Brucella, the causative agent of brucellosis, is a stealthy intracellular pathogen that is highly pathogenic to a range of mammals, including humans. The twin-arginine translocation (Tat) pathway transports folded proteins across the cytoplasmic membrane and has been implicated in virulence in many bacterial pathogens. However, the roles of the Tat system and related substrates in Brucella remain unclear. We report here that disruption of Tat increases the sensitivity of Brucella melitensis M28 to the membrane stressor sodium dodecyl sulfate (SDS), indicating cell envelope defects, as well as to EDTA.
KEYWORDS: Brucella melitensis, stress, translocated substrates, twin-arginine protein translocation, virulence
ABSTRACT
Brucella, the causative agent of brucellosis, is a stealthy intracellular pathogen that is highly pathogenic to a range of mammals, including humans. The twin-arginine translocation (Tat) pathway transports folded proteins across the cytoplasmic membrane and has been implicated in virulence in many bacterial pathogens. However, the roles of the Tat system and related substrates in Brucella remain unclear. We report here that disruption of Tat increases the sensitivity of Brucella melitensis M28 to the membrane stressor sodium dodecyl sulfate (SDS), indicating cell envelope defects, as well as to EDTA. In addition, mutating Tat renders M28 bacteria more sensitive to oxidative stress caused by H2O2. Further, loss of Tat significantly attenuates B. melitensis infection in murine macrophages ex vivo. Using a mouse model for persistent infection, we demonstrate that Tat is required for full virulence of B. melitensis M28. Genome-wide in silico prediction combined with an in vivo amidase reporter assay indicates that at least 23 proteins are authentic Tat substrates, and they are functionally categorized into solute-binding proteins, oxidoreductases, cell envelope biosynthesis enzymes, and others. A comprehensive deletion study revealed that 6 substrates contribute significantly to Brucella virulence, including an l,d-transpeptidase, an ABC transporter solute-binding protein, and a methionine sulfoxide reductase. Collectively, our work establishes that the Tat pathway plays a critical role in Brucella virulence.
INTRODUCTION
Brucellosis, affecting many mammalian species worldwide, is one of the most notorious and economically significant zoonotic diseases. Infections are characterized by various clinical manifestations. Animal brucellosis usually leads to reproductive failures associated with epididymitis, orchitis, and abortions (1, 2). In humans, brucellosis is characterized by weakness, undulant fever, and chronic inflammation in various organs, including the spleen, liver, and bone (3). The causative agents of brucellosis, Brucella spp., are Gram-negative facultative intracellular bacteria. Among the 10 recognized species of Brucella, B. melitensis is the most pathogenic to humans (4). Entry of Brucella into the human body usually occurs via inhalation or consumption of contaminated milk products. Upon entry, these bacteria can penetrate mucosal surfaces and be further delivered to lymph nodes via macrophages. Brucella has the ability to survive and replicate within macrophages in compartments called Brucella-containing vacuoles (BCVs) (5, 6). They can infect various host cells, such as epithelial cells and reproductive tissue cells (7). Despite being a highly pathogenic bacterial pathogen, Brucella rarely contains classical bacterial virulence factors, e.g., exotoxins, fimbria, and prophages. Well-studied virulence factors of Brucella include lipopolysaccharide (LPS) and type IV secretion systems (T4SS), which are employed to evade or suppress the host immune system, along with various enzymes utilized to promote fitness in vivo (8–11). However, current understanding of the contributing factors for Brucella's intracellular replication and its persistence in the host is limited. Identification of novel virulence-associated traits is desperately needed to better understand Brucella pathogenesis and define potential antimicrobial targets.
The twin-arginine protein translocation (Tat) system occurs in bacteria, archaea, and plants, and it generally comprises three membrane-bound components, i.e., TatABC (12). The Tat pathway targets folded proteins containing an N-terminal signal peptide with an S/TRRXFLK consensus motif for transport across the cytoplasmic membrane (13). TatC usually consists of six transmembrane helices (14); TatA and TatB are sequence-related proteins that share a structure containing an N-terminal transmembrane segment and a neighboring amphipathic helix (15). During the process of translocation, TatC and TatB coordinate transport by binding to signal peptides. TatA subunits oligomerize into a channel and are then recruited by TatC to assemble a secretion complex that is involved in protein export (16). Many of the Tat substrates are complexed with metal cofactors such as molybdenum or iron-sulfur clusters, thus requiring complete folding prior to their secretion. Nonetheless, Tat substrates with no cofactors have also been identified (17).
In many pathogenic bacteria, disruption of the Tat pathway leads to attenuation of virulence. Changes in virulence can be attributed to pleiotropic phenotypes, e.g., cell envelope defects, reduced motility, and impaired ability to resist stresses (18–20). In some cases, a phenotypic change is the result of multiple Tat substrates. For instance, triple deletion of amiA, amiC, and sufI in Salmonella results in a marked defect in the cell envelope, leading to virulence attenuation (21). In other cases, a single virulence factor transported by Tat directly contributes to pathogenicity; for example, the phospholipase toxins in Pseudomonas aeruginosa (19) and Legionella pneumophila (22), Shiga toxin 1 in enterohemorrhagic E. coli (EHEC) (23), and SufI in Yersinia pseudotuberculosis (24). Hence, a wide collection of studies on Tat suggest that Tat can contribute to pathogenicity directly by executing secretion of diverse virulence–associated factors or indirectly by exerting multiple effects.
Although B. abortus possesses a functional Tat translocase (25), the role of the Tat pathway in Brucella has not been defined, and Tat substrates in Brucella have yet to be experimentally studied at the system level. In this study, using the highly virulent strain B. melitensis M28 (26), we show that mutation in Tat disrupts cell envelope integrity, decreases the antioxidant capacity of B. melitensis, and dramatically reduces infection ex vivo and in vivo. Furthermore, Tat substrates were systematically identified, and the contribution of each substrate to virulence was explored.
RESULTS
Sequence analysis of TatABC in Brucella melitensis M28.
It has been shown that B. abortus possesses a functional Tat translocase (25). A search of the B. melitensis M28 genome revealed a Tat locus composed of tatA (A0889), tatB (A0890), and tatC (A0891) on chromosome I, which is the same genetic localization of Tat in B. abortus (Fig. 1A). No paralogs of TatABC were found in the B. melitensis M28 genome. The TatABC proteins in M28 containing 72, 188, and 274 amino acids are 100%, 97%, and 100% identical to those of B. abortus, respectively, but they have much lower identity with E. coli TatABC (42%, 27%, and 36%, respectively). Sequence alignment of well-characterized tatA genes from various Gram-positive and -negative bacteria showed that M28 TatA carries two important conserved domains: a transmembrane segment and an amphiphilic helix, which are also shared by M28 TatB (Fig. 1B) (27).
FIG 1.
Sequence analysis of the Tat loci in B. melitensis M28. (A) Genetic organization of the Tat loci in B.melitensis M28. tatABC in M28 is found on chromosome I and has been designated BM28_A0889, BM28_A0890, and BM28_A0891. Similar to B. abortus 2308, M28 tatABC are flanked by ScpB (encoding an SMC-Scp complex subunit) and serS (encoding a serine-tRNA ligase). Genes represented by arrows are drawn to scale. (B) Sequence alignment of TatA from M28 and other bacteria, as well as TatB of M28, illustrates the conservation of a transmembrane segment (TMS) and an amphiphilic helix (APH). The graph was created by ClustalW. Color scheme illustrates the different types of amino acids: blue, hydrophobic; red, positively charged; magenta, negatively charged; green, polar; orange, glycine; pink, cysteine; yellow, proline; cyan, aromatic; white, unconserved.
Disruption of Tat leads to cell envelope defects.
To examine the effects of tat mutation on M28, we constructed a deletion mutant ΔtatA (referred to as M28ΔtatA) by allelic replacement and its complemented strain which contains a plasmid-borne tatA locus plus its native promoter. The successful replacement of tatA by a kanamycin-resistance cassette was confirmed by PCR (Fig. 2A) and DNA sequencing. Quantitative reverse transcription-PCR (qPCR) analysis showed that expression levels of tatB and tatC were only slightly affected in the M28ΔtatA mutant (less than 2-fold, P < 0.05), compared to the wild-type strain (Fig. 2B). The M28ΔtatA mutant grew slightly slower than the wild type during the exponential phase, yet it caught up with the wild type as these strains entered the stationary phase (Fig. 2C). It has been reported that loss of Tat leads to cell envelope defects in many bacterial species (21, 28) and that increased sensitivity to hydrophobic drugs or the detergent SDS is indicative of disrupted cell envelope integrity in Tat mutants (29). To test whether cell envelope integrity was compromised due to Tat deficiency, we subjected M28 and its derivatives to sensitivity tests using a few stressors. All strains tested exhibited similar sensitivities to polymyxin B, ampicillin, and high concentrations of NaCl (data not shown). Using a classical disk diffusion assay, we found that M28ΔtatA was more sensitive to SDS treatment, as it showed a significantly larger inhibition zone than the wild type (Fig. 2D; P < 0.01); this phenotypic change was reversed when tatA was expressed from a plasmid in the tatA mutant strain. These data suggest that the Tat system contributes to the resistance to SDS treatment, and is important for maintaining cell envelope integrity in B. melitensis. EDTA is a metal chelating agent, and it can have various physiological effects on a range of microorganisms, including cell envelope damage and metal utilization defects (30, 31). Upon treatment with 2 mM EDTA on tryptic soy agar (TSA) plates, bacterial CFU of M28ΔtatA were decreased by ∼104-fold, whereas the wild type decreased by only ∼10-fold (P < 0.001; Fig. 2E), indicating that the M28ΔtatA mutant is more sensitive to EDTA than the wild type.
FIG 2.
Mutant construction and growth and sensitivity of various Brucella melitensis M28 strains to stressors. (A) Positions of PCR oligonucleotides used and gel images showing PCR amplicons of the wild type and M28ΔtatA mutant. HA, homologous arm. Lanes: 1 and 2, wild type and M28ΔtatA mutant using del-tatA-F and del-tatA-R primers, respectively; 3 and 4, wild type and M28ΔtatA mutant using tatA-IN-F and tatA-IN-R primers, respectively; M, DNA size marker. (B) Relative transcriptional levels of tatBC in the wild type and M28ΔtatA mutant. Total RNA was extracted from the wild type and the M28ΔtatA mutant, followed by genomic DNA removal, reverse transcription, and qPCR analysis. The expression of target genes in the wild type was normalized to 1, and a 2-ΔΔCt method was employed to indicate expression changes in the mutant. The dashed line denotes a transcriptional reduction of 2-fold. *, P < 0.05. Experiments were repeated 3 times, and values are means ± standard error of the mean (SEM). (C) Bacteria were grown in minimal medium and the optical density was measured at the indicated time points. Data were collected from three replicates, and error bars were too small to be visible on the graph. (D) SDS sensitivity assay. Bacteria (∼1.5 × 107 CFU) were plated onto TSA plates with an SDS-soaked disk placed in the center. The results are plotted as average diameters of the zones of inhibition around the disks. (E) EDTA stress survival assay. Serially diluted cultures of the wild type, M28ΔtatA mutant, and the complemented strain were spotted on plain TSA plates or TSA plates containing EDTA (2 mM). After 72 h of growth at 37°C under 5% CO2, CFU were counted and the ratio of CFU on the TSA plain plates (CFUunstressed) to CFU on the EDTA-supplemented TSA plates (CFUstressed) was calculated for each strain. (F) Survival of bacterial strains at various concentrations of H2O2. Bacterial suspensions containing approximately 5 × 105 CFU were mixed with H2O2 at final concentrations of 5 mM, 2.5 mM, and 1 mM. A control experiment was performed by adding H2O2-free PBS to the bacterial suspensions. After 1 h of treatment at 37°C with shaking, cells were plated onto TSA for counting. The survival of each bacterial strain was determined as the percentage of the control. Experiments were independently performed three times; the data are shown as the mean ± standard error of the mean. Student’s t test was used to evaluate significant differences between the M28ΔtatA mutant and the wild type. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Tat system contributes to the resistance of B. melitensis M28 to oxidative stress.
To survive and replicate within macrophages, Brucella needs to resist intracellular oxidative and acidic stress (32). We therefore evaluated whether deletion of tatA affects the resistance of M28 to H2O2 and acidic conditions. The percentage of survival for the wild type and M28ΔtatA strains was similar when cultured at pH 5.5, 4.5, and 3.8 (data not shown). In contrast, when exposed to different concentrations of H2O2, M28ΔtatA exhibited a lower survival rate than the wild type (Fig. 2F; P < 0.001), suggesting that the M28ΔtatA mutant is more sensitive to H2O2. However, the survival rate of the complemented strain was comparable to that of the wild-type strain (Fig. 2F). These findings indicate that the Tat pathway significantly contributes to the resistance of B. melitensis to H2O2.
Tat pathway is important for infection in a cell model and for full virulence in a mouse model.
Tat mutations render high H2O2 sensitivity in M28 cells. Thus, we further tested the ability of the wild type, M28ΔtatA mutant, and the complemented strain to infect in a murine macrophage cell line and in primary bone marrow-derived macrophages (BMDMs). Macrophages were infected with B. melitensis strains at an MOI of 100, followed by a gentamicin protection assay to evaluate intracellular bacterial counts. At an early stage of infection (6 h) in murine RAW264.7 cells, deletion of tatA resulted in a 2-fold reduction in the number of intracellular bacterial CFU, which became more severe at 24 h and 48 h postinfection (1-log and 2-log differences, respectively). Over time, the wild type and the complementation strain grew within macrophages, whereas intracellular M28ΔtatA mutants did not show any sign of growth (Fig. 3A). Similar results were obtained with BMDMs, except that as infection time increased, a reduction in the number of intracellular CFU was observed in M28ΔtatA, indicating an attenuation of infection (Fig. 3B). Altogether, these results demonstrate that the Tat system is important for B. melitensis infection in macrophages ex vivo.
FIG 3.
Tat contributes to virulence of B. melitensis in ex vivo and in vivo models. Intracellular CFU of M28 and its derivatives in murine RAW264.7 cells (A) and bone marrow-derived macrophages (BMDMs) (B). Monolayers of RAW264.7 cells (1.5 × 106 cells per well) and BMDM cells (2 × 105 cells per well) were seeded onto 24-well tissue culture plates and inoculated with bacterial cultures at a multiplicity of infection (MOI) of 100:1. After 3 h of infection, a gentamicin protection assay was performed at 37°C/5% CO2 for the indicated times. The intracellular CFU counts were determined by plating serial dilutions of the cultures on TSA. Asterisks denote statistically significant differences between the M28ΔtatA mutant group and the parental strain M28 group based on the Student’s t test (**, P < 0.01; ***, P < 0.001). All tests were done 3 times. (C to D) In vivo virulence assay. Six-week-old BALB/c mice were challenged intraperitoneally with the wild type, M28ΔtatA mutant, and the complemented strain M28ΔtatA pBBRtatA. Splenic bacterial burden (C) and spleen weights (D) were measured at 1, 3, and 7 weeks postinfection. The short lines amid data points in the graph represent the mean (n = 5 mice per bacterial strain per time point). The blank indicates a control group of mice inoculated with PBS. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (one-way ANOVA followed by Tukey's test). This experiment was repeated at least twice.
To characterize the role of the Tat system in B. melitensis virulence in vivo, we tested the virulence of various strains in a mouse model. Groups of 5 BALB/c mice were intraperitoneally challenged with M28 and its derivative strains. At 1, 3, and 7 weeks after infection, mice were euthanized, and the bacterial load in the spleen was measured by plating serial dilutions of spleen lysates. Compared to the wild-type strain, deletion of tatA caused a drastic reduction (10- to 100-fold) in bacterial load in the spleen by 1, 3, and 7 weeks postinfection (Fig. 3C). Complementation of the ΔtatA strain with a plasmid-borne tatA gene restored the levels of bacterial splenic colonization to those of the wild type. Additionally, mice infected with the wild type strain exhibited apparent splenomegaly and had larger and heavier spleens, whereas mice infected with the M28ΔtatA mutant had smaller and lighter spleens and displayed no noticeable splenomegaly (Fig. 3D; P < 0.001). The spleen weights of mice infected with M28ΔtatA were similar to that of the uninfected mice at 7 weeks postinfection (Fig. 3D), suggesting severe attenuation of virulence. These results indicate that the B. melitensis Tat system is required for infection in mice.
To compare the respective pathology induced by the wild type and M28ΔtatA mutant, spleens from infected mice were harvested at 1 week postinfection and stained with hematoxylin and eosin and anti-Brucella antibody, followed by microscopic observation. Compared to the naive (uninfected) mice, murine spleens infected with the wild type and the complemented strain displayed reduced white to red pulp ratio and higher extramedullary hematopoiesis, contained conspicuous granuloma and infiltrated neutrophils (Fig. 4A), and exhibited extensive Brucella immunoreactivities, whereas those infected with M28ΔtatA showed diminished pathological features, including unappreciable granuloma and neutrophil infiltration, minimally changed extramedullary hematopoiesis, and rare Brucella immunoreactivities (Fig. 4B). Pathology scores of murine spleens are presented in Table S1 in the supplemental material. These results, therefore, support the notion that the Tat system is necessary for full virulence of B. melitensis in a mouse model of infection.
FIG 4.
Histology of murine spleens at 1 week postinfection. Spleens (n = 3) were harvested, fixed, and stained with hematoxylin and eosin (A) or immunostained with goat anti-Brucella antibody (B). Images of spleens from uninfected mice (naive) and mice infected with the wild-type strain, M28ΔtatA mutant, or the complemented strain are presented from the left to the right columns; boxed areas represent specific regions that are shown at increased magnification below each spleen. Conspicuous granuloma and infiltrated neutrophils are indicated by black arrows. Brucella antigens were detected by immunohistochemistry with an anti-Brucella antibody (brown area). Short white lines indicate scale bars. Shown are representative images.
Bioinformatics prediction of Tat substrates in B. melitensis M28.
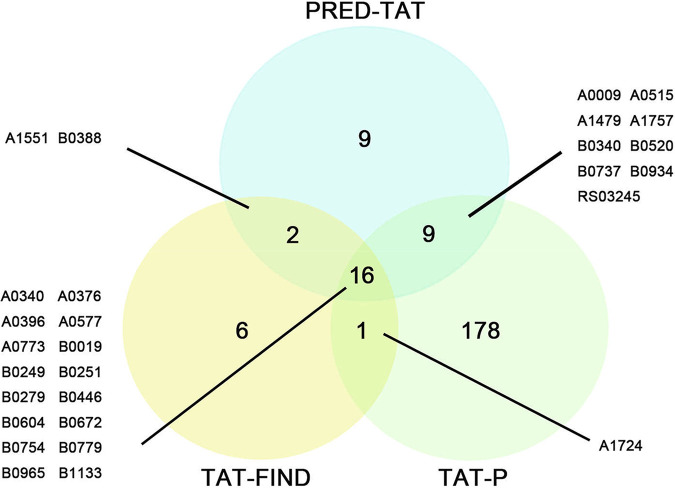
Next, we sought to identify the Tat substrates of B. melitensis M28 in a genome-wide scale. Using three popular Tat signal prediction programs, i.e., TAT-P (33), TAT-FIND (34), and PRED-TAT (35), we searched the entire B. melitensis M28 annotated genome for putative Tat signal peptides. This search led to the prediction of 204 Tat signal peptides by TAT-P (Table S2), 25 by TAT-FIND (Table S3), and 36 by PRED-TAT (Table S4). Among these, 16 Tat signal peptides were identified by all three programs (Fig. 5). In this study, we used a stringent criterion, by which a putative Tat signal peptide is considered a candidate only when it was predicted by at least two of the three programs. This criterion has also been adopted by other researchers (25, 36). A full list of candidate signal peptides is shown in Table 1. Note that two proteins, BM28_RS02740 and BM28_RS11470, were each predicted to have signal peptides of different lengths by two different programs and thus deemed candidates as well. Therefore, a total of at least 30 Tat signal peptides corresponding to 28 proteins were identified in B. melitensis M28 using bioinformatics approaches.
FIG 5.
Venn diagram depicting comparisons between outputs generated by three Tat signal peptide prediction programs. Outputs by TAT-FIND, TAT-P, and PRED-TAT are represented by yellow, green, and blue circles, respectively. Numbers displayed for each section of the circles represent the total number of proteins identified with one, two, or three prediction programs. A line links the substrate number indicative of each overlapping region to the list of the putative Tat candidates tested in this study.
TABLE 1.
Tat substrates identified by bioinformatics and experimental approaches in B. melitensis M28
| Gene ID | Predicted gene product | N-terminal signal sequence | Verified by amidase reporter assay |
|---|---|---|---|
| Substrate/solute-binding protein | |||
| A0009 | ABC transporter substrate-binding protein | MTFLCMNRRHFMGLSGSAALVACLPGQAFA | + |
| A0376 | TRAP transporter substrate-binding protein | MKETLSRRSFLTKGAAIGAAAATSGAALATPAIA | + |
| B0279 | ABC transporter substrate-binding protein | MTTTRRDILLGAAAFGLASIATRLPAYA | + |
| B0446 | ABC transporter substrate-binding protein | MHENNEIGQLDETGAALAEENTHMKFSLNRRQALLGMGAAGAAVAFGFPARA | + |
| B0520 | Periplasmic substrate-binding protein | MNIANIFDRMNFRRAVMFTVAAIGLTASPLFTAPAQA | + |
| B0754 | ABC transporter substrate-binding protein | MSGFVASRRAFLVGSTSLAALAFWPKATWA | + |
| B0779 | ABC transporter substrate-binding protein | MRLTRRQALGGMAALAAFPASRVWA | + |
| B0934 | MetQ/NlpA family ABC transporter substrate-binding protein | MSSVLSRYALTRRAGLKALLFTAAALTVGFASAPSHA | + |
| B1133 | D-xylose ABC transporter substrate-binding protein | MKRRNFLAGALVAAALGIGAMGATPAFA | _ |
| Oxidoreductases | |||
| A0396 | Disulfide bond reductase DsbA family protein | MMNRRQILAATAAGAVKFALSGGSGLA | + |
| A0515 | DsbA family protein | MPVALNRRHVISLAGAAAAGLAFTRGANA | _ |
| A1551 | Ubiquinol-cytochrome c reductase iron-sulfur subunit | MSAHDTAEPTRRDFLYIATGMAGVVGVGG | _ |
| LAWPFIDQMRPDASTLAAA | |||
| A1757 | Thiamine/thiamine pyrophosphate ABC transporter permease ThiP | MTATPARRTSLASPATKPVAGGLALAFLATL | _ |
| AGGALLALA | |||
| B0019 | Methionine sulfoxide reductase catalytic subunit YedY | MTGPRLSRRRFLTFTGMAGSAVLLSGCDA | + |
| B0251 | Nitrite reductase, copper-containing | TAT-P: MADQIQVNRRTILAGAALAGALGPVLSATSAWG | + |
| PRED-TAT: MADQIQVNRRTILAGAALAGALGPVLSATSAWGQGTMRKASA | + | ||
| B0672 | Multicopper oxidase CueO | MTGITRRRLLALGASAACVAALRPLGAFA | + |
| B0965 | Methionine sulfoxide reductase catalytic subunit MsrP | MSSFKPSRFSTARLTGDAVTPKSIYLRRREFMIGLGAIATTGAASSAFA | + |
| Cell envelope biosynthesis | |||
| A0577 | L,d-transpeptidase | PRED-TAT: MQTTLTRRSFLTAMTATAATGLAGCAQLG | + |
| TAT-P: MQTTLTRRSFLTAMTATA | _ | ||
| A0773 | Murein L,d-transpeptidase | MTKTDRPANAFRADRRRFLRSAATAGLSVAVSAMVSSAYA | + |
| A1479 | Linear amide C-N hydrolase | METKSSLWKSSRRVLAHGAATVLVAAGLIVPQAAMA | + |
| B0388 | O-antigen/exopolysaccharide biosynthesis protein | MIISDVSSANGRQPVADLLETIKRRKLMLTLPIIAGVGVGFAGYLTAPVSY | + |
| Others | |||
| A0340 | CAP domain-containing protein | MQQKHGIRLSRRGFLMLAGGAMALSALPVDWAQA | + |
| A1724 | 5-Formyltetrahydrofolate cyclo-ligase | METAGQGRSDEKQALRRVVLA | _ |
| B0340 | P1 family peptidase | MDRRTFAKSLAGLGAMPFAVNTASAGA | + |
| B0737 | Hint domain-containing protein | MSSELNGHSPRNRARRHFLGVAAAAAARVAILGALVSSSLPARA | + |
| RS03245 | TIGR02301 family protein | MKDYRRLFRLAAGAAALWLAVCPPVLA | + |
| B0249 | Lipase | MNRRSLLLGLLAGAALVLPSLPAEA | + |
| B0604 | Amidohydrolase | MCLMCGPAAKTILNITRRNLLKGLGAASLTAAIPFRAES | + |
Experimental verification of Tat substrates.
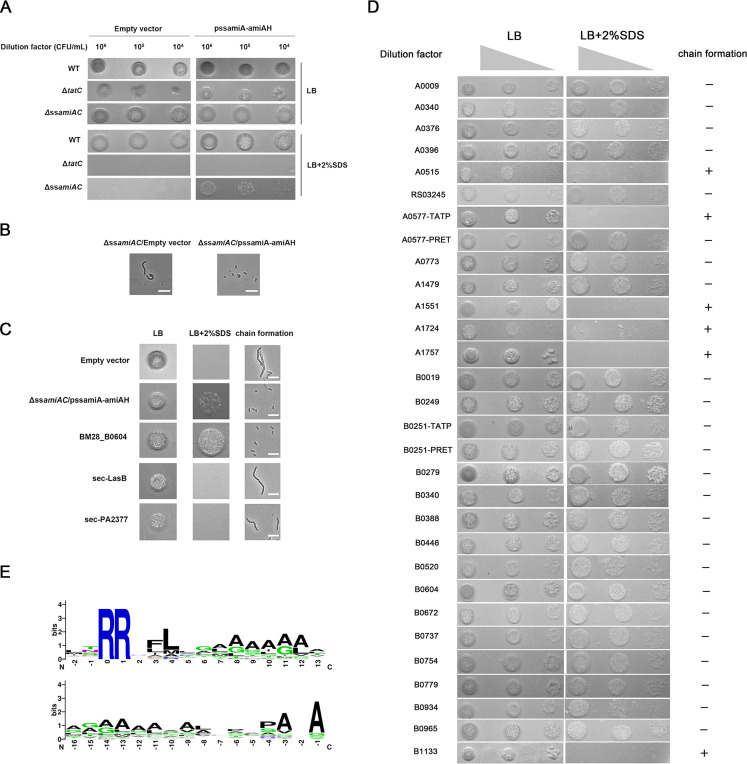
We went on to verify the predicted Tat substrates using an E. coli-based amidase reporter assay, which was developed for confirming Tat substrates in E. coli and other bacteria (36, 37). The Tat signal sequences of amiA and amiC, encoding amidases, were deleted in E. coli MC4100, resulting in the ΔssamiAC strain that is defective in translocating AmiA and AmiC, sensitive to SDS stress, and forms chains of cells (29). We demonstrated that the E. coli ΔssamiAC strain, with a plasmid carrying the AmiA signal sequence fused to amiA (pssAmiA-AmiAH), could recover the transport of AmiA, thus allowing the bacteria to form single cells and to tolerate SDS treatment; and this recovery depended on the Tat system (Fig. 6A and B). Furthermore, this system was able to distinguish Tat from Sec signal peptides, as signal peptides of Sec substrates LasB and PA2377 could not restore the phenotype (Fig. 6C), which is consistent with data from other reports (36). To determine Tat engagement of the 30 candidate Tat signal peptides of M28, we constructed recombinant plasmids by fusing each candidate signal peptide sequence to the sequence encoding the mature AmiAH protein and then tested whether expression of these fusion proteins could restore SDS tolerance and the single-cell phenotype of the E. coli ΔssamiAC strain. Figure 6D shows that 24 constructs can clearly complement the ΔssamiAC mutation, indicating that these are real Tat signal peptides. Of note, for the two A0577 signal peptides predicted by TAT-P and PRED-TAT, only one could successfully restore the phenotype. However, for the two B0251 signal peptides predicted by TAT-P and PRED-TAT, both versions restored the phenotype (Fig. 6D). Of these 23 proteins in M28, at least two (CueO and YedY) appear to be conserved Tat substrates that are shared in other bacterial species, including E. coli, Salmonella spp., and Pseudomonas aeruginosa (Table S5). Based on the functions and conserved domains, we categorized the 23 Tat substrates into four classes, namely, ABC transporters, oxidoreductases, cell wall biogenesis enzymes, and others (Table 1).
FIG 6.
Verification of the putative Tat candidates by an amidase reporter assay and the creation of Tat signal peptide logos from signal peptides identified in this work. (A) ssAmiA-AmiAH (His6-tagged AmiA with its own signal peptide sequence) is translocated properly, thus providing SDS resistance, and the process depends on Tat. Various E. coli MC4100 strains were grown aerobically and serial dilutions of bacterial cultures were spotted onto the LB agar plates containing 2% (wt/vol) SDS to evaluate SDS resistance. (B) Light micrographs indicate that possession of pssAmiA-AmiAH restores the single-cell phenotype. The scale bar represents 10 μm. (C) The amidase reporter assay discriminates between Tat- and Sec-dependent signal peptides. SDS sensitivity and chain formation phenotype were examined for the ΔssamiAC strain carrying the empty vector, pssAmiA-AmiAH, or derivatives of pssAmiA-AmiAH where the AmiA signal sequence has been replaced by the putative signal sequences of Tat-dependent (B0604) and Sec-dependent known substrates (LasB and PA2377). (D) SDS sensitivity and chain formation of the ΔssamiAC strain carrying derivatives of the pssAmiA-AmiAH construct where the AmiA signal sequence (ssAmiA) was replaced by the 30 candidate Tat signal peptides identified in silico. Authentic Tat signal peptides would allow the Tat-dependent translocation of amidase AmiA and enable the ΔssamiAC strain to form single cells and grow in the presence of SDS. +, cell chains formed; −, no cell chains formed. Experiments were repeated twice, and representative images are presented. (E) Sequence logos of Tat signal peptides aligned from two amino acids prior to the conserved twin-arginine (upper logo) or from the peptidase cleavage sites (lower logo). Amino acids are colored based on their chemical properties: green letters indicate polar amino acids (i.e., G, C, S, N, T, Y, and Q), blue indicates basic amino acids (i.e., R, K, and H), red indicates acidic amino acids (D and E), and black indicates hydrophobic amino acids (i.e., A, F, V, P, L, I, W, and M). The overall height of the stack demonstrates the sequence conservation at that position; the heights of symbols within each stack represent the relative frequency of the amino acids at that position.
To establish the sequence pattern and amino acid frequencies of the 23 signal peptides of B. melitensis M28, two sequence logos were created from two sites prior to the RR motif and from the putative peptidase cleavage site, respectively. As shown in Fig. 6E, the RRXFL motif and the classical signal peptidase cleavage site AXA occur in high frequency and are relatively conserved (13). Taken together, using a systemic approach, we confirmed a total of 23 Tat substrates and established the sequence pattern of Tat signal peptides in B. melitensis M28.
Contribution of each substrate to the virulence of B. melitensis M28 in a mouse model.
To evaluate the role of each substrate in virulence and to determine whether deletion of any individual substrate could recapitulate the decreased virulence of tatA deletion, we constructed deletion mutants of each individual substrate and used them to challenge mice via the intraperitoneal route. Spleen weight and splenic colonization of mice were assessed at 1 and 4 weeks postinfection. Of the 22 substrate mutants, 16 were found to be equally virulent in comparison to the parental strain. Compared to the wild type, (i) three mutants with deletions in genes encoding a lipase ΔB0249, a cell envelope-related amide hydrolase A1479, and a peptidase A0340 displayed lower bacterial loads but had comparable spleen weights (Fig. 7A); (ii) two mutants with deletions in genes encoding an ABC transporter substrate-binding protein ΔB0279 and the methionine sulfoxide reductase YedY (ΔB0019) exhibited reduced splenomegaly but indistinguishable bacterial burden (Fig. 7B); and (iii) deletion of A0577, encoding an L,d-transpeptidase, presented significantly reduced splenomegaly and bacterial burden at 4 weeks postinfection relative to the wild type (Fig. 7A and B). Collectively, we conclude that 6 Tat substrates contribute to different degrees of Brucella virulence in mice and that no single substrate can fully account for the observed virulence attenuation in tatA mutants.
FIG 7.
Role of each Tat substrate in B. melitensis virulence in a murine model. Six-week-old BALB/c mice were challenged intraperitoneally with wild-type B. melitensis M28 or each Tat substrate mutant. Splenic bacterial burden (A) and spleen weights (B) were measured at 1 and 4 weeks postinfection. The short lines amid data points in the graph represent the mean (n = 5 mice per bacterial strain per time point); colors of the short line indicate the functional group (see main text for detail): purple, ABC transporter solute-binding proteins; blue, oxidoreductases; red, cell envelope biogenesis enzymes; and black, others. Data were analyzed by one-way ANOVA followed by Tukey’s test. *, P < 0.05.
DISCUSSION
Tat translocase is an important and widespread protein translocation system that is responsible for secretion of folded proteins across the inner membrane. In B. abortus, a transposon sequencing study indicates that tatA and tatB insertional mutants can be obtained, but not a tatC mutant, suggesting that TatC may be critically important to B. abortus (38). In the present study, we took advantage of a tatA mutant and demonstrated the importance of the Tat system in stress resistance and virulence of B. melitensis M28. To date, only four bacteria, E. coli (∼30 substrates), Salmonella enterica (∼30 substrates) (39), Streptomyces coelicolor (∼27 substrates) (40), and P. aeruginosa (∼34 substrates) (36), have had their Tat substrates systematically predicted and verified with reporter and/or protein translocation assays. The various numbers of Tat substrates suggest the Tat system is utilized by different bacterial species to different extents. To our knowledge, we are the first to identify 23 Tat substrates and define at least the core set of Tat substrates in B. melitensis, which we accomplished through the use of both bioinformatics and experimental approaches (Fig. 5 and 6D). We were unable to experimentally validate six of the predicted signal peptides with the amidase reporter assay, suggesting that these might not be authentic Tat substrates. However, the failure could also be attributed to incompatibility due to species differences (36). Two sequence logos were created for the signal peptides identified in the M28 strain (Fig. 6E), which could help predict and identify more Tat signal peptides in Brucella, as well as engineer Tat signal peptides for purposeful use.
We showed that loss of Tat results in cell envelope defects and significantly attenuates Brucella virulence in both cell and mouse models (Fig. 2 to 4). The Tat system is crucial for virulence in many important bacterial pathogens (19, 20). Tat mutants of Salmonella fail to translocate two amidases, leading to envelope defects and, consequently, virulence attenuation (21). An extensive bioinformatics search did not identify any amidase as a Tat substrate in B. melitensis M28; nevertheless, the M28 Tat substrate set contains two putative L,d-transpeptidases, namely, A0577 and A0773 (Table 1). A0577 and A0773 both contain transpeptidase (TP) domains and are homologs of E. coli L,d-transpeptidase ErfK and YcbB, respectively. ErfK is responsible for cross-linking lipoproteins to the peptidoglycans (PGs), while YcbB forms direct meso-diaminopimelate (DAPDAP, or 3-3) cross-links within the PG. Amidases and transpeptidases are hydrolases that play important roles in cell wall integrity (41). In E. coli, the deletion of L,d-transpeptidase genes disrupts the cell envelope and induces leakage of periplasmic proteins (42), whereas in Mycobacterium tuberculosis, mutations in the L,d-transpeptidase gene alter cell size and morphology and attenuate virulence (43). Our results show that deletion of A0577 significantly reduces both splenomegaly and bacterial load at 28 days after infection (Fig. 7). Thus, a potential failure to properly translocate L,d-transpeptidases in tatA mutants, which can lead to cell envelope defects, could partly explain the virulence defect resulting from Tat disruption.
M28ΔtatA mutants also displayed increased sensitivity to H2O2 (Fig. 2F). Although a similar phenotype was reported in Tat mutants of Campylobacter jejuni, the underlying mechanism remains elusive (44). In our study, B0019 and B0965, both encoding methionine sulfoxide reductases (Msrs), were verified as Tat substrates (Fig. 6D). Homologs of these two Msrs are involved in resistance to H2O2 and/or HClO (45–47). The ability to resist H2O2 killing is correlated with survival and replication in macrophages, and subsequently determines the capacity of Brucella to establish chronic infections (48, 49). Indeed, deletion of B0019 significantly reduced splenomegaly compared to the parental strain (Fig. 7B). Brucella may employ multiple strategies to counter the stress conditions within macrophages, and translocation of Msrs into the periplasm by Tat likely constitutes an important protective mechanism.
The Tat substrate set of M28 contains more ABC transporter proteins than any other bacteria (Table S5) (36, 39, 40). The six ABC transporter proteins are well conserved in Brucella species. In a mouse model, B0279, encoding periplasmic substrate-binding protein of an ABC transporter, is involved in full virulence of M28. B0279 is somewhat similar to TauA, which is responsible for the transport of nitrate/sulfonate/bicarbonate (50). Based on the general roles of ABC transporters, we infer that these transporter proteins might function in the acquisition of nutrients for Brucella. However, their actual ligands remain unknown and thus warrant further studies. A0340 is predicted to be a surface-localized CAP domain-containing protein. It is similar to BB0689 of Borrelia burgdorferi, which is a surface-localized protein that is inducible during tick feeding. The CAP domain has the ability to bind to cholesterol, lipids, and proinflammatory leukotrienes (51). B0249 has a thioesterase domain (IPR001031), belonging to the α/β hydrolase superfamily; and proteins carrying this domain are often involved in the synthesis of secondary metabolites, such as polyketides. In Mycobacterium tuberculosis, many thioesterase domain-containing enzymes are associated with lipid metabolism, particularly in the degradation of host lipid material (52). Therefore, A0340 and B0249 might possibly contribute to virulence through direct Brucella-host interactions. A1479 encodes a predicted penicillin acylase with linear amide C-N hydrolase activities, and may have potential to modify Brucella peptidoglycan. Surprisingly, we were unable to generate a deletion mutant of B0388, despite repeated attempts. This protein was annotated as an O-antigen/exopolysaccharide biosynthesis protein (53); nonetheless, we found that the LPS profile of tatA mutants is similar to that of the wild type (data not shown).
Overall, the present study demonstrates the importance of Tat translocase and its substrates for B. melitensis M28 to cope with various stresses, and for infections ex vivo and in vivo. Our work has opened a new avenue for understanding Brucella pathobiology and virulence strategies. The severe virulence defects caused by Tat deficiency highlight that the Tat system may serve as a potential anti-Brucella target for developing antimicrobials or live attenuated vaccines. Future work should be directed toward identifying more Tat substrates and defining the functions of the substrates.
MATERIALS AND METHODS
Bacterial strains, cell cultures, and media.
All strains and plasmids used in the study are listed in Table S6. B. melitensis M28 and its derivatives were cultured on tryptic soy agar (TSA) or in tryptic soy broth (TSB) (Difco) at 37°C in a 5% CO2 atmosphere. E. coli strains (DH5α, MC4100, and their derivatives) were grown in Luria-Bertani (LB) medium at 37°C. When needed, appropriate antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; gentamicin, 5 μg/ml; and chloramphenicol, 25 μg/ml. RAW264.7 (ATCC) murine macrophage cell line was cultured in Dulbecco’s minimal essential medium (DMEM) basic (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) without antibiotics, and was incubated at 37°C in a 5% CO2 atmosphere.
Construction of deletion mutants and the complemented strains in M28.
All gene IDs were based on NC_017244.1 (Chromosome I) and NC_017245.1 (Chromosome II) from the NCBI database, and can be traced accordingly. In-frame deletion mutants of tatA (BM28_A0889) and each gene encoding Tat substrates in B. melitensis M28 were constructed by homologous recombination (26, 54). Briefly, 1,000-bp upstream and downstream regions of the target gene were PCR-amplified from B. melitensis M28, which was followed by cloning the two fragments so that they flanked a kanamycin resistance cassette in a pSP72 suicide vector (AmpR) using a MultiS one-step cloning kit (Vazyme, China). The resultant plasmid construct was transformed into M28 competent cells by electroporation, and the transformants were selected in TSA containing kanamycin. The candidate deletion mutants, which were resistant to kanamycin but sensitive to ampicillin, were further subjected to PCR amplification and DNA sequencing for verification. For complementation, the coding sequence of tatA plus its promoter region was amplified from M28 and cloned into pBBR1MCS4 (AmpR) using EcoRI and BamHI restriction sites. The resulting construct was confirmed by DNA sequencing and introduced into M28ΔtatA for complementation experiments. The empty vector did not affect the experiments we performed in this study (55–57). All primers used in this study are listed in Table S7.
RNA isolation and quantitative reverse transcription-PCR.
RNA extraction and quantitative reverse transcription-PCR (qPCR) analysis were performed as previously described (26, 54). B. melitensis M28 and the M28ΔtatA mutant were cultured in TSB medium at 37°C until they reached mid-log phase, and were then subjected to RNA extraction by TRIzol reagent according to the instructions (Invitrogen). RNA samples were treated to remove genomic DNA and subjected to reverse transcription using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Clontech). cDNA was used as the template for SYBR green-based qPCRs using TB Green Premix Ex Taq II reagent (TaKaRa, Clontech) and an ABI Quant 5 thermocycler. Melting curve analyses were performed after each reaction to ensure amplification specificity. Fold changes in transcript levels were calculated using the threshold cycle (2−ΔΔCt) method (58), and levels were normalized according to 16S rRNA expression.
Envelope stress assay.
A bacterial suspension from a fresh single colony was plated onto TSA plates and allowed to grow for 72 h. Cells were then harvested and resuspended in sterile phosphate-buffered saline (PBS) at an optical density at 600 nm (OD600) of ∼0.02 (∼1 × 108 CFU/ml) before performing a 10-fold serial dilution. Five microliters of each dilution was spotted onto plain TSA plates and TSA plates supplemented with 5 μg ml−1 ampicillin, 200 mM NaCl, or 2 mM EDTA. The plates were incubated at 37°C in a 5% CO2 atmosphere for 3 days to allow bacterial growth, which was followed by counting the CFU of each dilution.
Ratio of CFU on the TSA plain plates (CFUunstressed) to CFU on the EDTA-supplemented TSA plates (CFUstressed) was calculated for each dilution of the bacteria.
For the SDS sensitivity assay, a bacterial suspension from a fresh single colony was plated onto TSA plates and allowed to grow for 48 h. Cells were then harvested and resuspended in TSB containing 0.5% agar (maintained at 54°C) at a concentration of 1.5 × 107 CFU/ml. Twenty milliliters of this suspension was overlaid onto Brucella agar plates, and after the overlay solidified, a sterile 7-mm Whatman disk was placed in the center of each plate. Seven microliters of 10%, 5%, or 1% SDS, or 10 mg/ml polymyxin B solution, was applied to each filter disk. After incubation for 72 h at 37°C in a 5% CO2 atmosphere, the zone of inhibition around each disk was measured in millimeters.
Resistance to acidic environments and oxidative stress.
A bacterial suspension from a fresh single colony was plated onto TSA plates and allowed to grow for 48 h. Cells were then harvested and resuspended at a concentration of 107 CFU/ml in HCl-acidified TSB with a pH of 7.3, 5.5, 4.5, or 3.8. After 2 h of incubation at 37°C, bacterial cultures were serially diluted 10-fold and plated on TSA to quantify bacterial CFU. The bacterial survival percentages were calculated with respect to the numbers of CFU obtained from bacteria incubated in TSB at pH 7.3 (100% survival).
Resistance to oxidative stress was assayed according to previously described protocols with some modifications. M28 and its derivatives were grown on TSA for 48 h and adjusted to a concentration of 5 × 105 CFU/ml in PBS. Then, 100 μl of the bacterial suspension was mixed with the same volume of H2O2 (freshly prepared in PBS) at final concentrations of 5 mM, 2.5 mM, and 1 mM. A control experiment was performed by adding 100 μl of H2O2-free PBS to the same bacterial suspension. After 1 h of treatment at 37°C with shaking, the cells were swiftly serially diluted with PBS and plated onto TSA. After growth for 72 h at 37°C, CFU on each plate were counted, and the survival of each bacterial strain was determined as a percentage of the control.
In silico prediction of Tat substrates.
Potential Tat signal peptides in the B. melitensis M28 proteome were searched using three programs, TatP (http://www.cbs.dtu.dk/services/TatP/), TATFIND (http://signalfind.org/tatfind.html) and PRED-TAT (http://www.compgen.org/tools/PRED-TAT/), as previously described (25, 36). The output of each program is shown in Table S2-4. A protein containing Tat signal peptides predicted by at least two programs was considered a positive target and examined further for experimental verification.
The amidase reporter assay.
An amidase reporter assay based on the E. coli MC4100ΔssamiAC strain was performed essentially as previously described (36), but with minor modifications for plasmid construction. A double signal peptide deletion strain MC4100ΔssamiAC lacking residues 2 to 33 of AmiA and residues 2 to 32 of AmiC was constructed using the Lambda-Red recombinase system. The antibiotic resistance cassette was removed by a flippase encoded on the plasmid pCP20, and the strain was further confirmed by DNA sequencing (59). The resultant ΔssamiAC strain is defective in translocating AmiA and AmiC, sensitive to SDS stress, and forms chains of cells (29). Fragments containing a promoter region of M28 tatA and each predicted signal peptide-coding sequence were individually synthesized by Genewiz (China). The expression plasmid constructs were generated by linking each fragment mentioned above and the amiAH sequence into the linearized pSP72 vector using a MultiS one-step cloning kit (Vazyme, China). Each plasmid construct therefore allows PtatABC-driven expression of an N-terminal signal peptide fusion to mature His-tagged AmiA lacking a signal peptide. Each of these clones carries the tatA ribosome-binding site, with identical spacing between the ribosome-binding site and the start codon. The plasmid constructs of the experimental group and the control group were transferred into MC4100ΔssamiAC. The chain-forming phenotype and restoration of outer membrane integrity were determined by microscopy (Zeiss inverted microscope Primovert) and SDS sensitivity assays. For the signal peptides that yielded negative results in the reporter assay, Western blotting was used to detect the proper expression of the fusion AmiA with a His-tag. Western blotting was performed as described in previously described protocols (36) with a mouse anti-His6 primary antibody (Abcam, ab18184) and a KPL DyLight 800 labeled mouse IgG (KPL, 042-07-18-06).
Infection of murine macrophage cells.
BMDMs were isolated from femurs of 7- to 8-week-old BALB/c female mice, and subsequently differentiated into bone marrow-derived macrophages (BMDMs) as described previously (60). Evaluation of the intracellular survival of B. melitensis strains in BMDMs and RAW264.7 murine macrophage-like cell line was performed as previously described (54, 61). Briefly, monolayers of BMDM cells (2 × 105 cells per well) and RAW264.7 cells (1.5 × 106 cells per well) were seeded onto 24-well tissue culture plates and inoculated with bacterial culture diluted in DMEM (5% FBS) at an MOI (multiplicity of infection) of 100:1. After a 10-min centrifugation at 250 × g, the plates were placed in a 5% CO2 atmosphere at 37°C for 3 h. Then, cells were washed with DMEM three times and incubated in DMEM (5% FBS) containing 5 μg/ml gentamicin at 37°C/5% CO2 until the indicated times, when infected cells in each well were washed three times with PBS and lysed with 1 ml of 0.1% Triton X-100 in PBS. The intracellular CFU counts were determined by plating serial dilutions on TSA with the appropriate antibiotics.
Mouse infection assay.
Evaluation of B. melitensis virulence in BALB/c mice was performed in a BSL3 facility as described previously (26, 54). Briefly, groups of five 6-week-old mice with similar weights were inoculated intraperitoneally with 100 μl of a bacterial suspension containing 1 × 106 CFU of the indicated strain. At 1, 3, and 7 weeks postinfection (wpi), or at 1 wpi and 4 wpi for Tat substrate mutants, mice were sacrificed by cervical dislocation, and spleens were aseptically removed, weighed, and homogenized in PBS containing 0.1% Triton X-100. Each spleen homogenate was serially diluted in PBS and plated on TSA with the appropriate antibiotics to determine bacterial load, which is expressed as log10CFU per spleen.
Spleen histology.
Spleen histological analysis was performed according to previously described protocols with some modifications (62). At 1 week postinfection, spleens (n = 3 per strain) were removed and fixed with formalin for 7 days. Further, used formalin was replaced with fresh formalin, and fixation continued for another 7 days before washing and subsequent soaking in 70% ethanol. The fixed spleens were submitted to an in-house pathology sector for tissue embedding, section preparation, and hematoxylin and eosin (H & E) staining. For immunohistochemistry analysis, goat anti-Brucella antibodies (Bioss, bs-2229G) were applied, followed by detection with rabbit anti-goat IgG horseradish peroxidase (HRP)-conjugated antibodies (Abcam, ab6741). Observations were made and photographed using a Leica DM4000 B microscope. Pathological changes were scored by a pathologist in a blinded manner.
Statistical analysis.
One-way ANOVA (followed by Tukey’s test) analysis was used in mouse infection assays; for all other experiments, the Student’s t test was used to analyze differences between M28ΔtatA and the wild type (GraphPad, Prism). A P value of <0.05 was considered statistically significant.
Ethics statement.
Handling and care of laboratory animals were in accordance with the Beijing Administration Guidelines for the Use of Laboratory Animals. The complete protocol regarding animal studies was approved by the Review Board of Harbin Veterinary Research Institute and by the Animal Care and Use Committee of Heilongjiang Province (SYXK[H]2006-032). Infection of BALB/c mice by Brucella was conducted in a biosecurity level 3 laboratory approved by the Chinese Ministry of Agriculture.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erik Petersen from Gary Splitter’s lab for providing protocols and recipes for culturing Brucella in minimal medium, and Zhe Ma and Jiaoer Zhang for critical reviewing and helpful discussions. We would like to express our gratitude to all the staff members in the BSL3 facility for their assistance and daily service.
This work was supported in part by The National Key Research and Development Program of China (2018YFD0500501). The funders played no roles in study design, data collection and interpretation, or submission for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Banai M. 2002. Control of small ruminant brucellosis by use of Brucella melitensis Rev.1 vaccine: laboratory aspects and field observations. Vet Microbiol 90:497–519. doi: 10.1016/s0378-1135(02)00231-6. [DOI] [PubMed] [Google Scholar]
- 2.Byndloss MX, Tsolis RM. 2016. Brucella spp. virulence factors and immunity. Annu Rev Anim Biosci 4:111–127. doi: 10.1146/annurev-animal-021815-111326. [DOI] [PubMed] [Google Scholar]
- 3.Buzgan T, Karahocagil MK, Irmak H, Baran AI, Karsen H, Evirgen O, Akdeniz H. 2010. Clinical manifestations and complications in 1028 cases of brucellosis: a retrospective evaluation and review of the literature. Int J Infect Dis 14:e469. doi: 10.1016/j.ijid.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 4.Dean AS, Crump L, Greter H, Schelling E, Zinsstag J. 2012. Global burden of human brucellosis: a systematic review of disease frequency. PLoS Negl Trop Dis 6:e1865. doi: 10.1371/journal.pntd.0001865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starr T, Ng TW, Wehrly TD, Knodler LA, Celli J. 2008. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic 9:678–694. doi: 10.1111/j.1600-0854.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 6.Atluri VL, Xavier MN, de Jong MF, den Hartigh AB, Tsolis RM. 2011. Interactions of the human pathogenic Brucella species with their hosts. Annu Rev Microbiol 65 65:523–541. doi: 10.1146/annurev-micro-090110-102905. [DOI] [PubMed] [Google Scholar]
- 7.de Figueiredo P, Ficht TA, Rice-Ficht A, Rossetti CA, Adams LG. 2015. Pathogenesis and immunobiology of brucellosis: review of Brucella-host interactions. Am J Pathol 185:1505–1517. doi: 10.1016/j.ajpath.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barquero-Calvo E, Mora-Cartín R, Arce-Gorvel V, de Diego JL, Chacón-Díaz C, Chaves-Olarte E, Guzmán-Verri C, Buret AG, Gorvel J-P, Moreno E. 2015. Brucella abortus induces the premature death of human neutrophils through the action of its lipopolysaccharide. PLoS Pathog 11:e1004853. doi: 10.1371/journal.ppat.1004853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitzer JE, Zeczycki TN, Baumgartner JE, Martin DW, Roop RM. 2018. The manganese-dependent pyruvate kinase PykM is required for wild-type glucose utilization by Brucella abortus 2308 and its virulence in C57BL/6 mice. J Bacteriol 200:e00471-18. doi: 10.1128/JB.00471-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanna N, Jiménez de Bagüés MP, Ouahrani-Bettache S, El Yakhlifi Z, Köhler S, Occhialini A. 2011. The virB operon is essential for lethality of Brucella microti in the BALB/c murine model of infection. J Infect Dis 203:1129–1135. doi: 10.1093/infdis/jiq163. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz-Ranwez V, Posadas DM, Van der Henst C, Estein SM, Arocena GM, Abdian PL, Martín FA, Sieira R, De Bolle X, Zorreguieta A. 2013. BtaE, an adhesin that belongs to the trimeric autotransporter family, is required for full virulence and defines a specific adhesive pole of Brucella suis. Infect Immun 81:996–1007. doi: 10.1128/IAI.01241-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer T, Berks BC. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol 10:483–496. doi: 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 13.Berks BC. 1996. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol 22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 14.Ki JJ, Kawarasaki Y, Gam J, Harvey BR, Iverson BL, Georgiou G. 2004. A periplasmic fluorescent reporter protein and its application in high-throughput membrane protein topology analysis. J Mol Biol 341:901–909. doi: 10.1016/j.jmb.2004.05.078. [DOI] [PubMed] [Google Scholar]
- 15.Sargent F, Bogsch EG, Stanley NR, Wexler M, Robinson C, Berks BC, Palmer T. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J 17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cline K. 2015. Mechanistic aspects of folded protein transport by the twin arginine translocase (Tat). J Biol Chem 290:16530–16538. doi: 10.1074/jbc.R114.626820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer T, Sargent F, Berks BC. 2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol 13:175–180. doi: 10.1016/j.tim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Wagley S, Hemsley C, Thomas R, Moule MG, Vanaporn M, Andreae C, Robinson M, Goldman S, Wren BW, Butler CS, Titball RW. 2014. The twin arginine translocation system is essential for aerobic growth and full virulence of Burkholderia thailandensis. J Bacteriol 196:407–416. doi: 10.1128/JB.01046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochsner UA, Snyder A, Vasil AI, Vasil ML. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc Natl Acad Sci U S A 99:8312–8317. doi: 10.1073/pnas.082238299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavander M, Ericsson SK, Broms JE, Forsberg A. 2006. The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis. Infect Immun 74:1768–1776. doi: 10.1128/IAI.74.3.1768-1776.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craig M, Sadik AY, Golubeva YA, Tidhar A, Slauch JM. 2013. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol Microbiol 89:887–902. doi: 10.1111/mmi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossier O, Cianciotto NP. 2005. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect Immun 73:2020–2032. doi: 10.1128/IAI.73.4.2020-2032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pradel N, Ye CY, Livrelli V, Xu HG, Joly B, Wu LF. 2003. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect Immun 71:4908–4916. doi: 10.1128/iai.71.9.4908-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avican U, Doruk T, Ostberg Y, Fahlgren A, Forsberg A. 2017. The Tat substrate SufI is critical for the ability of Yersinia pseudotuberculosis to cause systemic infection. Infect Immun 85:e00867-16. doi: 10.1128/IAI.00867-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunez PA, Soria M, Farber MD. 2012. The twin-arginine translocation pathway in alpha-proteobacteria is functionally preserved irrespective of genomic and regulatory divergence. PLoS One 7:e33605. doi: 10.1371/journal.pone.0033605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F, Qiao Z, Hu S, Liu W, Zheng H, Liu S, Zhao X, Bu Z. 2013. Comparison of genomes of Brucella melitensis M28 and the B. melitensis M5-90 derivative vaccine strain highlights the translation elongation factor Tu gene tuf2 as an attenuation-related gene. Infect Immun 81:2812–2818. doi: 10.1128/IAI.00224-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walther TH, Gottselig C, Grage SL, Wolf M, Vargiu AV, Klein MJ, Vollmer S, Prock S, Hartmann M, Afonin S, Stockwald E, Heinzmann H, Nolandt OV, Wenzel W, Ruggerone P, Ulrich AS. 2013. Folding and self-assembly of the TatA translocation pore based on a charge zipper mechanism. Cell 152:316–326. doi: 10.1016/j.cell.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Caldelari I, Mann S, Crooks C, Palmer T. 2006. The Tat pathway of the plant pathogen Pseudomonas syringae is required for optimal virulence. Mol Plant Microbe Interact 19:200–212. doi: 10.1094/MPMI-19-0200. [DOI] [PubMed] [Google Scholar]
- 29.Ize B, Stanley NR, Buchanan G, Palmer T. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol Microbiol 48:1183–1193. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- 30.Finnegan S, Percival SL. 2015. EDTA: an antimicrobial and antibiofilm agent for use in wound Ccare. Adv Wound Care (New Rochelle) 4:415–421. doi: 10.1089/wound.2014.0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dos Santos MH, Da Costa AF, Da Silva Santos G, Dos Santos AL, Nagao PE. 2009. Effect of chelating agents on the growth, surface polypeptide synthesis and interaction of Streptococcus agalactiae with human epithelial cells. Mol Med Rep 2:81–84. doi: 10.3892/mmr_00000065. [DOI] [PubMed] [Google Scholar]
- 32.Teixeira-Gomes AP, Cloeckaert A, Zygmunt MS. 2000. Characterization of heat, oxidative, and acid stress responses in Brucella melitensis. Infect Immun 68:2954–2961. doi: 10.1128/iai.68.5.2954-2961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dilks K, Rose RW, Hartmann E, Pohlschroder M. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol 185:1478–1483. doi: 10.1128/jb.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. 2010. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 26:2811–2817. doi: 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- 36.Gimenez MR, Chandra G, Van Overvelt P, Voulhoux R, Bleves S, Ize B. 2018. Genome wide identification and experimental validation of Pseudomonas aeruginosa Tat substrates. Sci Rep 8:11950. doi: 10.1038/s41598-018-30393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ize B, Coulthurst SJ, Hatzixanthis K, Caldelari I, Buchanan G, Barclay EC, Richardson DJ, Palmer T, Sargent F. 2009. Remnant signal peptides on non-exported enzymes: implications for the evolution of prokaryotic respiratory chains. Microbiology 155:3992–4004. doi: 10.1099/mic.0.033647-0. [DOI] [PubMed] [Google Scholar]
- 38.Sternon JF, Godessart P, Goncalves de Freitas R, Van der Henst M, Poncin K, Francis N, Willemart K, Christen M, Christen B, Letesson JJ, De Bolle X. 2018. Transposon sequencing of Brucella abortus uncovers essential genes for growth in vitro and inside macrophages. Infect Immun 86:e00312-18. doi: 10.1128/IAI.00312-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer T, Sargent F, Berks BC. 2010. The Tat protein export pathway. EcoSal Plus 4. doi: 10.1128/ecosalplus.4.3.2. [DOI] [PubMed] [Google Scholar]
- 40.Widdick DA, Dilks K, Chandra G, Bottrill A, Naldrett M, Pohlschroder M, Palmer T. 2006. The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc Natl Acad Sci U S A 103:17927–17932. doi: 10.1073/pnas.0607025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vermassen A, Leroy S, Talon R, Provot C, Popowska M, Desvaux M. 2019. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol 10:331. doi: 10.3389/fmicb.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders AN, Pavelka MS. 2013. Phenotypic analysis of Eschericia coli mutants lacking L,D-transpeptidases. Microbiology 159:1842–1852. doi: 10.1099/mic.0.069211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoonmaker MK, Bishai WR, Lamichhane G. 2014. Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, the cytosolic matrix, protein localization, virulence, and resistance to beta-lactams. J Bacteriol 196:1394–1402. doi: 10.1128/JB.01396-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajashekara G, Drozd M, Gangaiah D, Jeon B, Liu Z, Zhang Q. 2009. Functional characterization of the twin-arginine translocation system in Campylobacter jejuni. Foodborne Pathog Dis 6:935–945. doi: 10.1089/fpd.2009.0298. [DOI] [PubMed] [Google Scholar]
- 45.Singh VK, Singh K, Baum K. 2018. The role of methionine sulfoxide reductases in oxidative stress tolerance and virulence of Staphylococcus aureus and other bacteria. Antioxidants (Basel) 7:128. doi: 10.3390/antiox7100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao C, Hartke A, La Sorda M, Posteraro B, Laplace JM, Auffray Y, Sanguinetti M. 2010. Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence. Infect Immun 78:3889–3897. doi: 10.1128/IAI.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alamuri P, Maier RJ. 2004. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol Microbiol 53:1397–1406. doi: 10.1111/j.1365-2958.2004.04190.x. [DOI] [PubMed] [Google Scholar]
- 48.Ahmed W, Zheng K, Liu ZF. 2016. Establishment of chronic infection: Brucella's stealth strategy. Front Cell Infect Microbiol 6:30. doi: 10.3389/fcimb.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steele KH, Baumgartner JE, Valderas MW, Roop RM 2nd, 2010. Comparative study of the roles of AhpC and KatE as respiratory antioxidants in Brucella abortus 2308. J Bacteriol 192:4912–4922. doi: 10.1128/JB.00231-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Javaux C, Joris B, De Witte P. 2007. Functional characteristics of TauA binding protein from TauABC Escherichia coli system. Protein J 26:231–238. doi: 10.1007/s10930-006-9064-x. [DOI] [PubMed] [Google Scholar]
- 51.Brangulis K, Jaudzems K, Petrovskis I, Akopjana I, Kazaks A, Tars K. 2015. Structural and functional analysis of BB0689 from Borrelia burgdorferi, a member of the bacterial CAP superfamily. J Struct Biol 192:320–330. doi: 10.1016/j.jsb.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 52.Maan P, Kumar A, Kaur J, Kaur J. 2018. Rv1288, a two domain, cell wall anchored, nutrient stress inducible carboxyl-esterase of Mycobacterium tuberculosis, modulates cell wall lipid. Front Cell Infect Microbiol 8:421. doi: 10.3389/fcimb.2018.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalynych S, Cherney M, Bostina M, Rouiller I, Cygler M. 2015. Quaternary structure of WzzB and WzzE polysaccharide copolymerases. Protein Sci 24:58–69. doi: 10.1002/pro.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu D, Song J, Li G, Cai W, Zong S, Li Z, Liu W, Hu S, Bu Z. 2018. A novel small RNA Bmsr1 enhances virulence in Brucella melitensis M28. Vet Microbiol 223:1–8. doi: 10.1016/j.vetmic.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Rajashekara G, Covert J, Petersen E, Eskra L, Splitter G. 2008. Genomic island 2 of Brucella melitensis is a major virulence determinant: functional analyses of genomic islands. J Bacteriol 190:6243–6252. doi: 10.1128/JB.00520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patey G, Qi Z, Bourg G, Baron C, O'Callaghan D. 2006. Swapping of periplasmic domains between Brucella suis VirB8 and a pSB102 VirB8 homologue allows heterologous complementation. Infect Immun 74:4945–4949. doi: 10.1128/IAI.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delrue RM, Deschamps C, Leonard S, Nijskens C, Danese I, Schaus JM, Bonnot S, Ferooz J, Tibor A, De Bolle X, Letesson JJ. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol 7:1151–1161. doi: 10.1111/j.1462-5822.2005.00543.x. [DOI] [PubMed] [Google Scholar]
- 58.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai W, Wannemuehler Y, Dell'anna G, Nicholson B, Barbieri NL, Kariyawasam S, Feng Y, Logue CM, Nolan LK, Li G. 2013. A novel two-component signaling system facilitates uropathogenic Escherichia coli's ability to exploit abundant host metabolites. PLoS Pathog 9:e1003428. doi: 10.1371/journal.ppat.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conde-Álvarez R, Arce-Gorvel V, Iriarte M, Manček-Keber M, Barquero-Calvo E, Palacios-Chaves L, Chacón-Díaz C, Chaves-Olarte E, Martirosyan A, von Bargen K, Grilló M-J, Jerala R, Brandenburg K, Llobet E, Bengoechea JA, Moreno E, Moriyón I, Gorvel J-P. 2012. The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition. PLoS Pathog 8:e1002675. doi: 10.1371/journal.ppat.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F, Hu S, Liu W, Qiao Z, Gao Y, Bu Z. 2011. Deep-sequencing analysis of the mouse transcriptome response to infection with Brucella melitensis strains of differing virulence. PLoS One 6:e28485. doi: 10.1371/journal.pone.0028485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herrou J, Willett JW, Fiebig A, Czyż DM, Cheng JX, Ultee E, Briegel A, Bigelow L, Babnigg G, Kim Y, Crosson S. 2019. Brucella periplasmic protein EipB is a molecular determinant of cell envelope integrity and virulence. J Bacteriol 201:e00134-19. doi: 10.1128/JB.00134-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.