Abstract
The HIV-1 Gag protein is responsible for genomic RNA (gRNA) packaging and immature viral particle assembly. Although the presence of gRNA in virions is required for viral infectivity, in its absence, Gag can assemble around cellular RNAs and form particles resembling gRNA-containing particles. When gRNA is expressed, it is selectively packaged despite the presence of excess host RNA, but how it is selectively packaged is not understood. Specific recognition of a gRNA packaging signal (Psi) has been proposed to stimulate the efficient nucleation of viral assembly. However, the heterogeneity of Gag–RNA interactions renders capturing this transient nucleation complex using traditional structural biology approaches challenging. Here, we used native MS to investigate RNA binding of wild-type (WT) Gag and Gag lacking the p6 domain (GagΔp6). Both proteins bind to Psi RNA primarily as dimers, but to a control RNA primarily as monomers. The dimeric complexes on Psi RNA require an intact dimer interface within Gag. GagΔp6 binds to Psi RNA with high specificity in vitro and also selectively packages gRNA in particles produced in mammalian cells. These studies provide direct support for the idea that Gag binding to Psi specifically promotes nucleation of Gag–Gag interactions at the early stages of immature viral particle assembly in a p6-independent manner.
Keywords: human immunodeficiency virus, HIV, virus assembly, mass spectrometry, MS, RNA binding protein, RNA, RNA-protein interaction, native mass spectrometry, genomic RNA packaging, Gag, Gag p6 domain
The ability to specifically select the viral genomic RNA (gRNA) for packaging into the assembling virus particle is absolutely necessary for specific replication of HIV-1 and other retroviruses. This selection is critical as the gRNA is surrounded by a great excess of cellular RNAs, and these RNAs can also be packaged under certain conditions. Approximately 2500 copies of the viral structural protein (“Gag”) assemble around a gRNA dimer forming an immature virion (1–5). The full-length 9.4-kb viral RNA that is packaged into virions serves both as gRNA and as the mRNA for translation of all structural and enzymatic viral proteins including Gag (6). Remarkably, when expressed in cells lacking viral gRNA, Gag still forms virus-like particles (VLPs) (7). Thus, despite the critical role of gRNA for the infectivity of viral particles, Gag VLP assembly does not depend on its presence. Although there are many therapeutics in clinical use that target various steps of the viral life cycle, including entry, reverse transcription, and integration, there are currently no antiviral therapies that inhibit gRNA packaging or virion assembly (8).
The mechanism of selective packaging of HIV-1 gRNA is not well understood. The selection depends upon its “packaging signal” (Psi), a region of ∼100 bases near the 5′ end of the RNA. We have recently found that at physiological ionic strengths in vitro, Gag binds with roughly equal affinity to Psi-containing and control RNAs (9, 10). In light of these observations, we and others (7, 11, 12) have suggested that gRNA is selectively packaged because it initiates assembly more rapidly or more efficiently than other RNAs. Understanding the initial stages of this process, i.e. nucleation of Gag–Gag interactions, is an essential step in developing new therapeutics that can interfere with immature particle assembly.
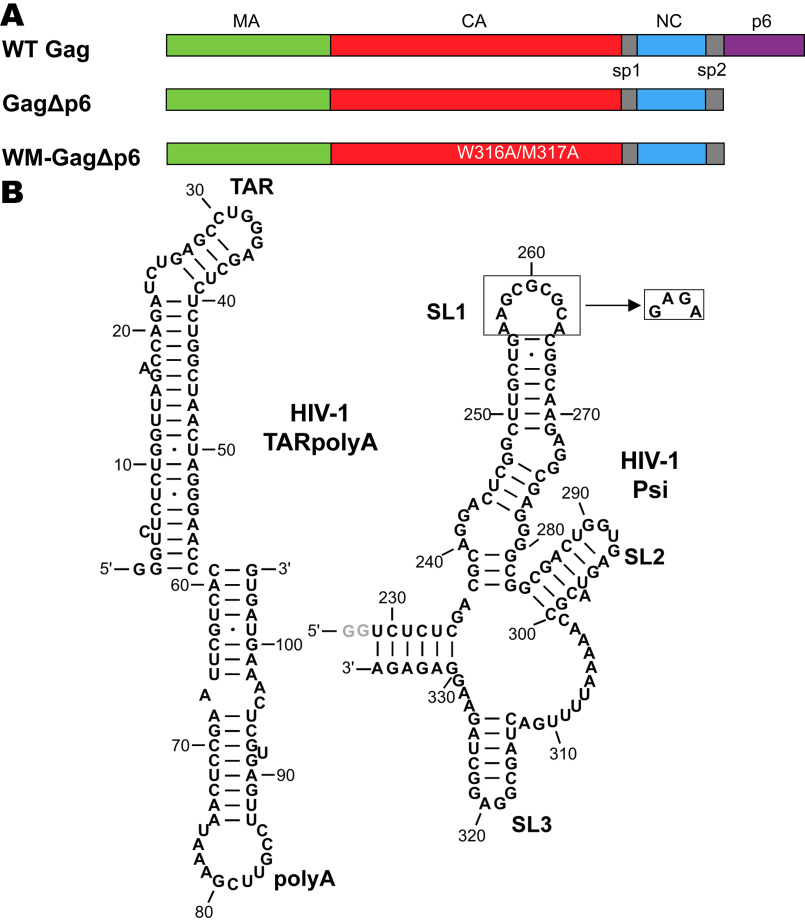
One complication in the attempt to reconcile the selective packaging observed in vivo with the in vitro binding data is that the recombinant Gag protein used in vitro has, with very few exceptions (13–16), lacked the C-terminal p6 domain. The Gag protein is composed of four major functional domains from N terminus to C terminus: matrix (MA), capsid (CA), nucleocapsid (NC), and p6, as well as the short spacer peptides SP1 and SP2 (Fig. 1A). MA is cotranslationally myristoylated (17), and during virion assembly, this domain becomes anchored to the plasma membrane via its myristoyl group (18, 19). MA is also highly basic and capable of binding RNA (18, 20–25). Gag–Gag interactions are mediated by CA and SP1, and the NC domain is primarily responsible for specific gRNA recognition (26, 27). The p6 domain facilitates the release of viral particles from the surface of virus-producing cells and is not, as far as is known, involved in the formation of the virus (2). It has frequently been omitted from the Gag protein produced in bacteria largely for reasons of convenience. However, it has recently been suggested that p6 may, in fact, function in the selective packaging of gRNA or Gag–Gag oligomerization during viral assembly (14, 15). The first goal of the experiments presented here is to determine whether the p6 domain is dispensable in the selective packaging of gRNA by Gag.
Figure 1.
A, domain structure of WT and mutant Gag proteins investigated in this work. B, sequence and predicted secondary structures of TARpolyA (left) and Psi (right) RNAs. To ensure a monomeric state, the SL1 loop of Psi was mutated to a GAGA tetraloop as indicated. The gray nucleotides at the 5′ end of Psi (GG) are not encoded by HIV-1 but were added to improve the yield of in vitro transcription.
The second major goal of this work is to investigate the role of Psi RNA binding in nucleating higher-order Gag multimerization. In the cytosol, HIV-1 Gag exists as monomers or lower-order oligomers and only forms higher-order multimers at the plasma membrane (28–30). Packaging of gRNA is initiated in the cytoplasm through specific, nonelectrostatic interactions between the NC domain of Gag and Psi (7, 22, 31, 32). The 5′ UTR regulates many stages of the viral life cycle, including genome dimerization, splicing, and initiation of reverse transcription, as well as packaging. In addition to Psi, the 5′ UTR is composed of several structural elements: the transactivation response hairpin (TAR), the poly(A) hairpin (poly(A)), and the primer binding site. Psi is composed of three stem loops (SL1, SL2, SL3) with a conserved GC-rich palindromic dimerization initiation site located within SL1 (6, 7, 33). Our previous studies showed that wild-type (WT) dimeric Psi RNA and a monomeric Psi variant were bound by Gag with similar specificity in vitro (9, 10). Thus, to simplify the data analysis, monomeric Psi RNA was used in this work. Because Gag can assemble around non-gRNA in the cell, we hypothesize that Gag–Gag interactions are facilitated by NC domain binding to Psi in a manner that is distinct from binding to non-Psi RNA sequences.
In this work, native MS (nMS) was carried out to test this hypothesis. Native MS can be used to determine the stoichiometry of RNA–protein complexes if Kd values range from low nanomolar to micromolar, with exact values depending on the mass spectrometer and the particular complex under investigation. The stoichiometry of transient RNA–protein interactions can also be determined by nMS (34); covalent crosslinking may also be used to capture such transient interactions (35–37). Here, we used a 109-nt Psi construct (Fig. 1B) that was previously characterized to display high Gag binding specificity, and the transactivation response–poly(A) hairpin element (TARPolyA or TpA) as the non-Psi sequence (Fig. 1B), as this RNA was previously shown to be characterized by low Gag binding specificity (10). The results of in vitro binding assays, nMS, and cell-based RNA packaging assays strongly support the hypothesis that Gag–Gag interactions are facilitated by specific binding to Psi RNA and that this phenomenon is independent of the p6 domain.
Results and discussion
Gag oligomeric state in the absence of RNA
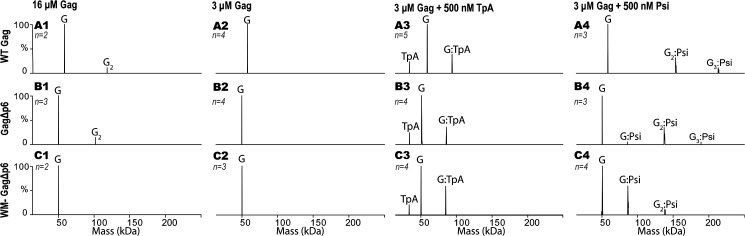
Native MS was first used to analyze the purity of the Gag constructs studied in this work (Fig. 1A). The experimental molecular masses for the monomeric and dimeric forms of each Gag construct closely match the theoretical molecular masses, confirming the high purity of the preparations and the binding of two zinc ions (Figs. S1–S3 and Table S1). We next examined the oligomeric state of free Gag proteins at multiple concentrations in the absence of any nucleic acids. The presence of the p6 domain had a modest effect on the oligomeric states observed; a lower amount of dimer was observed in the case of GagΔp6 at 12 μm relative to WT Gag at 9 μm (Figs. S1 and S2). This is consistent with a previous report showing that WT Gag has a greater propensity to oligomerize relative to GagΔp6 (14). At 16 μm, a similar extent of dimerization was observed for both proteins (Fig. 2, A1 and B1). All subsequent experiments were performed at 3 μm Gag, where both proteins alone were exclusively monomeric (Fig. 2, A2 and B2). The presence of the p6 domain influenced the charge state distribution (Figs. S1 and S2). The multimodal charge state distribution observed for WT Gag is attributed to the presence of the intrinsically disordered p6 domain (38). As expected, dimerization depended on the well-characterized dimer interface in the C-terminal domain of CA, as WM-GagΔp6 was exclusively monomeric (Fig. 2, C1 and C2, and Fig. S3) (39, 40).
Figure 2.
Zero-charge mass spectra of WT Gag (A1), GagΔp6 (B1), and WM-GagΔp6 (C1) at 16 μm; WT Gag (A2), GagΔp6 (B2), and WM-GagΔp6 (C2) at 3 μm; 500 nm TARpolyA in the presence of 3 μm WT Gag (A3), GagΔp6 (B3), and WM-GagΔp6 (C3); 500 nm Psi in the presence of 3 μm WT Gag (A4), GagΔp6 (B4), and WM-GagΔp6 (C4). Notation is as follows: G = Gag, G2 = two Gag, G3 = three Gag, TpA = TARpolyA. Molecular masses for all analytes are listed in Table S1. The number of replicates (n) per experiment are indicated on each spectrum with additional repeats shown in Figs. S1–S6.
Gag oligomeric state in the presence of RNA
We also analyzed the oligomeric state of Gag in the presence of Psi RNA and a control RNA, TARpolyA (Fig. 1B). Although nMS is a sensitive analytical platform, the analyses reported here used submicromolar concentrations of RNA and micromolar concentrations of Gag. Lower concentrations of Gag did not result in observable TpA binding under the ionic strength used here. At these protein concentrations, VLP formation was avoided by using 500 mm ammonium acetate, a salt concentration in which nonspecific Gag–RNA interactions are minimized. Gag–Psi interactions are more salt resistant than Gag–TpA interactions, because of the presence of nonelectrostatic binding modes (9, 10). Thus, the high-salt concentrations used in the nMS assays allowed viral assembly nucleation complexes to be observed while preventing VLP assembly. As expected, Gag, which is present at 6-fold molar excess over RNA in our experiments, is the dominant species detected for all Gag:RNA mixtures investigated (Fig. 2, A3, B3, C3 and A4, B4, C4, and Figs. S4–S6). Both WT Gag and GagΔp6 formed complexes with TARpolyA. These complexes were found to contain mainly one copy of Gag and a single copy of TARpolyA (Fig. 2, A3 and B3, and Figs. S4 and S5). In contrast, in the presence of Psi these proteins formed complexes comprised predominantly of two copies of Gag bound to one copy of Psi RNA, with minor formation of 3:1 Gag:RNA complexes (Fig. 2, A4 and B4, and Figs. S4 and S5).
The formation of Gag:RNA complexes with 2:1 stoichiometry may be explained either by Gag binding to RNA as a dimer or, alternatively, by two Gag molecules binding to two separate RNA-binding sites. The dimerization of Gag is mediated by an interface within CA. To determine whether complexes comprised of two Gag and one RNA molecule were formed via Gag–Gag as well as Gag–RNA interactions, we tested a Gag mutant (WM-GagΔp6) lacking this interface. WM-GagΔp6 formed complexes containing predominantly one copy of WM-GagΔp6 and one copy of RNA for both TARpolyA and Psi RNAs (Fig. 2, C3 and C4, and Fig. S6), supporting the conclusion that Psi RNA binding promotes dimerization of Gag rather than cooperative Gag binding to two independent binding sites in Psi RNA. Collectively, these observations demonstrate that the primarily monomeric WT Gag protein dimerizes by protein–protein interaction, using the WM interface, in the presence of RNA. The increased abundance of Gag dimer in the presence of Psi RNA may result from changes in the Gag conformation upon NC domain binding to Psi, which exposes dimerization interfaces although we did not examine conformational changes in our study. Previous studies have demonstrated that the mature NC domain can adopt different conformations upon binding to different RNAs (41, 42). Thus, it is feasible that the identity of the RNA dictates the conformation NC adopts when bound, which may allosterically regulate the formation of a dimerization competent Gag conformation.
Similar Psi RNA binding specificity for GagΔp6 and WT Gag in vitro
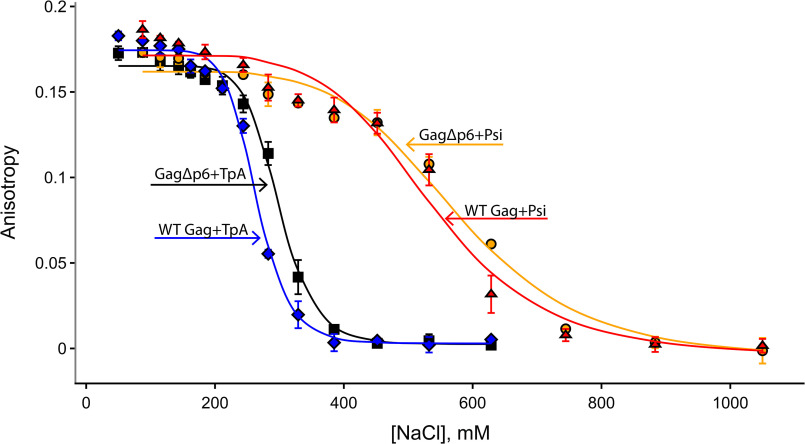
To further investigate any differences in binding specificity between WT Gag and GagΔp6, we performed fluorescence anisotropy (FA)–based salt titration binding assays (43). A nonspecific interaction between protein and RNA, mediated primarily by electrostatic interactions, will dissociate at a lower salt concentration than one that also involves specific nonelectrostatic interactions (Fig. 3 and Figs. S7 and S8 for ammonium acetate). Two parameters can be determined from this analysis: Kd,1M and Zeff. The extrapolated dissociation constant at 1 M salt (Kd,1M) reflects the strength of nonelectrostatic contributions to binding, while Zeff is a measure of the number of Na+ ions displaced from RNA upon protein binding (44, 45). Using this assay, we previously reported that the binding of GagΔp6 to Psi is far more salt-resistant than its binding to TARpolyA (10). In good agreement with these previous studies, the Kd,1M values for GagΔp6 binding to Psi and TARpolyA RNAs are 3.9 × 10−5 M and 1.5 × 10−1 M, respectively, and the values of Zeff are 6.1 and 10.8, respectively. In the case of WT Gag, the Kd,1M values for Psi and TARpolyA are 7.7 × 10−5 M and 3.4 × 10−1 M, respectively, and the Zeff values are 6.3 and 10.4, respectively. Thus, we conclude that the presence of the p6 domain does not significantly affect the specificity of Gag toward Psi RNA (Fig. 3). Importantly, titrations using ammonium acetate in place of the typical NaCl were also performed, which verified that significant binding differences between Psi and TARPolyA RNA are observed under nMS-compatible solution conditions (Figs. S7 and S8).
Figure 3.
Fluorescence anisotropy results for WT Gag and GagΔp6 binding to Psi and TARPolyA RNAs as a function of NaCl concentration (n = 3, error bars reflect the S.D. of three independent measurements). See Figs. S7 and S8 for data acquired in ammonium acetate.
Selective packaging of gRNA in cells is independent of p6
We also tested the ability of Gag with and without the p6 domain to selectively package gRNA in human cells. 293T cells were transiently transfected with our Gag expression plasmid and a plasmid expressing an HIV-derived GFP vector. This vector contains Psi and will therefore be selectively packaged by WT Gag. Packaging of the vector RNA in released virus particles was quantitated by real-time RT-PCR. Parallel experiments were performed using a mutant Gag expression plasmid in which the Gag coding region lacked the p6 domain.
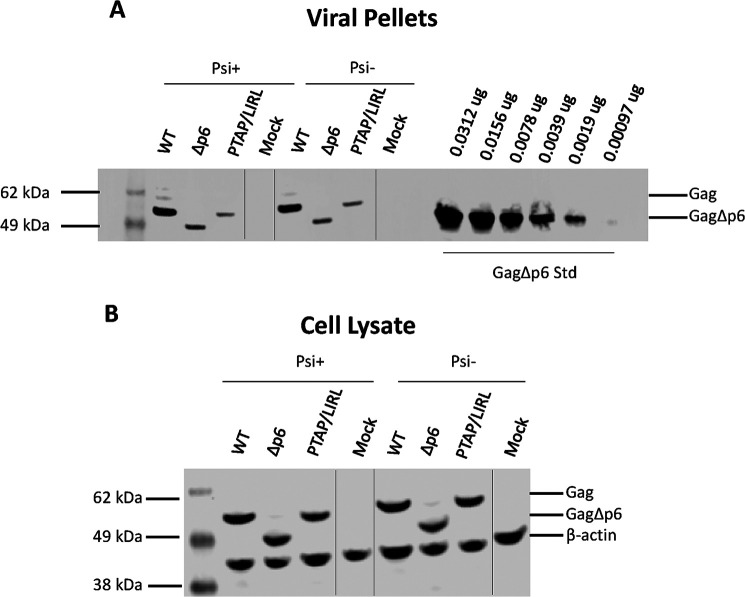
The effects of p6 removal upon virus particle production were quantified by Western blot analysis of viral pellets from the medium of the transfected cultures. GagΔp6 produced a lower level of VLPs than WT Gag. The average yield of VLPs over several experiments with GagΔp6 was 45% of that obtained in parallel transfections of WT Gag. Similar levels of WT Gag and GagΔp6 were present in lysates of the transfected cells (Fig. 4), showing that the expression of the two proteins was similar. Transmission electron micrographs of cells expressing GagΔp6 (Fig. S9) showed many partially formed and malformed VLPs, as well as assembled particles arrested at the cell surface. All of these observations are fully consistent with the known function of p6 in virus particle release (27, 46, 47).
Figure 4.
A and B, viral pellets (A) and cell lysates (B) from a representative transfection experiment were analyzed for Gag by Western blot analysis as described in “Experimental Procedures.” The cell lysates (B) were also probed for β-actin. A dilution series of recombinant GagΔp6 protein was loaded adjacent to the viral pellets (A) so that a standard curve could be constructed (Fig. S12). In the “mock” samples, no Gag plasmid was transfected, but the vector (Psi+ or Psi−, as indicated) was. The vertical lines in panels A and B indicate where lanes unrelated to this study were spliced out of the gel photo.
RNA was also extracted from the pellets and assayed for the vector RNA; the results of these measurements, corrected for the difference in virus particle content, are shown in Fig. 5A. It is evident that the ratio of vector RNA to Gag protein in the viral pellets from GagΔp6 is only ∼4% of the ratio in the WT Gag pellets.
Figure 5.
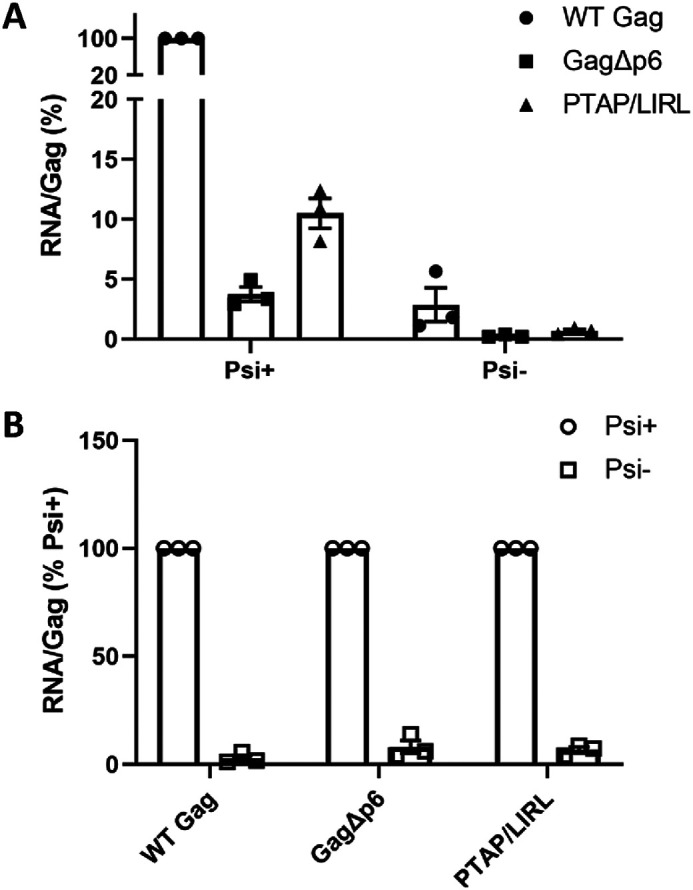
RNA packaging by mutant Gag proteins. The figure shows the results of three separate transfections. In each case, viral pellets were assayed for copies of the vector RNA and for Gag content as described in “Experimental Procedures.” A, the ratio of these two quantities in the WT control was set to 100. B, for each Gag, the ratio of these two quantities for Psi+ was set to 100.
The low level of vector RNA in the GagΔp6 pellets might indicate that GagΔp6 has completely lost the ability to selectively package Psi-containing RNA; perhaps this level represents nonspecific packaging, as seen with nearly any cellular mRNA in the absence of Psi-specific packaging of gRNA (48). Alternatively, the ability to selectively package Psi-containing RNA might be retained but somewhat diminished in GagΔp6. To distinguish between these possibilities, we measured the packaging of a derivative of the vector lacking Psi (Psi−). As shown in Fig. 5B, the deletion of Psi profoundly reduced the packaging of the vector RNA by GagΔp6 (as well as by WT Gag), so that the RNA:Gag ratio in the pellets was <10% of that seen with the Psi+ vector. These data show that the GagΔp6 still retains, to a very significant extent, the ability to preferentially package Psi-containing RNA.
The p6 domain functions in the interactions of Gag with the cellular ESCRT machinery during virus particle budding from virus-producing cells. Thus, it is conceivable that the ablation of this interaction in GagΔp6 is somehow responsible for the reduction in packaging of Psi+ RNA. Indeed, as noted above, malformed or partial virions can be seen in electron micrographs of transfected cells (Fig. S9); perhaps some of these are released without packaging gRNA, or perhaps the malformed GagΔp6 particles fail to protect their RNA cargo from degradation. To explore this possibility, we also measured packaging of the vector RNA by full-length Gag in which the PTAP motif, the site in p6 that interacts with the ESCRT component Tsg101, was replaced with LIRL (47). This mutation, like the removal of p6, modestly reduced the level of particle production (Fig. 4) and resulted in the formation of misshapen particles (Fig. S9). As shown in Fig. 5A, these pellets also show a far lower ratio of vector RNA to Gag protein than those formed by WT Gag. As with WT and GagΔp6, we also found that removal of Psi from the vector significantly reduced its packaging by the PTAP mutant Gag, showing that the packaging of the intact vector by this mutant Gag is Psi-specific (Fig. 5B). Taken together, the data suggest that the reduction in Psi-specific RNA packaging by GagΔp6 is a result of its defective interaction with ESCRT machinery, and that it has not lost specificity in its interactions with RNA. It has previously been reported that virions assembled from GagΔp6 or PTAP–Gag are deficient in reverse transcriptase and integrase (47); it seems likely that gRNA, like these internal viral proteins, is lost from or degraded within the budding mutant virions during the extended period between assembly and release from the cell.
Conclusions
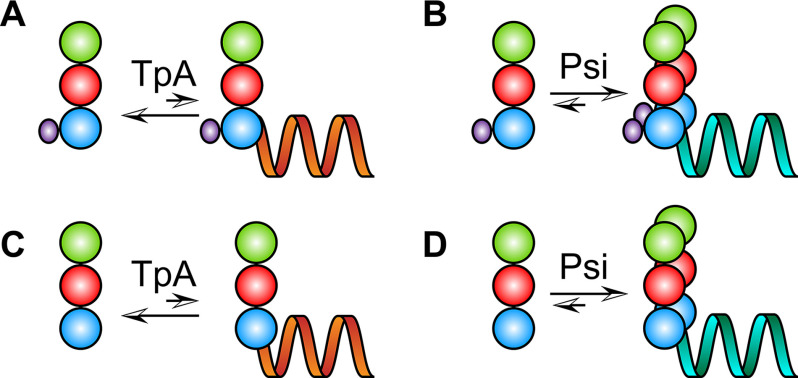
The data presented here lead to two main conclusions regarding the interactions between Gag, the structural protein of HIV-1 virus particles, and RNA. First, the presence or absence of the p6 domain has minimal effect on Psi interactions in in vitro binding assays and does not appear to significantly affect the packaging of viral RNA in virus-producing cells, except for an effect attributable to the “late domain” mutant phenotype seen with this truncated Gag protein. Second, Gag (with or without p6) dimerization is promoted when it binds the Psi RNA packaging signal, but far less dimer is observed when it binds a control RNA (Fig. 2).
A previous report used short RNA oligonucleotides to mimic RNA interactions that may occur during viral assembly (14). Differences in the binding thermodynamic parameters between WT Gag and GagΔp6 were observed in the case of A-rich RNAs, whereas similar parameters were obtained for both proteins when binding to GU-rich RNA and to the SL3 Psi hairpin. These data suggest a role for p6 in driving Gag–Gag oligomerization (see also Figs. S1 and S2) and assembly but not in initial Psi RNA selection, in good agreement with our data (Figs. 2–5).
Another recent report indicated that p6 plays a role in selective Psi RNA binding in vitro (15). The latter work used different RNA constructs and different analytical techniques from those employed here, and one or more of these differences is presumably responsible for the discrepancy. However, we found no evidence supporting a role for p6 in the interactions of Gag with RNAs, either in vitro or in vivo.
A significant effect of the p6 domain on Gag/RNA interactions, if it had been found, would have important implications for our understanding of virus assembly. This is because the vast majority of in vitro studies with recombinant Gag protein have used a truncated protein lacking p6. The findings presented here thus provide important support for the relevance of these studies to the virus-assembly problem.
Multimerization of Gag upon nucleic acid binding has been previously investigated both in vitro and in cells (49, 50). However, the stronger tendency for Gag to multimerize on Psi RNA relative to non-Psi RNA has, to our knowledge, not been previously reported. The dimerization of Gag protein on Psi RNA would appear to have major ramifications regarding virus particle assembly. Gag normally packages Psi-containing RNA with very high selectivity, despite the presence of a substantial excess of cellular mRNA species that can also be packaged. The mechanism of this selective packaging is not fully understood. It has previously been reported (9, 10) that at physiological ionic strengths, Gag binds with similar high affinities to RNAs with or without the packaging signal. Thus, the selection of Psi+ RNA during virus assembly is evidently not because of a uniquely high affinity of Gag for this RNA. We and others have suggested that binding to Psi induces a nucleating event, such as formation of a small Gag oligomer, more efficiently than binding to other RNAs, and that this difference might underlie the selective packaging of Psi-containing RNA (7, 11, 12, 51–53). The present results provide very strong support for this fundamental concept.
A model for the nucleation of HIV-1 viral assembly has previously been proposed (10, 31, 40, 54, 55). In this model, Gag exists in an equilibrium between bent and extended conformations. We previously hypothesized that different Gag conformations are adopted while binding to different RNAs (10). When Gag binds Psi, the extended conformation is favored, which allows rapid dimerization of Gag via CA–CA (39, 40), SP1–SP1 (56, 57), and potentially NC–NC interactions (50). In contrast, the bent conformation is preferred for binding to TARpolyA, requiring a rate-limiting conformational switch before dimerization can occur. In the bent conformation, MA binding to the nucleic acid may also further stabilize this nonproductive complex.
Although the work presented here does not test the Gag conformational changes, we demonstrate that Gag (with or without p6) dimerizes more efficiently on Psi RNA than on a similarly sized non-Psi RNA (Fig. 6). The difference in Gag–RNA binding stoichiometry may be because of the conformational differences discussed above; however, additional studies will be required to determine exactly how specific RNAs modulate the conformation of Gag.
Figure 6.
Model showing WT Gag and GagΔp6 binding to Psi and TARpolyA based on the results of this work. In the presence of TARpolyA (orange), WT Gag (panel A) and GagΔp6 (panel C) bind primarily as monomers, whereas the presence of Psi (teal) promotes dimerization of both proteins (panels B and D). Gag domains are colored as follows: MA (green), CA (red), NC (blue), and p6 (purple).
The capability of nMS to characterize disordered and heterogeneous systems has allowed us to gain new insights into viral assembly. In the future, additional MS-based technologies such as ion-mobility MS, RNA–protein crosslinking approaches, and covalent labeling may provide additional insight into RNA-induced Gag conformational changes and virion assembly. The versatility of nMS should also be helpful in the future investigation of Gag binding to the full-length gRNA 5′ UTR.
Experimental Procedures
Protein purification
WT HIV-1 BH10 Gag, GagΔp6, and WM-GagΔp6 (Fig. 1A) were expressed in Escherichia coli (BL21(DE3)pLysS) and purified as described previously (58), with the following alterations. Soluble lysate fractions were treated with polyethylenimine to precipitate endogenous nucleic acids prior to the ammonium sulfate precipitation step. The pellet was resuspended in 100 mm NaCl, 20 mm Tris-HCl, pH 7.4, 10 mm 2-mercaptoethanol (βME) and 1 μm ZnCl2, loaded onto a HiTrap Heparin HP affinity column (GE Healthcare) equilibrated in the same buffer, and eluted with 0.75–1 M NaCl as described (59).
The purification of WT HIV-1 Gag was identical to that of GagΔp6 except for the addition of a cleavable C-terminal His6-tag. The Gag-His6 protein was loaded onto a HIS-select affinity column (Sigma), washed (20 mm Tris-HCl, pH 7.4, 500 mm NaCl, 5 mm βME, 1 μm ZnCl2, 0.1% Tween 20, 5 mm imidazole) and eluted in the same buffer using a step gradient of 10 mm, 20 mm, 50 mm, 75 mm, and 150 mm imidazole. Gag eluted in the 20–75 mm imidazole fractions. These fractions were pooled and dialyzed overnight into the same buffer without imidazole using a Slide-A-Lyzer Dialysis Cassette (20,000 MWCO, Thermo Scientific). During dialysis, the His-tag was cleaved using 1 mg tobacco etch virus (TEV) protease/20 mg of Gag. The TEV protease was purified in house as described (60).
WT Gag was further purified by elution through a HiTrap Heparin HP affinity column as described above. All steps were carried out at 4°C and in all cases, the purest protein fractions were pooled for use in salt-titration binding and nMS studies (Fig. S10). For nMS studies, purified Gag proteins were dialyzed (3500 MWCO dialysis tubing, BioDesign Inc., New York) into 500 mm ammonium acetate, pH 6.8, and concentrated using an Amicon-ultra 30,000 MWCO ultrafiltration device (Millipore-Sigma). For salt-titration binding assays, the proteins were dialyzed using Slide-A-Lyzer Dialysis Cassette (20,000 MWCO) into 20 mm Tris-HCl, pH 7.4, 500 mm NaCl, 10% glycerol, 10 mm βME, and 1 μm ZnCl2. Protein concentrations were determined by measuring the absorbance at 280 nm using an extinction coefficient of 63,223 m−1cm−1.
RNA preparation
HIV-1 NL4-3 TARPolyA and Psi RNAs shown in Fig. 1B were generated via T7 RNA polymerase-directed in vitro transcription as described (61) from either a TARPolyA-encoding pUC19 plasmid or a Psi-encoding pIDTsmart plasmid (Integrated DNA Technologies).Plasmids were linearized via FOK1 digestion prior to transcription. TARPolyA was preceded by a hammerhead ribozyme designed to cleave during the transcription reaction, resulting in the desired 5′ end. A hammerhead ribozyme construct was only used in the case of TARpolyA as it significantly improved the homogeneity of this RNA, as analyzed by nMS. The Psi RNA was engineered with two additional guanosines at the 5′ end to improve the yield of in vitro transcription. This Psi construct was shown previously to be recognized by Gag with high specificity (10). RNAs were purified by electrophoresis on 6% polyacrylamide/8 m urea denaturing gels followed by elution and concentration via the crush and soak method (61). RNA quality was checked on 6% polyacrylamide/8 m urea denaturing gels with ethidium bromide staining (not shown) and by MS (Fig. S11).
Fluorescence anisotropy binding assays
RNAs used in FA-based salt titration binding assays were labeled with fluorescein-5-thiosemicarbazide (FTSC) at the 3′ end as described (43). The labeling efficiency of the RNAs was determined by measuring the absorbance at 260 nm and 495 nm and using the extinction coefficients ε495 nm = 8.5 × 104 m−1·cm−1 for FTSC and ε260 nm = 9.3 × 105 m−1·cm−1 and 9.7 × 105 m−1·cm−1 for TARpolyA and Psi RNAs, respectively. A correction factor (ε260 nm/ε495 nm = 0.3266) was used to correct the RNA concentration for the absorbance of the dye at 260 nm. The NaCl salt titration binding assays were performed and the data analyzed as described (43). A very similar approach was used to determine whether Gag:RNA binding was affected by nMS-compatible solution conditions. For these measurements, all nonvolatile salts were replaced with ammonium acetate in both the RNA and the Gag samples. FTSC-labeled RNAs were refolded in 50 mm ammonium acetate (pH 6.8) and 1 mm magnesium acetate by heating (90°C, 2 min) and snap-cooling (ice, 2 min) followed by incubation at 37°C for 30 min. The GagΔp6 protein was dialyzed overnight in 500 mm ammonium acetate, pH 6.8, prior to carrying out the ammonium acetate salt titration binding assays and data analysis as previously described (43).
Native MS
Analysis of the three Gag constructs (WT Gag, GagΔp6, and WM-GagΔp6) in the absence of RNA was carried out at both 3 and 16 μm protein in 500 mm ammonium acetate, pH 6.8. Prior to nMS analysis, RNAs were refolded as described above. Gag:RNA complexes were formed by mixing 3 μm Gag with 0.5 μm refolded RNA at room temperature. Gag samples were stored on ice prior to analysis whereas reactions containing Gag and RNA were incubated at room temperature for 15–30 min prior to analysis. An aliquot (1–3 μl) was loaded into a borosilicate glass nano-electrospray ionization (nESI) emitter prepared in house on a Sutter p-97 micropipette puller and directly injected into a Thermo Fisher Scientific Q ExactiveTM UHMR Hybrid Quadrupole-OrbitrapTM mass spectrometer. Separate nESI emitters were used for each measurement. The instrument settings were as follows: capillary temperature, 225°C; S-lens RF level, 200; high-energy collision dissociation, 0 V; in-source fragmentation, 0 V; in-source trapping, −100 V; trap gas, 8 (arbitrary units); m/z range, 500–80,000. Nano-electrospray ionization was initiated by applying voltage (0.9–1.1 kV) to the sample via a platinum wire that was inserted into the nESI emitter. It was observed that signal intensity of all species requires several minutes of nESI to stabilize, as shown in Figs. S4–S6. Backing pressure on the electrospray decreased the initial spray differences caused by starting the spray by voltage change only; this suggests different electrophoretic mobilities of RNA and protein, as might be expected, or differences in adsorption of the RNA and protein to the borosilicate capillaries used. Once the spray is stabilized, the dominant species in all spectra is the species in concentration excess, as would be expected (Gag or Gag mutant). Because of possible differences in ionization efficiencies of RNA and protein, we focus on our main question of interest, the stoichiometries of the RNA–protein complexes. A minimum of three technical repeats were performed for each combination of Gag and RNA. Mass spectra were deconvolved using UniDec v4.1 (62, 63). Theoretical molecular weights were calculated based on protein and RNA sequences (provided in Table S2) using the protein/RNA molecular weight calculator tool in UniDec.
Constructs for analysis of RNA packaging
Gag was expressed in mammalian cells from pCMV55M1-10 (64), a Rev-independent version of the HXB2 Gag gene. This plasmid does not contain Psi. We measured packaging of the RNA from an HIV-1–derived GFP vector constructed from pLenti6/V5-DEST (Thermo Fisher). A Psi− version of this vector was constructed, using inverse PCR, by deleting nucleotides corresponding to 214–365 of NL4-3 RNA; the deleted stretch encompasses the stem loops SL1–SL4. Mutants of the Gag gene in pCMV55M1-10 were generated by inverse PCR. All constructs were confirmed by sequencing of the entire coding region.
Transfection
HEK293T/17 cells were seeded at a density of 4 × 106 cells in a 10-cm cell culture dish. The following day they were transfected with 6 μg Gag plasmid + 6 μg vector plasmid + 3 μg pCMV-Rev (to support nuclear export of vector RNA) using Transit-293 (Mirus) following the manufacturer's protocol. Cell culture supernatants were collected after 48 h and 72 h and filtered through 0.22-μm syringe filters (Merck Millipore). Filtered supernatants were stored at −80 °C until further analysis. The transfected cells were lysed at 72 h for immunoblotting as described below. The results presented represent three independent transfections. Transfected cultures were also analyzed by transmission EM. Cells were fixed 48 h after transfection and processed as described previously (55).
RNA analysis
For extracting RNA from VLPs, the filtered cell culture fluid was ultracentrifuged (25,000 rpm, 4°C, SW55Ti Rotor, Beckman Coulter) through a 20% sucrose cushion prepared in TNE buffer (10 mm Tris-HCl, 100 mm NaCl, and 1 mm EDTA, pH 7.5). The VLPs in the pellet were lysed by adding PK Lysis buffer (50 mm Tris-HCl, 100 mm NaCl, 10 mm EDTA, 1% SDS, 100 μg/ml proteinase K, pH 7.5) and incubating at 37°C for 30 min. RNA was extracted from the lysed VLPs using TriReagent (Ambion) following the manufacturer's protocol. Glycoblue (Ambion) was used as a carrier in the precipitation step. The RNA pellet was resuspended in nuclease-free water and stored at −80 °C until further analysis.
Copies of sequences in the RNA preparations were then enumerated by real-time RT-PCR. RNA representing ∼350 μl of culture fluid was treated with DNaseI (RNase free, Ambion) at 37°C for 30 min in a total reaction volume of 10 μl. The DNaseI was inactivated by adding 1 μl of 50 mm EDTA to the reaction followed by heat inactivation at 75°C for 10 min. First-strand synthesis was performed using an iScript cDNA synthesis kit (Bio-Rad) in a total reaction volume of 20 μl following the manufacturer's protocol. This DNA was then analyzed by SYBR Green–based (FastStart Essential DNA Green Master, Roche) real-time PCR. Standard curves were generated from serial dilutions of a transcript containing the target sequence. These transcripts were digested with DNaseI and the transcripts were cleaned up twice using RNeasy spin columns (Qiagen) after in vitro transcription. RNA transcripts were quantitated using A260 and Ribogreen (Thermo Fisher). In the real-time RT-PCR assays, HIV-1–derived packageable RNA was amplified using primers: 556 forward: 5′-AAACAAAAGTAAGACCACCGCAC-3′ and 556 reverse: 5′-ACCACTCTTCTCTTTGCCTTGG-3′, spanning nucleotides 891–1047 of pLenti6/V5-DEST RNA. All real-time RT-PCR assays also included a no-RT control, which gave extremely low RNA copy numbers, indicating that the experimental values represent RNA copy numbers without significant DNA contamination.
Immunoblotting
Gag in VLPs was quantitated as follows. VLPs were pelleted through a 20% sucrose cushion in TNE buffer. Pellets were resuspended in 1× NuPAGE sample dye containing reducing agent and 1× HALT protease inhibitors (Invitrogen) and stored at −80°C until further analysis. For detecting intracellular HIV-1 Gag, cells were lysed using the same 1× NuPAGE sample dye mixture (Invitrogen), sonicated for complete lysis and stored at −80°C. Prior to electrophoresis, samples were thawed and heated at 90°C for 5 min. Electrophoresis was carried out using NuPAGE 4 to 12% Bis-Tris polyacrylamide gels (Invitrogen) in 1× NuPAGE buffer followed by transfer to Immobilon-FL polyvinylidene difluoride membrane (Millipore). Membrane was blocked using Intercept Blocking Buffer (LI-COR); this was followed by the addition of primary antibodies (goat anti-p24 from National Institutes of Health, and mouse anti-actin, Santa Cruz Biotechnology, C4 sc-47778) diluted in the blocking buffer and incubated overnight at 4°C. Membranes were washed thrice with TBS (20 mm Tris-HCl, pH 7.5, 500 mm NaCl) and probed with secondary antibodies conjugated to Dylight 800 or 700 (LI-COR). Blots were imaged using the LI-COR Odyssey imaging system and images were analyzed using ImageStudioLite. Absolute quantities of Gag protein were obtained by reference to a standard curve prepared using recombinant GagΔp6 (a kind gift of S. Datta, NCI), an example of which is in Fig. S12. Sample band intensities were within the linear range of the standard curve.
Data Availability
All data are contained within the article.
Supplementary Material
Acknowledgments
We thank Drs. Zac VanAernum and Florian Busch for mass spectrometry assistance and discussion and Drs. Ioulia Rouzina and Siddhartha Datta for helpful discussions. We also thank Ferri Soheilian and Kunio Nagashima for electron microscopy and Dr. Barbara Felber (NCI-Frederick) for the gift of the pCMV55M1-10 and pCMV-Rev plasmids.
This article contains supporting information.
Author contributions—S. S., B. B., E. D. O., A. R., V. H. W., and K. M.-F. conceptualization; S. S., B. B., S. L., E. D. O., and J. P. K. data curation; S. S., B. B., S. L., A. R., and V. H. W. formal analysis; S. S., B. B., and S. L. validation; S. S., B. B., S. L., A. R., V. H. W., and K. M.-F. investigation; S. S., B. B., and S. L. visualization; S. S., B. B., A. R., and K. M.-F. writing-original draft; S. S., B. B., S. L., E. D. O., J. P. K., A. R., V. H. W., and K. M.-F. writing-review and editing; B. B., S. L., and E. D. O. methodology; A. R., V. H. W., and K. M.-F. resources; A. R., V. H. W., and K. M.-F. supervision; A. R., V. H. W., and K. M.-F. funding acquisition; A. R., V. H. W., and K. M.-F. project administration; J. P. K. preparation of RNAs for forming Gag-RNA complexes and help writing and editing manuscript.
Funding and additional information—This work was supported by the National Institutes of Health Grants P41 GM128577 (to V. H. W.) for driving technology development for RNA:protein complexes, RO1 AI153216 (to K. M.-F.) for FA binding and native MS studies, U54 AI150472 (to K. M.-F.) for protein and RNA preparation, and T32 GM118291 (to J. P. K.). This study was also supported by the Intramural Research Program of the Center for Cancer Research, NCI, National Institutes of Health, and in part with funds from the Intramural AIDS Targeted Antiviral Therapy Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- gRNA
- genomic RNA
- VLP
- virus-like particle
- Psi
- packaging signal
- MA
- matrix domain
- CA
- capsid domain
- NC
- nucleocapsid domain
- TAR
- transactivation response hairpin
- poly(A)
- poly(A) hairpin
- nMS
- native MS
- FA
- fluorescence anisotropy
- βME
- 2-mercaptoethanol
- TEV
- tobacco etch virus
- MWCO
- molecular weight cutoff
- FTSC
- fluorescein-5-thiosemicarbazide
- nESI
- nano-electrospray ionization.
References
- 1. Bieniasz P., and Telesnitsky A. (2018) Multiple, switchable protein:RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu. Rev. Virol. 5, 165–183 10.1146/annurev-virology-092917-043448 [DOI] [PubMed] [Google Scholar]
- 2. Sundquist W. I., and Kräusslich H.-G. (2012) HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2, a006924 10.1101/cshperspect.a006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turner B. G., and Summers M. F. (1999) Structural biology of HIV. J. Mol. Biol. 285, 1–32 10.1006/jmbi.1998.2354 [DOI] [PubMed] [Google Scholar]
- 4. Wilk T., and Fuller S. D. (1999) Towards the structure of the human immunodeficiency virus: Divide and conquer. Curr. Opin. Struct. Biol. 9, 231–243 10.1016/S0959-440X(99)80033-5 [DOI] [PubMed] [Google Scholar]
- 5. Chen Y., Wu B., Musier-Forsyth K., Mansky L. M., and Mueller J. D. (2009) Fluorescence fluctuation spectroscopy on viral-like particles reveals variable Gag stoichiometry. Biophys. J. 96, 1961–1969 10.1016/j.bpj.2008.10.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuzembayeva M., Dilley K., Sardo L., and Hu W.-S. (2014) Life of Psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454–455, 362–370 10.1016/j.virol.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rein A. (2019) RNA Packaging in HIV. Trends Microbiol. 27, 715–723 10.1016/j.tim.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puhl A. C., Garzino Demo A., Makarov V. A., and Ekins S. (2019) New targets for HIV drug discovery. Drug Discov. Today 24, 1139–1147 10.1016/j.drudis.2019.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comas-Garcia M., Datta S. A., Baker L., Varma R., Gudla P. R., and Rein A. (2017) Dissection of specific binding of HIV-1 Gag to the 'packaging signal' in viral RNA. eLife 6, e27055 10.7554/eLife.27055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Webb J. A., Jones C. P., Parent L. J., Rouzina I., and Musier-Forsyth K. (2013) Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: Implications for viral genomic RNA packaging. RNA 19, 1078–1088 10.1261/rna.038869.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dilley K. A., Nikolaitchik O. A., Galli A., Burdick R. C., Levine L., Li K., Rein A., Pathak V. K., and Hu W.-S. (2017) Interactions between HIV-1 Gag and viral RNA genome enhance virion assembly. J. Virol. 91, e02319–16 10.1128/JVI.02319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nikolaitchik O. A., Dilley K. A., Fu W., Gorelick R. J., Tai S.-H. S., Soheilian F., Ptak R. G., Nagashima K., Pathak V. K., and Hu W.-S. (2013) Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 9, e1003249 10.1371/journal.ppat.1003249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abd El-Wahab E. W., Smyth R. P., Mailler E., Bernacchi S., Vivet-Boudou V., Hijnen M., Jossinet F., Mak J., Paillart J.-C., and Marquet R. (2014) Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat. Commun. 5, 4304 10.1038/ncomms5304 [DOI] [PubMed] [Google Scholar]
- 14. Tanwar H. S., Khoo K. K., Garvey M., Waddington L., Leis A., Hijnen M., Velkov T., Dumsday G. J., McKinstry W. J., and Mak J. (2017) The thermodynamics of Pr55Gag-RNA interaction regulate the assembly of HIV. PLoS Pathog. 13, e1006221 10.1371/journal.ppat.1006221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dubois N., Khoo K. K., Ghossein S., Seissler T., Wolff P., McKinstry W. J., Mak J., Paillart J.-C., Marquet R., and Bernacchi S. (2018) The C-terminal p6 domain of the HIV-1 Pr55Gag precursor is required for specific binding to the genomic RNA. RNA Biol. 15, 923–936 10.1080/15476286.2018.1481696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bernacchi S., El-Wahab E. W. A., Dubois N., Hijnen M., Smyth R. P., Mak J., Marquet R., and Paillart J.-C. (2017) HIV-1 Pr55Gag binds genomic and spliced RNAs with different affinity and stoichiometry. RNA Biol. 14, 90–103 10.1080/15476286.2016.1256533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schultz A. M., Henderson L. E., and Oroszlan S. (1988) Fatty acylation of proteins. Annu. Rev. Cell Biol. 4, 611–647 10.1146/annurev.cb.04.110188.003143 [DOI] [PubMed] [Google Scholar]
- 18. Wang C. T., and Barklis E. (1993) Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J. Virol. 67, 4264–4273 10.1128/JVI.67.7.4264-4273.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bryant M., and Ratner L. (1990) Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. U. S. A. 87, 523–527 10.1073/pnas.87.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hearps A. C., Wagstaff K. M., Piller S. C., and Jans D. A. (2008) The N-terminal basic domain of the HIV-1 matrix protein does not contain a conventional nuclear localization sequence but is required for DNA binding and protein self-association. Biochemistry 47, 2199–2210 10.1021/bi701360j [DOI] [PubMed] [Google Scholar]
- 21. Kutluay S. B., Zang T., Blanco-Melo D., Powell C., Jannain D., Errando M., and Bieniasz P. D. (2014) Global changes in the RNA binding specificity of HIV-1 Gag regulate virion genesis. Cell 159, 1096–1109 10.1016/j.cell.2014.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rye-McCurdy T., Olson E. D., Liu S., Binkley C., Reyes J.-P., Thompson B. R., Flanagan J. M., Parent L. J., and Musier-Forsyth K. (2016) Functional equivalence of retroviral MA domains in facilitating Psi RNA binding specificity by Gag. Viruses 8, 256 10.3390/v8090256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parent L. J., and Gudleski N. (2011) Beyond plasma membrane targeting: Role of the MA domain of Gag in retroviral genome encapsidation. J. Mol. Biol. 410, 553–564 10.1016/j.jmb.2011.04.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alfadhli A., McNett H., Tsagli S., Bächinger H. P., Peyton D. H., and Barklis E. (2011) HIV-1 matrix protein binding to RNA. J. Mol. Biol. 410, 653–666 10.1016/j.jmb.2011.04.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chukkapalli V., Oh S. J., and Ono A. (2010) Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U. S. A. 107, 1600–1605 10.1073/pnas.0908661107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bell N. M., and Lever A. M. L. (2013) HIV Gag polyprotein: Processing and early viral particle assembly. Trends Microbiol. 21, 136–144 10.1016/j.tim.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 27. Freed E. O. (2015) HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 13, 484–496 10.1038/nrmicro3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hendrix J., Baumgärtel V., Schrimpf W., Ivanchenko S., Digman M. A., Gratton E., Kräusslich H.-G., Müller B., and Lamb D. C. (2015) Live-cell observation of cytosolic HIV-1 assembly onset reveals RNA-interacting Gag oligomers. J. Cell Biol. 210, 629–646 10.1083/jcb.201504006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fogarty K. H., Berk S., Grigsby I. F., Chen Y., Mansky L. M., and Mueller J. D. (2014) Interrelationship between cytoplasmic retroviral Gag concentration and Gag–membrane association. J. Mol. Biol. 426, 1611–1624 10.1016/j.jmb.2013.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kutluay S. B., and Bieniasz P. D. (2010) Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 6, e1001200 10.1371/journal.ppat.1001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olson E. D., and Musier-Forsyth K. (2019) Retroviral Gag protein–RNA interactions: Implications for specific genomic RNA packaging and virion assembly. Semin. Cell Dev. Biol. 86, 129–139 10.1016/j.semcdb.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu K., Heng X., and Summers M. F. (2011) Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 410, 609–633 10.1016/j.jmb.2011.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keane S. C., Heng X., Lu K., Kharytonchyk S., Ramakrishnan V., Carter G., Barton S., Hosic A., Florwick A., Santos J., Bolden N. C., McCowin S., Case D. A., Johnson B. A., Salemi M., et al. (2015) Structure of the HIV-1 RNA packaging signal. Science 348, 917–921 10.1126/science.aaa9266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen L., Tanimoto A., So B. R., Bakhtina M., Magliery T. J., Wysocki V. H., and Musier-Forsyth K. (2019) Stoichiometry of triple-sieve tRNA editing complex ensures fidelity of aminoacyl-tRNA formation. Nucleic Acids Res. 47, 929–940 10.1093/nar/gky1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dybkov O., Urlaub H., Lührmann R., (2014) Protein–RNA crosslinking in native ribonucleoprotein particles. In Handbook of RNA Biochemistry (Hartmann R. K., Bindereif A., and Schön A., and Westhof E., eds.), pp. 1029–1054, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 36. Kramer K., Sachsenberg T., Beckmann B. M., Qamar S., Boon K.-L., Hentze M. W., Kohlbacher O., and Urlaub H. (2014) Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods 11, 1064–1070 10.1038/nmeth.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harris M. E., (2014) Probing RNA solution structure by photocrosslinking: Incorporation of photoreactive groups at RNA termini and determination of crosslinked sites by primer extension. In Handbook of RNA Biochemistry (Hartmann R. K., Bindereif A., and Schön A., and Westhof E., eds.), pp. 231–254, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 38. Li J., Lyu W., Rossetti G., Konijnenberg A., Natalello A., Ippoliti E., Orozco M., Sobott F., Grandori R., and Carloni P. (2017) Proton dynamics in protein mass spectrometry. J. Phys. Chem. Lett. 8, 1105–1112 10.1021/acs.jpclett.7b00127 [DOI] [PubMed] [Google Scholar]
- 39. Gamble T. R., Yoo S., Vajdos F. F., Schwedler U. K., von, Worthylake D. K., Wang H., McCutcheon J. P., Sundquist W. I., and Hill C. P. (1997) Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278, 849–853 10.1126/science.278.5339.849 [DOI] [PubMed] [Google Scholar]
- 40. Datta S. A. K., Zhao Z., Clark P. K., Tarasov S., Alexandratos J. N., Campbell S. J., Kvaratskhelia M., Lebowitz J., and Rein A. (2007) Interactions between HIV-1 Gag molecules in solution: An inositol phosphate-mediated switch. J. Mol. Biol. 365, 799–811 10.1016/j.jmb.2006.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. De Guzman R. N., Wu Z. R., Stalling C. C., Pappalardo L., Borer P. N., and Summers M. F. (1998) Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science 279, 384–388 10.1126/science.279.5349.384 [DOI] [PubMed] [Google Scholar]
- 42. Amarasinghe G. K., De Guzman R. N., Turner R. B., Chancellor K. J., Wu Z. R., and Summers M. F. (2000) NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301, 491–511 10.1006/jmbi.2000.3979 [DOI] [PubMed] [Google Scholar]
- 43. Rye-McCurdy T., Rouzina I., and Musier-Forsyth K. (2015) Fluorescence anisotropy-based salt-titration approach to characterize protein-nucleic acid interactions. Methods Mol. Biol. 1259, 385–402 10.1007/978-1-4939-2214-7_23 [DOI] [PubMed] [Google Scholar]
- 44. Record M. T., Lohman T. M., and Haseth P. D. (1976) Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 107, 145–158 10.1016/S0022-2836(76)80023-X [DOI] [PubMed] [Google Scholar]
- 45. Rouzina I., and Bloomfield V. A. (1997) Competitive electrostatic binding of charged ligands to polyelectrolytes: Practical approach using the non-linear Poisson-Boltzmann equation. Biophys. Chem. 64, 139–155 10.1016/S0301-4622(96)02231-4 [DOI] [PubMed] [Google Scholar]
- 46. Freed E. O. (1998) HIV-1 gag proteins: Diverse functions in the virus life cycle. Virology 251, 1–15 10.1006/viro.1998.9398 [DOI] [PubMed] [Google Scholar]
- 47. Demirov D. G., Orenstein J. M., and Freed E. O. (2002) The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76, 105–117 10.1128/jvi.76.1.105-117.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rulli S. J., Hibbert C. S., Mirro J., Pederson T., Biswal S., and Rein A. (2007) Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 81, 6623–6631 10.1128/JVI.02833-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. El Meshri S. E., Dujardin D., Godet J., Richert L., Boudier C., Darlix J. L., Didier P., Mély Y., and de Rocquigny H. (2015) Role of the nucleocapsid domain in HIV-1 Gag oligomerization and trafficking to the plasma membrane: A fluorescence lifetime imaging microscopy investigation. J. Mol. Biol. 427, 1480–1494 10.1016/j.jmb.2015.01.015 [DOI] [PubMed] [Google Scholar]
- 50. Zhao H., Datta S. A. K., Kim S. H., To S. C., Chaturvedi S. K., Rein A., and Schuck P. (2019) Nucleic acid–induced dimerization of HIV-1 Gag protein. J. Biol. Chem. 294, 16480–16493 10.1074/jbc.RA119.010580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Comas-Garcia M., Kroupa T., Datta S. A., Harvin D. P., Hu W.-S., and Rein A. (2018) Efficient support of virus-like particle assembly by the HIV-1 packaging signal. eLife 7, e38438 10.7554/eLife.38438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jouvenet N., Simon S. M., and Bieniasz P. D. (2009) Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. U. S. A. 106, 19114–19119 10.1073/pnas.0907364106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Floderer C., Masson J.-B., Boilley E., Georgeault S., Merida P., El Beheiry M., Dahan M., Roingeard P., Sibarita J.-B., Favard C., and Muriaux D. (2018) Single molecule localisation microscopy reveals how HIV-1 Gag proteins sense membrane virus assembly sites in living host CD4 T cells. Sci. Rep. 8, 16283 10.1038/s41598-018-34536-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Munro J. B., Nath A., Färber M., Datta S. A. K., Rein A., Rhoades E., and Mothes W. (2014) A conformational transition observed in single HIV-1 Gag molecules during in vitro assembly of virus-like particles. J. Virol. 88, 3577–3585 10.1128/JVI.03353-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Datta S. A. K., Heinrich F., Raghunandan S., Krueger S., Curtis J. E., Rein A., and Nanda H. (2011) HIV-1 Gag extension: Conformational changes require simultaneous interaction with membrane and nucleic acid. J. Mol. Biol. 406, 205–214 10.1016/j.jmb.2010.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Datta S. A. K., Temeselew L. G., Crist R. M., Soheilian F., Kamata A., Mirro J., Harvin D., Nagashima K., Cachau R. E., and Rein A. (2011) On the role of the SP1 domain in HIV-1 particle assembly: A molecular switch? J. Virol. 85, 4111–4121 10.1128/JVI.00006-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Datta S. A. K., Clark P. K., Fan L., Ma B., Harvin D. P., Sowder R. C., Nussinov R., Wang Y.-X., and Rein A. (2016) Dimerization of the SP1 region of HIV-1 Gag induces a helical conformation and association into helical bundles: Implications for particle assembly. J. Virol. 90, 1773–1787 10.1128/JVI.02061-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Datta S. A. K., and Rein A. (2009) Preparation of recombinant HIV-1 Gag protein and assembly of virus-like particles in vitro. Methods Mol. Biol. 485, 197–208 10.1007/978-1-59745-170-3_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen Z., and Cheng W. (2017) Reversible aggregation of HIV-1 Gag proteins mediated by nucleic acids. Biochem. Biophys. Res. Commun. 482, 1437–1442 10.1016/j.bbrc.2016.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tropea J. E., Cherry S., and Waugh D. S. (2009) Expression and purification of soluble His6-tagged TEV protease. In High Throughput Protein Expression and Purification: Methods and Protocols (Doyle S. A., ed.), pp. 297–307, Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 61. Milligan J. F., Groebe D. R., Witherell G. W., and Uhlenbeck O. C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798 10.1093/nar/15.21.8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reid D. J., Diesing J. M., Miller M. A., Perry S. M., Wales J. A., Montfort W. R., and Marty M. T. (2019) MetaUniDec: High-throughput deconvolution of native mass spectra. J. Am. Soc. Mass Spectrom. 30, 118–127 10.1007/s13361-018-1951-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Marty M. T., Baldwin A. J., Marklund E. G., Hochberg G. K. A., Benesch J. L. P., and Robinson C. V. (2015) Bayesian deconvolution of mass and ion mobility spectra: From binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 10.1021/acs.analchem.5b00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schneider R., Campbell M., Nasioulas G., Felber B. K., and Pavlakis G. N. (1997) Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71, 4892–4903 10.1128/JVI.71.7.4892-4903.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fabris D., Zaia J., Hathout Y., and Fenselau C. (1996) Retention of thiol protons in two classes of protein zinc ion coordination centers. J. Am. Chem. Soc 118, 12242–12243 10.1021/ja962863r [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.