Abstract
Objective:
MiR-140 and miR-196a were known to be correlated with cancer diagnosis and prognosis. The current study aimed at the analysis of miR-140 and miR-196a expression patterns and their clinical significance for breast cancer (BC) patients.
Methods:
Differentially expressed miR-140 and miR-196a were examined via quantitative PCR in 110 cases of BC and their adjacent non-tumor (ANT) tissues.
Results:
The results indicated that miR-140 and miR-196a, respectively, notably decreased and increased expression in BC samples in comparison with ANT (p<0.001). Reduced miR-140 expression was also related to Lymph node metastasis (LNM, P= 0.023) and stage (P = 0.009). Additionally, Receiver Operating Characteristics (ROC) analysis illustrated that miR-140 had a significant diagnostic accuracy for stage and LNM of BC patients. We also discovered a strong negative correlation between miR-196a expression with histological grade (P = 0.038), LNM (P = 0.012) and stage (P = 0.001).
Conclusion:
Overall, exploring the miR-140 and miR-196a profiles not only can statistically different among BC and ANT samples, but it is also expected to become potential BC biomarkers.
Key Words: Biomarkers, miR-140, miR-196a, quantitative PCR, breast cancer
Introduction
As the most predominant malignancies in females, breast cancer (BC) accounts for the number one cause of cancer mortality worldwide among women (Jafari-Gharabaghlou et al., 2018; Javidfar et al., 2018). The relationship between genes and environmental risk factors associated with the etiopathology of various tumors (Rudolph et al., 2016). Recently, many studies have been made aiming to identify the connection between genetic alterations of protein-coding genes and mechanisms of carcinogenesis (Chakravarthi et al., 2016). But latest studies have supported the potential role of noncoding RNAs in tumorigenesis, in which miRNAs recognized as new players in cancer pathobiology (Sheervalilou et al., 2016; Mohammadian et al., 2017a). microRNAs (miRNAs/miRs) are short (about 18–23 nucleotides), conserved, noncoding regulatory RNAs that negatively regulate gene expression based on targeting of the mRNA 3′-untranslated regions (3′-UTR) (Mohammadian et al., 2016b; Norouzi et al., 2019). Computational approaches report that miRNA-mediated the activity of about 30% of all human genomes, in which each miRNA on average potentially interact with >100 of mRNA targets (Mohammadian et al., 2016a; Sheervalilou et al., 2020). The biological function of miRNAs as either tumor suppressors or oncogenes related to their target mRNA (Bertoli et al., 2015; Shahabi et al., 2019). Given that the particular miRNA signatures reported in many tumor types, so, logically, the alterations of miRNA profiles would be promising diagnostics and prognostic cancer biomarkers (Mohammadian et al., 2017b; Rupaimoole and Slack, 2017). The dysregulation of miRNA profiles may contribute to the malignant transformation such as cancer initiation, progression, and therapeutic resistance (O’Bryan et al., 2017). At the moment, numerous investigations have illustrated that miR-140 and miR-196a expression frequently are altered in human cancers tissues, which proposes that they play key roles in tumorigenesis (Schimanski et al., 2009; Green et al., 2015).
Recent studies have demonstrated that miR-140 expression mediated phenotypic alterations of tumor cells (Fang et al., 2017). As an example, miR-140 expression significantly represses the growth and BC stem cell self-renewal potential (Li et al., 2013) and effectively prevents tumorigenicity of glioblastoma, osteosarcoma and non-small cell lung carcinoma (NSCLC) (Liu et al., 2016; Ji et al., 2018; Yang et al., 2018). Indeed, numerous published studies have displayed that the miR-140 may serve as tumor suppressor miRNA and its replacement has been indicated as a novel therapeutic strategy among different cancer types (Fang et al., 2017; Li et al., 2018). miR-196a belongs to the homeotic genes (HOX) family which has different function in various types of cell lines and tissues (Chen et al., 2011). Several recent profiling studies of miRNAs reported that upregulated miR-196a expression was significantly linked to tumor progression (Sun et al., 2012; Jin et al., 2016). Besides, numerous studies illustrated that miR-196a has significant effects on the growth, differentiation, and metastasis of tumor cells based on the regulation of specific genes (Papaconstantinou et al., 2012; Jiang et al., 2018). These investigations permit us to simultaneously evaluate the miR–196a and miR–140 expression profiles in BC tissues and their matched adjacent non-tumor (ANT) tissues, and to demonstrate their clinicopathological significance. Our results support the interpretation that upregulation of tumor suppressor miR–140 and downregulation of oncogene miR-196a may be an intriguing possibility for future BC treatment.
Materials and Methods
Patient and clinical specimens
All specimens (BC and ANT tissues) were acquired from 110 patients admitted in Noor Nejat Hospital (Tabriz, Iran) and then transferred to −80°C before further analysis. Current research underwent ethical review and was applied through the ethics committee of Tabriz University of Medical Sciences.
Real-time qRT–PCR for miRNAs
The relative levels of miR–140 and miR–196a in both BC and ANT samples were analyzed by Real-time quantitative PCR (qPCR) method (Jeddi et al., 2019). The tissues were homogenized using a tissue homogenizer and total RNA was isolated from the homogenized tissue following the Exiqon’s miRCURY™ RNA Isolation Kit (Exiqon, Denmark) and Phenol/Chloroform methods. Total RNA concentrations were investigated by using NanoDrop 2000 (Thermo Scientific, Waltham, MA). Also, the integrity of total RNA was investigated by agarose gel electrophoresis on 1.5% agarose gel containing GelRedTM (Biotium). PCR reactions for quantifying miR-140 and miR-196a were performed in triplicate by LNA Universal RT miRNA PCR kit (Exiqon, Denmark). qPCR protocol was: 95°C pre-denaturation step for 10 min, 95°C 40 cycles of denaturation step for 15 seconds, annealing, and synthesis steps at 60°C for 1 min. The relative expression levels of miR-140 and miR-196a were normalized by U6 and the results were calculated based on the 2−ΔΔCt method (Livak and Schmittgen, 2001). The sequence and details of PCR primer for mature miR-140, miR-196a, and U6 are shown in Table 1.
Table 1.
Primer Sequences
| Objective genes | Primer sequence |
|---|---|
| miR-140 | Forward: 5′-GAGTGTCAGTGGTT TTACCCT-3′ |
| Reverse: 5′-GCAGGGTCCGAGGTATTC-3′ | |
| miR-196a | Forward: 5′-CGTCAGAAGGAATGATGCACAG-3′ |
| Reverse: 5′-ACCTGCGTAGGTAGTTTCATGT-3′ | |
| U6 | Forward: 5'-CTCGCTTCGGCAGCACATATACT-3' |
| Reverse: 5'-ACGCTTCACGAATTTGCGTGTC-3' |
Data analysis
Mann-Whitney U test was applied for the evaluation of statistical differences in tissue miRNAs expression between BC and ANT groups. Chi-square test (χ2) was to measure the relationship among both miR-140 and miR196a expression levels and clinical parameters. Receiver operating characteristic curves (ROC curves) were considered for investigation mentioned miRNAs as two biomarkers. Statistical analyses were investigated by the SPSS® software (SPSS, Version 16.0; SPSS Inc, Chicago, IL) or GraphPad Prism Software (version 6, GraphPad software, lnc., San Diego, CA, USA).
Results
Patient characteristics
From April 2016 to July 2018, 110 female BC patients were recruited for the present study. The clinicopathological information of all participants is indicated in Table 2. The mean age was 46 years and the range of 27–65 years.
Table 2.
Relationship between miR-140 and miR-196a Expression with Clinicopathological Parameters of BC Patients. (n = 110). BC, Breast cancer
| Clinical pathological criteria | miR–140 Expression |
miR–196a Expression |
||||||
|---|---|---|---|---|---|---|---|---|
| Low 53 (%) | High 57 (%) | P-value | Low 49 (%) | High 61 (%) | P-value | |||
| Age | <50 | 75 | 32 | 43 | NS | 29 | 46 | NS |
| >50 | 35 | 21 | 14 | 20 | 15 | |||
| histological grade | I | 21 | 9 | 12 | NS | 8 | 13 | 0.038 |
| II | 79 | 38 | 41 | 35 | 44 | |||
| III | 10 | 6 | 4 | 6 | 4 | |||
| Progesterone receptor | negative | 34 | 20 | 14 | NS | 18 | 16 | NS |
| positive | 76 | 33 | 43 | 31 | 45 | |||
| Estrogen receptors | negative | 27 | 12 | 15 | NS | 9 | 18 | NS |
| positive | 83 | 41 | 42 | 40 | 43 | |||
| HER2 | negative | 99 | 50 | 49 | NS | 45 | 54 | NS |
| positive | 11 | 3 | 8 | 4 | 7 | |||
| lymph node metastasis | negative | 46 | 21 | 25 | 0.023 | 18 | 28 | 0.012 |
| positive | 64 | 32 | 32 | 31 | 33 | |||
| Stage | I | 16 | 7 | 9 | 0.009 | 7 | 9 | 0.001 |
| II | 72 | 37 | 35 | 35 | 37 | |||
| III | 18 | 8 | 10 | 7 | 11 | |||
| IV | 4 | 1 | 3 | 0 | 4 | |||
| Tumor size | <20mm | 39 | 16 | 23 | NS | 14 | 25 | NS |
| 20-49mm | 61 | 32 | 29 | 31 | 30 | |||
| >50mm | 10 | 5 | 5 | 4 | 6 | |||
Differential miR-140 and miR-196a expression levels in BC and ANT tissues
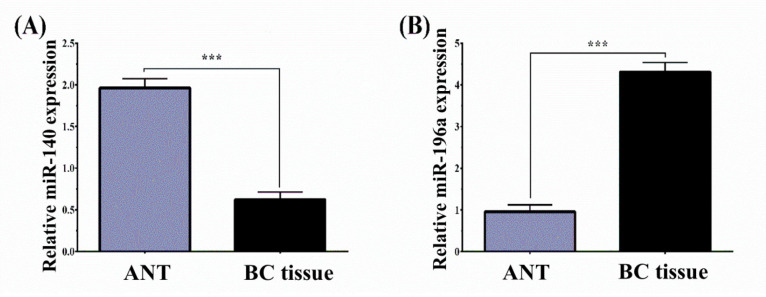
Using the RT-qPCR method, the relative level of miR-140 and miR-196a expression was assessed in BC and ANT tissues. Our findings illustrated that the relative level of miR-140 in BC tissues significantly was significantly downregulated than that of the ANT tissues (BC vs. ANT: 0.623 ± 1.07 vs 1.74 ±1.393, P< 0.001) (Figure 1A). However, the expression of miR-196a was higher in BC patients than that in ANT (BC vs ANT: 4.31 ± 2.55 vs 2.77 ± 2.45, P< 0.001) (Figure 1B). To investigate the correlation of these miRNAs with clinicopathologic features, we classified BC patients into miR-140-low/high and miR-196a-low/high expression groups according to the median value of miR-140 and miR-196a expression.
Figure 1.
MiR-140 and miR-196a Expression Levels of 110 BC and ANT Tissues Normalized to U6 Analyzed by Using qRT-PCR. The results represent that (A) miR-140 was downregulated in BC compared to that in ANT group (BC vs. ANT: 0.623 ± 1.07 vs 1.74 ±1.393, P< 0.001) and (B) miR-196a was upregulated in BC compared to that in ANT group (BC vs ANT: 4.31 ± 2.55 vs 2.77 ± 2.45, ***P< 0.001). BC, Breast cancer; ANT, Adjacent non‐tumor; qRT-PCR, Real-Time Quantitative Reverse Transcription PCR
Correlation between downregulated miR-140 expression and clinical characteristics
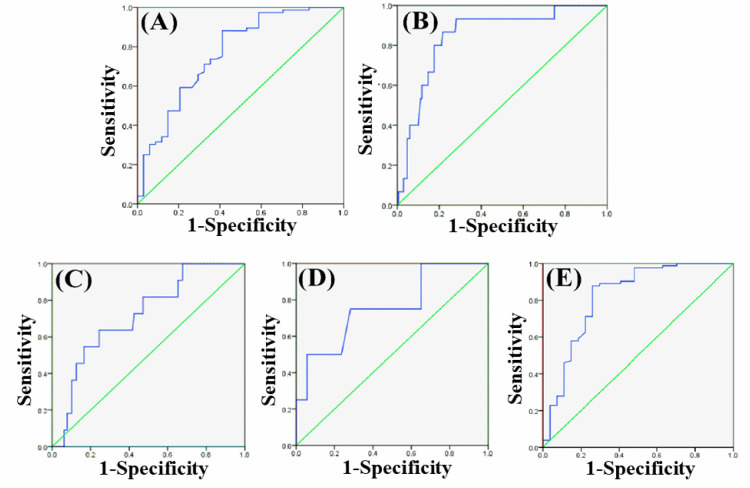
Correlation of miR-140 relative expression level with clinical criteria is presented in Table 2. Low miR-140 expression was positively associated with Lymph node metastasis (LNM, P= 0.023) and stage (P = 0.009) in BC patients. But, correlations between miR-140 expression and age, histological grade, estrogen receptor (ER) expression, progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) were not statistically significant. Additionally, the ROC curve was plotted to investigate the miRNA diagnostic power cancer detection. According to the ROC curve, miR-140 had the ability only to predict stage and LNM occurrence in BC patients (Table 3). At an appropriate cut-off value, miR-140 superior performance in predicting the LNM occurrence (AUC=0.765, P=0.001) and stage (AUC=0.847, P<0.001). The specificity and sensitivity values, respectively, were 77% and %, 68, and 95%, and 87% (Figure 2A, B).
Table 3.
AUC of ROC Curve Corresponding to the Diagnostic miR-140 and miR-196a Values in BC. AUC, Area under the curve; ROC, Receiver operating characteristics; BC, Breast cancer
|
miR–140 Expression |
miR–196a Expression |
|||||||
|---|---|---|---|---|---|---|---|---|
| Parameter | AUC | Standard Error | 95 % C.I | P-value | AUC | Standard Error | 95 % C.I | P-value |
| Histological grade | 0.635 | 0.09 | 0.458–0.812 | 0.143 | 0.718 | 0.072 | 0.576–0.859 | 0.017 |
| lymph node metastasis | 0.765 | 0.051 | 0.665–0.866 | 0.001 | 0.758 | 0.061 | 0.671–0.845 | 0.01 |
| Stage | 0.847 | 0.049 | 0.751–0.942 | <0.001 | 0.82 | 0.054 | 0.714–0.927 | <0.001 |
Figure 2.
ROC Curve Analysis of the Expression miR-140 and miR-196a to Detect BC. Expression of miR-140 was an applicable biomarker for prediction of occurrence of LNM (A), and stage (B). Moreover, ROC curve analysis revealed the potential of the miR-196a level to predict histological grade (C), the occurrence of LNM (D) and stage (E), and in BC. BC, Breast cancer; LNM, Lymph node metastasis; ROC, Receiver operating characteristics
Correlation between upregulated miR-196a expression and clinical characteristics
As showing in Table 2, high miR-196a expression was closely linked with histological grade (P = 0.038), LNM (P = 0.012) and stage (P = 0.001) in BC patients. However, no remarkable associations between the miR-196a expression and age, ER/PR, and Her2 status were evident. Based on the ROC findings, miR-196a was an important predictor of histological grade, LNM, and stage in BC patients (Table 3). At an appropriate cut-off value, miR-196a superior performance in predicting the histological grade (AUC=0.718, P=0.017), LNM (AUC=0.758, P=0.010) and stage (AUC=0.820, P<0.001). The specificity and sensitivity values, respectively, were 72% and 65%, 74% and 70%, and 88% and 81% (Figures 2C, D, E).
Discussion
Tumor markers are highly important for early detection, outcome prediction, and selection of effective treatment strategies (Mehta et al., 2010). As involved in different cancer biological processes, miRNA studies have become imperative for the detection of miRNA profiles-based prognosis, diagnosis, and theranostics biomarkers (Sheervalilou et al., 2020; Wang et al., 2018). Altered expression of miRNAs provides crucial information about molecular signatures of disease status without the performance of biopsy (Lu et al., 2005; Ayub et al., 2015). The objective of our research was to clarify the prognostic significance of miR-140 and miR-196a in 110 Iranian women with BC and to describe their possible relationship with clinicopathological criteria. Our data exhibited that the miR-140 and miR-196a levels in BC samples were notably lower and higher than those in corresponding ANT tissues. The reduced level of miR–140 was significantly relevant to LNM and stage. Besides, ROC results illustrated that the expression of miR–140 was an applicable molecular biomarker for the prediction of LNM occurrence and stage in BC patients. Based on prior studies results, miR-140 considered being one of the importantly reduced miRNAs in human cancers such as BC. For instance, Zhou et al., (2019) found that high levels of miR-140 expression could significantly suppress BC cell growth and migration. In another study, Lu et al., (2017) also found similar results that the level of miR–140 was negatively related to the patient’s clinical-grade and tumor metastasis. Furthermore, both gain and loss of function studies by Yuan et al., (2013) revealed that miR-140 represses the NSCLC metastasis. Also, the high expression of miR-145 significantly reduced lung cancer growth in nude mice models. Other studies also reported that miR-140 was downregulated in cell lines and human tumor tissues (Liu et al., 2016; Yan et al., 2017). According to our study, an inverse association also found between the miR-196ea expression profile and histological grade, LNM, and stage. Besides, ROC curve analysis indicated the potential of miR–196a level to predict histological grade, LNM occurrence, and stage in the BC. Based on the tumor tissue and the cell type, miR-196a has oncogenic or tumor-suppressor functions. Few reports have revealed the miR-196 family function as a tumor suppressor through negatively regulating the expression of oncogenes. For instance, Cao et al., (2018) indicated that miR-196a level was considerably lower in clinical osteosarcoma tissues than normal controls, and the osteosarcoma cell progression and migration abilities were inhibited by miR-196 overexpression. Likewise, in melanoma, miR-196a inhibits the processes of proliferation and metastasis through the negative regulation of transcription factor HOXC8 (Mueller and Bosserhoff, 2011). However, several other studies (e.g. Liu et al. (Liu et al., 2012); Hou et al., (2014); Yang et al., (2016); Milevskiy et al., (2019) have examined and reported the significant roles of miR-196a in promoting proliferation, migration and invasion of NSCLC, cervical, epithelial ovarian and ER+ BC cell lines and malignant tissues in comparison with healthy tissues.
Our findings recommend that both miR-140 and miR-196a are promising biomarkers for the diagnosis and management of BC patients. Nevertheless, the shortcomings of the present study were that we could not investigate the correlation between miRNA expression and the survival status because it needs a 5-year follow-up of BC patients.
In conclusion, briefly, the results exhibited that both miR-140 and miR-196a may be potential biomarkers for determining the BC diagnosis and prognosis, which may serve, respectively, as a tumor-suppressing and -promoting gene. However, more research is required to determine the miR-140 and miR-196a molecular mechanisms in the BC progression.
Acknowledgments
The authors would like to grateful the Infectious and Tropical Diseases Research Center, Tabriz University of Medical Sciences, for financially supporting this study.
Ethical approval
All procedures performed in studies involving human participants were under the ethical standards of the Ethics Committee of Tabriz University of Medical Science (NO. IR.TBZMED.REC.1396.55) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Author’s contribution
NZ and BN designed and directed the project; AS and VM processed the experimental data and performed the analysis, drafted the manuscript, and designed the figures; NZ, BN, and KA contributed to the final version of the manuscript. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Informed consent
Informed consent was obtained from all individual participants included in the study.
References
- Ayub SG, Kaul D, Ayub T. Microdissecting the role of microRNAs in the pathogenesis of prostate cancer. Cancer Genet. 2015;208:289–302. doi: 10.1016/j.cancergen.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Wu B, Wang D, et al. miR-196 acts as a tumor suppressor in osteosarcoma by targeting HOXA9. Int J Clin Exp Patho. 2018;11:4579–84. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chakravarthi BV, Nepal S, Varambally S. Genomic and epigenomic alterations in cancer. Am J Pathol. 2016;186:1724–35. doi: 10.1016/j.ajpath.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang Y, Zhang L, et al. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2011;15:14–23. doi: 10.1111/j.1582-4934.2010.01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Yin S, Sun R, et al. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16:139. doi: 10.1186/s12943-017-0708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D, Dalmay T, Fraser WD. Role of miR-140 in embryonic bone development and cancer. Clin Sci. 2015;129:863–73. doi: 10.1042/CS20150230. [DOI] [PubMed] [Google Scholar]
- Hou T, Ou J, Zhao X, et al. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27 Kip1. Br J Cancer. 2014;110:1260. doi: 10.1038/bjc.2013.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari-Gharabaghlou D, Pilehvar-Soltanahmadi Y, Dadashpour M, et al. Combination of metformin and phenformin synergistically inhibits proliferation and hTERT expression in human breast cancer cells. Iran J Basic Med Sci. 2018;21:1167. doi: 10.22038/IJBMS.2018.30460.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javidfar S, Pilehvar-Soltanahmadi Y, Farajzadeh R, et al. The inhibitory effects of nano-encapsulated metformin on growth and hTERT expression in breast cancer cells. J Drug Deliv Sci Technol. 2018;43:19–26. [Google Scholar]
- Jeddi F, Alipour S, Najafzadeh N, et al. Reduced Levels of miR–28 and miR–200a Act as Predictor Biomarkers of Aggressive Clinicopathological Characteristics in Gastric Cancer Patients. GMJ. 2019;8:1329. doi: 10.31661/gmj.v8i0.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Wang E, Tian F. MicroRNA-140 suppresses osteosarcoma tumor growth by enhancing anti-tumor immune response and blocking mTOR signaling. Biochem Biophys Res Commun. 2018;495:1342–8. doi: 10.1016/j.bbrc.2017.11.120. [DOI] [PubMed] [Google Scholar]
- Jiang C-F, Shi Z-M, Li D-M, et al. Estrogen-induced miR-196a elevation promotes tumor growth and metastasis via targeting SPRED1 in breast cancer. Mol Cancer. 2018;17:83. doi: 10.1186/s12943-018-0830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zhang Y, Li J. Upregulation of MiR-196a promotes cell proliferation by downregulating p27kip1 in laryngeal cancer. Biol Res. 2016;49 doi: 10.1186/s40659-016-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zou K, Yu L, et al. MicroRNA-140 inhibits the epithelial-mesenchymal transition and metastasis in colorectal cancer. Mol Ther Nucleic Acids. 2018;10:426–37. doi: 10.1016/j.omtn.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li Q, Yao Y, Eades G, et al. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene. 2013;33:2589. doi: 10.1038/onc.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-h, Lu K-h, Wang K-m, et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012;12:348. doi: 10.1186/1471-2407-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang S, Yuan A, et al. MicroRNA-140 represses glioma growth and metastasis by directly targeting ADAM9. Oncol Rep. 2016;36:2329–38. doi: 10.3892/or.2016.5007. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu Y, Qin T, Li J, et al. MicroRNA-140-5p inhibits invasion and angiogenesis through targeting VEGF-A in breast cancer. Cancer Gene Ther. 2017;24:386. doi: 10.1038/cgt.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Shelling A, Muthukaruppan A, et al. Predictive and prognostic molecular markers for cancer medicine. Ther Adv Med Oncol. 2010;2:125–48. doi: 10.1177/1758834009360519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milevskiy MJ, Gujral U, Marques CDL, et al. MicroRNA-196a is regulated by ER and is a prognostic biomarker in ER+ Breast Cancer. Br J Cancer. 2019;120:621. doi: 10.1038/s41416-019-0395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadian F, Abhari A, Dariushnejad H, et al. Effects of chrysin-PLGA-PEG nanoparticles on proliferation and gene expression of miRNAs in gastric cancer cell line. Iran J Cancer Prev. 2016a;9:e4190. doi: 10.17795/ijcp-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadian F, Pilehvar-Soltanahmadi Y, Alipour S, et al. Chrysin alters microRNAs expression levels in gastric cancer cells: possible molecular mechanism. Drug Res (Stuttg) 2017a;67:509–14. doi: 10.1055/s-0042-119647. [DOI] [PubMed] [Google Scholar]
- Mohammadian F, Pilehvar-Soltanahmadi Y, Mofarrah M, et al. Down regulation of miR-18a, miR-21 and miR-221 genes in gastric cancer cell line by chrysin-loaded PLGA-PEG nanoparticles. Artif Cells Nanomed Biotechnol. 2016b;44:1972–8. doi: 10.3109/21691401.2015.1129615. [DOI] [PubMed] [Google Scholar]
- Mohammadian F, Pilehvar-Soltanahmadi Y, Zarghami F, et al. Upregulation of miR-9 and Let-7a by nanoencapsulated chrysin in gastric cancer cells. Artif Cells Nanomed Biotechnol. 2017b;45:1201–6. doi: 10.1080/21691401.2016.1216854. [DOI] [PubMed] [Google Scholar]
- Mueller DW, Bosserhoff AK. MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. Int J Cancer. 2011;129:1064–74. doi: 10.1002/ijc.25768. [DOI] [PubMed] [Google Scholar]
- Norouzi M, Yasamineh S, Montazeri M, et al. Recent advances on nanomaterials-based fluorimetric approaches for microRNAs detection. Mater Sci Eng C. 2019;104:110007. doi: 10.1016/j.msec.2019.110007. [DOI] [PubMed] [Google Scholar]
- O’Bryan S, Dong S, Mathis JM, et al. The roles of oncogenic miRNAs and their therapeutic importance in breast cancer. Eur J Cancer. 2017;72:1–11. doi: 10.1016/j.ejca.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Papaconstantinou IG, Lykoudis PM, Gazouli M, et al. A review on the role of microRNA in biology, diagnosis, and treatment of pancreatic adenocarcinoma. Pancreas. 2012;41:671–7. doi: 10.1097/MPA.0b013e31823c9d21. [DOI] [PubMed] [Google Scholar]
- Rudolph A, Chang-Claude J, Schmidt MK. Gene–environment interaction and risk of breast cancer. Br J Cancer. 2016;114:125. doi: 10.1038/bjc.2015.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- Schimanski CC, Frerichs K, Rahman F, et al. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahabi A, Naghili B, Ansarin K, et al. The relationship between microRNAs and Rab family GTPases in human cancers. J Cell Physiol. 2019;234:12341–52. doi: 10.1002/jcp.28038. [DOI] [PubMed] [Google Scholar]
- Sheervalilou R, Ansarin K, Fekri Aval S, et al. An update on sputum Micro RNA s in lung cancer diagnosis. Diagn Cytopathol. 2016;44:442–9. doi: 10.1002/dc.23444. [DOI] [PubMed] [Google Scholar]
- Sheervalilou R, Shahraki O, Hasanifard L, et al. Electrochemical nano-biosensors as novel approach for the detection of lung cancer-related MicroRNAs. Curr Mol Med. 2020;20:13–35. doi: 10.2174/1566524019666191001114941. [DOI] [PubMed] [Google Scholar]
- Sun M, Liu X-h, Li J-h, et al. MiR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27kip1. Mol Cancer Ther. 2012;11:842–52. doi: 10.1158/1535-7163.MCT-11-1015. [DOI] [PubMed] [Google Scholar]
- Wang H, Peng R, Wang J, et al. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics. 2018;10:59. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Zhu Z, Xu S, et al. MicroRNA-140-5p inhibits hepatocellular carcinoma by directly targeting the unique isomerase Pin1 to block multiple cancer-driving pathways. Sci Rep. 2017;7:45915. doi: 10.1038/srep45915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Li S-Z, Ma L, et al. Expression and mechanism of action of miR-196a in epithelial ovarian cancer. Asian Pac J Trop Dis. 2016;9:1105–10. doi: 10.1016/j.apjtm.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Yang P, Xiong J, Zuo L, et al. miR1405p regulates cell migration and invasion of nonsmall cell lung cancer cells through targeting VEGFA. Mol Med Rep. 2018;18:2866–72. doi: 10.3892/mmr.2018.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Shen Y, Xue L, et al. miR-140 suppresses tumor growth and metastasis of non-small cell lung cancer by targeting insulin-like growth factor 1 receptor. PLoS One. 2013;8:e73604. doi: 10.1371/journal.pone.0073604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang B, Wang Y, et al. miR1403p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol Lett. 2019;17:3835–41. doi: 10.3892/ol.2019.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]