Abstract
Among the multiple antiviral defense mechanisms found in prokaryotes, CRISPR-Cas systems stand out as the only known RNA-programmed pathways for detecting and destroying bacteriophages and plasmids. Class 1 CRISPR-Cas systems, the most widespread and diverse of these adaptive immune systems, use an RNA-guided multiprotein complex to find foreign nucleic acids and trigger their destruction. In this review, we describe how these multisubunit complexes target and cleave DNA and RNA and how regulatory molecules control their activities. We also highlight similarities to and differences from Class 2 CRISPR-Cas systems, which use a single-protein effector, as well as other types of bacterial and eukaryotic immune systems. We summarize current applications of the Class 1 CRISPR-Cas systems for DNA/RNA modification, control of gene expression, and nucleic acid detection.
Keywords: CRISPR-Cas, crRNA, DNA endonuclease, ribonuclease, enzyme mechanism, bacteriophage, cyclic nucleotide, gene regulation, bacteria, archaea, prokaryotic adaptive immunity, genome editing
All cells must defend against infection by harmful genetic elements, like viruses or transposons. Prokaryotes use a multitude of different strategies to combat their viruses, which are called phages. These include, but are not limited to, adsorption and injection blocking, abortive infection, toxin-antitoxin, restriction-modification, and CRISPR-Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated) systems (1). CRISPR-Cas loci constitute the only known adaptive immune system in bacteria and archaea. They typically include an array of repeat sequences (CRISPRs) with intervening “spacers,” matching sequences of DNA or RNA from viruses or other mobile genetic elements, and a set of genes encoding Cas proteins (Fig. 1A). Transcription across the CRISPR array produces a precursor crRNA (pre-crRNA) that is processed by nucleases into small, noncoding CRISPR RNAs (crRNAs) (Fig. 1B). Each crRNA molecule assembles with one or more Cas proteins into an effector complex that binds crRNA-complementary regions in foreign DNA or RNA (Fig. 1, C–E). The effector complex then triggers degradation of the targeted DNA or RNA using either an intrinsic nuclease activity or a separate nuclease in trans (Fig. 1, A and C–E).
Figure 1.
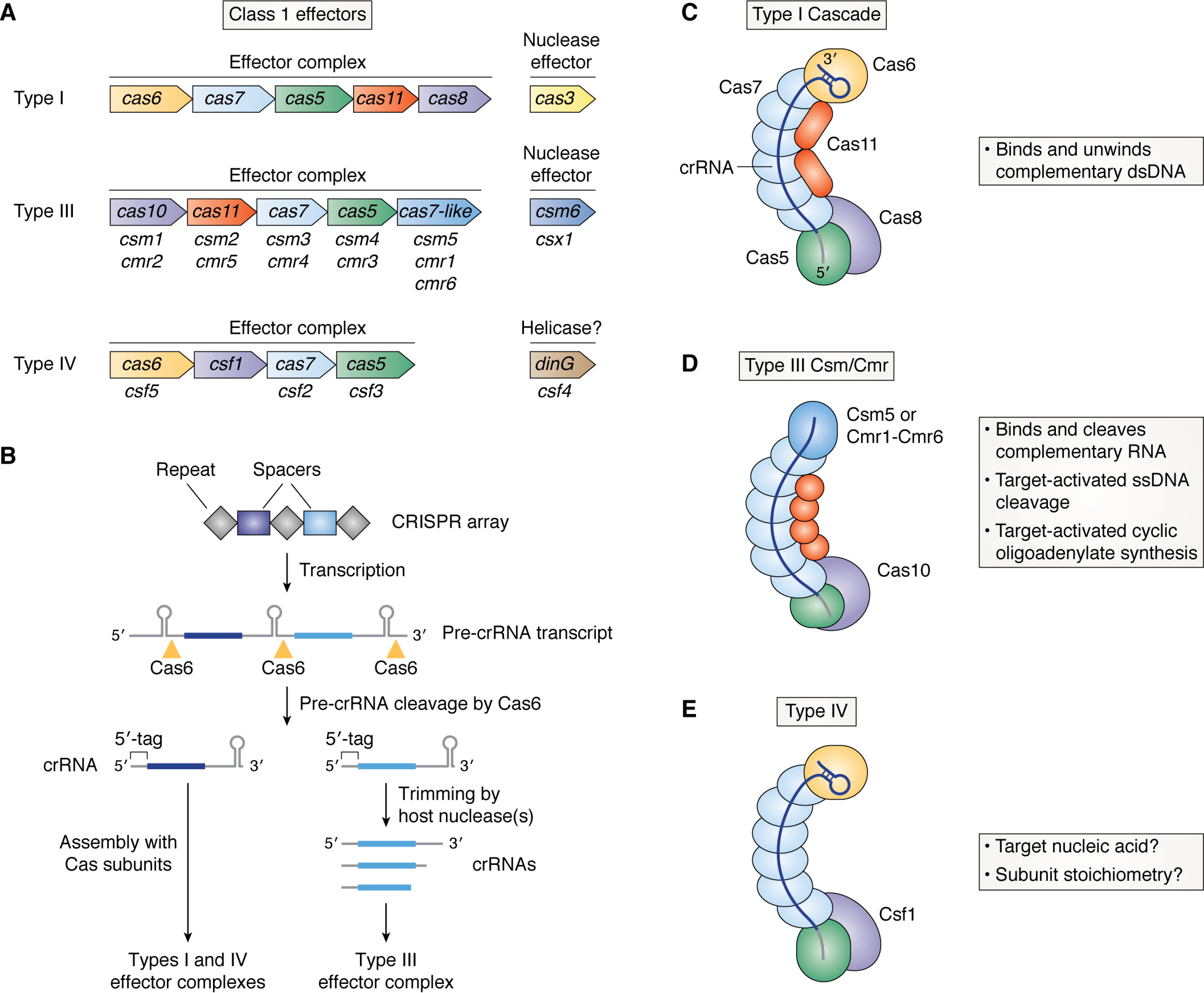
Effector complex architecture and crRNA biogenesis in Class 1 CRISPR-Cas systems. A, Cas protein composition of Class 1 effector complexes and their associated nuclease effectors. Subunits that are analogous between the different types are shown with the same color. Type III– and IV–specific names for Cas7, Cas5, Cas6, and Cas11 subunits are also shown below the canonical subunit names. B, biogenesis of Class 1 crRNAs. Transcription across a CRISPR array (repeat sequences are shown as gray diamonds, and unique spacers are shown as dark and light blue rectangles) leads to production of a pre-crRNA transcript that is then cleaved by Cas6 into individual guide molecules. Each crRNA has a 5′-tag that is derived from the repeat sequence. Individual guides are then directly incorporated into Type I and IV complexes or trimmed at their 3′ end by host nucleases before assembly with Type III effector subunits. C, architecture and enzymatic activities of the Type I crRNA-guided effector complex, Cascade. Subunits are shown with the same color scheme as in A. The crRNA is shown with the same color scheme as in B. D, as in C but for the Type III effector complex, Csm (subtypes III-A/D/E/F) or Cmr (subtypes III-B/C). Subunits unique to Type III systems (Cas10 and either Csm5 or Cmr1/6) are labeled. E, as in C but for the Type IV effector complex. The subunit unique to the Type IV system (Csf1) is labeled. Type IV complexes contain a crRNA assembled with Cas7, Cas6, and Csf1, but their enzymatic activity, precise stoichiometry, and structure are not yet known.
CRISPR-Cas systems have been classified into two groups comprising three types each (Class 1 includes Types I, III, and IV; Class 2 includes Types II, V, and VI) (2). Class 1 systems use multisubunit complexes that contain multiple different Cas proteins, whereas Class 2 effectors contain only a single protein (Fig. 1, C–E) (2). To date, much attention has focused on the mechanism of Class 2 effectors, such as Cas9, Cas12, and Cas13, given their practical applications in genome editing and manipulation (3, 4). Class 1 systems, although less well-studied, are far more abundant in nature, comprising about 90% of CRISPR-Cas systems in bacteria and archaea (2, 5, 6). They are also present across diverse bacterial and archaeal phyla and likely evolved earlier than Class 2 systems (2). Class 1 CRISPR-Cas systems harbor a number of different enzymatic activities, including cleavage of dsDNA, ssDNA, and RNA, and synthesis of a second messenger molecule, cyclic oligoadenylate (cOA). These functions could be harnessed for genome or transcriptome manipulation and control of cellular outcomes. Here, we review the interference mechanisms of effector complexes from Class 1 systems and their regulation, focusing on new paradigms of adaptive immunity from recent studies of Type I and III systems, and emerging applications of these systems in genome and transcriptome engineering.
Class 1 surveillance complex architecture and crRNA biogenesis
Surveillance complex architecture and activity
Class 1 systems use RNA-guided surveillance complexes that are composed of multiple different subunits assembled around a single crRNA molecule (2). Type I systems are the most abundant, and they include nine different subtypes (I-A to I-E, I-F1, I-F2, I-F3, and I-G) (2). Structures of Type I “Cascade” (CRISPR-associated complex for antiviral defense) complexes show that they adopt a seahorse-like architecture (7). They typically contain a single copy of Cas8, the large subunit, and Cas5 at the 5′ end of the crRNA (“foot”), a helical “backbone” filament composed of Cas7 subunits that assembles along the crRNA spacer region, a “belly” filament composed of Cas11 subunits, and a Cas6 subunit that binds to the 3′ end of the crRNA (“head”) and caps the backbone (Fig. 1C) (8–10). Structural studies of Type III effector complexes, “Csm” (Cas subtype Mtube; subtypes III-A/D/E/F) and “Cmr” (Cas module RAMP; subtypes III-B/C), indicate that they adopt a similar architecture but have a more extended, wormlike shape (11–13). Type III complexes also include Cas10 as the large subunit, instead of Cas8 (Fig. 1D). Although Cas10 and Cas8 occupy similar positions in the complex, they are highly divergent by amino acid sequence and play different roles in target interference (2). Another notable difference between Type I and III complexes is that Type III complexes lack Cas6, instead employing specialized Cas7-like subunits (Csm5 in Type III-A and Cmr1 and Cmr6 in Type III-B) to bind the 3′ end of the crRNA (Fig. 1D) (11–14). Type IV CRISPR-Cas complexes, which include three subtypes (IV-A to IV-C), have a unique large subunit, Csf1, as well as subunits homologous to Cas7, Cas6, and Cas5 (Fig. 1E) (15, 16). The subunit assembly and architecture of Type IV complexes is not clear, as structures of the entire complex have not yet been determined.
Homology between Cas7, Cas6, and Cas5 and the similarity of complex architectures across Class 1 systems point to a common evolutionary origin for these effector complexes. Both Type I and III systems also have Cas11, known as the “small subunit.” Cas11 proteins do not share significant sequence similarity across types, but they exhibit structural homology and occupy analogous positions in Type I and III complexes (Fig. 1, C and D) (2, 7, 17). The divergence of large subunits, Cas10 and Cas8, on the other hand, suggests that they may be under greater evolutionary pressure from phage counterdefense strategies. This is consistent with the roles that Cas8 and Cas10 play in activating and regulating immunity, which we will discuss in subsequent sections of this review.
Several subtypes of Type I CRISPR-Cas also lack genes encoding for the large and/or small subunits (2). In these systems, other subunits typically take over the functional roles and positions of Cas8 and/or Cas11 in the complex. For example, in the Type I-F2 system from Shewanella putrefaciens, Cas5 and Cas7 substitute for the lack of Cas8 and Cas11, respectively (18). In addition, many Class 1 complexes include fusions of subunits into a single protein (2). The newly identified Type III-E locus lacks the large subunit, Cas10, and encodes a predicted fusion of Cas7 with Cas11 (2). Whether one of the subunits replaces Cas10, or if this system has a novel function compared with other Type III systems, is not known. Future biochemical and structural studies of diverse Type I and III variants will likely uncover unexpected functional and structural versatility of Class 1 CRISPR-Cas complexes and identify minimal complexes that could be more easily expressed and assembled in heterologous systems for genome engineering.
Class 1 CRISPR-Cas systems also encode for effector nucleases and/or helicases that cooperate with the surveillance complex for RNA-guided immunity (Fig. 1A). The Type I Cascade has no intrinsic enzymatic activity and relies on recruitment of Cas3, a helicase-nuclease, to degrade dsDNA in trans (Fig. 1A) (19, 20). Type III CRISPR-Cas Csm/Cmr possess intrinsic DNase and RNase activities but also synthesize a second messenger, cOA, which binds and stimulates RNA cleavage by Csm6/Csx1, a separate nuclease effector (Fig. 1A) (21). Type IV systems are associated with Csf4, a DinG family helicase (Fig. 1A) (2). Csf4 is required for in vivo plasmid interference by Type IV systems, but its role in the process is unclear (22). These “partner” enzymes illustrate how a common RNA-guided surveillance complex architecture can be adapted to perform diverse functions.
crRNA biogenesis
In contrast to the CRISPR-Cas9 effector, which requires both a crRNA and a tracrRNA (trans-acting crRNA) for activity, all Class 1 complexes contain only a single crRNA molecule (2, 23). Processing of the crRNA in Class 1 systems typically requires the Cas6 RNase, which cuts pre-crRNA transcripts into individual crRNA molecules containing a repeat-derived 5′-tag, a spacer region, and a 3′-stem loop hairpin (Fig. 1B). In Type I and IV systems, the crRNA retains the 3′ hairpin structure (Fig. 1B) (15, 16). In Type III systems, host nucleases trim the 3′ end to variable lengths corresponding to the number of Cas7 subunits in the complex (Fig. 1B) (24–27). Genetic and biochemical studies of a Type III-A system from Staphylococcus epidermidis suggest that Csm5, the subunit that caps the Cas7 helical filament, recruits polynucleotide phosphorylase (PNP), to trim the exposed 3′ end of the crRNA (28, 29). However, deletion of the pnp gene did not result in complete loss of mature, trimmed crRNAs, suggesting that other host RNases may also contribute to processing (29). A recent study also showed that a Type III-Bv system, which lacks Cas6, uses a host RNase E enzyme for crRNA maturation (30). This highlights the importance of studying different subtypes to understand how Class 1 crRNAs are specifically assembled with Cas proteins into RNA-guided effector complexes. Understanding the minimal requirements for guide maturation and complex assembly would also facilitate introduction of these complexes into eukaryotic cells for DNA and RNA detection and editing.
Class 1 CRISPR-Cas interference
Type I and III CRISPR-Cas systems together comprise the most abundant types of CRISPR-Cas systems and encompass diverse subtypes (2, 5). In this section, we review recent advances in our mechanistic understanding of nucleic acid targeting by Type I and III surveillance complexes and their modes of regulation.
DNA targeting and regulation by type I CRISPR-Cas systems
Type I CRISPR-Cas systems target homologous regions of dsDNA in phages or plasmids for degradation (2). The overall mechanism of targeting involves two major steps—recognition of a complementary target in foreign DNA by the surveillance complex and cleavage of the target by Cas3, a protein with an SF2 (Superfamily 2) helicase and HD (histidine-aspartate) nuclease domain, that is recruited in trans (31–33). Target recognition requires complementarity between the crRNA and the target, as well as the presence of a protospacer-adjacent motif (PAM), which allows the host to avoid self-immunity (34).
RNA-guided DNA binding and cleavage by type I CRISPR-Cas systems
During an infection, the Type I complex first scans the viral genome for the PAM, a 2–5-bp motif flanking the target sequence (Fig. 2A) (34). Because the PAM is not present in the repeat sequences flanking the CRISPR spacers, this protects the host's own DNA from being targeted for degradation (35). The mechanism of PAM recognition has been most well-studied for the Type I-E Cascade complex from Escherichia coli, which recognizes a trinucleotide 5′-A-(T/C/A)-G-3′ (36). The PAM is recognized in a double-stranded form through minor groove contacts with a lysine finger, glutamine wedge, and glycine loop in Cas8 (also known as Cse1 in Type I-E systems) (36). Local bending of the DNA combined with insertion of the wedge motif into the DNA duplex following the PAM initiates DNA unwinding (36, 37). Wedge and loop motifs are functionally and structurally conserved across different Type I systems, but sequence variability enables recognition of distinct PAM sequences (36–39). Some Type I complexes have also evolved to use other subunits for PAM recognition. The Type I-F2 complex, which lacks Cas8, uses Cas5 to recognize a 5′-GG-3′ PAM through major groove interactions (18). Studies of different Type I variants will likely reveal further diversity in the mechanisms and protein motifs used to recognize PAM sequences. This information could be used to engineer more flexible PAM recognition or nearly PAM-less variants of Type I effector complexes for genome engineering, similar to those that have been developed for Cas9 (40, 41).
Figure 2.
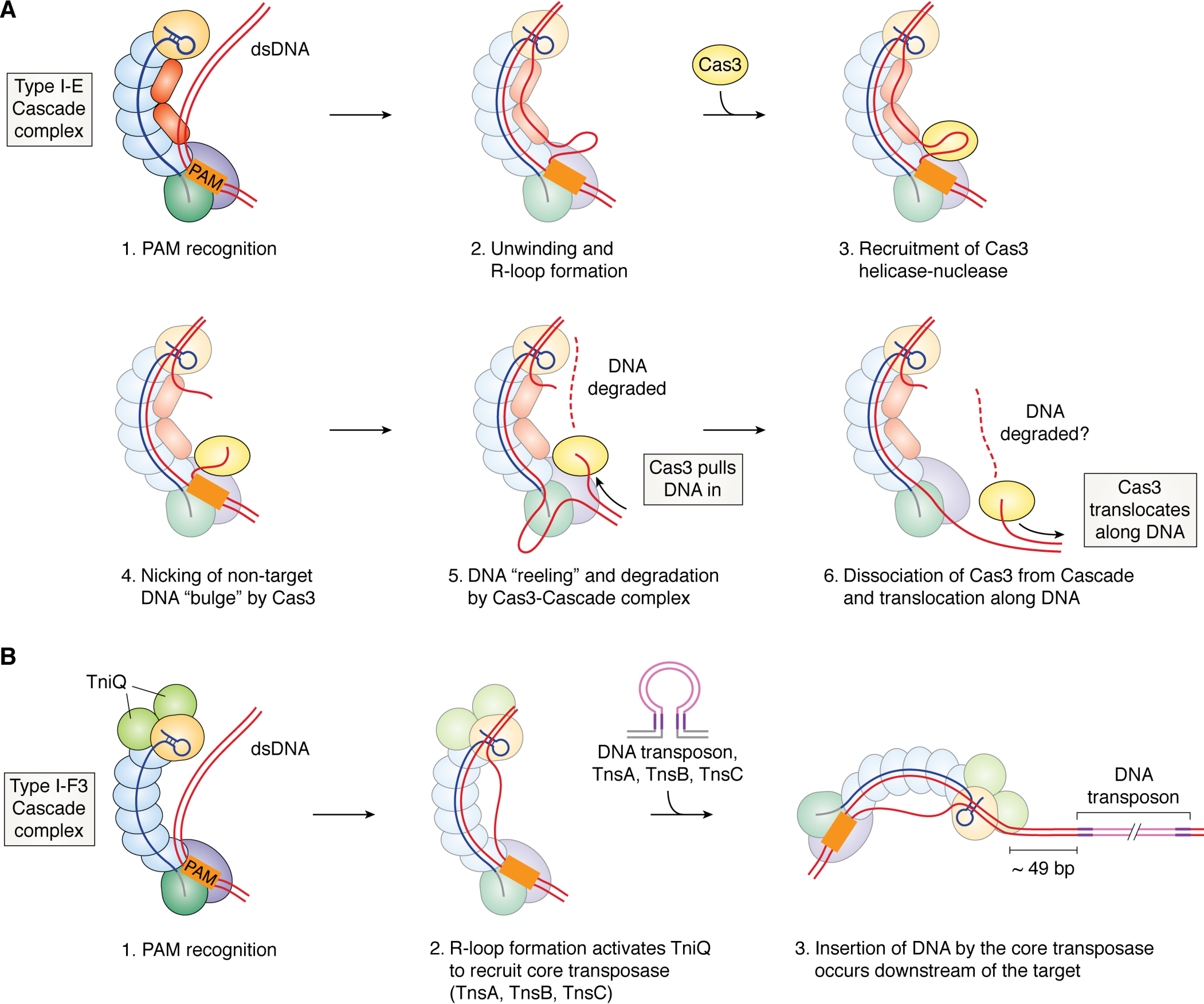
Type I CRISPR-Cas interference mechanism. A, target binding and degradation by the Type I-E Cascade complex. 1, recognition of the PAM (shown as an orange rectangle) by Cas8 leads to DNA bending and initiation of unwinding; 2, hybridization of the crRNA with the target strand of DNA leads to displacement of the nontarget DNA strand and formation of an R-loop; 3, a conformational change in Cascade that accompanies R-loop formation leads to recruitment of Cas3, a helicase-nuclease protein, to a small bulge in the nontarget strand; 4, the nontarget strand bulge is cleaved by Cas3, and the ssDNA is loaded into the helicase; 5, Cas3's helicase domain unwinds DNA upstream of the PAM, and “reels in” ssDNA toward its nuclease active site; 6, Cas3 dissociates from Cascade and continues to translocate; whether or not degradation occurs during translocation is unclear. B, cooperation of Type I-F3 Cascade with a Tn7 transposase to mediate RNA-guided DNA insertion. Recognition of the PAM (1) and base-pairing with a complementary DNA sequence (2) leads to R-loop formation, which activates the TniQ dimer to recruit the core transposase (TnsA, TnsB, TnsC) and transposon DNA with paired ends (violet lines) to the Cascade-bound target site. 3, integration of the transposon's DNA cargo (pink lines) occurs ∼49 bp downstream of the DNA target. In all panels, individual Cas proteins are colored as in Fig. 1, but with colors muted for greater clarity of the nucleic acid strands.
Once DNA unwinding initiates at the PAM, crRNA hybridization with the target strand of DNA leads to displacement of the nontarget DNA strand, forming a three-stranded nucleic acid structure known as an R-loop (Fig. 2A) (42, 43). Structural, biochemical, and single-molecule experiments on purified Type I complexes have led to a detailed understanding of R-loop formation. Complementarity at a seed region (positions 1–5 and 7–8 following the PAM) is required for target binding and interference (37, 44). In the Thermobifida fusca Type I-E Cascade, binding of the PAM first leads to bending of DNA and unwinding of an ∼11-nucleotide “seed loop” intermediate (37). Further base-pairing along the crRNA then expands the seed loop into a full R-loop, which is then locked in place by interactions with Cas7 and/or Cas11 (37, 45). In Type I-E Cascade, positively charged residues on the surface of Cas8 and Cas5 guide the displaced nontarget strand away from target strand toward the back of the Cas11 subunits, where it is locked (Fig. 2A) (18, 36–38). In Type I-F2 complexes, which lack Cas11 and Cas8, the nontarget strand winds through a positively charged “trench” formed by Cas5 and Cas7 subunits (18, 37). The Cas7 “backbone” and Cas11 “belly” filaments are also involved in “locking” the target DNA once it hybridizes with the crRNA (18, 37, 38). Single-molecule studies indicate that R-loop formation serves as a step for rejecting off-target DNA, as mismatches between the crRNA and the target increase the likelihood of R-loop collapse before it reaches the locked state (45). Bacterial RNA polymerases, which unwind DNA without energy input, also bend the path of the DNA duplex, suggesting that this may be a conserved mechanism to facilitate DNA unwinding (46, 47). Studies of DNA unwinding and R-loop stabilization in subtypes of Type I systems may reveal further similarities with other protein families that unwind or bend DNA, including helicases, RNA polymerases, transcription factors, and RecA.
Formation of the R-loop induces a conformational change in the complex that enables recruitment of Cas3, a helicase-nuclease protein that is required for target DNA degradation (Fig. 2A) (19, 20, 48). Current evidence supports a model in which Cas3 nicks the DNA at the R-loop, loads onto the ssDNA, and processively unwinds and degrades DNA in a unidirectional and ATP-dependent manner (Fig. 2A) (19, 33, 49). Structures of T. fusca Cascade bound to a DNA target and Cas3 revealed that a protruding “bubble” in the nontarget strand of the R-loop is required for Cas3 to nick the DNA (Fig. 2A) (50). Cascade bound to a partial R-loop lacking the protruding bubble could recruit Cas3, but it did not induce DNA cleavage by Cas3 (50, 51). Single-molecule FRET and bulk fluorescence experiments indicate that after ssDNA loading, Cas3 first stays associated with Cascade and cleaves ssDNA by a “reeling” mechanism; in this model, Cas3 uses its SF2 helicase domain to repeatedly pull in and present ssDNA to its HD nuclease domain for cleavage (Fig. 2A) (52–54). Previous studies also showed that Cas3 can break free of Cascade and translocate on its own, but no evidence of DNA degradation during translocation was observed (Fig. 2A) (53). Thus, it is unclear whether Cas3 would degrade DNA during translocation. In addition, whereas structural and biochemical studies have shed light on how Cas3 degrades the nontarget strand, how it nicks and degrades the target strand of DNA is less well-understood. One possibility is that once the nontarget strand has been degraded, the exposed ssDNA of the target strand would become a substrate for a second Cas3 molecule to nick and degrade.
Anti-CRISPR inhibition of type I CRISPR-Cas effector complexes
Several phage and prophage genomes encode for “anti-CRISPRs,” small proteins (∼50–300 amino acids) that inhibit CRISPR-mediated immunity (55). Identification and characterization of anti-CRISPRs that inhibit Type I-E or I-F systems show that they block DNA interference using diverse mechanisms (55). Some anti-CRISPRs mimic duplex DNA or induce a conformational change in Cas8 to interfere with PAM binding (9, 38). Structural studies of Type I-F anti-CRISPRs indicate that some can bind to the Cas7 backbone or the crRNA to prevent crRNA:ssDNA base-pairing and R-loop formation (9, 38). In addition, Type I-E and I-F anti-CRISPRs, AcrIE1 and AcrIF3, bind and inhibit the recruitment of Cas3 by Cascade for DNA cleavage (56–58). Structural comparison of AcrIF3 with Cas8 revealed that AcrIF3 resembles a helical bundle in Cas8 that binds Cas3, which indicates that mimicry of host proteins could be a common strategy for phages to evade CRISPR-Cas interference (48). Binding of the Type I-F anti-CRISPR, AcrIF9, to the surveillance complex also induces nonspecific DNA binding, which could sequester the complex away from its target (59, 60). These discoveries highlight not only the diversity of anti-CRISPRs but also the importance of PAM recognition, R-loop formation, and DNA cleavage in Type I CRISPR-Cas immunity. The steps at which anti-CRISPRs inhibit Class 1 complexes are analogous to steps at which anti-CRISPRs inhibit DNA-targeting Class 2 systems, Cas9 and Cas12, illustrating a remarkable evolutionary convergence of counterdefense strategies by phages (61, 62). Further discovery and characterization of anti-CRISPRs against other Type I subtypes could reveal new insights into the host-virus evolutionary arms race and lead to new strategies to control Type I CRISPR-Cas effectors for genome manipulation.
RNA-guided DNA transposition by type I CRISPR-Cas systems
Bioinformatic analyses revealed that some Type I-B and I-F systems lacking Cas3 have been co-opted by mobile genetic elements (63). The transposons in which these systems are found also lack a key protein involved in directing site-specific transposition (63). Thus, it was hypothesized that the Type I effector in these systems used crRNAs to direct DNA insertion by the transposase to specific sites (63). This concept was recently demonstrated by experiments showing that a transposon-encoded Type I-F effector complex from Vibrio cholerae can mediate targeted insertion of cargo DNA sequences when expressed in E. coli (Fig. 2B) (64). Biochemical and genetic experiments indicate that the Type I-F effector specifically interacts directly with TniQ, a transposition protein, and that this interaction is required for RNA-guided transposition (64, 65). Cryo-EM structures also showed that the Type I-F complex associates with a dimer of TniQ through contacts with Cas6 and a Cas7 subunit at the 3′ end of the crRNA (65). Interestingly, RNA-guided transposition is sensitive to mismatches not only in the PAM-proximal seed region, but also in a four-nucleotide region near the TniQ-binding site (64, 65). Further structural and biochemical studies are needed to determine how target DNA binding and unwinding leads to recruitment of the core transposase, comprising TnsA, TnsB, and TnsC, for RNA-guided DNA insertion (Fig. 2B). Such studies could facilitate the targeted insertion of large DNA elements into the genomes of microbes and eukaryotic cells without requiring homologous recombination, which is often inefficient and only occurs in dividing cells.
Dual DNA and RNA targeting by type III CRISPR-Cas systems
Type III CRISPR-Cas systems are the most evolutionarily ancient CRISPR-Cas systems and are widespread across bacteria and archaea (66). Their effector complexes include enzymatic domains that cleave RNA and ssDNA and that synthesize second messenger molecules to activate antiviral nucleases in trans (Fig. 3A). Their effector complexes coordinate a sophisticated, multipronged defense against invasive genetic elements, including DNA and RNA phages, plasmids, and jumbo phages (67–71). They are divided into six subtypes (III-A to III-F), but the most common are Type III-A and III-B systems (2, 5). The study of these systems has offered insights into the evolutionary origins of CRISPR-Cas immunity and surprising parallels with other prokaryotic and eukaryotic immune systems.
Figure 3.
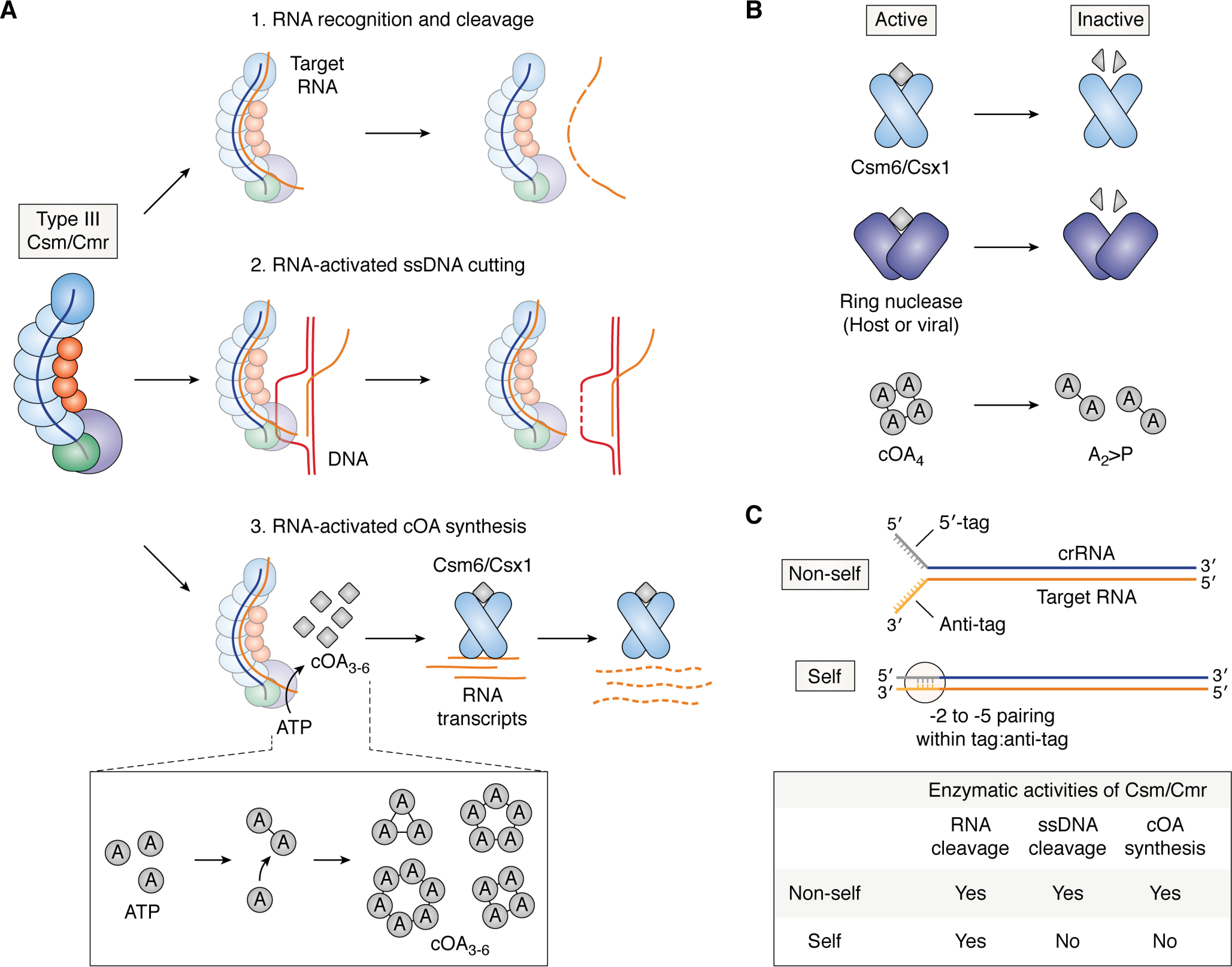
Type III CRISPR-Cas interference mechanism. A, multipronged interference mechanism of Type III effector complexes. Type III Csm or Cmr complexes recognize and degrade complementary target RNA (orange lines) (1), cleave ssDNA (red lines) nonspecifically when bound to an RNA target, possibly at exposed R-loops (2), and synthesize a second messenger molecule, cyclic oligoadenylate (cOA, gray diamonds), when bound to an RNA target (3). cOA binds to and activates an accessory nuclease, Csm6 or Csx1, to nonspecifically cleave host and viral transcripts. The bottom inset shows the mechanism of cOA synthesis by Cas10. ATP molecules (gray circles labeled with A) are polymerized into linear oligoadenylates with 3′,5′-phosphodiester linkages. This is then followed by ring closure to produce cOA with 3–6 AMPs per ring (cOA3-6). cOA4 and cOA6 molecules bind Csm6/Csx1 and activate its RNase activity. B, cOA signaling is regulated by Csm6/Csx1 (top row) and dedicated ring nucleases (middle row), which use their CARF domains to degrade cOA4 or cOA6. Ring nucleases may be derived from the host (e.g. Crn-1) or from phages (e.g. AcrIII-1). The bottom row illustrates how cOA4 is degraded by ring nucleases or Csm6/Csx1 into linear diadenylates with 2′,3′-cyclic phosphates (A2>P). cOA6 is degraded in a similar manner. C, self versus nonself discrimination in Type III CRISPR-Cas systems depends on 5′-tag:anti-tag complementarity. Base-pairing at the −2 to −5 positions (+1 is the first nucleotide of the spacer region) inhibits ssDNA cleavage and cOA synthesis, but not complementary RNA degradation by Csm/Cmr. In all panels, Cas proteins are colored as in Fig. 1 but with colors muted for clarity of nucleic acid substrates.
RNA-guided RNA cleavage by type III CRISPR-Cas complexes
Type III CRISPR-Cas effector complexes recognize RNA through base-pairing interactions with their crRNAs and cleave it using their Cas7-like subunits, Csm3 (III-A/D/E/F) or Cmr4 (III-B/C) (Fig. 3A). As in Type I complexes, the crRNA is presented by the effector complex in discontinuous segments for base-pairing with the target nucleic acid (11–13, 72). In addition, each Csm3/Cmr4 inserts a “finger” loop into the duplex, flipping out every 6th base pair (11–14). This closely resembles the mechanism of ssDNA binding in the R-loops of Type I effector complexes (10, 38). Type III effector complexes do not have a clear seed region for RNA binding, unlike Type I Cascade and other RNA-guided RNA nucleases, like Argonaute and CRISPR-Cas13 (73, 74). Each segment of target RNA is recognized and cleaved independently by Csm3 or Cmr4; crRNA:target mismatches or deoxynucleotide modifications that disrupt cleavage at one site do not inhibit cleavage at other sites (25, 75). Complete guide:target complementarity is not required for RNA cleavage, although reduced base-pairing results in a slower rate of cleavage (68, 76). A seed or “target capture” motif has been reported at the 5′ end of the target in Type III-B Cmr, but truncation of this region did not entirely inhibit RNA binding or cleavage (77, 78).
Unlike in Type I effectors, the Cas7-like subunits of Type III effector complexes (Csm3/Cmr4) are catalytically active. Cleavage requires a conserved Asp residue in Csm3/Cmr4 and occurs 3′ to every flipped base, resulting in a characteristic six-nucleotide cleavage periodicity (11–14, 72). RNA cleavage requires a 2′-OH in the target RNA for cleavage, and the resulting termini of the reaction products have a 5′-OH and either a 3′-phosphate or 2′,3′-cyclic phosphate (14, 79). This suggests a metal-independent cleavage mechanism, but experiments show that divalent metal ions are required for RNA cleavage (24–26, 68, 77, 79, 80). Atomic-resolution structures of Type III complexes show how RNA targets are positioned prior to cleavage, but the use of active-site mutants or a noncleavable ssDNA target in these structures has prevented determination of the cleavage mechanism (12–14). Thus, structural studies of the complex in cleavage-competent and post-cleavage states will be required to understand the catalytic mechanism of RNA cleavage by Type III effectors.
RNA-activated DNA cleavage by type III CRISPR-Cas effector complexes
Biochemical experiments showed that Csm and Cmr recognize and cleave complementary single-stranded RNA in vitro (Fig. 3A) (81). However, Type III-A and III-B CRISPR-Cas systems exhibit transcription-dependent DNA targeting in vivo (82, 83). This was a puzzle, until the discovery that recognition of RNA allosterically activates a latent ssDNA endonuclease activity in Cas10, the large subunit (Fig. 3A) (26, 75, 84–86). DNA cleavage is catalyzed by Cas10's HD nuclease domain and requires RNA binding but not RNA cleavage by the effector complex (26, 75, 85–87). The HD domain cleaves random sequences of ssDNA and generally requires transition metals (Ni2+ or Mn2+) for maximal activity, similar to Cas3 (26, 72, 75, 84–86). How Cas10 is activated to bind and cleave ssDNA is not clear, as ssDNA could not be visualized in structures of Csm/Cmr, and few conformational changes were observed in the HD domain upon RNA binding (12, 13, 72). The identification of Cas10 mutations that constitutively activate ssDNA cleavage indicate that RNA binding may relieve an autoinhibited state (13). Elucidation of the conformational changes and dynamics involved in activation are needed to fully understand how crRNA-guided RNA binding activates the Cas10 subunit for ssDNA cleavage.
Due to the ability of Type III effector complexes to cleave both ssDNA and RNA, it was proposed that the Type III complex would co-transcriptionally bind the growing transcript and trigger cleavage of both the RNA and the unwound ssDNA in transcription elongation complexes (26, 75, 84, 85). However, experiments in which Csm was added to in vitro transcription reactions or stalled transcription complexes indicated that Type III-A Csm prefers to cut RNA transcripts, rather than ssDNA (72, 88). This preference is likely because the transcript is more accessible than ssDNA bound by RNA polymerase during transcription. This is supported by the finding that Csm can cleave ssDNA in free R-loops that are not bound by RNA polymerase (72). Similarly, the Cas3 nuclease in Type I systems only nicks the ssDNA of the R-loop when it forms an exposed loop that protrudes above the surface of Cascade (50). Further studies are required to identify the DNA target of Type III CRISPR-Cas effector complexes in the cell. Potential targets may include unbound R-loops (Fig. 3A) or DNA replication intermediates (43, 89). Type III effectors could also cleave DNA during transcription initiation, when longer lengths of ssDNA are exposed by RNA polymerase during a process known as “scrunching” (90). Thus, future studies may reveal an unanticipated level of coordination between CRISPR-Cas immunity and other DNA processes in the cell.
DNA cleavage is important to help clear a phage infection, but it could also be deleterious to the host's genomic integrity if it persists indefinitely. Thus, Cas10 is gradually inactivated over time by cleavage and dissociation of the RNA from the effector complex (Fig. 3A) (75, 84). Consistent with this, use of a modified, noncleavable RNA or a cleavage-deficient Csm3/Cmr4 mutant prevented inactivation of ssDNA cleavage (75, 84). In the cell, RNA cleavage by the Csm3/Cmr4 subunits in the effector complex is likely important for turning off Cas10's DNase activity once viral transcripts have been cleared.
RNA-guided cOA synthesis in Type III complexes
In addition to DNA cleavage, RNA binding also activates the Cas10 subunit for cOA synthesis and activation of signaling effectors (Fig. 3A) (91–95). Binding of cOA to accessory nucleases, Csm6 (III-A) or Csx1 (III-B), dramatically stimulates their enzymatic activity (Fig. 3A) (91, 93, 94). This leads to degradation of viral and host transcripts, induces a growth arrest in the host cell, and promotes plasmid clearance (70, 96, 97). Target RNA cleavage and dissociation from the crRNA-guided effector complex eventually inactivates the Palm domains of Cas10 for cOA synthesis, which would prevent persistent degradation of host transcripts after the phage infection has been cleared (91, 95). Genomic analyses of CRISPR-Cas loci suggest that the ancestral function of these systems was a nucleotide-based stress-signaling pathway, similar to the Type III cOA-signaling pathway (98). Thus, further studies into RNA-guided cOA signaling could reveal unexpected connections between RNA-guided CRISPR-Cas immunity and other nucleotide-signaling pathways.
Biochemical and structural studies of Type III-A complexes suggest the following mechanism for cOA synthesis. In addition to the HD nuclease domain, Cas10 also has two Palm domains that form a composite active site for cOA synthesis (12, 91, 93, 99). The catalytic motif for cOA synthesis, GGDD, is present in only one of these domains (91, 94). Cooperative binding of two ATP molecules by the Palm domains positions the 3′-OH of one ATP molecule for attack of the 5′-α-phosphate of the second molecule to generate a 3′–5′ phosphodiester bond (12, 91, 93, 99). Specific recognition of ATP is mediated through a network of hydrogen-bonding interactions (99). Further reaction of this substrate with incoming ATP molecules leads to extension of the oligoadenylate chain and eventually ring closure through intramolecular attack of the terminal nucleotide's 5′-α-P by the first nucleotide's 3′-OH (Fig. 3A, bottom inset) (91, 99). Release of the cyclic oligoadenylates occurs through a channel formed by Cas10 and Csm4 (99). The lengths of the cOA species range from 3 to 6 AMP molecules per ring (Fig. 3A, bottom inset), but how the size of the ring is determined is not well-understood. In addition, how exactly RNA binding allosterically activates Cas10's Palm domains for cOA synthesis is also not understood.
RNA-guided cOA signaling in Type III CRISPR-Cas systems
Type III CRISPR-Cas loci are frequently associated with genes encoding for Csm6 or Csx1, which contain an N-terminal CARF (CRISPR-associated Rossman fold) “sensor” domain and a C-terminal HEPN (higher eukaryotes and prokaryotes nucleotide-binding) “effector” domain (5, 100, 101). The RNase activity of Csm6/Csx1's HEPN domain is allosterically activated by binding of either cOA4 or cOA6 to the CARF domains (Fig. 3A) (91, 93, 94, 102). The HEPN domains of Csm6 and Csx1 exhibit a base cleavage preference that varies by ortholog, with most cleaving adjacent to either purines or cytosines (91, 97, 100, 103). Structures of Csm6 and Csx1 reveal that they typically form dimers, but some orthologs also exhibit the ability to form higher-order oligomers (101, 102, 104, 105). Binding of cOA does not appear to induce large conformational changes in their HEPN active sites, suggesting that conformational activation may involve subtle changes or transient sampling of an activated state (102, 104).
In addition to Csm6 and Csx1, several DNases fused to CARF domains also respond to cOA molecules. For example, binding of cOA3 to NucC, an enzyme whose gene is associated with Type III CRISPR-Cas loci and other prokaryotic defense modules, activates it for dsDNA cleavage (106). A recent study also identified can1 (CRISPR-associated nuclease 1) in a genome with a Type III-A CRISPR-Cas system (107). Binding of cOA4 to Can1 activates it for nicking at random sequences of dsDNA (107). These cOA-activated nucleases may promote immunity by triggering degradation of viral DNA during replication or induce host death before the phage can replicate and infect other cells. Bioinformatic studies have also identified additional CARF domain proteins with transmembrane or other nuclease domains (108, 109). Further characterization of cOA-regulated effectors is needed to determine the full effects of cOA signaling by Type III systems in cells.
Nonspecific RNA or DNA degradation by nucleases can have deleterious effects on the host cell. Thus, cells have evolved dedicated enzymes called “ring nucleases” that degrade cOA and switch off the signaling pathway (110) (Fig. 3B). Ring nucleases degrade cOA4 or cOA6 using a catalytically active CARF domain (Fig. 3B) (110). Some Csm6/Csx1 orthologs also harbor an intrinsic ring nuclease activity in their CARF domains that leads to slow self-inactivation over time (Fig. 3B) (102, 105, 111). Cleavage proceeds in two steps, with the first step generating a linear oligoadenylate with a 2′,3′-cyclic phosphate, followed by a second step in which it is split into two halves (Fig. 3B) (102, 105, 110). Several Csm6 orthologs are still activated by linear A4 or A6 with 2′,3′-cyclic phosphates at their 3′ termini, suggesting that the second cleavage event is crucial for complete inactivation (94). Interestingly, anti-CRISPR proteins that inactivate Type III CRISPR-Cas systems are either highly active ring nucleases (e.g. AcrIII-1) or proteins that bind Cas10 and inhibit its cyclic oligoadenylate activity (e.g. AcrIIIB1) (112, 113). This highlights the importance of the cOA signaling pathway in bacterial immunity. How the opposing activities of Csm6/Csx1 and ring nucleases are effectively coordinated to mount a defense against phages is still not well-understood. In vitro kinetic studies of substrate binding and cleavage by the effector complex and its associated nucleases have been used to model the dynamics of Type III CRISPR-Cas immunity in cells and illustrate the distinct effects that host and viral ring nucleases have on immunity (114). Further studies that measure actual cellular concentrations of cOA and both host and viral transcript levels during an infection will reveal whether these kinetic models accurately describe how the Type III cOA signaling pathway protects cells from invaders.
Regulation of Type III Cas10 by tag:anti-tag pairing
All immune systems must distinguish between self versus nonself. Antisense transcription across the CRISPR array produces RNA molecules that are complementary to the crRNA (115, 116). To prevent these antisense transcripts from triggering autoimmunity, complementarity between the 5′-tag of the crRNA and the “anti-tag” sequence flanking the 3′ end of the target RNA inhibits Cas10's enzymatic activities (Fig. 3C) (26, 69, 91). In Type III-A systems, base-pairing between positions −2 to −5 in the crRNA 5′-tag (+1 is the first nucleotide of the spacer region) and the corresponding positions in the anti-tag is crucial for recognition of self RNA (Fig. 3C) (12, 13, 69, 84, 91). In the Type III-B system from Thermotoga maritima, inhibition is similarly mediated by tag:anti-tag complementarity but is additionally enhanced by the presence of a guanine opposite position −1 in the crRNA tag (117). How this guanine promotes Cas10 inhibition is unclear. The Type III-B CRISPR-Cas system from Pyrococcus furiosus also recognizes a “protospacer-flanking sequence” in the first three nucleotides flanking the 3′ end of the target (opposite positions −1 to −3 of the crRNA) to license ssDNA cleavage and cOA synthesis by Cas10 (85, 118). It is unclear how the protospacer-flanking sequence is recognized by the P. furiosus Cmr and whether tag:anti-tag complementarity still plays a role. Further analysis of how Type III complexes bind different anti-tag sequences will advance our understanding of how RNA binding regulates both ssDNA cleavage and cOA signaling by Cas10. Interestingly, the inhibition of Type III complexes by complementarity between the crRNA 5′-tag and the anti-tag has also been reported in an RNA-targeting Class 2 system, Type VI Cas13 (119). Thus, further insights into self versus nonself discrimination by Type III systems could reveal concepts that apply to other RNA-guided RNA nucleases.
Comparison of type III CRISPR-Cas systems with other nucleotide-based immune systems in bacteria and eukaryotes
Recent studies show that Type III CRISPR-Cas systems are not the only bacterial immune systems that use cyclic nucleotides for signaling. For instance, a cyclic GMP-AMP synthase (cGAS)-like enzyme in a bacterial defense module synthesizes a cyclic GMP-AMP in response to phage infection, which leads to membrane degradation by a phospholipase and cell death (120). A recent study also reported that a cGAS/DncV-like nucleotidyltransferase (CD-NTase) could synthesize cOA3, which in turn could bind and activate NucC, a DNA nuclease (121). Thus, insights into the cOA signaling pathway in Type III CRISPR-Cas systems may reveal concepts that are broadly applicable to other cyclic nucleotide–based antiphage signaling systems in prokaryotes.
The Type III cOA signaling pathway also bears similarities to the oligoadenylate synthase (OAS)-RNase L and cGAS-stimulator of interferon genes (STING) pathways in eukaryotes (122). OAS-RNase L constitutes a eukaryotic innate immune system, in which sensing of viral dsRNA activates OAS to synthesize 2′,5′-linked oligoadenylates. These oligoadenylates in turn bind and activate RNase L to cleave viral transcripts. Like Csm6, RNase L also has a HEPN domain that catalyzes RNA cleavage. In the cGAS-STING pathway, cytosolic DNA activates the cGAS enzyme to synthesize a cyclic GMP-AMP, which binds to the STING receptor and ultimately activates transcription of antiviral genes. Thus, studies of the cOA signaling pathway in Type III systems may reveal evolutionary connections between bacterial adaptive immunity and eukaryotic innate immunity.
Editing and applications of Class 1 CRISPR-Cas effectors
Although Class 1 systems are less widely used than Class 2 systems in genome editing, they are emerging as tools for genome and transcriptome manipulation in both microbial and eukaryotic cells (Fig. 4, A–C). In bacteria, targeting of Type I CRISPR-Cas effectors to DNA sequences in the absence of Cas3 or with a Cas3-inhibiting anti-CRISPR leads to transcriptional repression (Fig. 4A) (56, 123). Transcriptional inhibition is strongest when guide RNAs target the promoter rather than the ORF, similar to dCas9, a cleavage-deficient mutant of Cas9 engineered to repress transcription in cells (123, 124). Fusion of a transcriptional activator or repressor domain to Type I CRISPR-Cas complex subunits also enables the complex to modulate gene expression in plant or mammalian cells, illustrating its utility across different kingdoms (Fig. 4A) (125, 126).
Figure 4.
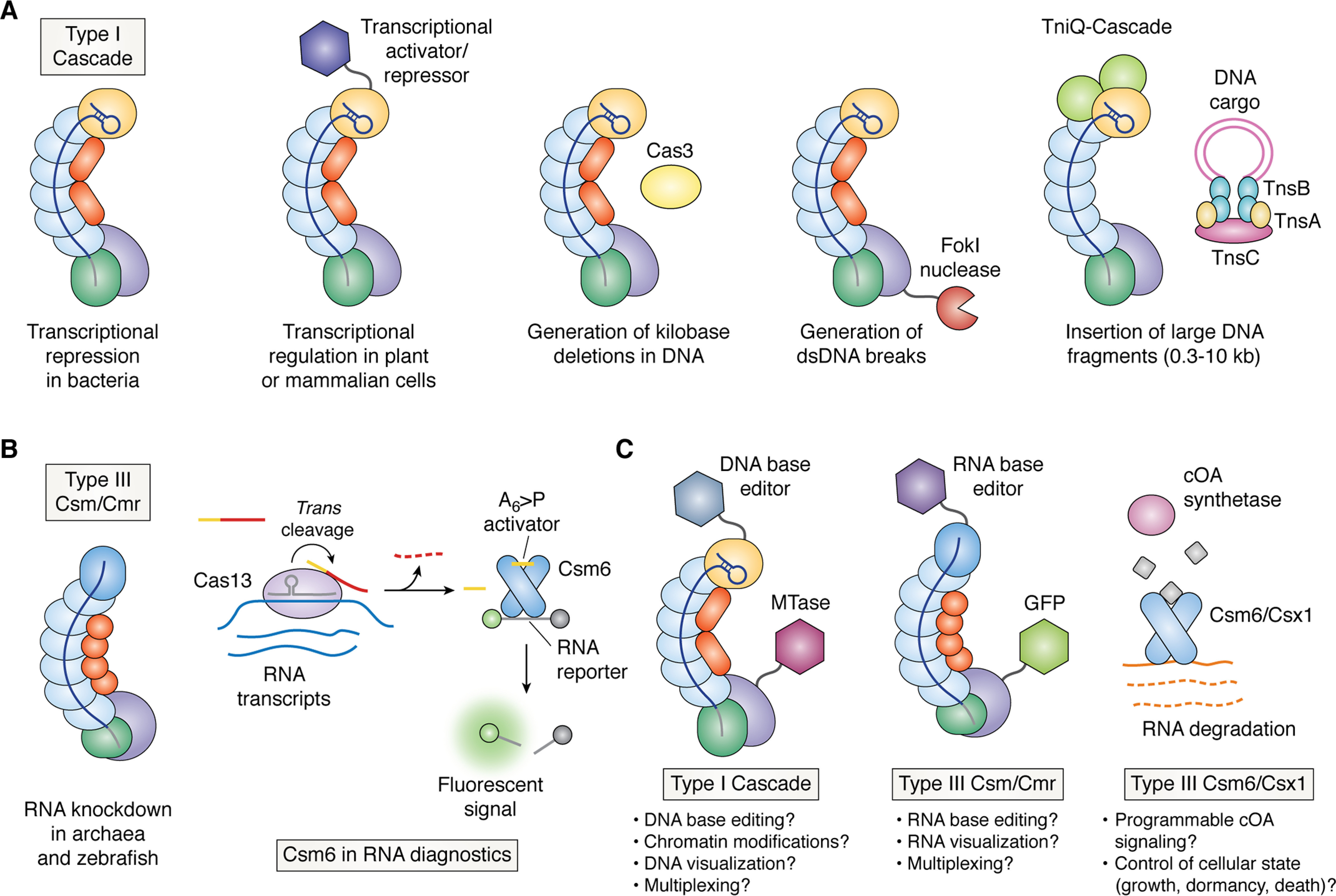
Current and future applications of Class 1 systems. A, applications of Type I CRISPR-Cas effector complexes include transcriptional repression/activation, generation of long-range genomic deletions, generation of dsDNA breaks, and insertion of large DNA fragments (left to right). Subunits are colored as in Fig. 1. B, applications of Type III effectors include RNA knockdown by Csm or Cmr (left) and use of the Csm6 RNase in RNA diagnostics (right). Subunits are colored as in Fig. 1. Cas13 is an RNA-guided RNA nuclease from the Type VI CRISPR-Cas system. Trans cleavage of an RNA substrate containing adenosines (yellow) and uridines (red) by Cas13 leads to release of a linear hexaadenylate with a 2′,3′-cyclic phosphate (A6>P activator). A6>P can bind and stimulate Csm6 to cleave a fluorescent RNA reporter. C, future applications of Class 1 CRISPR-Cas systems. The multisubunit Type I and III complexes could be fused to diverse functional domains for DNA and RNA editing. Multiple subunits per effector complex could also facilitate multiplexing. The cOA signaling pathway of Type III systems could be harnessed for control of cellular states in prokaryotes or eukaryotes by coupling cOA-binding nucleases to a cOA synthetase. MTase, methyltransferase.
Type I systems have also been introduced into various cell types for DNA modification (127–130). Introduction of the Type I effector complex and the Cas3 helicase-nuclease into mammalian cells results in long-range chromosomal deletions in DNA (Fig. 4A) (127–129). These deletions are unidirectional, consistent with biochemical studies of Cas3 degradation. Type I-E effectors fused to the FokI nuclease can also be programmed with a pair of guide RNAs to induce dsDNA breaks, triggering both small deletions and templated repair in mammalian cells (Fig. 4A) (129). Endogenous Type I systems have been harnessed for faster genome manipulation of the archaeon, Sulfolobus islandicus (130). Lastly, transposase-associated Type I-F systems have been shown to specifically insert synthetic “cargo” DNA up to 10 kb in length in E. coli with high fidelity, and thus holds promise as a technique to knock-in genes without requiring homologous recombination (Fig. 4A) (64). Indeed, recent preprints have reported the use of transposase-associated Type I-F systems for targeted insertion of antibiotic resistance genes in members of a microbial community and multiplexed gene insertion in several medically and industrially important bacterial species (131, 132).
Type III CRISPR-Cas effectors have been repurposed for RNA knockdown in archaea and in zebrafish, which lack an RNAi pathway (Fig. 4B) (133–135). They have also been used to assist in phage DNA editing (136). In addition, the cOA-regulated enzyme, Csm6, has been repurposed for viral diagnostics in conjunction with Cas13, a Type IV CRISPR-Cas effector that is activated to cleave RNA in trans upon crRNA-guided recognition of an RNA target (Fig. 4B) (103). Trans cleavage of an RNA oligonucleotide bearing an A6 at its 5′ end and multiple uridines at its 3′ end by Leptotrichia wadei Cas13 leads to production of a linear A6 with a 2′,3′-cyclic phosphate, which can bind to and activate certain Csm6 orthologs (103). This led to an ∼3.5-fold boost in RNA detection sensitivity over Cas13 alone (103). Further exploration of Csm6 and Csx1 orthologs from different organisms could lead to improved kinetics and sensitivity, expanded multiplexing, and greater thermostability of RNA diagnostic technologies. There may also be additional opportunities for reprogramming endogenous Type III systems in individual bacteria or bacterial communities by delivery of crRNA guides. The RNA-sensing function of Type III CRISPR-Cas effector complexes coupled with nonspecific RNA degradation by Csm6 could also be used to modify cell state (e.g. induce cell death or inhibit cell growth) in response to transcription of specific genes (Fig. 4C). This may be useful in the context of antimicrobials or for developing disease therapeutics that target cells with aberrant gene expression.
A major challenge for the widespread use of these systems in eukaryotic cells has been the delivery of these large complexes to the site of editing in cells. Several methods have now been established for introducing Type I and III complexes into eukaryotic cells, including nucleofection of preformed ribonucleoprotein complexes and expression from multiple DNA vectors (125–129). Further discovery of minimal Type I and III complexes and advances in RNA and protein delivery methods will simplify delivery and facilitate the continued development of these systems as tools for DNA and RNA manipulation in diverse cell types.
Class 1 CRISPR-Cas effectors extend the toolbox for genome engineering beyond the capabilities of Class 2 systems. The distinct and flexible PAM sequence requirements of Type I systems, which differ from the PAM sequences recognized by Cas9 and Cas12, broaden the array of DNA targets that can be recognized (34). The generation of long-range deletions by Type I Cascade also contrasts with the smaller deletions (“indels”) that result from Cas9 or Cas12 editing (Fig. 4A) (127, 128). The multisubunit composition of Class 1 effectors would also facilitate multiplexed fusion of domains that perform DNA or RNA base editing, epigenetic modifications, visualization of genomic loci or RNA sequences, and/or transcriptional regulation (Fig. 4C). Transposase-associated Type I-F systems also integrate DNA cargo with fewer off-target events than a transposase-associated Type V (Cas12k) system (64, 132, 137). Lastly, the unique, cOA-regulated RNases of Type III systems could also be repurposed for control of cellular growth or behavior, in response to an upstream signal generated by a cOA synthetase (Fig. 4C). Further study of Class 1 systems and their mechanism will likely continue to broaden the array of tools available for investigation of genome and transcriptome function in cells and for in vitro nucleic acid detection.
Summary and outlook
Class 1 CRISPR-Cas systems are the most common adaptive immunity pathways in prokaryotes. The diverse functions and activities of Type I and III crRNA-guided complexes and enzymes illustrate their versatility, and recent studies highlight their promise and development as tools for genome and transcriptome manipulation. These complexes resemble each other in subunit assembly and use a conserved mechanism for crRNA-mediated target recognition. Type I and III systems have evolved distinct mechanisms to recognize dsDNA and RNA, respectively. Discrimination between self and nonself occurs through interactions with sequences flanking their DNA or RNA targets; Type I Cascade recognizes a dsDNA PAM, whereas Type III Csm/Cmr probes for noncomplementarity between an RNA anti-tag and the 5′-tag of its crRNA. DNA recognition and R-loop formation by Type I complexes licenses foreign DNA degradation by Cas3. In Type III systems, RNA recognition triggers nonspecific ssDNA cleavage and initiation of a cOA signaling pathway that activates additional nucleases for DNA or RNA degradation in trans. Type III systems also include a timer for self-inactivation by slow cleavage of the RNA target and degradation of cOA, the second messenger. Critical steps of interference are inhibited by anti-CRISPR proteins against both systems. Fundamental studies of Class 1 enzymes have enabled the application of Type I systems for transcriptional regulation and genome editing and Type III systems for RNA degradation and diagnostics. Future studies on Class 1 CRISPR-Cas complexes will expand our understanding of the mechanism and evolution of prokaryotic RNA-guided immunity and reveal unexpected connections with other cellular DNA processes, nucleotide signaling pathways, and eukaryotic innate immunity. Such insights will also open new avenues for using CRISPR-Cas systems to interrogate genome and transcriptome function, control gene expression, and detect DNA or RNA for disease diagnostics.
Acknowledgments
We thank Patrick Pausch and Haridha Shivram for critical reading and comments on the manuscript and other members of the Doudna laboratory for helpful suggestions. We apologize to those investigators whose primary work we could not cite due to length considerations.
Funding and additional information—T. Y. L. was supported by the Howard Hughes Medical Institute (HHMI). J. A. D. is an Investigator of HHMI. This work was supported by HHMI, National Science Foundation Grant 1817593, and NIGMS, National Institutes of Health, Grant P01GM051487. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The Regents of the University of California have patents issued for CRISPR technologies on which J. A. D. is an inventor and patents pending for CRISPR technologies on which both J. A. D. and T. Y. L. are inventors. J. A. D. is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences. J. A. D. is a scientific advisor to Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Algen Biotechnologies, Felix Biosciences, Mammoth Biosciences, and Inari. J. A. D. is a Director at Johnson & Johnson and has research projects sponsored by Pfizer, Roche, AppleTree Partners, and Biogen.
- CRISPR
- clustered regularly interspaced short palindromic repeats
- pre-crRNA
- precursor crRNA
- crRNA
- CRISPR RNA
- cOA
- cyclic oligoadenylate
- PAM
- protospacer adjacent motif
- CARF
- CRISPR-associated Rossman fold
- HEPN
- higher eukaryotes and prokaryotes nucleotide-binding
- cGAS
- cyclic GMP-AMP synthase
- OAS
- oligoadenylate synthase
- STING
- stimulator of interferon genes.
References
- 1. Dy R. L., Richter C., Salmond G. P. C., and Fineran P. C. (2014) Remarkable mechanisms in microbes to resist phage infections. Annu. Rev. Virol. 1, 307–331 10.1146/annurev-virology-031413-085500 [DOI] [PubMed] [Google Scholar]
- 2. Makarova K. S., Wolf Y. I., Iranzo J., Shmakov S. A., Alkhnbashi O. S., Brouns S. J. J., Charpentier E., Cheng D., Haft D. H., Horvath P., Moineau S., Mojica F. J. M., Scott D., Shah S. A., Siksnys V., et al. (2020) Evolutionary classification of CRISPR–Cas systems: a burst of Class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 10.1038/s41579-019-0299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knott G. J., and Doudna J. A. (2018) CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 10.1126/science.aat5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murugan K., Babu K., Sundaresan R., Rajan R., and Sashital D. G. (2017) The revolution continues: newly discovered systems expand the CRISPR-Cas toolkit. Mol. Cell 68, 15–25 10.1016/j.molcel.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Makarova K. S., Wolf Y. I., Alkhnbashi O. S., Costa F., Shah S. A., Saunders S. J., Barrangou R., Brouns S. J. J., Charpentier E., Haft D. H., Horvath P., Moineau S., Mojica F. J. M., Terns R. M., Terns M. P., et al. (2015) An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13, 722–736 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burstein D., Sun C. L., Brown C. T., Sharon I., Anantharaman K., Probst A. J., Thomas B. C., and Banfield J. F. (2016) Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat. Commun. 7, 10613 10.1038/ncomms10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jackson R. N., and Wiedenheft B. (2015) A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol. Cell. 58, 722–728 10.1016/j.molcel.2015.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jackson R. N., Golden S. M., van Erp P. B. G., Carter J., Westra E. R., Brouns S. J. J., van der Oost J., Terwilliger T. C., Read R. J., and Wiedenheft B. (2014) Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345, 1473–1479 10.1126/science.1256328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chowdhury S., Carter J., Rollins M. C. F., Golden S. M., Jackson R. N., Hoffmann C., Nosaka L. L., Bondy-Denomy J., Maxwell K. L., Davidson A. R., Fischer E. R., Lander G. C., and Wiedenheft B. (2017) Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell 169, 47–57.e11 10.1016/j.cell.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mulepati S., Héroux A., and Bailey S. (2014) Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 345, 1479–1484 10.1126/science.1256996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taylor D. W., Zhu Y., Staals R. H. J., Kornfeld J. E., Shinkai A., van der Oost J., Nogales E., and Doudna J. A. (2015) Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science 348, 581–585 10.1126/science.aaa4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. You L., Ma J., Wang J., Artamonova D., Wang M., Liu L., Xiang H., Severinov K., Zhang X., and Wang Y. (2019) Structure studies of the CRISPR-Csm complex reveal mechanism of co-transcriptional interference. Cell 176, 239–253.e16 10.1016/j.cell.2018.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jia N., Mo C. Y., Wang C., Eng E. T., Marraffini L. A., and Patel D. J. (2019) Type III-A CRISPR-Cas Csm complexes: assembly, periodic RNA cleavage, DNase activity regulation, and autoimmunity. Mol. Cell 73, 264–277.e5 10.1016/j.molcel.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Osawa T., Inanaga H., Sato C., and Numata T. (2015) Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol. Cell 58, 418–430 10.1016/j.molcel.2015.03.018 [DOI] [PubMed] [Google Scholar]
- 15. Özcan A., Pausch P., Linden A., Wulf A., Schühle K., Heider J., Urlaub H., Heimerl T., Bange G., and Randau L. (2019) Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum. Nat. Microbiol. 4, 89–96 10.1038/s41564-018-0274-8 [DOI] [PubMed] [Google Scholar]
- 16. Taylor H. N., Warner E. E., Armbrust M. J., Crowley V. M., Olsen K. J., and Jackson R. N. (2019) Structural basis of Type IV CRISPR RNA biogenesis by a Cas6 endoribonuclease. RNA Biol. 16, 1438–1447 10.1080/15476286.2019.1634965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Venclovas Č. (2016) Structure of Csm2 elucidates the relationship between small subunits of CRISPR-Cas effector complexes. FEBS Lett. 590, 1521–1529 10.1002/1873-3468.12179 [DOI] [PubMed] [Google Scholar]
- 18. Pausch P., Müller-Esparza H., Gleditzsch D., Altegoer F., Randau L., and Bange G. (2017) Structural variation of type I-F CRISPR RNA guided DNA surveillance. Mol. Cell 67, 622–632.e4 10.1016/j.molcel.2017.06.036 [DOI] [PubMed] [Google Scholar]
- 19. He L., St. John James M., Radovcic M., Ivancic-Bace I., and Bolt E. L. (2020) Cas3 protein—a review of a multi-tasking machine. Genes (Basel) 11, 208 10.3390/genes11020208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jackson R. N., Lavin M., Carter J., and Wiedenheft B. (2014) Fitting CRISPR-associated Cas3 into the helicase family tree. Curr. Opin. Struct. Biol. 24, 106–114 10.1016/j.sbi.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pyenson N. C., and Marraffini L. A. (2017) Type III CRISPR-Cas systems: when DNA cleavage just isn't enough. Curr. Opin. Microbiol. 37, 150–154 10.1016/j.mib.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 22. Crowley V. M., Catching A., Taylor H. N., Borges A. L., Metcalf J., Bondy-Denomy J., and Jackson R. N. (2019) A type IV-A CRISPR-Cas system in Pseudomonas aeruginosa mediates RNA-guided plasmid interference in vivo. CRISPR J. 2, 434–440 10.1089/crispr.2019.0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., and Charpentier E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Staals R. H. J. H. J., Agari Y., Maki-Yonekura S., Zhu Y., Taylor D. W. W., van Duijn E., Barendregt A., Vlot M., Koehorst J. J. J., Sakamoto K., Masuda A., Dohmae N., Schaap P. J. J., Doudna J. A. A., Heck A. J. R. J. R., et al. (2013) Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of Thermus thermophilus. Mol. Cell 52, 135–145 10.1016/j.molcel.2013.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Staals R. H. J., Zhu Y., Taylor D. W., Kornfeld J. E., Sharma K., Barendregt A., Koehorst J. J., Vlot M., Neupane N., Varossieau K., Sakamoto K., Suzuki T., Dohmae N., Yokoyama S., Schaap P. J., et al. (2014) RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol. Cell 56, 518–530 10.1016/j.molcel.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu T. Y., Iavarone A. T., and Doudna J. A. (2017) RNA and DNA targeting by a reconstituted Thermus thermophilus type III-A CRISPR-Cas system. PLoS ONE 12, e0170552 10.1371/journal.pone.0170552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hatoum-Aslan A., Samai P., Maniv I., Jiang W., and Marraffini L. A. (2013) A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J. Biol. Chem. 288, 27888–27897 10.1074/jbc.M113.499244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker F. C., Chou-Zheng L., Dunkle J. A., and Hatoum-Aslan A. (2016) Molecular determinants for CRISPR RNA maturation in the Cas10–Csm complex and roles for non-Cas nucleases. Nucleic Acids Res. 45, 2112–2123 10.1093/nar/gkw891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chou-Zheng L., and Hatoum-Aslan A. (2019) A type III-A CRISPR-Cas system employs degradosome nucleases to ensure robust immunity. eLife 8, 1–25 10.7554/eLife.45393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Behler J., Sharma K., Reimann V., Wilde A., Urlaub H., and Hess W. R. (2018) The host-encoded RNase E endonuclease as the crRNA maturation enzyme in a CRISPR–Cas subtype III-Bv system. Nat. Microbiol. 3, 367–377 10.1038/s41564-017-0103-5 [DOI] [PubMed] [Google Scholar]
- 31. Hochstrasser M. L., Taylor D. W., Bhat P., Guegler C. K., Sternberg S. H., Nogales E., and Doudna J. A. (2014) CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc. Natl. Acad. Sci. U. S. A. 111, 6618–6623 10.1073/pnas.1405079111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mulepati S., and Bailey S. (2013) In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem. 288, 22184–22192 10.1074/jbc.M113.472233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Westra E. R., van Erp P. B. G., Künne T., Wong S. P., Staals R. H. J., Seegers C. L. C., Bollen S., Jore M. M., Semenova E., Severinov K., de Vos W. M., Dame R. T., de Vries R., Brouns S. J. J., and van der Oost J. (2012) CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by cascade and Cas3. Mol. Cell. 46, 595–605 10.1016/j.molcel.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gleditzsch D., Pausch P., Müller-Esparza H., Özcan A., Guo X., Bange G., and Randau L. (2019) PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 16, 504–517 10.1080/15476286.2018.1504546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Westra E. R., Semenova E., Datsenko K. A., Jackson R. N., Wiedenheft B., Severinov K., and Brouns S. J. J. (2013) Type I-E CRISPR-Cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 9, e1003742 10.1371/journal.pgen.1003742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hayes R. P., Xiao Y., Ding F., van Erp P. B. G., Rajashankar K., Bailey S., Wiedenheft B., and Ke A. (2016) Structural basis for promiscuous PAM recognition in type I–E Cascade from E. coli. Nature 530, 499–503 10.1038/nature16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao Y., Luo M., Hayes R. P., Kim J., Ng S., Ding F., Liao M., and Ke A. (2017) Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas system. Cell 170, 48–60.e11 10.1016/j.cell.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo T. W., Bartesaghi A., Yang H., Falconieri V., Rao P., Merk A., Eng E. T., Raczkowski A. M., Fox T., Earl L. A., Patel D. J., and Subramaniam S. (2017) Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell 171, 414–426.e12 10.1016/j.cell.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rollins M. F., Schuman J. T., Paulus K., Bukhari H. S. T., and Wiedenheft B. (2015) Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 43, 2216–2222 10.1093/nar/gkv094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walton R. T., Christie K. A., Whittaker M. N., and Kleinstiver B. P. (2020) Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296 10.1126/science.aba8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu J. H., Miller S. M., Geurts M. H., Tang W., Chen L., Sun N., Zeina C. M., Gao X., Rees H. A., Lin Z., and Liu D. R. (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aguilera A., and García-Muse T. (2012) R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 46, 115–124 10.1016/j.molcel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 43. Santos-Pereira J. M., and Aguilera A. (2015) R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16, 583–597 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- 44. Wiedenheft B., van Duijn E., Bultema J. B., Waghmare S. P., Zhou K., Barendregt A., Westphal W., Heck A. J. R., Boekema E. J., Dickman M. J., Doudna J. A., Bultema J. B., Waghmare S. P., Waghmare S. P., Zhou K., et al. (2011) RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U. S. A. 108, 10092–10097 10.1073/pnas.1102716108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rutkauskas M., Sinkunas T., Songailiene I., Tikhomirova M. S., Siksnys V., and Seidel R. (2015) Directional R-loop formation by the CRISPR-cas surveillance complex cascade provides efficient off-target site rejection. Cell Rep. 10, 1534–1543 10.1016/j.celrep.2015.01.067 [DOI] [PubMed] [Google Scholar]
- 46. Zuo Y., and Steitz T. A. A. (2015) Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol. Cell 58, 534–540 10.1016/j.molcel.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bae B., Feklistov A., Lass-Napiorkowska A., Landick R., and Darst S. A. (2015) Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife 4, e08504 10.7554/eLife.08504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rollins M. F., Chowdhury S., Carter J., Golden S. M., Miettinen H. M., Santiago-Frangos A., Faith D., Lawrence C. M., Lander G. C., and Wiedenheft B. (2019) Structure reveals a mechanism of CRISPR-RNA-guided nuclease recruitment and anti-CRISPR viral mimicry. Mol. Cell 74, 132–142.e5 10.1016/j.molcel.2019.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sinkunas T., Gasiunas G., Fremaux C., Barrangou R., Horvath P., and Siksnys V. (2011) Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30, 1335–1342 10.1038/emboj.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xiao Y., Luo M., Dolan A. E., Liao M., and Ke A. (2018) Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science 361, eaat0839 10.1126/science.aat0839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huo Y., Nam K. H., Ding F., Lee H., Wu L., Xiao Y., Farchione M. D., Zhou S., Rajashankar K., Kurinov I., Zhang R., and Ke A. (2014) Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat. Struct. Mol. Biol. 21, 771–777 10.1038/nsmb.2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Loeff L., Brouns S. J. J., and Joo C. (2018) Repetitive DNA reeling by the Cascade-Cas3 complex in nucleotide unwinding steps. Mol. Cell 70, 385–394.e3 10.1016/j.molcel.2018.03.031 [DOI] [PubMed] [Google Scholar]
- 53. Redding S., Sternberg S. H., Marshall M., Gibb B., Bhat P., Guegler C. K., Wiedenheft B., Doudna J. A., and Greene E. C. (2015) Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell 163, 854–865 10.1016/j.cell.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nimkar S., and Anand B. (2020) Cas3/I-C mediated target DNA recognition and cleavage during CRISPR interference are independent of the composition and architecture of Cascade surveillance complex. Nucleic Acids Res. 48, 2486–2501 10.1093/nar/gkz1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wiegand T., Karambelkar S., Bondy-Denomy J. and Wiedenheft B. (2020) Structures and strategies of Anti-CRISPR-mediated immune suppression. Annu. Rev. Microbiol. 74, 21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pawluk A., Shah M., Mejdani M., Calmettes C., Moraes T. F., Davidson A. R., and Maxwell K. L. (2017) Disabling a Type I-E CRISPR-Cas nuclease with a bacteriophage-encoded anti-CRISPR protein. mBio 8, 1–12 10.1128/mBio.01751-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bondy-Denomy J., Garcia B., Strum S., Du M., Rollins M. F., Hidalgo-Reyes Y., Wiedenheft B., Maxwell K. L., and Davidson A. R. (2015) Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 10.1038/nature15254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang X., Yao D., Xu J. G., Li A. R., Xu J., Fu P., Zhou Y., and Zhu Y. (2016) Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat. Struct. Mol. Biol. 23, 868–870 10.1038/nsmb.3269 [DOI] [PubMed] [Google Scholar]
- 59. Hirschi M., Lu W.-T., Santiago-Frangos A., Wilkinson R., Golden S. M., Davidson A. R., Lander G. C., and Wiedenheft B. (2020) AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex. Nat. Commun. 11, 2730 10.1038/s41467-020-16512-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang K., Wang S., Li S., Zhu Y., Pintilie G. D., Mou T.-C., Schmid M. F., Huang Z., and Chiu W. (2020) Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR–Cas complex revealed by cryo-EM. Proc. Natl. Acad. Sci. U. S. A. 117, 7176–7182 10.1073/pnas.1922638117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bondy-Denomy J. (2018) Protein Inhibitors of CRISPR-Cas9. ACS Chem. Biol. 13, 417–423 10.1021/acschembio.7b00831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Knott G. J., Thornton B. W., Lobba M. J., Liu J. J., Al-Shayeb B., Watters K. E., and Doudna J. A. (2019) Broad-spectrum enzymatic inhibition of CRISPR-Cas12a. Nat. Struct. Mol. Biol. 26, 315–321 10.1038/s41594-019-0208-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Peters J. E., Makarova K. S., Shmakov S., and Koonin E. V. (2017) Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc. Natl. Acad. Sci. U.S.A. 114, E7358–E7366 10.1073/pnas.1709035114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Klompe S. E., Vo P. L. H. H., Halpin-Healy T. S., and Sternberg S. H. (2019) Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 10.1038/s41586-019-1323-z [DOI] [PubMed] [Google Scholar]
- 65. Halpin-Healy T. S., Klompe S. E., Sternberg S. H., and Fernández I. S. (2020) Structural basis of DNA targeting by a transposon-encoded CRISPR–Cas system. Nature 577, 271–274 10.1038/s41586-019-1849-0 [DOI] [PubMed] [Google Scholar]
- 66. Mohanraju P., Makarova K. S., Zetsche B., Zhang F., Koonin E. V., and van der Oost J. (2016) Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147 10.1126/science.aad5147 [DOI] [PubMed] [Google Scholar]
- 67. Marraffini L. A., and Sontheimer E. J. (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tamulaitis G., Kazlauskiene M., Manakova E., Venclovas Č., Nwokeoji A. O. O., Dickman M. J. J., Horvath P., and Siksnys V. (2014) Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol. Cell 56, 506–517 10.1016/j.molcel.2014.09.027 [DOI] [PubMed] [Google Scholar]
- 69. Marraffini L. A., and Sontheimer E. J. (2010) Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571 10.1038/nature08703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jiang W., Samai P., and Marraffini L. A. (2016) Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164, 710–721 10.1016/j.cell.2015.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Malone L. M., Warring S. L., Jackson S. A., Warnecke C., Gardner P. P., Gumy L. F., and Fineran P. C. (2020) A jumbo phage that forms a nucleus-like structure evades CRISPR–Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat. Microbiol. 5, 48–55 10.1038/s41564-019-0612-5 [DOI] [PubMed] [Google Scholar]
- 72. Liu T. Y., Liu J.-J., Aditham A. J., Nogales E., and Doudna J. A. (2019) Target preference of Type III-A CRISPR-Cas complexes at the transcription bubble. Nat. Commun. 10, 3001 10.1038/s41467-019-10780-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. O'Connell M. R. (2019) Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J. Mol. Biol. 431, 66–87 10.1016/j.jmb.2018.06.029 [DOI] [PubMed] [Google Scholar]
- 74. Jinek M., and Doudna J. A. (2009) A three-dimensional view of the molecular machinery of RNA interference. Nature 457, 405–412 10.1038/nature07755 [DOI] [PubMed] [Google Scholar]
- 75. Estrella M. A., Kuo F.-T., and Bailey S. (2016) RNA-activated DNA cleavage by the type III-B CRISPR–Cas effector complex. Genes Dev. 30, 460–470 10.1101/gad.273722.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pan S., Li Q., Deng L., Jiang S., Jin X., Peng N., Liang Y., She Q., and Li Y. (2019) A seed motif for target RNA capture enables efficient immune defence by a type III-B CRISPR-Cas system. RNA Biol. 16, 1166–1178 10.1080/15476286.2019.1618693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hale C. R., Cocozaki A., Li H., Terns R. M., and Terns M. P. (2014) Target RNA capture and cleavage by the Cmr type III-B CRISPR–Cas effector complex. Genes Dev. 28, 2432–2443 10.1101/gad.250712.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li Y., Zhang Y., Lin J., Pan S., Han W., Peng N., Liang Y. X., and She Q. (2017) Cmr1 enables efficient RNA and DNA interference of a III-B CRISPR–Cas system by binding to target RNA and crRNA. Nucleic Acids Res. 45, 11305–11310 10.1093/nar/gkx791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hale C. R., Zhao P., Olson S., Duff M. O., Graveley B. R., Wells L., Terns R. M., and Terns M. P. (2009) RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956 10.1016/j.cell.2009.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang W. (2011) Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 44, 1–93 10.1017/S0033583510000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tamulaitis G., Venclovas Č., and Siksnys V. (2017) Type III CRISPR-Cas immunity: major differences brushed aside. Trends Microbiol. 25, 49–61 10.1016/j.tim.2016.09.012 [DOI] [PubMed] [Google Scholar]
- 82. Goldberg G. W., Jiang W., Bikard D., and Marraffini L. A. (2014) Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514, 633–637 10.1038/nature13637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Deng L., Garrett R. A., Shah S. A., Peng X., and She Q. (2013) A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 87, 1088–1099 10.1111/mmi.12152 [DOI] [PubMed] [Google Scholar]
- 84. Kazlauskiene M., Tamulaitis G., Kostiuk G., Venclovas Č., and Siksnys V. (2016) Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell 62, 295–306 10.1016/j.molcel.2016.03.024 [DOI] [PubMed] [Google Scholar]
- 85. Elmore J. R., Sheppard N. F., Ramia N., Deighan T., Li H., Terns R. M., and Terns M. P. (2016) Bipartite recognition of target RNAs activates DNA cleavage by the type III-B CRISPR–Cas system. Genes Dev. 30, 447–459 10.1101/gad.272153.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Han W., Li Y., Deng L., Feng M., Peng W., Hallstrøm S., Zhang J., Peng N., Liang Y. X., White M. F., and She Q. (2017) A type III-B CRISPR-Cas effector complex mediating massive target DNA destruction. Nucleic Acids Res. 45, 1983–1993 10.1093/nar/gkw1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Aravind L., and Koonin E. V. (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23, 469–472 10.1016/S0968-0004(98)01293-6 [DOI] [PubMed] [Google Scholar]
- 88. Samai P., Pyenson N., Jiang W., Goldberg G. W., Hatoum-Aslan A., and Marraffini L. A. (2015) Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell 161, 1164–1174 10.1016/j.cell.2015.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. O'Donnell M., Langston L., and Stillman B. (2013) Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 5, a010108 10.1101/cshperspect.a010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kapanidis A. N., Margeat E., Ho S. O., Kortkhonjia E., Weiss S., and Ebright R. H. (2006) Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314, 1144–1147 10.1126/science.1131399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kazlauskiene M., Kostiuk G., Venclovas Č., Tamulaitis G., and Siksnys V. (2017) A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609 10.1126/science.aao0100 [DOI] [PubMed] [Google Scholar]
- 92. Nasef M., Muffly M. C., Beckman A. B., Rowe S. J., Walker F. C., Hatoum-Aslan A., and Dunkle J. A. (2019) Regulation of cyclic oligoadenylate synthesis by the Staphylococcus epidermidis Cas10-Csm complex. RNA 25, 948–962 10.1261/rna.070417.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Han W., Stella S., Zhang Y., Guo T., Sulek K., Peng-Lundgren L., Montoya G., and She Q. (2018) A Type III-B Cmr effector complex catalyzes the synthesis of cyclic oligoadenylate second messengers by cooperative substrate binding. Nucleic Acids Res. 46, 10319–10330 10.1093/nar/gky844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Niewoehner O., Garcia-do, Val C., Rostøl J. T., Berk C., Schwede F., Bigler L., Hall J., Marraffini L. A., and Jinek M. (2017) Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548 10.1038/nature23467 [DOI] [PubMed] [Google Scholar]
- 95. Rouillon C., Athukoralage J. S., Graham S., Grüschow S., and White M. F. (2018) Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife 7, e36734 10.7554/elife.36734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rostøl J. T., and Marraffini L. A. (2019) Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR–Cas immunity. Nat. Microbiol. 4, 656–662 10.1038/s41564-018-0353-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Foster K., Kalter J., Woodside W., Terns R. M., and Terns M. P. (2019) The ribonuclease activity of Csm6 is required for anti-plasmid immunity by Type III-A CRISPR-Cas systems. RNA Biol. 16, 449–460 10.1080/15476286.2018.1493334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Koonin E. V., and Makarova K. S. (2018) Discovery of oligonucleotide signaling mediated by CRISPR-associated polymerases solves two puzzles but leaves an enigma. ACS Chem. Biol. 13, 309–312 10.1021/acschembio.7b00713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jia N., Jones R., Sukenick G., and Patel D. J. (2019) Second messenger cA4 formation within the composite Csm1 palm pocket of type III-A CRISPR-Cas Csm complex and its release path. Mol. Cell 75, 933–911 10.1016/j.molcel.2019.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sheppard N. F., Glover C. V. C., Terns R. M., and Terns M. P. (2016) The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA 22, 216–224 10.1261/rna.039842.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Niewoehner O., and Jinek M. (2016) Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 22, 318–329 10.1261/rna.054098.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jia N., Jones R., Yang G., Ouerfelli O., and Patel D. J. (2019) CRISPR-Cas III-A Csm6 CARF domain is a ring nuclease triggering stepwise cA4 cleavage with ApA>p formation terminating RNase activity. Mol. Cell 75, 944–956.e6 10.1016/j.molcel.2019.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gootenberg J. S., Abudayyeh O. O., Kellner M. J., Joung J., Collins J. J., and Zhang F. (2018) Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 10.1126/science.aaq0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Molina R., Stella S., Feng M., Sofos N., Jauniskis V., Pozdnyakova I., López-Méndez B., She Q., and Montoya G. (2019) Structure of Csx1-cOA4 complex reveals the basis of RNA decay in Type III-B CRISPR-Cas. Nat. Commun. 10, 4302 10.1038/s41467-019-12244-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Garcia-Doval C., Schwede F., Berk C., Rostøl J. T., Niewoehner O., Tejero O., Hall J., Marraffini L. A., and Jinek M. (2020) Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6. Nat. Commun. 11, 1596 10.1038/s41467-020-15334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lau R. K., Ye Q., Birkholz E. A., Berg K. R., Patel L., Mathews I. T., Watrous J. D., Ego K., Whiteley A. T., Lowey B., Mekalanos J. J., Kranzusch P. J., Jain M., Pogliano J., and Corbett K. D. (2020) Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol. Cell 77, 723–733.e6 10.1016/j.molcel.2019.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. McMahon S. A., Zhu W., Graham S., Rambo R., White M. F., and Gloster T. M. (2020) Structure and mechanism of a type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate. Nat. Commun. 11, 500 10.1038/s41467-019-14222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Shah S. A., Alkhnbashi O. S., Behler J., Han W., She Q., Hess W. R., Garrett R. A., and Backofen R. (2019) Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR-cas gene cassettes reveals 39 new cas gene families. RNA Biol. 16, 530–542 10.1080/15476286.2018.1483685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shmakov S. A., Makarova K. S., Wolf Y. I., Severinov K. V., and Koonin E. V. (2018) Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc. Natl. Acad. Sci. 115, E5307–E5316 10.1073/pnas.1803440115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Athukoralage J. S., Rouillon C., Graham S., Grüschow S., and White M. F. (2018) Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature 562, 277–280 10.1038/s41586-018-0557-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Athukoralage J. S., Graham S., Grüschow S., Rouillon C., and White M. F. (2019) A type III CRISPR ancillary ribonuclease degrades its cyclic oligoadenylate activator. J. Mol. Biol. 431, 2894–2899 10.1016/j.jmb.2019.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bhoobalan-Chitty Y., Johansen T. B., Di Cianni N., and Peng X. (2019) Inhibition of type III CRISPR-Cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell 179, 448–458.e11 10.1016/j.cell.2019.09.003 [DOI] [PubMed] [Google Scholar]
- 113. Athukoralage J. S., McMahon S. A., Zhang C., Grüschow S., Graham S., Krupovic M., Whitaker R. J., Gloster T. M., and White M. F. (2020) An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577, 572–575 10.1038/s41586-019-1909-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Athukoralage J. S., Graham S., Rouillon C., Grüschow S., Czekster C. M., and White M. F. (2020) The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling. eLife 9, 1–16 10.7554/eLife.55852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hale C. R., Majumdar S., Elmore J., Pfister N., Compton M., Olson S., Resch A. M., Glover C. V. C., Graveley B. R., Terns R. M., and Terns M. P. (2012) Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 45, 292–302 10.1016/j.molcel.2011.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lillestøl R. K., Shah S. A., Brügger K., Redder P., Phan H., Christiansen J., and Garrett R. A. (2009) CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 72, 259–272 10.1111/j.1365-2958.2009.06641.x [DOI] [PubMed] [Google Scholar]
- 117. Johnson K., Learn B. A., Estrella M. A., and Bailey S. (2019) Target sequence requirements of a type III-B CRISPR-Cas immune system. J. Biol. Chem. 294, 10290–10299 10.1074/jbc.RA119.008728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Foster K., Grüschow S., Bailey S., White M. F., and Terns M. P. (2020) Regulation of the RNA and DNA nuclease activities required for Pyrococcus furiosus Type III-B CRISPR–Cas immunity. Nucleic Acids Res. 48, 4418–4434 10.1093/nar/gkaa176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Meeske A. J., and Marraffini L. A. (2018) RNA guide complementarity prevents self-targeting in type VI CRISPR systems. Mol. Cell 71, 791–801.e3 10.1016/j.molcel.2018.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Cohen D., Melamed S., Millman A., Shulman G., Oppenheimer-Shaanan Y., Kacen A., Doron S., Amitai G., and Sorek R. (2019) Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574, 691–377 10.1038/s41586-019-1605-5 [DOI] [PubMed] [Google Scholar]
- 121. Ye Q., Lau R. K., Mathews I. T., Birkholz E. A., Watrous J. D., Azimi C. S., Pogliano J., Jain M., and Corbett K. D. (2020) HORMA domain proteins and a Trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Mol. Cell 77, 709–722.e7 10.1016/j.molcel.2019.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hornung V., Hartmann R., Ablasser A., and Hopfner K.-P. (2014) OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 14, 521–528 10.1038/nri3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Luo M. L., Mullis A. S., Leenay R. T., and Beisel C. L. (2015) Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 43, 674–681 10.1093/nar/gku971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., and Lim W. A. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pickar-Oliver A., Black J. B., Lewis M. M., Mutchnick K. J., Klann T. S., Gilcrest K. A., Sitton M. J., Nelson C. E., Barrera A., Bartelt L. C., Reddy T. E., Beisel C. L., Barrangou R., and Gersbach C. A. (2019) Targeted transcriptional modulation with type I CRISPR–Cas systems in human cells. Nat. Biotechnol. 37, 1493–1501 10.1038/s41587-019-0235-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Young J. K., Gasior S. L., Jones S., Wang L., Navarro P., Vickroy B., and Barrangou R. (2019) The repurposing of type I-E CRISPR-Cascade for gene activation in plants. Commun. Biol. 2, 383 10.1038/s42003-019-0637-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Morisaka H., Yoshimi K., Okuzaki Y., Gee P., Kunihiro Y., Sonpho E., Xu H., Sasakawa N., Naito Y., Nakada S., Yamamoto T., Sano S., Hotta A., Takeda J., and Mashimo T. (2019) CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nat. Commun. 10, 5302 10.1038/s41467-019-13226-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Dolan A. E., Hou Z., Xiao Y., Gramelspacher M. J., Heo J., Howden S. E., Freddolino P. L., Ke A., and Zhang Y. (2019) Introducing a spectrum of long-range genomic deletions in human embryonic stem cells using type I CRISPR-Cas. Mol. Cell 74, 936–950.e5 10.1016/j.molcel.2019.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cameron P., Coons M. M., Klompe S. E., Lied A. M., Smith S. C., Vidal B., Donohoue P. D., Rotstein T., Kohrs B. W., Nyer D. B., Kennedy R., Banh L. M., Williams C., Toh M. S., Irby M. J., et al. (2019) Harnessing type I CRISPR–Cas systems for genome engineering in human cells. Nat. Biotechnol. 37, 1471–1477 10.1038/s41587-019-0310-0 [DOI] [PubMed] [Google Scholar]
- 130. Li Y., Pan S., Zhang Y., Ren M., Feng M., Peng N., Chen L., Liang Y. X., and She Q. (2016) Harnessing type I and type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 44, e34–e34 10.1093/nar/gkv1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Vo P. L. H., Ronda C., Klompe S. E., Chen E. E., Acree C., Wang H. H., and Sternberg S. H. (2020) CRISPR RNA-guided integrases for high-efficiency and multiplexed bacterial genome engineering. bioRxiv 10.1101/2020.07.17.209452 [DOI] [PMC free article] [PubMed]
- 132. Rubin B. E., Diamond S., Cress B. F., Crits-Christoph A., He C., Xu M., Zhou Z., Smock D. C., Tang K., Trenton K., Krishnappa N., Jillian F., and Doudna J. A. (2020) Targeted genome editing of bacteria within microbial communities. bioRxiv 10.1101/2020.07.17.209189 [DOI]
- 133. Zebec Z., Zink I. A., Kerou M., and Schleper C. (2016) Efficient CRISPR-mediated post-transcriptional gene silencing in a hyperthermophilic archaeon using multiplexed crRNA expression. G3 (Bethesda) 6, 3161–3168 10.1534/g3.116.032482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zebec Z., Manica A., Zhang J., White M. F., and Schleper C. (2014) CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 42, 5280–5288 10.1093/nar/gku161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Fricke T., Tamulaitis G., Smalakyte D., Pastor M., Kolano A., Siksnys V., and Bochtler M. (2017) Targeted RNA knockdown by crRNA guided Csm in zebrafish. bioRxiv 10.1101/228775 [DOI] [PMC free article] [PubMed]
- 136. Nayeemul Bari S. M., and Hatoum-Aslan A. (2019) CRISPR–Cas10 assisted editing of virulent staphylococcal phages. Methods Enzymol. 616, 385–409 10.1016/bs.mie.2018.10.023 [DOI] [PubMed] [Google Scholar]
- 137. Strecker J., Ladha A., Gardner Z., Schmid-Burgk J. L., Makarova K. S., Koonin E. V., and Zhang F. (2019) RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 10.1126/science.aax9181 [DOI] [PMC free article] [PubMed] [Google Scholar]


