mRNA levels (Fig. 3, C and D), because both were more modest in P2C-stimulated monocytes than in LPS-stimulated macrophages.
The kinase activity of TBK1 was required for inducing IFN-I downstream of TLR2: in response to P2C, monocytes pretreated with the TBK1/IKKε inhibitor MRT67307 (48) induced significantly less IFNβ and ISG54 mRNA, whereas elevated levels of the NF-κB–regulated gene IL8 were detected (Fig. 4C). This was in line with observations by others for a different TLR2 ligand, lipoteichoic acid (LTA) (49). In addition, MRT67307 dose-dependently inhibited TLR2-induced, but not IFNα-induced, ISG15 protein expression (Fig. 4, D and E), confirming the involvement of TBK1/IKKε kinase activity in productive IFN-I signaling downstream of human TLR2.
In view of the contributions of upstream TBK1 (Fig. 4, B–E) and downstream STAT1 (Fig. 4B), we were surprised that IRF3 phosphorylation was not detected in our experimental set-up (Fig. 4B). Examples have been reported where the role of IRF3 only became evident from depletion experiments, potentially related to (in)sensitivity of the IRF3 activation assays used. For instance, siRNA experiments in human fibroblasts showed that IRF3 was required for the induction of ISGs in response to UV-inactivated Newcastle disease virus (NDV) and human cytomegalovirus, despite a lack of detectable levels of IRF3 dimerization or Ser-396 phosphorylation (50).
To conclusively determine whether IRF3 is involved in TLR2-induced IFN-I signaling in human monocytes, we made use of IRF3 knockout THP1 Dual cells (ΔIRF3). Incubation with the TLR2 ligand P2C, P3C (TLR2/1), or FSL-1 (TLR2/6) yielded only marginal IRF reporter activity in ΔIRF3 monocytes, as opposed to the 9–14-fold increase in their WT counterparts (Fig. 4F, left). As a control, ΔIRF3 cells robustly induced the IRF-Lucia reporter following exposure to IFNα, indicating that IFNAR signaling was not disrupted in these cells. The TLR2 ligands triggered strong NF-κB activity both in WT and ΔIRF3 cells, implying that IRF3 has no major effect on NF-κB activation (Fig. 4F, right). In line with the IRF reporter data, ΔIRF3 cells were also hampered in ISG15 protein induction in response to diverse TLR2 ligands (P2C, P3C, and FSL-1), whereas the response to IFNα treatment remained intact (Fig. 4G). This substantiates that the major IRF driving TLR1/2- and TLR2/6-induced IFN-I signaling in human monocytes is IRF3.
Because a weak TLR2-induced IRF activity remained detectable upon genetic depletion of IRF3 (2–4-fold; Fig. 4F, left), other IRF family members may play an additional (minor) role. In addition to IRF3, monocytes express the myeloid-specific IRF8 and the IFN-I–inducible IRF7 transcription factors, of which the latter was only detectable in Western blots of IFN-I–treated cells (data not shown). To assess the contribution of these IRFs to TLR2-dependent IFN-I signaling, expression of IRF7 and IRF8 was silenced in THP1 monocytes (Fig. S3, A and B). Without affecting IRF3 protein levels (Fig. S3B), knockdown of IRF7 or IRF8 reduced—but did not completely abolish—TLR2-induced ISG15 expression (Fig. S3C). Therefore, both IRF7 and IRF8 may contribute to the TLR2-induced IFN-I response in human monocytes.
Combined, our findings support a model for human monocytes wherein TLR2 depends on TBK1 to modestly activate IRF3, which, together with other IRF family members, drives the expression of IFNβ (Fig. 4A).
Downstream of TAK1, the NF-κB, but not AP-1, signaling axis contributes to TLR2-dependent IFN-I induction in human monocytes
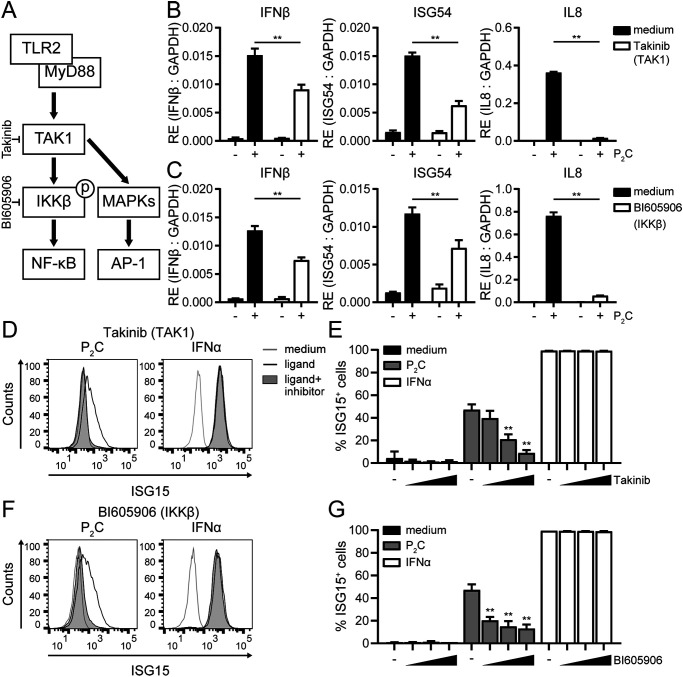
Besides IRF family members, the transcription factors NF-κB and AP-1 (ATF2/c-Jun) are activated upon TLR2 stimulation (Fig. 1A, right) (51). Because both NF-κB and AP-1 can also bind and regulate the IFNβ promoter (52), one could envisage that signaling modules driving these transcription factors contribute to TLR2-dependent induction of IFNβ in human monocytes in conjunction with the TBK1/IRF3 axis, particularly when IRF3 is modestly activated. To test this, we performed experiments using inhibitors targeting kinases that act in the NF-κB or AP-1 signaling pathways downstream of TLR2 (Fig. 5A).
Figure 5.
The drivers of NF-κB activation TAK1 and IKKβ contribute to IFN-I signaling downstream of human TLR2. A, schematic overview of TLR2-induced signaling events under study that trigger NF-κB and AP-1 activation, highlighting the branching of pathways downstream of TAK1 and the level at which the inhibitors used (in B–G) act. To determine the role of TAK1 kinase activity in TLR2 signaling outcome, THP1 monocytes were (pre)treated with the inhibitor Takinib (10 μm in B and D; doses of 1, 5, or 10 μm in E); to determine the role of the downstream kinase IKKβ, THP1 monocytes were (pre)treated with the inhibitor BI605906 (12.5 μm in C and F; doses of 3.1, 6.2, or 12.5 μm in G). B and C, at 1 h (IFNβ) and 4 h (ISG54 and IL8) after stimulation with P2C (100 nm), RE of the indicated transcripts was assessed by qRT-PCR and normalized to GAPDH. Data are presented as means ± S.D. (error bars) from a representative of three independent experiments (unpaired Student's t test). D–G, cellular ISG15 levels 24 h after stimulation with P2C (100 nm) or IFNα (1000 units/ml) were measured. Data are depicted as fluorescence intensity histograms (D and F) and as percentage of ISG15-positive cells ± S.D. (error bars). E and G, of three independent experiments (one-way ANOVA with Dunnett's multiple-comparison test). **, p < 0.01.
TAK1 forms a central signaling hub in the proinflammatory signaling cascade, leading to the activation of both NF-κB and AP-1 (27). A role for this kinase in regulating secretion of IFN-I besides TNFα was implicated in LTA– or S. aureus–treated cells based on the use of a broadly acting kinase inhibitor, 5Z-7-oxozeaenol (49). We confirmed the role of TAK1 in human TLR2 signaling as follows: pretreatment of THP1 monocytes with the recently published, highly specific inhibitor of TAK1, Takinib (53), virtually abolished P2C-induced expression of the NF-κB–regulated gene product IL8 and significantly reduced TLR2-dependent expression of IFNβ and ISG54 mRNAs (Fig. 5B). At the highest dose tested, Takinib treatment almost completely blocked ISG15 production (Fig. 5, D and E). At the same time, IFNα-induced ISG15 levels were not affected, excluding broadly toxic effects of the inhibitor at the concentrations used. Thus, TAK1 activity is required for TLR2-dependent synthesis of IFNβ to levels that support productive IFN-I signaling.
Downstream of TAK1, signaling diverges, and either cascade may effectuate the observed role of TAK1 in IFN-I induction (Fig. 5A). On the one hand, TAK1 recruits and activates an IκB kinase complex, consisting of IKKα, IKKβ, and IKKγ, which ultimately leads to activation and nuclear translocation of NF-κB. On the other hand, TAK1 triggers several mitogen-activated protein kinase (MAPK) pathways that drive AP-1 activation. We did not find alterations in TLR2-induced ISG15 protein levels in THP1 monocytes pretreated with inhibitors of various MAPKs (Jun N-terminal kinases 1 and 2 (JNK1/2), MAPK/ERK kinases 1 and 2 (MEK1/2), and p38α; Fig. S4). IFN-I induction in monocytes, therefore, does not appear to rely on the activation of AP-1. In contrast, in line with recent observations for other TLR2 ligands (49), the inhibitor BI605906, which interferes with IKKβ activity and thereby selectively blocks NF-κB activation (48), recapitulated the phenotypic effects of the TAK1 inhibitor: in response to P2C, inhibitor-treated cells expressed significantly lower levels of IFNβ, ISG54, and IL8 transcripts (Fig. 5C) and displayed a dose-dependent reduction in induced ISG15 levels (Fig. 5, F and G). From these findings, we conclude that TAK1-induced activation of NF-κB through IKKβ, rather than activation of AP-1 through MAPKs, is required—in conjunction with the TBK1/IRF3 signaling axis—to optimally induce IFNβ expression and subsequent IFN-I signaling in human monocyte-like cells following TLR2 stimulation.
TLR2 ligand engagement at the cell surface of human monocytes suffices to drive an IFN-I response
Having defined downstream signaling components that relay IFNβ expression in human monocytes, we next addressed whether TLR2 internalization is required to initiate these signaling cascades. In LPS-stimulated macrophages, internalization of TLR4 molecules is a prerequisite for promoting IFN-I expression (29), which coincides with surface TLR4 down-regulation. In contrast, TLR2 surface levels on THP1 monocytes remained constant following P2C or P3C stimulation (data not shown). This implies that a majority of receptors stays at the plasma membrane after stimulation but does not rule out the possibility that a small fraction of TLR2 molecules is internalized and responsible for initiating IFN-I signaling.
In the case of TLR4, one route of internalization is via macropinocytosis, a process depending on the co-receptor CD14 (54). CD14 also acts as a co-receptor for TLR2, in particular for capturing triacylated ligands (55) that are recognized by TLR2/1 heterodimers. As THP1 monocytes do not express CD14 (Fig. S1A), CD14-dependent processes were not involved in the observed TLR2-induced IFNβ expression. When stably introduced into THP1 cells (Fig. S5A), CD14 expression did not substantially increase P2C- or P3C-induced levels of IFNβ and ISG54 transcripts (Fig. S5B) or ISG15 proteins (Fig. S5, C and D). Thus, the absence of CD14 did not negatively impact TLR2-dependent IFN-I responses in human monocytes, rendering receptor internalization via macropinocytosis an unlikely requirement.
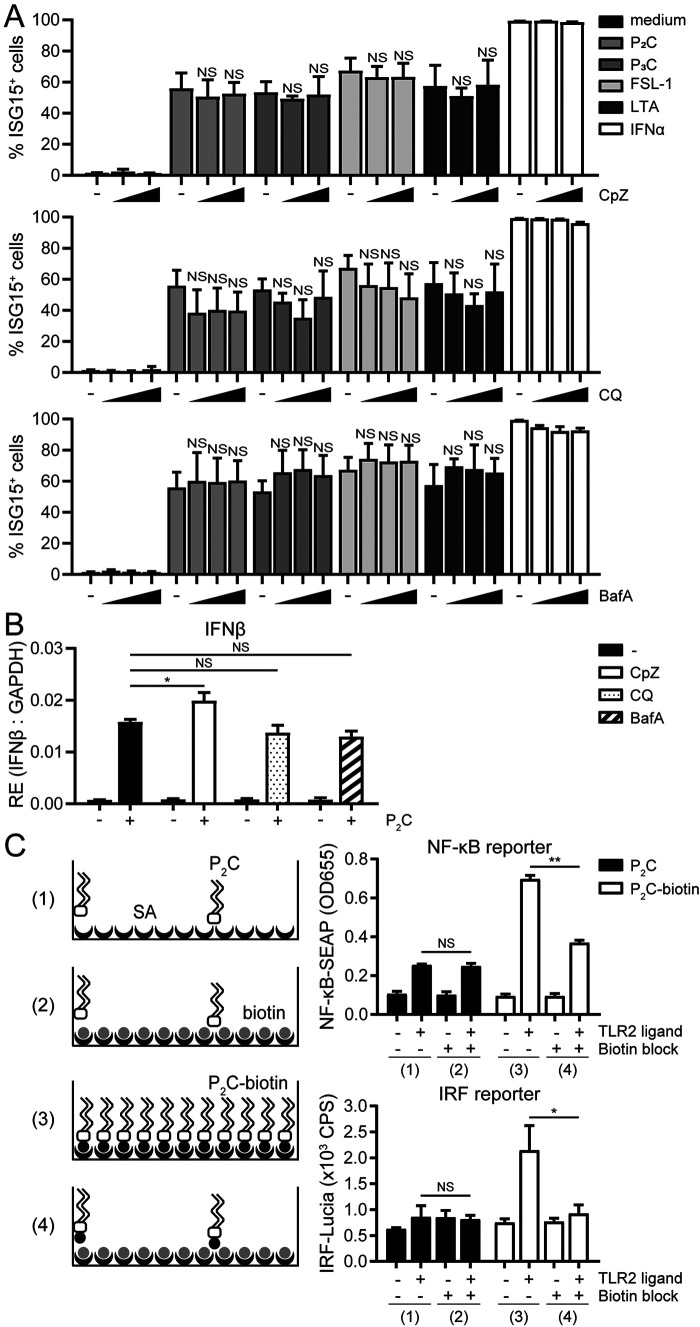
Surface TLRs, including TLR2 and TLR4, can be internalized through clathrin-dependent endocytosis (54, 56, 57). To examine whether endocytosis is necessary for TLR2-induced IFN-I signaling, THP1 monocytes were (pre)incubated with different chemical inhibitors, over concentration ranges that were not toxic, as they did not interfere with ISG15 induction by IFNα (Fig. 6A and Fig. S6). Chlorpromazine (CpZ) inhibits the formation of clathrin-coated pits at the cell surface, whereas chloroquine (CQ) and bafilomycin A1 (BafA) block endosome maturation by preventing endosomal acidification (58, 59). CpZ, CQ, and BafA did not prevent up-regulation of ISG15 in response to four well-defined ligands for TLR1/2 and/or TLR2/6 heterodimers (P2C, P3C, FSL-1, and LTA; Fig. 6A and Fig. S6). In line with this, the endocytosis inhibitors did not significantly reduce P2C-induced IFNβ mRNA levels (Fig. 6B). This strongly suggests that TLR2 can induce IFN-I signaling independently of receptor endocytosis and implies that the signaling events leading to IFN-I production originate either from the plasma membrane or from a preexisting intracellular pool of receptors.
Figure 6.
TLR2 can drive IFN-I expression from the surface of human monocyte-like cells, irrespective of receptor internalization. A and B, to evaluate the role of endocytosis in TLR2 signaling outcome, THP1 monocytes were (pre)treated with the inhibitor of clathrin-mediated endocytosis CpZ (dose range 6.2–12.5 μm (A) or 25 μm (B)) or the inhibitors of endosome maturation CQ (doses of 7.5, 15, or 30 μm (A) or 15 μm (B)) or BafA (doses of 25, 50, or 100 nm (A) or 50 nm (B)). A, cellular ISG15 levels 24 h after stimulation with TLR2 ligand P2C (100 nm), TLR2/1 ligand P3C (1 μm), TLR2/6 ligand FSL-1 (100 nm) or LTA (2.5 μg/ml), or IFNα (1000 units/ml) were measured. Data are shown as percentage of ISG15-positive cells ± S.D. (error bars) from three independent experiments (one-way ANOVA with Dunnett's multiple-comparison test). Representative fluorescence intensity histograms are presented in Fig. S6. B, at 1 h after stimulation with P2C (100 nm), RE of the IFNβ transcript was assessed by qRT-PCR and normalized to GAPDH. Data are presented as means ± S.D. (error bars) from a representative of four independent experiments (one-way ANOVA with Dunnett's multiple-comparison test). C, to assess whether surface TLR engagement was sufficient to induce an IFN-I response, P2C-biotin was immobilized on the surface of streptavidin (SA)-coated plates (condition 3). Unconjugated P2C was taken along as a negative control that does not bind SA (condition 1). Control SA plates were preblocked with D-biotin to prevent immobilization of the biotinylated TLR2 ligand (conditions 2 and 4). After culturing THP1 Dual reporter cells in these plates for 24 h, levels of the reporter proteins reflecting NF-κB (SEAP) and IRF (Lucia) activation were determined in the culture supernatants. Data are presented as means ± S.D. (error bars) from a representative of three independent experiments (unpaired Student's t test). NS, not significant; *, p < 0.05; **, p < 0.01.
To specifically assess the outcome of TLR2 signaling from the cell surface, we immobilized a biotin-conjugated derivative of P2C on streptavidin-coated culture wells to prevent cells from internalizing these ligands (Fig. 6C). In parallel, unconjugated P2C served as a negative control, as it cannot bind to streptavidin. After removal of unbound ligand through washing steps, THP1 Dual monocytes were incubated onto the coated wells, and after 24 h, levels of NF-κB-SEAP and IRF-Lucia reporter proteins were determined in the supernatants. Cells incubated in wells treated with unconjugated TLR2 ligand produced low levels of both reporter proteins, likely caused by minor, nonspecific adherence of the ligand to the wells (Fig. 6C, condition 1). Markedly higher levels of both NF-κB-SEAP and IRF-Lucia were detected in the culture supernatants of cells incubated on immobilized P2C-biotin (Fig. 6C, condition 3). Moreover, when we preblocked streptavidin-coated wells with D-biotin to prevent immobilization of P2C-biotin on streptavidin, reporter induction triggered by P2C-biotin was significantly reduced to levels comparable with those induced by remaining unconjugated P2C (Fig. 6C, condition 4 compared with conditions 1 and 2). These combined findings show that the immobilized fraction of P2C-biotin induced both NF-κB and IRF activity in THP1 Dual monocytes, thereby demonstrating that TLR2 signaling from the cell surface of monocytes can initiate both a proinflammatory and an IFN-I response.
Discussion
In this study, we found that diverse TLR2 ligands evoke an IFN-I response in human monocyte-like cells, which is lost upon differentiation into macrophages. Stimulated monocytes transiently express modest levels of IFNβ resulting from a signaling cascade (Fig. 7) that 1) occurs upon exposure to TLR2/1 and TLR2/6 ligands, 2) critically depends on the signaling adaptor MyD88, 3) involves TBK1 and IRF3 activity, 4) is complemented by robust TAK1/IKKβ-induced activation of NF-κB, rather than of AP-1, and 4) does not require TLR2 internalization. The levels of IFNβ produced by TLR2-activated monocytes supported productive downstream IFN-I signaling. In contrast, the TLR2 response in macrophages was limited to NF-κB–dependent proinflammatory cytokine production. Thus, monocytes and macrophages employ distinct molecular mechanisms to regulate the quality of the innate response following human TLR2 engagement, in particular with respect to IFN-I induction.
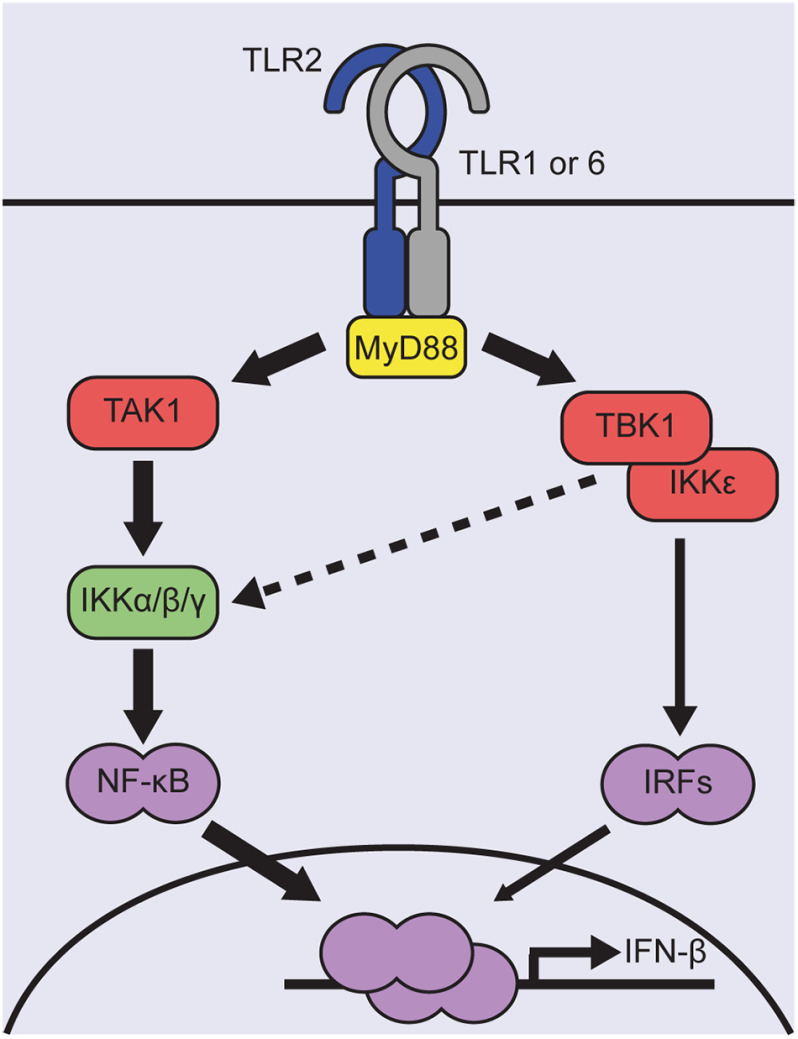
Figure 7.
Model for the TLR2-induced signaling events leading to IFNβ transcription in human monocyte-like cells. Ligand binding induces TLR2 heterodimerization with either TLR1 or TLR6. Activated receptor dimers recruit the adaptor MyD88 to transduce downstream signaling events. This results in modest and transient IFNβ expression, relying on the kinase activities of TAK1, IKKβ, and TBK1/IKKε. Via TAK1 and an IKK complex containing IKKβ, NF-κB is robustly activated. TBK1/IKKε, instead, regulate the activity of IRFs. IRF3 was identified as the dominant IRF for driving IFNβ expression, which may be aided by IRF7 and IRF8. Due to cross-talk with the canonical IKK complex, TBK1 may additionally be involved in modulating NF-κB activity to appropriate levels (striped line). Overall, TLR2-induced IFNβ expression is governed by a low level of IRF3 in conjunction with NF-κB activity.
Differentiation-dependent outcomes of TLR2 signaling in human cells may well have contributed to the inconsistent findings in studies addressing whether the IFN-I response observed downstream of murine TLR2 could be extrapolated to human TLR2. Our findings are in line with, extend, and complement a very recent report by Musilova et al. (49), who reported that primary human monocytes as well as undifferentiated THP1 cells secreted relatively low levels of IFN-I when they were cocultured with live S. aureus or LTA, which is an S. aureus-derived TLR2 ligand. They did not study differentiated THP1 cells, for which we observed that the TLR2-induced IFN-I response was selectively lost (Figs. 1 (A and B) and 2 (B–D) and Fig. S2B); this was also the case for primary M1 and M2 macrophages (Fig. S7). Cumulative evidence now indicates that human TLR2 signaling in monocytes can yield an IFN-I response in addition to the well-established proinflammatory one but that this response is not induced after macrophage differentiation.
The molecular basis for why the monocyte IFN-I response downstream of human TLR2 is selectively lost upon differentiation is unclear. Before and after PMA differentiation, cells mount comparable TLR2-dependent proinflammatory responses (Figs. 1 (A and B) and 2B), and levels of TLRs and signaling adaptors are not reduced upon differentiation (Fig. S1). THP1 macrophages have up-regulated CD36 and CD14 (Fig. S1), two co-receptors for TLR2/6 and TLR2/1 heterodimers, respectively (55, 60), but elevated co-receptor expression has been associated with increased TLR2 signaling rather than reduced (61). We therefore conclude that the inability of human macrophages to mount an IFN-I response via TLR2 is not due to loss of (co)-receptor expression. Further research is warranted to decipher at what level signaling downstream of TLR2 diverges between monocytes and macrophages to culminate in distinct signaling outcomes. Additionally, it is as yet unclear what the biological meaning of this would be.
With respect to the differentiation-dependent signaling outcome, human TLR2 differs from mouse TLR2, as the latter yields IFN-I responses in both monocytes and macrophages (32, 33), yet, under the right conditions, IFN-I synthesis can be initiated by TLR2 ligands in myeloid cells from both species. Combined with the recent observation that TLR2-stimulated human pDCs produce IFNα (62), it demonstrates that the outcome of TLR2 signaling in some human cells includes not only a proinflammatory response but also an additional IFN-I response.
One way to regulate TLR signaling outcome is by the use of alternative signaling adaptors. The prototypic switch downstream of TLR4 from proinflammatory signaling to IFN-I is explained by displacement of TIRAP/MyD88 by TRAM/TRIF (28, 63). Accordingly, whereas LPS-stimulated peritoneal macrophages lacking MyD88 no longer produced proinflammatory cytokines, TRAM/TRIF-dependent IFN-I signaling remained (64). Also, human ΔMyD88 THP1 macrophages were still capable of producing IFNβ and ISGs following TLR4 stimulation (Fig. 3, A and B (right panels) and D). In contrast, MyD88 was essential downstream of TLR2 activation: IFN-I and ISG expression by human THP1 monocytes was ablated by knockout (Fig. 3 (A and B (left panels) and C) and Fig. S2C) or reduced by silencing (49) of MyD88. From this, we conclude that TLR2 activation on human monocytes does not appear to yield IFN-I output through displacement of MyD88 from the receptor by other adaptors. The ability to induce both proinflammatory and IFN-I signaling through a single adaptor protein is not unique, because TLR7 and TLR9 also confer both of these responses through MyD88 in pDCs (65). In the mouse, several examples support MyD88 dependence of IFN-I induction downstream of TLR2. Murine cytomegalovirus and VV triggered IFN-I via TLR2 in WT but not in ΔMyD88 inflammatory monocytes (32), and synthetic TLR2 ligands failed to induce proinflammatory cytokines (TNFα and IL12) and IFNβ in ΔMyD88 bone marrow–derived macrophages (33). Without displacing MyD88, other adaptor molecules expressed by monocytes (Fig. S1) could still aid in the induction of IFN-I. Roles for TRAM and TRIF have been proposed in murine TLR2 signaling (37, 66), where TRAM appeared to interact with both TLR2 and MyD88 (37). Although we did not investigate the contributions of TRAM and TRIF, silencing of these adaptor molecules did not have a major impact on human TLR2-induced IFN-I signaling (49). Based on these combined observations, MyD88 adaptor switching does not appear to be a mechanism by which human monocyte-like cells regulate IFN-I production in response to TLR2 ligands.
Whereas human TLR2 initiated both IFN-I and proinflammatory cytokines responses via MyD88, the minimal ligand dose for IFN-I induction might be higher than that for a proinflammatory response (Fig. 2A). The question then arises of how higher ligand doses could affect the downstream signaling cascade. Therefore, we analyzed which signaling intermediates are involved in the IFN-I response.
In general, IRFs are the transcription factors activated to induce IFN-I following TLR stimulation (67), whereas proinflammatory cytokines are instead produced from the NF-κB pathway. The IRF family comprises nine members, which are differentially expressed among cell types. IRF1, IRF3, IRF5, and IRF7 have been implicated in IFN-I transcription (52), with certain IRFs dominating in specific cells and depending on the TLR activated. For instance, TLR4 strongly activates IRF3 in macrophages, through the TRIF-dependent pathway (28), and IRF3-IFNβ promoter interactions are stabilized by the myeloid-specific transcription factor IRF8 (68). Downstream of TLR7 and TLR9 in pDCs, IRF7 mainly drives the IFN-I response (69). For TLR2 in murine cells, multiple IRF family members, including IRF1, IRF3, IRF7, and IRF8, have been implicated in regulating IFNβ expression (32, 33, 37, 66, 70), but their relative contributions remain unclear. In human cells, we now show that TLR2-dependent IFN-I signaling is virtually lost in IRF3 knockout THP1 monocytes (Fig. 4, F and G), whereas NF-κB signaling is unaltered; this reveals the important role IRF3 plays in facilitating IFN-I synthesis. To a lesser extent, IRF7 and IRF8 also contribute (Fig. S3). The limited role of IRF7 is likely explained by the kinetics of TLR2-induced IFNβ transcription, which peaks at times when IRF7 levels are low (Fig. 1C). IRF8 could serve to enhance IRF3 effects, as described above for TLR4 signaling. Of note, IRF8 may also act indirectly, as it regulates transcription of developmental genes in addition to specific ISGs (71). The major role we found for IRF3 in the induction of IFN-I downstream of human TLR2 is reminiscent of that downstream of TLR4 (64). Unlike the latter, the indispensability of IRF3 for TLR2 was—to our surprise—not accompanied by common hallmarks of IRF3 activation, such as its phosphorylation (Fig. 4B) or nuclear translocation (data not shown). We hypothesize that this reflects limited IRF3 activity, which would underlie the more modest IFN-I response in TLR2-stimulated monocytes compared with TLR4-stimulated macrophages (Figs. 1A and 3 (C and D)).
How then is this limited IRF3 activity achieved? The noncanonical IKKs TBK1 and IKKε regulate IRF3 through phosphorylation (30). Robust activation of TBK1 occurred upon TLR2 ligation on the monocytes (Fig. 4B) and was essential for the IFN-I response (Fig. 4, C–E). Although efficient phosphorylation of IRF3 requires TBK1 to interact with adaptor proteins containing a conserved pLXIS motif, such as TRIF (72, 73), in human monocytes, modest IFN-I levels were induced via TLR2 irrespective of TRIF expression (49). In vitro, TBK1 can (inefficiently) phosphorylate IRF3 in the absence of TRIF (74), and therefore, this kinase might by itself impose low-level phosphorylation onto IRF3 in TLR2-stimulated human monocytes, ensuring a limited IFN-I response.
Having only low levels of IRF activity might not be sufficient to fully drive TLR2-dependent IFNβ expression and appears to be complemented by NF-κB. Two kinases involved in the NF-κB signaling cascade, TAK1 and IKKβ, contributed to the TLR2-induced IFN-I response in human monocytes (Fig. 5); TAK1-induced events that trigger AP-1 activity did not appear to be involved (Fig. S4). Cross-talk between TBK1/IKKε and IKKβ has been reported (48), suggesting that TBK1 activity may add to proper NF-κB activation downstream TAK1. A precedent of concerted actions of IRF3 and NF-κB in initiating IFN-I synthesis is provided by NDV-infected cells (75). Late in infection, NDV evokes robust IRF3 activity and IFNβ expression, but during the first 6 h, limited IRF3 activation is not sufficient for detection in a dimerization assay. It is at these early times that complementation by NF-κB signaling is crucial for modest IFNβ expression. Thus, NF-κB becomes particularly important for IFN-I synthesis when IRF activity is limited. In summary (Fig. 7), combined activities of kinases like TBK1, IKKβ, and TAK1 culminate in activation of IRF3 and NF-κB to jointly effectuate a modest level of IFNβ transcription downstream of TLR2 in human monocytes.
In addition to the use of distinct signaling adaptors and intermediates to regulate IFN-I, differential outcomes of TLR signaling could originate from distinct cellular locations. A striking observation we made is that, in human monocytes, the MyD88-dependent signaling events driving IFNβ expression in response to the TLR2 ligand P2C did not require receptor internalization (Fig. 6, A and B) and were induced from the cell surface (Fig. 6C). This contrasts with macrophages requiring TLR4 to be internalized through endocytosis to support TRIF-dependent signaling pathways toward IFN-I (28) yet is in line with continued signaling via the MyD88 signaling adaptor (Fig. 3 and Fig. S2C). Earlier studies yielded inconsistent conclusions on human TLR2 internalization as a requirement for inducing signaling events. Using microscopy, TLR2 fusion proteins as well as fluorescent TLR2 ligands have been observed at the plasma membrane and in endosomes (57, 66, 76). Increased internalization kinetics upon TLR2 stimulation suggested a role for endocytosis in TLR2 signal transduction (33, 37, 66) but did not prove from which location signaling toward IFN-I originated; internalization could also serve to end signaling via receptor degradation. To functionally address the impact of blocking receptor internalization, chemical inhibitors of endocytosis are widely used. In our experiments, we used dose ranges of chlorpromazine, chloroquine, and bafilomycin A1 that did not interfere with IFNAR signaling and found that blocking endocytosis did not alter IFN-I signaling in response to multiple TLR2 ligands, with no detectable differences related to the receptor heterodimers that can be bound (Fig. 6A and Fig. S6). Circumventing potential off-target effects that chemical inhibitors have on overall cell functioning, we showed that when P2C was restrained extracellularly, monocytes still mounted an IFN-I response (Fig. 6C), thereby conclusively demonstrating that IFN-I transcription can result from TLR2 signaling events induced at the plasma membrane. Although our results seem to be in contradiction with the reduced secretion of IFN-I from endocytosis inhibitor–treated THP1 monocytes infected with S. aureus (49), the use of entire pathogens might not be one-on-one comparable with an isolated small synthetic TLR2 ligand, as even stimulation with the S. aureus–derived TLR2 ligand LTA yielded IFN-I signaling in treated cells (Fig. 6). Our data do not exclude the possibility that TLR2 can signal to IFN-I from intracellular vesicles in addition to from the cell surface. Finally, by restricting the TLR2 ligand P2C to the cell surface, we did not separate the spatial dependences of TLR2/1 and TLR2/6 heterodimers for signaling to IFN-I, although we did not find indications that the IFN-I responsiveness of P3C (TLR2/1) or FSL-1/LTA (TLR2/6) differed from P2C in other assays (Figs. 2, 3 (A and B), 4 (F and G), and 6A and Fig. S2). In the future, chemical approaches to generate diverse TLR2 ligands that function exclusively intracellularly may provide definitive insight into the signals originating selectively from TLR2 in endosomes (25). Our observation that human TLR2 can induce IFN-I signaling from the cell surface of monocytes (Fig. 6) at least indicates that the outcome of TLR2 signaling is not as strictly regulated through TLR trafficking events in these cells as seen for TLR4 in macrophages.
Our combined results reveal that cells resembling two myeloid differentiation stages differentially respond to TLR2 ligands, with monocytes producing both proinflammatory cytokines and modest levels of IFN-I, whereas macrophages only induce a proinflammatory response. Intriguingly, also TLR4 responses differ in human macrophages versus monocytes. Compared with macrophages, monocytes induce much lower levels of IFNβ following LPS stimulation, which has been observed both for THP1 cells1 and primary cells (77, 78). Thus, TLR2 and TLR4 ligands both induce relatively modest levels of IFNβ in human monocytes, whereas TLR4 signaling in macrophages is tailored to induce higher levels of IFN-I. Aside from playing a key role in the innate defense against infections (8, 9, 79), IFN-I also pose the risk of inflammatory damage (80). Monocytes circulate in the blood, and therefore, the production of high levels of IFN-I systemically might do more harm than good. However, low levels of IFN-I may be beneficial for effector functions, such as the polarization of innate and adaptive immune cells (81). The localized detrimental effects of high TLR4-induced IFN-I levels by macrophages in infected tissues may, instead, be outweighed by the beneficial effects of clearing an infection. The finding that TLR2 (and TLR4) signaling and outcome differ not only between cell types, but even between differentiation states, may therefore reflect the importance of tailoring IFN-I output depending on cellular context. Specific mechanisms acting to limit TLR-induced IFN-I production in monocytes may have been overlooked earlier, because monocytes and macrophages were often assumed to similarly transduce TLR signals. Delineation of such mechanisms is essential for the development of preventive or therapeutic intervention strategies aimed at achieving balanced IFN-I responses.
Experimental procedures
Cells
The human monocytic cell line THP1 (ATCC, TIB-202) was maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. WT, ΔMyD88, ΔIFNAR2, and ΔIRF3 THP1 Dual NF-κB/IRF reporter cells were obtained from Invivogen and were cultured according to the manufacturer's instructions. Cells were maintained at 37 °C and 5% CO2. To generate differentiated macrophages, THP1 cells were treated with 150 nm PMA (Sigma–Aldrich) for 24 h and were left to rest for another 24 h before use in experiments, as described previously (82).
Lentiviral transductions
For knockdown experiments, pLKO.1-Puro lentiviral vectors from the Sigma–Aldrich Mission shRNA library were used, two targeting IRF7 (TRCN0000014861 and TRCN0000014858), two for IRF8 (TRCN0000020984 and TRCN0000020985), a nontargeting control (SH002), and a control targeting β2m (SH008). To construct a lentiviral vector encoding hCD14, the coding sequence of hCD14 was excised from pUNO-CD14 (Invivogen) and cloned under the control of the hybrid EF1α-HTLV promoter in the pSicoR backbone (Addgene, catalog no. 11579); the final construct, pSicoR.EF1α-HTLV.hCD14, was sequence-verified. Third-generation replication-deficient SIN recombinant lentiviruses were generated by polyethylenimine transfection of 293T cells with pCMV-VSVG, pMDLg-RRE, and pRSV-REV and the above lentiviral vectors. THP1 Dual cells were transduced with the lentivirus-containing culture supernatants by spin infection (2000 rpm, 33 °C, 90 min) in the presence of 8 µg/ml Polybrene. To obtain pure cell populations, cells transduced with shRNA constructs were selected with puromycin (0.6 µg/ml; Invivogen). Transduced CD14-expressing cells were stained with anti-CD14-PerCP (MφP9, BD Biosciences), and CD14+ cells were sorted as described under flow cytometry.
Ligand stimulations
The following reagents were used to stimulate THP1 cells: TLR2/6 ligand FSL-1 (Invivogen), purified LTA from S. aureus (Invivogen; TLR2/6 ligand), TLR2/1 ligand Pam3CSK4 (Invivogen), TLR4 agonist LPS (from E. coli O55:B5; Invivogen), hIFNα-2a (PBL Assay Science), and hTNFα (Miltenyi Biotec). The TLR2 ligand Pam2CSK4 (P2C) and a biotin-conjugated derivative (P2C-biotin) were synthesized in-house (see Fig. S8 for further details on the chemical synthesis). Compounds 1–3 (Fig. S8) were synthesized in-house: compound 2 was synthesized using Stick's diazotransfer reagent and Fmoc-Lys-OH, and compounds 1 (83) and 3 (84, 85) were synthesized via literature procedures. Peptide resins were purchased from Rapp Polymere.
Inhibitor treatments
Inhibitors targeting TAK1 (Takinib, MedChem Express), TBK1/IKKε (MRT67307, Sigma), and IKKβ (BI605906, MedChem Express) were used. To assess the involvement of MAPK pathways in determining TLR2 signaling outcome, inhibitors for JNK (SP600125, Invivogen), MEK1/2 (PD0325901, MedChem Express), and p38α (losmapimod, kindly provided by Dr. J. Neefjes) were applied. Endocytosis was blocked at different stages using chlorpromazine (Sigma), chloroquine (kindly provided by Dr. A de Wilde), and bafilomycin A1 (Sigma). Cells were preincubated with inhibitors for 45–60 min, and subsequent ligand stimulations were conducted in the presence of the inhibitors.
NF-κB and IRF reporter assays
THP1 Dual reporter cells, plated in 96-well flat bottom plates, were stimulated for 24 h in a final volume of 200 μl. Three wells with 90,000 cells were tested for each experimental condition (ligand; dose). Culture supernatants were collected, and levels of the two secreted reporter proteins were determined: SEAP reflects NF-κB activation, whereas Lucia luciferase is produced upon IRF activation. Colorimetric enzyme assays to detect SEAP were performed using QUANTI-Blue reagent (Invivogen) according to the manufacturer's instructions. The enzyme-induced color change was detected by measuring the optical density at 655 nm using a Bio-Rad iMark microplate reader. The levels of Lucia luciferase in the samples were determined with a bioluminescent assay using QUANTI-Luc reagent (Invivogen). Light emitted upon reagent conversion was detected using a Victor X3 multilabel plate reader (PerkinElmer Life Sciences).
Selective stimulation of cell surface TLR2
P2C-biotin was immobilized on Pierce Streptavidin-coated high-sensitivity 8-well strips (Thermo Scientific) by incubating the ligand for 1 h at 4 °C in PBS, 2.5% BSA. Wells treated with unconjugated P2C served as a control for nonspecific binding to the wells. Wells preblocked with D-biotin (250 μm, 4 °C, overnight) served as negative controls, to which P2C-biotin was no longer able to bind. Strips were washed four times with PBS, 2.5% BSA, 0.5% Tween 20 and two times with PBS, 2.5% BSA to remove unbound ligand. THP1 Dual reporter cells were seeded into the culture wells, and after 24 h, culture supernatants were collected for NF-κB and IRF reporter assays.
RNA extraction and qRT-PCR
RNA was isolated from cells (300,000 cells/well; 24-well plates) at the indicated time points after stimulation using the NucleoSpin RNA kit (Machery–Nagel) or with TRIzol reagent (Invitrogen) according to the manufacturer's protocols. RNA was quantified by measuring the A260 on a Nanodrop ND-1000 (Isogen Life Science). cDNA was generated from 300–1000 ng of total RNA using anchored oligo(dT) primers (AB-1247 (Thermo Scientific) or PM-305L (Jena Bioscience)) and Superscript III reverse transcriptase (Invitrogen). For each cDNA sample, expression of genes of interest was measured by qRT-PCR in triplicate, except for the data in Fig. 1C and Fig. S5B, which were measured in duplicate, using iQ SYBR Green Supermix (Bio-Rad) on a CFx connect thermocycler (Bio-Rad). The primers used for qRT-PCR amplification are listed in Table 1. Relative expression (RE) levels were calculated using the ΔΔCt method (86), using GAPDH or β-actin (ACTB) expression levels as an internal reference. Where indicated, -fold mRNA inductions were calculated by normalizing the RE levels to that of the unstimulated control cells.
Table 1.
List of primers for qRT-PCR
| Gene | Primer | Sequence (5′–3′) |
|---|---|---|
| hIFNβ | Fw | TCTGGCACAACAGGTAGTAGGC |
| Rv | GAGAAGCACAACAGGAGAGCAA | |
| Fw_2 | AGTAGGCGACACTGTTCGTG | |
| Rv_2 | GTCTCATTCCAGCCAGTGCT | |
| hCXCL10 | Fw | GTGGCATTCAAGGAGTACCTC |
| Rv | TGATGGCCTTCGATTCTGGATT | |
| hISG15 | Fw | AGGCAGCGAACTCATCTTTG |
| Rv | CCAGCATCTTCACCGTCAG | |
| hISG54 | Fw | ATGTGCAACCTACTGGCCTAT |
| Rv | TGAGAGTCGGCCCATGTGATA | |
| hIRF7 | Fw | TGGTCCTGGTGAAGCTGGAA |
| Rv | GATGTCGTCATAGAGGCTGTTGG | |
| Fw_2 | TCAGAGTCTTCTTCCAAGAGCTG | |
| Rv_2 | GATGGTATAGCGTGGGGAGC | |
| hIRF8 | Fw | TGATCAAGGAGCCTTCTGTGG |
| Rv | CTGGAAGGAGCTGACTCCGA | |
| hViperin (RSAD2) | Fw | AGCAATGGAAGCCTGATCC |
| Rv | ACTTCCTCGTCAAAGCTGTC | |
| hTNFα | Fw | CTCTTCTGCCTGCTGCACTTTG |
| Rv | ATGGGCTACAGGCTTGTCACTC | |
| hIL8 | Fw | TTTTGCCAAGGAGTGCTAAAGA |
| Rv | AACCCTCTGCACCCAGTTTTC | |
| hCCL5 (RANTES) | Fw | TGCCTGTTTCTGCTTGCTCTTGTC |
| Rv | TGTGGTAGAATCTGGGCCCTTCAA | |
| hTLR1 | Fw | ATTCCGCAGTACTCCATTCCT |
| Rv | CTTTGCTTGCTCTGTCAGCTT | |
| hTLR6 | Fw | TGCCCATCTGTAAGGAATTTG |
| Rv | TGGGTGAAAAACAAGGTGAAG | |
| hMD2 (Ly96) | Fw | GCTCAGAAGCAGTATTGGGTCTG |
| Rv | CGCTTTGGAAGATTCATGGTG | |
| hMyD88 | Fw | ATGGTGGTGGTTGTCTCTGATG |
| Rv | GACAGTGATGAACCTCAGGATGC | |
| hMAL | Fw | AGCGATGCTTCACAGCCTACCT |
| Rv | GTCTTTGCTCCAGCGACTACTG | |
| hTRIF | Fw | ACGCCATAGACCACTCAGCTTTCA |
| Rv | AGGTTGCTCATCATGGCTTGGTTC | |
| hTRAM | Fw | AGATGAAGCCCTCAGAGTCCAG |
| Rv | CTGTAAATGCTGTCTGCCACATG | |
| hGAPDH | Fw | GCAAATTTCCATGGCACCGT |
| Rv | GCCCCACTTGATTTTGGAGG | |
| hActinB | Fw | ATTGCCGACAGGATGCAGAA |
| Rv | GCTGATCCACATCTGCTGGAA |
Flow cytometry
To assess the surface display of the indicated molecules, cells were treated with TruStain FcX (Biolegend) to block Fc receptors in all experiments, except for Fig. S5A, and subsequently stained with antibodies diluted in PBS supplemented with 0.5% BSA and 0.02% sodium azide. The following antibodies were used: anti-CD11b-PE (ICRF44, BD Biosciences), anti-TLR2-PE (TL2.1, eBioscience), anti-TLR4-A488 (HTA125, eBioscience), anti-CD36-PE (5-271, Biolegend), and anti-CD14-PE or anti-CD14-PerCP (both MφP9, BD Biosciences). ISG15 levels were determined using anti-hISG15-PE (R&D Systems) on cells fixed with 2% paraformaldehyde and permeabilized in saponin buffer (PBS, 2% fetal calf serum, 0.5% saponin).
Samples were subjected to flow cytometry by measuring 10,000 cells/sample on an LSR II (BD Biosciences), and data were analyzed using FlowJo software (V10, Treestar). Data were gated for live cells using FSC/SSC. For Fig. S1B, autofluorescence-corrected GeoMFI values were used to calculate relative surface levels as indicated in the figure legend. Quantification of percentages of ISG15-positive cells was based on gating, as shown in Fig. 1D. For cell sorting on an Aria III (BD Biosciences), cells were stained with antibodies diluted in sterile 0.2 μm filtered PBS containing 0.5% BSA.
Western blotting analysis
Total cell lysates were generated in Pierce radioimmune precipitation assay buffer (Thermo Fisher Scientific), supplemented with cOmplete mini protease inhibitor (Roche Applied Science), and 50 units/ml Benzonase (Santa Cruz Biotechnology, Inc.). For the detection of phosphoproteins, a 10 mm concentration of the phosphatase inhibitor NaF was added. Samples were subjected to 30 min of rotation at 4 °C and centrifuged to remove cellular debris (13,000 rpm, 4 °C, 5 min). Lysates were denatured by adding Laemmli sample buffer containing 20 mm DTT and heating for 5 min at 95 °C. Precision Plus All Blue prestained protein standard (Bio-Rad) was used as a molecular weight marker. Proteins were separated on 10% SDS-polyacrylamide gels and transferred onto 0.2-μm nitrocellulose membranes using the Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were blocked using 10% milk in TBS. For the detection of phosphoproteins, membranes were instead blocked using 5% BSA in TBS. Antibodies were diluted in immunobooster solution (TaKaRa) or 5% milk in TBS supplemented with 0.1% Tween 20 (TBS-T). Blots were incubated overnight at 4 °C with primary antibodies and for 30–45 min at room temperature with secondary antibodies. Blots were washed with TBS-T.
The following antibodies were used for staining: anti-IRF3 (Cell Signaling, catalog no. 11904), anti-IRF7 (Santa Cruz Biotechnology, sc-74471), anti-IRF8 (Santa Cruz Biotechnology, sc-365042), anti-RIG-I (Cell Signaling, catalog no. 3743), anti-TBK1 (Cell Signaling, catalog no. 3504), anti-phospho-TBK1 (Cell Signaling, catalog no. 5483), anti-phospho-IRF3 (Cell Signaling, catalog no. 4947), anti-STAT1 (Santa Cruz Biotechnology, sc-464), anti-phospho-STAT1 (Cell Signaling, catalog no. 9167), anti-vinculin (Sigma, V9131), and IRDye-conjugated secondary antibodies anti-rabbit 800CW (LI-COR Biosciences) and anti-mouse 680RD (LI-COR Biosciences).
Proteins were visualized using the Odyssey CLx Imaging System (LI-COR Biosciences). Stripping of blots prior to reprobing was performed using NewBlot IR stripping buffer (LI-COR Biosciences) according to the manufacturer's instructions.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software. For the comparison between two groups, data were analyzed using the unpaired Student's t test. For multiple comparisons, wherein samples were compared with a control sample, one-way analysis of variance with Dunnett's multiple comparison was performed. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01.
Data availability
Data described in this article are available from the corresponding author (Maaike E. Ressing, m.e.ressing@lumc.nl) upon reasonable request.
Supplementary Material
Acknowledgments
We thank Martijn Rabelink, Steve Cramer, and Kimberley Walburg for excellent experimental assistance and Drs. Rob Hoeben, Sjaak Neefjes, Tom Ottenhoff, and Annemarthe van der Veen for valuable discussions and critical reading of the manuscript.
This article contains supporting information.
Author contributions—T. O., M. J. v. d. G., S. v. K., and M. E. R. conceptualization; T. O. and M. J. v. d. G. formal analysis; T. O. and S. v. K. funding acquisition; T. O. and M. J. v. d. G. validation; T. O. and M. J. v. d. G. investigation; T. O. and M. E. R. visualization; T. O., M. J. v. d. G., M. C. H., S. v. K., and M. E. R. methodology; T. O. and M. E. R. writing-original draft; T. O., M. J. v. d. G., S. v. K., and M. E. R. writing-review and editing; M. C. H. resources; S. v. K. and M. E. R. supervision.
Funding and additional information—T. O. was supported by NWO Graduate Programme Grant 022.006.010. S. v. K. and M. J. v. d. G. were supported by ERC Starting Grant 639005.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
T. Oosenbrug, M. C. Haks, and M. E. Ressing, unpublished observations.
- PRR
- pattern recognition receptor
- PAMP
- pathogen-associated molecular pattern
- TLR
- Toll-like receptor
- Fmoc
- N-(9-fluorenylmethoxycarbonyl)
- IFN-I
- type I interferon(s)
- IFNAR
- IFN-α/β receptor
- ISG
- interferon-stimulated gene
- DC
- dendritic cell
- pDC
- plasmacytoid DC
- cDC
- conventional DC
- LPS
- lipopolysaccharide
- PMA
- phorbol 12-myristate 13-acetate
- SEAP
- secreted embryonic alkaline phosphatase
- qRT-PCR
- quantitative RT-PCR
- LTA
- lipoteichoic acid
- NDV
- Newcastle disease virus
- IRF
- IFN regulatory factor
- MAPK
- mitogen-activated protein kinase
- JNK
- c-Jun N-terminal kinase
- MEK
- MAPK/ERK kinase
- CpZ
- chlorpromazine
- CQ
- chloroquine
- BafA
- bafilomycin A1
- RE
- relative expression
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ANOVA
- analysis of variance
- SA
- streptavidin.
References
- 1. Brubaker S. W., Bonham K. S., Zanoni I., and Kagan J. C. (2015) Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290 10.1146/annurev-immunol-032414-112240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mogensen T. H. (2009) Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273, Table of Contents 10.1128/CMR.00046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barton G. M. (2007) Viral recognition by Toll-like receptors. Semin. Immunol. 19, 33–40 10.1016/j.smim.2007.01.003 [DOI] [PubMed] [Google Scholar]
- 4. Iwasaki A., and Medzhitov R. (2004) Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5, 987–995 10.1038/ni1112 [DOI] [PubMed] [Google Scholar]
- 5. Pasare C., and Medzhitov R. (2005) Toll-like receptors: linking innate and adaptive immunity. Adv. Exp. Med. Biol. 560, 11–18 10.1007/0-387-24180-9_2 [DOI] [PubMed] [Google Scholar]
- 6. Noppert S. J., Fitzgerald K. A., and Hertzog P. J. (2007) The role of type I interferons in TLR responses. Immunol. Cell Biol. 85, 446–457 10.1038/sj.icb.7100099 [DOI] [PubMed] [Google Scholar]
- 7. Crosse K. M., Monson E. A., Beard M. R., and Helbig K. J. (2018) Interferon-stimulated genes as enhancers of antiviral innate immune signaling. J. Innate Immun. 10, 85–93 10.1159/000484258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schoggins J. W., and Rice C. M. (2011) Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1, 519–525 10.1016/j.coviro.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boxx G. M., and Cheng G. (2016) The roles of type I interferon in bacterial infection. Cell Host Microbe 19, 760–769 10.1016/j.chom.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beilharz M. W., Cummins J. M., and Bennett A. L. (2007) Protection from lethal influenza virus challenge by oral type 1 interferon. Biochem. Biophys. Res. Commun. 355, 740–744 10.1016/j.bbrc.2007.02.019 [DOI] [PubMed] [Google Scholar]
- 11. Müller U., Steinhoff U., Reis L. F. L., Hemmi S., Pavlovic J., Zinkernagel R. M., and Aguet M. (1994) Functional-role of type-I and type-Ii interferons in antiviral defense. Science 264, 1918–1921 10.1126/science.8009221 [DOI] [PubMed] [Google Scholar]
- 12. Mancuso G., Midiri A., Biondo C., Beninati C., Zummo S., Galbo R., Tomasello F., Gambuzza M., Macrì G., Ruggeri A., Leanderson T., and Teti G. (2007) Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J. Immunol. 178, 3126–3133 10.4049/jimmunol.178.5.3126 [DOI] [PubMed] [Google Scholar]
- 13. Castiglia V., Piersigilli A., Ebner F., Janos M., Goldmann O., Damböck U., Kröger A., Weiss S., Knapp S., Jamieson A. M., Kirschning C., Kalinke U., Strobl B., Müller M., Stoiber D., et al. (2016) Type I interferon signaling prevents IL-1 β-driven lethal systemic hyperinflammation during invasive bacterial infection of soft tissue. Cell Host Microbe 19, 375–387 10.1016/j.chom.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 14. Apelbaum A., Yarden G., Warszawski S., Harari D., and Schreiber G. (2013) Type I interferons induce apoptosis by balancing cFLIP and caspase-8 independent of death ligands. Mol. Cell Biol. 33, 800–814 10.1128/MCB.01430-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schreiber G., and Piehler J. (2015) The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 36, 139–149 10.1016/j.it.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 16. Lee-Kirsch M. A. (2017) The type I interferonopathies. Annu. Rev. Med. 68, 297–315 10.1146/annurev-med-050715-104506 [DOI] [PubMed] [Google Scholar]
- 17. Lund J. M., Alexopoulou L., Sato A., Karow M., Adams N. C., Gale N. W., Iwasaki A., and Flavell R. A. (2004) Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 101, 5598–5603 10.1073/pnas.0400937101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., and Akira S. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 10.1038/35047123 [DOI] [PubMed] [Google Scholar]
- 19. Liu Y. J. (2005) IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23, 275–306 10.1146/annurev.immunol.23.021704.115633 [DOI] [PubMed] [Google Scholar]
- 20. Honda K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., and Taniguchi T. (2005) Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434, 1035–1040 10.1038/nature03547 [DOI] [PubMed] [Google Scholar]
- 21. Hoshino K., Sasaki I., Sugiyama T., Yano T., Yamazaki C., Yasui T., Kikutani H., and Kaisho T. (2010) Cutting edge: critical role of IκB kinase α in TLR7/9-induced type I IFN production by conventional dendritic cells. J. Immunol. 184, 3341–3345 10.4049/jimmunol.0901648 [DOI] [PubMed] [Google Scholar]
- 22. Klein D. C. G., Skjesol A., Kers-Rebel E. D., Sherstova T., Sporsheim B., Egeberg K. W., Stokke B. T., Espevik T., and Husebye H. (2015) CD14, TLR4 and TRAM show different trafficking dynamics during LPS stimulation. Traffic 16, 677–690 10.1111/tra.12274 [DOI] [PubMed] [Google Scholar]
- 23. Yurchenko M., Skjesol A., Ryan L., Richard G. M., Kandasamy R. K., Wang N. H., Terhorst C., Husebye H., and Espevik T. (2018) SLAMF1 is required for TLR4-mediated TRAM-TRIF-dependent signaling in human macrophages. J. Cell Biol. 217, 1411–1429 10.1083/jcb.201707027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Solis M., Romieu-Mourez R., Goubau D., Grandvaux N., Mesplede T., Julkunen I., Nardin A., Salcedo M., and Hiscott J. (2007) Involvement of TBK1 and IKKε in lipopolysaccharide-induced activation of the interferon response in primary human macrophages. Eur. J. Immunol. 37, 528–539 10.1002/eji.200636090 [DOI] [PubMed] [Google Scholar]
- 25. Oosenbrug T., van de Graaff M. J., Ressing M. E., and van Kasteren S. I. (2017) Chemical tools for studying TLR signaling dynamics. Cell Chem. Biol. 24, 801–812 10.1016/j.chembiol.2017.05.022 [DOI] [PubMed] [Google Scholar]
- 26. O'Neill L. A. J., and Bowie A. G. (2007) The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7, 353–364 10.1038/nri2079 [DOI] [PubMed] [Google Scholar]
- 27. Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., and Chen Z. J. J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 10.1038/35085597 [DOI] [PubMed] [Google Scholar]
- 28. Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., and Akira S. (2003) Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643 10.1126/science.1087262 [DOI] [PubMed] [Google Scholar]
- 29. Kagan J. C., Su T., Horng T., Chow A., Akira S., and Medzhitov R. (2008) TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat. Immunol. 9, 361–368 10.1038/ni1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., and Maniatis T. (2003) IKK ε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- 31. Lin R. T., Heylbroeck C., Pitha P. M., and Hiscott J. (1998) Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell Biol. 18, 2986–2996 10.1128/mcb.18.5.2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barbalat R., Lau L., Locksley R. M., and Barton G. M. (2009) Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat. Immunol. 10, U1200–U1287 10.1038/ni.1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dietrich N., Lienenklaus S., Weiss S., and Gekara N. O. (2010) Murine toll-like receptor 2 activation induces type I interferon responses from endolysosomal compartments. PLoS ONE 5, e10250 10.1371/journal.pone.0010250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoshimura A., Lien E., Ingalls R. R., Tuomanen E., Dziarski R., and Golenbock D. (1999) Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via toll-like receptor 2. J. Immunol. 163, 1–5 [PubMed] [Google Scholar]
- 35. Bieback K., Lien E., Klagge I. M., Avota E., Schneider-Schaulies J., Duprex W. P., Wagner H., Kirschning C. J., Ter Meulen V., and Schneider-Schaulies S. (2002) Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J. Virol. 76, 8729–8736 10.1128/jvi.76.17.8729-8736.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ge Y., Mansell A., Ussher J. E., Brooks A. E., Manning K., Wang C. J., and Taylor J. A. (2013) Rotavirus NSP4 triggers secretion of proinflammatory cytokines from macrophages via Toll-like receptor 2. J. Virol. 87, 11160–11167 10.1128/JVI.03099-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stack J., Doyle S. L., Connolly D. J., Reinert L. S., O'Keeffe K. M., McLoughlin R. M., Paludan S. R., and Bowie A. G. (2014) TRAM is required for TLR2 endosomal signaling to type I IFN induction. J. Immunol. 193, 6090–6102 10.4049/jimmunol.1401605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chanput W., Mes J. J., and Wichers H. J. (2014) THP-1 cell line: an in vitro cell model for immune modulation approach. Int. Immunopharmacol. 23, 37–45 10.1016/j.intimp.2014.08.002 [DOI] [PubMed] [Google Scholar]
- 39. Petes C., Mintsopoulos V., Finnen R. L., Banfield B. W., and Gee K. (2018) The effects of CD14 and IL-27 on induction of endotoxin tolerance in human monocytes and macrophages. J. Biol. Chem. 293, 17631–17645 10.1074/jbc.RA118.003501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsuchiya S., Kobayashi Y., Goto Y., Okumura H., Nakae S., Konno T., and Tada K. (1982) Induction of maturation in cultured human monocytic leukemia-cells by a phorbol diester. Cancer Res. 42, 1530–1536 [PubMed] [Google Scholar]
- 41. Toshchakov V., Jones B. W., Perera P. Y., Thomas K., Cody M. J., Zhang S. L., Williams B. R. G., Major J., Hamilton T. A., Fenton M. J., and Vogel S. N. (2002) TLR4, but not TLR2, mediates IFN-β-induced STAT1 α/β-dependent gene expression in macrophages. Nat. Immunol. 3, 392–398 10.1038/ni774 [DOI] [PubMed] [Google Scholar]
- 42. Andersen J., VanScoy S., Cheng T. F., Gomez D., and Reich N. C. (2008) IRF-3-dependent and augmented target genes during viral infection. Genes Immun. 9, 168–175 10.1038/sj.gene.6364449 [DOI] [PubMed] [Google Scholar]
- 43. Farhat K., Riekenberg S., Heine H., Debarry J., Lang R., Mages J., Buwitt-Beckmann U., Röschmann K., Jung G., Wiesmüller K.-H., and Ulmer A. J. (2008) Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J. Leukocyte Biol. 83, 692–701 10.1189/jlb.0807586 [DOI] [PubMed] [Google Scholar]
- 44. Omueti K. O., Beyer J. M., Johnson C. M., Lyle E. A., and Tapping R. I. (2005) Domain exchange between human Toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. J. Biol. Chem. 280, 36616–36625 10.1074/jbc.M504320200 [DOI] [PubMed] [Google Scholar]
- 45. Honda K., and Taniguchi T. (2006) IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6, 644–658 10.1038/nri1900 [DOI] [PubMed] [Google Scholar]
- 46. Clark K., Takeuchi O., Akira S., and Cohen P. (2011) The TRAF-associated protein TANK facilitates cross-talk within the IκB kinase family during Toll-like receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 108, 17093–17098 10.1073/pnas.1114194108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Panne D., McWhirter S. M., Maniatis T., and Harrison S. C. (2007) Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J. Biol. Chem. 282, 22816–22822 10.1074/jbc.M703019200 [DOI] [PubMed] [Google Scholar]
- 48. Clark K., Peggie M., Plater L., Sorcek R. J., Young E. R. R., Madwed J. B., Hough J., McIver E. G., and Cohen P. (2011) Novel cross-talk within the IKK family controls innate immunity. Biochem. J. 434, 93–104 10.1042/BJ20101701 [DOI] [PubMed] [Google Scholar]
- 49. Musilova J., Mulcahy M. E., Kuijk M. M., McLoughlin R. M., and Bowie A. G. (2019) Toll-like receptor 2-dependent endosomal signaling by Staphylococcus aureus in monocytes induces type I interferon and promotes intracellular survival. J. Biol. Chem. 294, 17031–17042 10.1074/jbc.RA119.009302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Noyce R. S., Collins S. E., and Mossman K. L. (2009) Differential modification of interferon regulatory factor 3 following virus particle entry. J. Virol. 83, 4013–4022 10.1128/JVI.02069-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Troutman T. D., Bazan J. F., and Pasare C. (2012) Toll-like receptors, signaling adapters and regulation of the pro-inflammatory response by PI3K. Cell Cycle 11, 3559–3567 10.4161/cc.21572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Honda K., Takaoka A., and Taniguchi T. (2006) Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 10.1016/j.immuni.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 53. Totzke J., Gurbani D., Raphemot R., Hughes P. F., Bodoor K., Carlson D. A., Loiselle D. R., Bera A. K., Eibschutz L. S., Perkins M. M., Eubanks A. L., Campbell P. L., Fox D. A., Westover K. D., Haystead T. A. J., et al. (2017) Takinib, a selective TAK1 inhibitor, broadens the therapeutic efficacy of TNF-α inhibition for cancer and autoimmune disease. Cell Chem. Biol. 24, 1029–1039.e7 10.1016/j.chembiol.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Płóciennikowska A., Hromada-Judycka A., Borzęcka K., and Kwiatkowska K. (2015) Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell Mol. Life Sci. 72, 557–581 10.1007/s00018-014-1762-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Manukyan M., Triantafilou K., Triantafilou M., Mackie A., Nilsen N., Espevik T., Wiesmüller K.-H., Ulmer A. J., and Heine H. (2005) Binding of lipopeptide to CD14 induces physical proximity of CD14, TLR2 and TLR1. Eur. J. Immunol. 35, 911–921 10.1002/eji.200425336 [DOI] [PubMed] [Google Scholar]
- 56. Watanabe S., Kumazawa Y., and Inoue J. (2013) Liposomal lipopolysaccharide initiates TRIF-dependent signaling pathway independent of CD14. PLoS ONE 8, e60078 10.1371/journal.pone.0060078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marre M. L., Petnicki-Ocwieja T., DeFrancesco A. S., Darcy C. T., and Hu L. T. (2010) Human integrin α3β1 regulates TLR2 recognition of lipopeptides from endosomal compartments. PLoS ONE 5, e12871 10.1371/journal.pone.0012871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dutta D., and Donaldson J. G. (2012) Search for inhibitors of endocytosis: intended specificity and unintended consequences. Cell. Logist. 2, 203–208 10.4161/cl.23967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoshimori T., Yamamoto A., Moriyama Y., Futai M., and Tashiro Y. (1991) Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266, 17707–17712 [PubMed] [Google Scholar]
- 60. Hoebe K., Georgel P., Rutschmann S., Du X., Mudd S., Crozat K., Sovath S., Shamel L., Hartung T., Zähringer U., and Beutler B. (2005) CD36 is a sensor of diacylglycerides. Nature 433, 523–527 10.1038/nature03253 [DOI] [PubMed] [Google Scholar]
- 61. Nilsen N. J., Deininger S., Nonstad U., Skjeldal F., Husebye H., Rodionov D., von Aulock S., Hartung T., Lien E., Bakke O., and Espevik T. (2008) Cellular trafficking of lipoteichoic acid and Toll-like receptor 2 in relation to signaling; role of CD14 and CD36. J. Leukocyte Biol. 84, 280–291 10.1189/jlb.0907656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Raieli S., Trichot C., Korniotis S., Pattarini L., and Soumelis V. (2019) TLR1/2 orchestrate human plasmacytoid predendritic cell response to gram+ bacteria. PLoS Biol. 17, e3000209 10.1371/journal.pbio.3000209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yamamoto M., Sato S., Hemmi H., Uematsu S., Hoshino K., Kaisho T., Takeuchi O., Takeda K., and Akira S. (2003) TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 4, 1144–1150 10.1038/ni986 [DOI] [PubMed] [Google Scholar]
- 64. Kawai T., Takeuchi O., Fujita T., Inoue J., Mühlradt P. F., Sato S., Hoshino K., and Akira S. (2001) Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol. 167, 5887–5894 10.4049/jimmunol.167.10.5887 [DOI] [PubMed] [Google Scholar]
- 65. Kawai T., and Akira S. (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 66. Nilsen N. J., Vladimer G. I., Stenvik J., Orning M. P. A., Zeid-Kilani M. V., Bugge M., Bergstroem B., Conlon J., Husebye H., Hise A. G., Fitzgerald K. A., Espevik T., and Lien E. (2015) A role for the adaptor proteins TRAM and TRIF in Toll-like receptor 2 signaling. J. Biol. Chem. 290, 3209–3222 10.1074/jbc.M114.593426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Uematsu S., and Akira S. (2007) Toll-like receptors and type I interferons. J. Biol. Chem. 282, 15319–15323 10.1074/jbc.R700009200 [DOI] [PubMed] [Google Scholar]
- 68. Li P., Wong J. J. Y., Sum C., Sin W. X., Ng K. Q., Koh M. B. C., and Chin K. C. (2011) IRF8 and IRF3 cooperatively regulate rapid interferon-β induction in human blood monocytes. Blood 117, 2847–2854 10.1182/blood-2010-07-294272 [DOI] [PubMed] [Google Scholar]
- 69. Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., and Taniguchi T. (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- 70. Park J. H., Ko R., and Lee S. Y. (2017) Reciprocal regulation of TLR2-mediated IFN-β production by SHP2 and Gsk3β. Sci. Rep. 7, 6807 10.1038/s41598-017-07316-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mancino A., Termanini A., Barozzi I., Ghisletti S., Ostuni R., Prosperini E., Ozato K., and Natoli G. (2015) A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev. 29, 394–408 10.1101/gad.257592.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhao B. Y., Shu C., Gao X. S., Sankaran B., Du F. L., Shelton C. L., Herr A. B., Ji J. Y., and Li P. W. (2016) Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 113, E3403–E3412 10.1073/pnas.1603269113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu S. Q., Cai X., Wu J. X., Cong Q., Chen X., Li T., Du F. H., Ren J. Y., Wu Y. T., Grishin N. V., and Chen Z. J. J. (2015) Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- 74. Shu C., Sankaran B., Chaton C. T., Herr A. B., Mishra A., Peng J. M., and Li P. W. (2013) Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure 21, 1137–1148 10.1016/j.str.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang J. M., Basagoudanavar S. H., Wang X. Y., Hopewell E., Albrecht R., García-Sastre A., Balachandran S., and Beg A. A. (2010) NF-κB RelA subunit is crucial for early IFN-β expression and resistance to RNA virus replication. J. Immunol. 185, 1720–1729 10.4049/jimmunol.1000114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Petnicki-Ocwieja T., Kern A., Killpack T. L., Bunnell S. C., and Hu L. T. (2015) Adaptor protein-3-mediated trafficking of TLR2 ligands controls specificity of inflammatory responses but not adaptor complex assembly. J. Immunol. 195, 4331–4340 10.4049/jimmunol.1501268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gessani S., Testa U., Varano B., Dimarzio P., Borghi P., Conti L., Barberi T., Tritarelli E., Martucci R., Seripa D., Peschle C., and Belardelli F. (1993) Enhanced production of LPS-induced cytokines during differentiation of human monocytes to macrophages: role of LPS receptors. J. Immunol. 151, 3758–3766 [PubMed] [Google Scholar]
- 78. Kato A., Ogasawara T., Homma T., Saito H., and Matsumoto K. (2004) Lipopolysaccharide-binding protein critically regulates lipopolysaccharide-induced IFN-β signaling pathway in human monocytes. J. Immunol. 172, 6185–6194 10.4049/jimmunol.172.10.6185 [DOI] [PubMed] [Google Scholar]
- 79. Auerbuch V., Brockstedt D. G., Meyer-Morse N., O'Riordan M., and Portnoy D. A. (2004) Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 200, 527–533 10.1084/jem.20040976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Davidson S., Maini M. K., and Wack A. (2015) Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J. Interferon Cytokine Res. 35, 252–264 10.1089/jir.2014.0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. González-Navajas J. M., Lee J., David M., and Raz E. (2012) Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 12, 125–135 10.1038/nri3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gram A. M., Sun C. L., Landman S. L., Oosenbrug T., Koppejan H. J., Kwakkenbos M. J., Hoeben R. C., Paludan S. R., and Ressing M. E. (2017) Human B cells fail to secrete type I interferons upon cytoplasmic DNA exposure. Mol. Immunol. 91, 225–237 10.1016/j.molimm.2017.08.025 [DOI] [PubMed] [Google Scholar]
- 83. Hida T., Hayashi K., Yukishige K., Tanida S., Kawamura N., and Harada S. (1995) Synthesis and biological activities of TAN-1511 analogues. J. Antibiot. (Tokyo) 48, 589–603 10.7164/antibiotics.48.589 [DOI] [PubMed] [Google Scholar]
- 84. van der Linden W. A., Li N., Hoogendoorn S., Ruben M., Verdoes M., Guo J., Boons G. J., van der Marel G. A., Florea B. I., and Overkleeft H. S. (2012) Two-step bioorthogonal activity-based proteasome profiling using copper-free click reagents: a comparative study. Bioorg. Med. Chem. 20, 662–666 10.1016/j.bmc.2011.06.037 [DOI] [PubMed] [Google Scholar]
- 85. Schultz M. K., Parameswarappa S. G., and Pigge F. C. (2010) Synthesis of a DOTA-Biotin conjugate for radionuclide chelation via Cu-free click chemistry. Org. Lett. 12, 2398–2401 10.1021/ol100774p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−ΔΔC) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 87. Korbee C. J., Heemskerk M. T., Kocev D., van Strijen E., Rabiee O., Franken K. L. M. C., Wilson L., Savage N. D. L., Džeroski S., Haks M. C., and Ottenhoff T. H. M. (2018) Combined chemical genetics and data-driven bioinformatics approach identifies receptor tyrosine kinase inhibitors as host-directed antimicrobials. Nat. Commun. 9, 358 10.1038/s41467-017-02777-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in this article are available from the corresponding author (Maaike E. Ressing, m.e.ressing@lumc.nl) upon reasonable request.