Abstract
The circadian clock regulates diverse physiological processes by maintaining a 24-h gene expression pattern. Genetic and environmental cues that disrupt normal clock rhythms can lead to cancer, yet the extent to which this effect is controlled by the cancer cells versus non-malignant cells in the tumor microenvironment (TME) is not clear. Here we set out to address this question, by selective manipulation of circadian clock genes in the TME. In two different mouse models of cancer we find that expression of the core clock gene Per2 in the TME is crucial for tumor initiation and metastatic colonization, whereas another core gene, Per1, is dispensable. We further show that loss of Per2 in the TME leads to significant transcriptional changes in response to cancer cell introduction. These changes may contribute to a tumor-suppressive microenvironment. Thus, our work unravels an unexpected protumorigenic role for the core clock gene Per2 in the TME, with potential implications for therapeutic dosing strategies and treatment regimens.
Keywords: circadian clock, Per2, tumor microenvironment, colon cancer, chronobiology, metastasis, liver, MC38 colon cancer cells
Introduction
The circadian clock is an endogenous, evolutionally conserved and ubiquitously expressed pacemaker, consisting of cell autonomous clocks and a central pacemaker located in the hypothalamus suprachiasmatic nucleus (SCN). Together these clocks synchronize numerous biological processes between the organism and its environment. Amongst these processes are DNA damage repair, metabolism, and cell cycle (Blakeman et al., 2016). The circadian clocks act as oscillators to drive 24-h rhythms in gene expression and protein function, and these rhythms help the organism maintain a homeostatic relationship with the environment (Padmanabhan and Billaud, 2017). Disruption of circadian rhythms has been associated with various forms of cancer in humans, and it has been shown that a disordered circadian clock, whether genetically or due to environmental signals (e.g., changes of dark/light exposure) accelerates tumor progression (Savvidis and Koutsilieris, 2012; Hadadi et al., 2019). Additionally, epidemiological studies show that women who work in irregular shift work may be at higher risk of developing breast cancer (Schernhammer et al., 2003; Hansen and Stevens, 2012; Blakeman et al., 2016). Moreover, the toxicity and efficacy of various cancer therapies is dictated by time-of-day (Kobayashi et al., 2002; Lauriola et al., 2014; Borniger et al., 2017). Cancer cells with a disrupted clock show increased growth in culture, and mice with a disrupted clock tend to develop more radiation-induced tumors than wild type (WT) mice (Shostak, 2017). Mutations in genes that regulate the molecular clock have been found in human cancer samples, indicating a gene-specific causal relation between the circadian clock and cancer (Kettner et al., 2014). At the cellular level, most cancer cell-lines show loss of rhythmicity, specifically of genes that are related to proliferation and apoptosis, thus enabling their uncontrolled proliferation (Relógio et al., 2014).
The currently held molecular model for circadian rhythmicity is based on a transcription-translation feedback loop. This mechanism includes the transcription factors BMAL1 and CLOCK that drive transcription of the Period (Per) and Cryptochrome (Cry) genes. Then, PER and CRY proteins dimerize, translocate to the nucleus and repress Clock-Bmal1 transcription. This orchestrated machinery results in periodic expression of approximately half of the mammalian genes (Zhang et al., 2014). Previous studies found that Per1 and Per2 are essential for the proper function of the circadian clock, while the contribution of Per3 is not as important (Bae et al., 2001). Per2 was shown to be essential for proper function of the circadian clock under conditions of constant dark or light, as mice lacking Per2 exhibit a faulty clock that is reflected by a shorter period and rhythmic instability (Zheng et al., 1999; Tamiya et al., 2016). In vitro, loss of Per1 or Per2 was suggested to have a much milder effect on cell rhythmicity (Ramanathan et al., 2014), compared to the effect observed in vivo.
It is well established that the non-malignant components of the tumor, also termed the tumor microenvironment (TME) are an integral and essential part of the tumor. The TME is composed of many cell types, including immune cells, cancer associated fibroblasts (CAF), endothelial cells, and extracellular matrix (ECM). The TME is essential for tumor formation, homeostasis, and progression (Quail and Joyce, 2013; Yuan et al., 2016; Jin et al., 2017). It is dynamically reshaped, thus certain cell populations in the TME can be detrimental for early stages of tumor initiation, while protumorigenic in later stages of tumor progression (Lu et al., 2012; Bercovici et al., 2019; Lin et al., 2019; Wei et al., 2020). Previous studies have shown that cancer cells within a tumor lose their rhythmicity, yet there is sparse data about rhythmicity of the TME. This is partially due to the fact that the vast majority of published studies on cancer and circadian clocks used mouse models in which both the TME and the cancer cells are affected by a disrupted clock, either by genetic mutations or by environmental cues. Recently it was shown that mice with environmentally disrupted clocks had a more protumorigenic immune microenvironment in breast cancer (Hadadi et al., 2019) and thoracic cancer (Yang et al., 2019). Pan-cancer analysis supported these findings, and suggested that circadian genes are involved in the regulation of cancer immunity (Zhou et al., 2020). Not only the immune microenvironment, but also CAFs were proposed to play a role in clock regulation in cancer, when shown that co-culturing colon cancer cells with CAFs improved the rhythmicity of cancer cells (Fuhr et al., 2019). Gaining a better understanding of how circadian clocks in the TME affect the development and progression of the tumor could lead to novel therapeutic advances and approaches. It is possible that synchronization, or lack of synchronization, between the TME and the cancer cells, plays a role in cancer progression. For example, tumors may rely on the TME maintaining intact circadian rhythms to compensate for imbalances resulting from loss of rhythmicity in the cancer cells. Alternatively, loss of rhythmicity may be essential for cells of the TME to shift from antitumorigenic to protumorigenic. In this study we set out to explore the role of the circadian clock in the TME. Using orthotopic injection of cancer cells into clock-deficient mice, we find that stromal Per2, but not Per1, is required for tumor growth and metastatic colonization. Loss of Per2 in the TME leads to transcriptional rewiring at early stages of metastases formation, and suppresses subsequent metastatic tumor progression, highlighting Per2 as an unexpected protumorigenic mediator in the TME.
Results
The Circadian Rhythm in Cancer Cells Is Disrupted, While Normal Fibroblasts Maintain a Robust Rhythm, in vitro
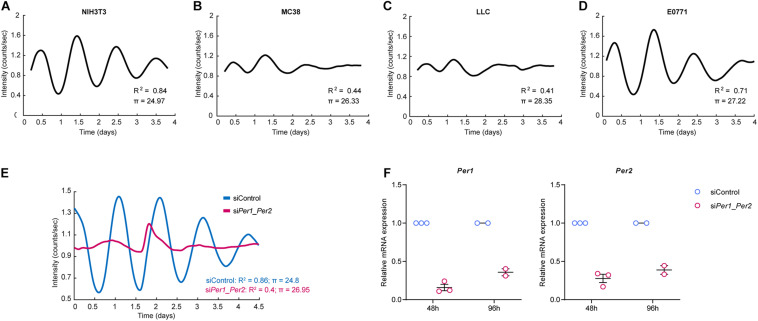
A disrupted circadian clock is associated with tumorigenesis, and cancer cells are thought to express a less robust clock compared to non-malignant cells (Welsh et al., 2004; Kiessling et al., 2017). To test this, we set out to evaluate the rhythmicity of NIH3T3, a non-malignant immortalized fibroblast cell line, and of three cancer cell lines – MC38 (colon), LLC (lung), and E0771 (breast) – using continuous bioluminescence monitoring of cells. Cells were transduced with a Per2 promotor-driven luciferase reporter (Yoo et al., 2004; Liu et al., 2008), synchronized by the synthetic glucocorticoid Dexamethasone, and longitudinally assessed using a LumiCycle luminometer (Yamazaki and Takahashi, 2005). To better understand the rhythmic behavior of cells, we developed a regression model that calculates the period length (π) of the curve (see “Materials and Methods”). The coefficient of determination R2 can be used as a readout for how closely our empiric data follows a sinusoidal pattern. As evident by the period and the coefficient of determination, NIH3T3 fibroblasts exhibited robust rhythmicity (Figure 1A), while all three cancer cell lines were less rhythmic than NIH3T3 (Figures 1B–D). E0771 cells exhibited the highest coefficient of determination of all three cancer cell lines, indicating that their rhythmicity is the most intact, while still less robust than that of non-malignant cells. The period length of all three cancer cell lines was longer than that of the fibroblasts, with LLC having the longest period (28.3 h), followed by E0771 (27.2 h), and MC38 (26.3 h). Next, we asked whether the rhythmicity of the NIH3T3 fibroblasts could be disrupted by loss of Per1/Per2. We knocked down Per1 and Per2 using siRNA and assessed their rhythmicity using bioluminescence. Indeed, we found that deletion of Per1 and Per2 impairs cell rhythmicity (Figures 1E,F). These results suggest that the rhythmicity of cancer cells is disrupted, in vitro, while normal cells maintain a functional clock, the activity of which is dependent on the expression of the core clock genes Per1 and Per2.
FIGURE 1.
Cancer cells show disrupted rhythmicity compared to normal fibroblasts. (A–D) Per2::luc expressing NIH3T3 fibroblasts or cancer cells, as indicated, were synchronized by dexamethasone and their luminescence signal was measured continuously (every 12 min) in a LumiCycle luminometer. The period and non-linear regression model coefficient of determination are presented (see “Materials and Methods”). The curves are an average of 3 technical repeats, and are representative of at least two independent experiments. (E,F) Per1 and Per2 were knocked down in Per2::luc-expressing NIH3T3 cells using siRNA. The cells were synchronized by dexamethasone and their luminescence was measured continuously (every 12 min) in a LumiCycle luminometer. (E) Period and the non-linear regression model coefficient of determination are presented. The curves are an average of 3 technical repeats, and are representative of two experiments. (F) Validation of Per gene knockdown efficiency at two time points by RT-PCR is presented as mean ± SEM.
Loss of Per1 and Per2 Inhibits Metastatic Colonization
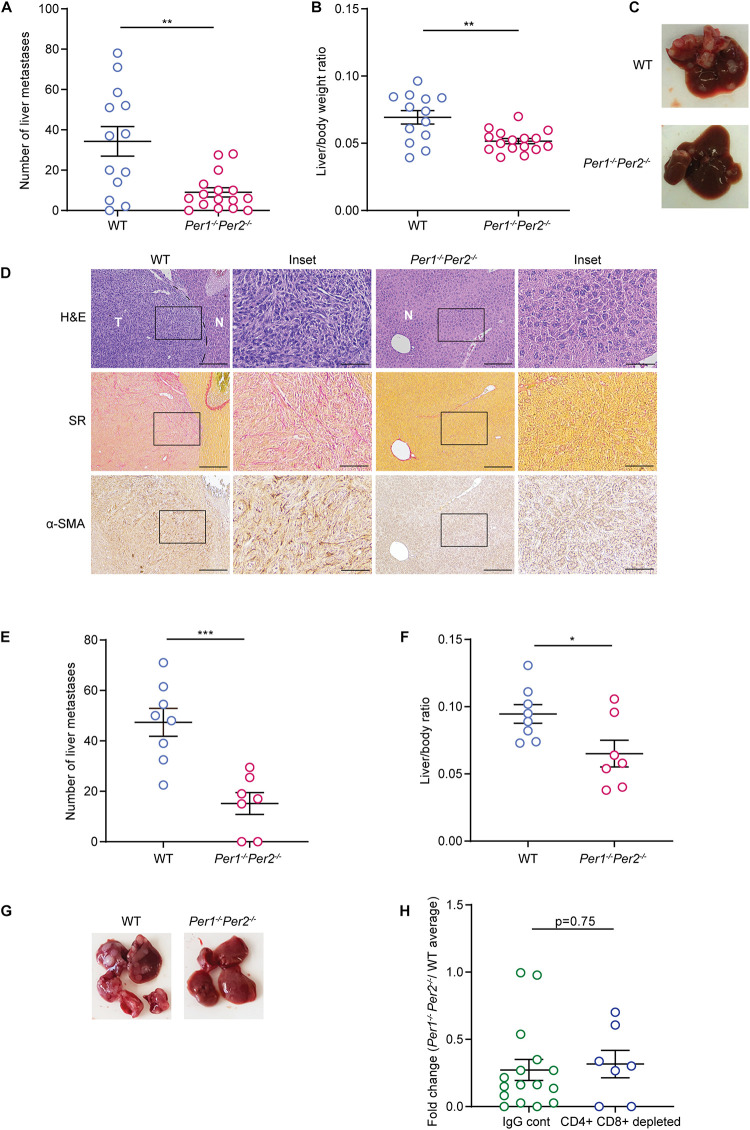
Several reports have shown that depletion of the Per family genes enhances cancer cell growth and tumor progression (Fu et al., 2002; Gery et al., 2006; Yang et al., 2009; Papagiannakopoulos et al., 2016). To understand how loss of rhythmicity in the TME affects tumor growth, we used a mouse model of liver metastasis, employing a syngeneic colorectal cancer cell line (MC38) in C57Bl/6 mice. Since the liver functions in a circadian manner (Reinke and Asher, 2016, 2019), and is affected directly by the feeding patterns of the organism, it serves as a good model to investigate the circadian clock in vivo (Tahara and Shibata, 2016). In our model, 20,000 cancer cells were injected to the hepatic portal vein of mice, producing liver metastases 21 days post injection. We applied this model to WT and Per1–/–Per2–/– mice to assess the effect of a clock-impaired TME on metastatic colonization. Surprisingly, Per1–/–Per2–/– mice had a lower metastatic burden than WT mice, as measured by the reduced number of metastases and lower liver/body weight ratio (Figures 2A–C and Supplementary Figure 1A). WT mice developed dozens of liver metastases, which exhibited substantial ECM rearrangements, as evident by Sirius red staining for collagen, and were heavily infiltrated by α-SMA-positive CAFs (Figure 2D). In stark contrast, the livers of Per1–/–Per2–/– mice had significantly fewer metastases, and stained mostly negative for α-SMA and Sirius Red (Figure 2D). These results suggest that loss of Per genes in the stroma inhibits metastatic colonization and ECM-remodeling of the metastatic niche.
FIGURE 2.
Clock deficient mice exhibit lower metastatic burden. (A–D) WT and Per1–/–Per2–/– mice were injected with MC38 cancer cells into the hepatic portal vein. At 21 days post injection macrometastases were quantified by visual inspection (A) and the ratio of liver weight to body weight was calculated (B). n = 13–16 mice per genotype combined from three independent experiments. P-value was calculated using unpaired Student’s t-test with Welch’s correction. Error bars represent SEM. (C) Representative images of livers from WT and Per1–/–Per2–/–mice. (D) Representative Hematoxylin and Eosin (H&E), Sirius red (SR), and α-SMA staining of livers from WT and Per1–/–Per2–/– mice. Scale bar – 200 μm; inset – 67 μm; T-tumor, N-normal tissue. (E–H) WT and Per1–/–Per2–/– mice were injected with antibodies of CD4+ and CD8+ 2 days prior to MC38 injection and every 5 days until the end point. At 21 days post injection macrometastases were quantified by visual inspection (E) and the ratio of liver weight to body weight was calculated (F). n = 7–8 per genotype combined from two independent experiments. P-value was calculated using unpaired Student’s t-test. (G) Representative images of livers from WT and Per1–/–Per2–/– mice with T cell depletion are shown. (H) The number of metastases in normal and T cell depleted Per1–/–Per2–/– mice was normalized to the average number of metastases in WT mice. P-value was calculated using unpaired Student’s t-test. In all figures error bars represent SEM, *p < 0.05, **p < 0.005, ***p < 0.0005.
T-cells are a major component of the TME, and have a key role in the cytotoxic activity against cancer cells. To assess whether T cells mediate the tumor growth inhibition observed in Per1–/–Per2–/– mice, as well as to exclude the possibility that Per1–/–Per2–/– mice reject the MC38 cancer cells due to their expression of Per1 and Per2 genes, we injected cancer cells into WT or Per1–/–Per2–/– mice in which T cells were depleted by CD4 and CD8 antibodies (Supplementary Figure 1B). CD4 and CD8 antibodies were injected 2 days prior to the injection of cancer cells, and every 5 days until the endpoint, and the efficiency of depletion was verified by FACS analysis (Supplementary Figure 1B). Per1–/–Per2–/– mice were significantly more resistant to metastatic colonization than WT mice, even upon T cell depletion (Figures 2E–G). The effect of Per1–/–Per2–/– loss in the TME was similar in the presence and absence of T cells (Figure 2H). Together, these results suggest that T cells do not mediate stromal Per1–/–Per2–/–-associated tumor inhibition.
Stromal Per2 Regulates Tumor Progression
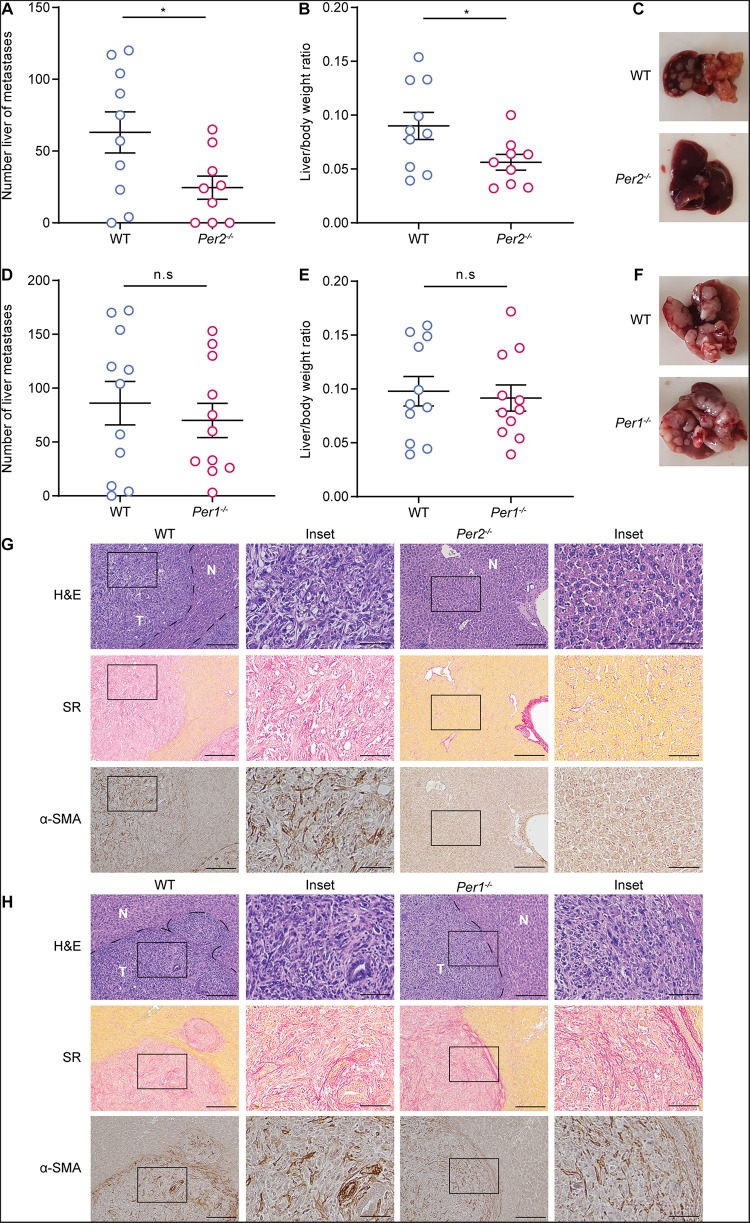
It is well established that both Per1 and Per2 are essential for maintaining proper circadian clock activity (Bae et al., 2001). To test whether both genes are also essential for the stromal regulation of tumor progression, we injected MC38 cancer cells to the portal vein of mice in which each of the Per genes (Per1, Per2) was knocked-out separately, and assessed metastatic colonization. WT mice were used as control. Per2–/– mice exhibited significantly lower metastatic burden than WT mice (Figures 3A–C). Loss of Per1, on the other hand, did not affect metastasis, and Per1–/– mice exhibited similar metastatic burden to that of WT (Figures 3D–F). Per2–/– livers also showed less collagen deposition and CAF infiltration than WT (Figure 3G), while the amount and the structure of collagen and infiltrating CAFs was similar between Per1–/– and WT livers (Figure 3H). These results suggest that stromal Per2 is essential for metastatic colonization, while Per1 is dispensable, thus implying that the effect of stromal Per2 knock-out on metastatic colonization may be clock-independent.
FIGURE 3.
Stromal Per2, and not Per1 promotes metastatic colonization. (A–C) WT and Per2–/– mice were injected with MC38 cancer cells into the hepatic portal vein. At 21 days post injection (A) macrometastases were quantified by visual inspection and (B) the ratio of liver weight to body weight was calculated. n = 9–10 mice per genotype combined from three independent experiments. (C) Representative images of livers from WT and Per2–/– mice are shown. (D–F) WT and Per1–/– mice were injected with MC38 cancer cells into the hepatic portal vein. At 21 days post injection macrometastases were quantified by visual inspection (D) and the ratio of liver weight to body weight was calculated (E). n = 11 mice per genotype combined from three independent experiments. (F) Representative images of livers from WT and Per1–/– mice are shown. (G,H) Representative H&E, Sirius red (SR), and α-SMA staining of livers from WT, Per2–/– and Per1–/– mice harvested at day 21 post injection. Scale bar – 200 μm, inset – 67 μm; T-tumor N-normal tissue. P-value was calculated using unpaired Student’s t-test. Error bars represent SEM, *p < 0.05.
Transcriptional Analysis of WT and Per2–/– Livers Suggests a Role for Per2 in Eliciting a Pre-metastatic Niche
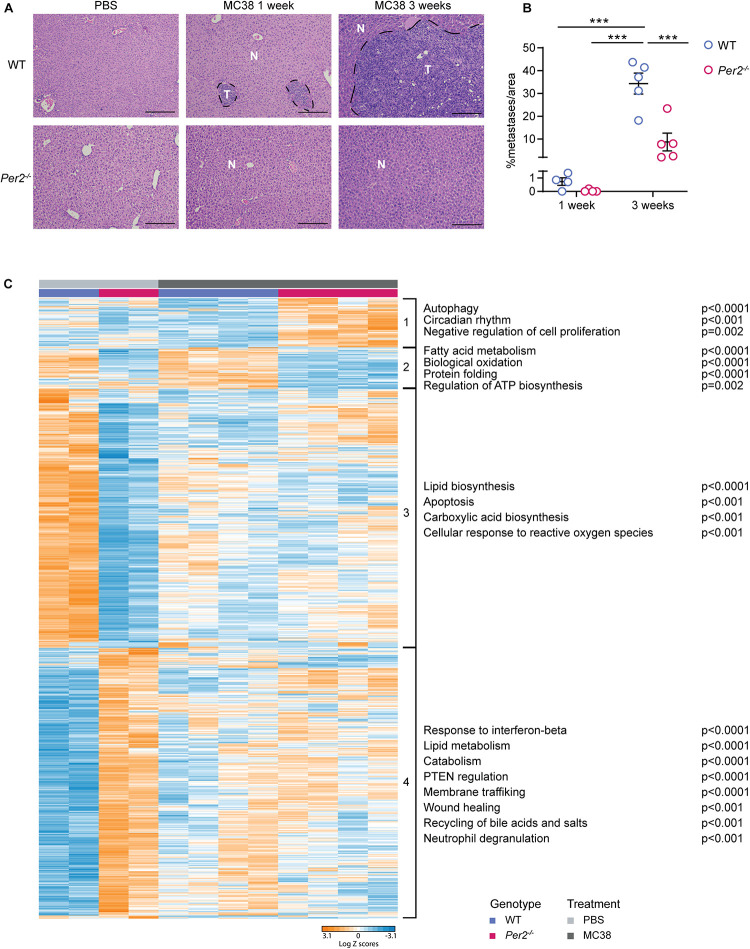
We hypothesized that stromal Per2 plays a role in preparing the hepatic niche for the initiation of metastasis. To test this hypothesis, we injected MC38 cancer cells (or PBS, as control) to the portal vein of Per2–/– and WT mice, harvested the livers 1 week following injection, and performed RNA-sequencing on total liver extracts (Supplementary Table 1). At this early stage only micrometastases (∼0.0005 mm3), and not macrometastases (∼5 mm3 at 3 weeks) were visible in WT livers (Figure 4A). The average area covered by metastases in the liver was 0.7% in WT mice and 0.05% in Per2–/– mice at this early time point (compared to 34.3% in WT mice and 8.7% in Per2–/– mice at 3 weeks post injection; Figure 4B). Therefore, we assume that the sequencing data represents mostly the normal liver and the pre-metastatic niche, and not cancer cells. Principal component analysis (PCA) showed that the basal transcriptional landscape of WT and Per2–/– livers is different, and further changes following injection of cancer cells (Supplementary Figure 2B). Hierarchical clustering highlighted 1222 differentially expressed genes between WT and Per2–/– livers injected with PBS or with MC38 cancer cells (Figure 4C and Supplementary Table 1). In the control mice (PBS), loss of Per2–/– led to upregulation of the response to interferon-beta (Ifit1, Stat1, Tgtp1, Eif2ak2), catabolism (Adam9, Pik3c2a, Tnfaip3, Rock1), wound healing (Cd36, Cldn1, Pdpn, Pdgfra, Pten), recycling of bile acids and salts (Slc10a2, Abcb11, Slco1b2), and neutrophil degranulation (Adam10, Ctsc, Degs1; Figure 4C and cluster 4 and Supplementary Table 2). Apoptosis (Akt1, Bad, Bcl2l1, Bnip3), carboxylic acid biosynthesis (Aldoa, Apoa1, Apoa4, Ccnd3), and the cellular response to reactive oxygen species (Sod3, Pdk2, Park7, Ccs) were downregulated in the Per2–/– livers (Figure 4C and cluster 3 and Supplementary Table 2). The same genes and pathways were also downregulated (though to a lesser extent) following injection of MC38 cells to either WT or Per2–/– mice, suggesting that these are genes involved in normal liver functions that fail to function properly upon loss of Per2–/– or in early cancer stages (Figure 4C, cluster 3). We could not identify pathways upregulated in WT livers 1 week after cancer cell injection. In Per2–/– livers, however, we observed upregulation of genes involved in autophagy (Ulk1, Plk3, Sesn2), circadian regulation (Ngfr, Klf10, Atf5), and inhibition of cell proliferation (Ngfr, Hes1, Tob1) following cancer cell injection (Figure 4C and cluster 1 and Supplementary Table 2). This analysis suggests that in early stages of metastases formation, house-keeping genes are downregulated in WT livers, while only minor transcriptional upregulation is observed. Loss of Per2, however, rearranges the premetastatic liver niche, in a manner that activates tumor suppressing pathways in response to the presence of cancer cells, resulting in attenuated metastatic spread.
FIGURE 4.
Stromal Per2–/– is essential for the formation of the pre-metastatic niche. WT and Per2–/– mice were injected with MC38 cancer cells into the hepatic portal vein and sacrificed 1 or 3 weeks post injection, as indicated. PBS was injected to control mice which were sacrificed 1 week post injection. n = 4–5 mice per genotype. (A) Representative H&E staining of livers from WT and Per2–/– mice post injection of PBS or MC38 cancer cells. Scale bar – 200 μm, inset – 67 μm; T-tumor, N-normal tissue. (B) The area covered by metastases in MC38-injected livers was calculated by QuPath based on H&E images. P-value was calculated using two way ANOVA. Error bars represent SEM, ***p < 0.0005. (C) RNA-sequencing was performed on livers of WT and Per2–/– mice injected with MC38 (n = 4 per genotype) and PBS (n = 2 per genotype). Heatmap showing hierarchical clustering of 1222 differentially expressed genes between all four conditions. Pathway analysis was performed using Metascape. Significant pathways are shown; p < 0.05, FDR < 0.05 (for details see Supplementary Table 2).
We further interrogated our RNA-sequencing data using CIBERSORT. This tool provides an estimation of the abundance of various cell types in a mixed cell population, based on expression of cell-type specific hallmark genes (Newman et al., 2019). We compiled several published single-cell datasets to create a reference dataset with which we examined the composition of our samples [Supplementary Table 3 and references (Heng and Painter, 2008; Dobie et al., 2019; Xiong et al., 2019)]. We found that the cellular composition is similar across samples (Supplementary Table 4), with one exception: cholangiocytes were significantly elevated in Per2–/– livers compared to WT livers (Supplementary Figure 2C and Supplementary Table 4). While this analysis did not highlight changes in cholangiocyte numbers between MC38 treated and non-treated livers, these findings suggest that Per2 plays a role in cholangiocyte homeostasis. These findings are in line with previous reports showing that deletion of Per2 exacerbates cholestatic liver injury and fibrosis (Chen et al., 2013). The majority of cells detected in all of our samples were hepatocytes (>95%, Supplementary Table 4), suggesting that transcriptional rewiring of these cells, rather than changes in cell composition, leads to the observed changes in gene expression.
Loss of Per2 in the Stroma Impairs Primary Tumor Growth
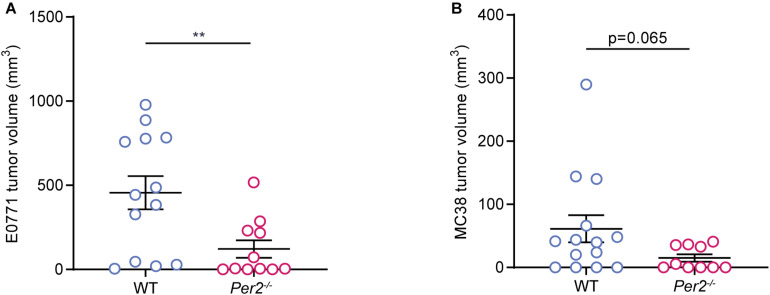
Next, we asked whether stromal Per2 is essential not only for metastatic colonization, but also for primary tumor formation. We used two orthotropic models: The E0771 triple negative breast cancer cell line injected to the mammary fat pad, and the MC38 colon cancer cell line (used for the liver metastasis model) injected into the colon submucosa by an endoscopy-guided procedure [as described in Zigmond et al. (2011)]. In both models, cancer cells were injected to Per2–/– and WT mice, and tumor volume was assessed at the end point. Per2–/– mice exhibited significantly smaller E0771 tumors compared to WT mice (Figure 5A), and a similar trend was seen for the MC38 colon model (Figure 5B). Taken together with our results from the liver metastasis model, these results suggest that stromal Per2 is required for tumor formation both in the primary site as well as in the metastatic site. These findings also imply that PER2 is required throughout the course of tumor progression and metastasis.
FIGURE 5.
Stromal Per2 controls primary tumor growth. (A) WT and Per2–/– mice were injected with E0771 cancer cells into the mammary fat pad. The mice were sacrificed at day 32, and tumor volume was calculated. n = 11–13 per genotype, combined from two independent experiments. (B) WT and Per2–/– mice were injected with MC38 cancer cells into the colon wall. Mice were sacrificed at day 14 and tumor volume was calculated. n = 10–14 per genotype combined from two independent experiments. P-value was calculated using unpaired student’s t-test with Welch’s correction. Error bars represent SEM, **p < 0.005.
Discussion
Disrupted circadian rhythms are generally thought to contribute to tumorigenesis and poor prognosis (Savvidis and Koutsilieris, 2012; Hadadi et al., 2019). However, much of the work performed on cancer and rhythmicity, to date, was done in a context in which both cancer cells and their microenvironment have disrupted clocks. Here we found that expression of the core clock gene Per2 in the TME supports different stages of cancer progression. Employing different orthotopic tumor models, we showed that loss of stromal Per2, but not Per1, inhibits tumorigenesis and metastasis.
The circadian clock is a major regulator of metabolism and cell cycle and its disruption is well known to promote cancer. In cancer cells, both PER1 and PER2 were shown to act as tumor suppressors (Fu et al., 2002, 2016; Yang et al., 2009; Su et al., 2017; Zhu et al., 2019; Hou et al., 2020; Liu et al., 2020) and mutations in these genes were associated with human cancer (Chen et al., 2005; Kettner et al., 2014; Wu et al., 2019). However, whether these effects are clock-dependent is not clear. Moreover, despite the expansive knowledge of Per1 and Per2 roles in cancer cells, their roles in the TME were largely overlooked. Here we show that selective loss of the core clock genes Per1 and Per2 in the TME not only does not promote cancer but actually inhibits tumor growth and metastatic colonization. These findings suggest that the tumor-suppressive effects of PER1 and PER2 are most likely restricted to the cancer cells themselves, whereas in the TME PER1 and PER2 play different roles, that may be clock-independent.
The TME is comprised of different cell types. Which of these cell types contributes to the observed inhibitory effect of Per2 loss on tumor growth and metastasis? Recent studies suggested an immune-regulatory role for Per genes in the TME, as mice with disrupted clocks have more protumorigenic immune microenvironments. Yet we find that T cell depletion does not compensate for the Per1–/–Per2–/–-driven inhibition of tumor growth, nor do we detect immune cell transcriptional signatures in early stages of metastases formation, suggesting that the adaptive immune microenvironment does not mediate this effect. Various other cell types play protumorigenic roles in the TME. We observed increased numbers of cholangiocytes in Per2–/– mice, which may contribute to the anti-tumorigenic TME. Additionally, we detect massive transcriptional changes in hepatocytes of the pre-metastatic niche. Indeed, hepatocytes were recently reported to drive the formation of a pro-metastatic niche in the liver (Lee et al., 2019). CAFs were also proposed to play a role in clock regulation of cancer (Fuhr et al., 2019), and are generally known to have key roles in cancer progression. We observed massive infiltration of α-SMA-positive CAFs and collagen secretion in WT tumors which were not seen in Per2–/– or Per1–/–Per2–/– mice. Whether this CAF infiltration is driven by Per2 in fibroblasts, or perhaps by Per2 in other cells of the TME, remains to be determined in future studies.
The liver is a circadian organ, which allows for the circadian regulation of different metabolic functions, such as the synthesis and metabolism of glucose, lipid, cholesterol, and bile acid (Bass and Takahashi, 2010; Ferrell and Chiang, 2015). These pathways are massively deregulated in Per2–/– mice. Injection of cancer cells leads to further deregulation of these pathways, and induces other pathways (autophagy and inhibition of proliferation) which may contribute to the tumor-suppressive environment that prevents subsequent metastatic tumor growth. These processes may explain why, while PER2 serves as a tumor suppressor in cancer cells, it serves as a protumorigenic factor in the TME. In circadian biology, PER1, and PER2 function together to regulate clock functions. Our results show that Per2, but not Per1, plays an important role in the stromal regulation of tumor progression. These findings may imply a non-circadian role for Per2, differentiating it from Per1, and offering a new attractive therapeutic target in the TME.
Materials and Methods
Ethics Statement
All animal studies were conducted in accordance with the regulations formulated by the Institutional Animal Care and Use Committee (IACUC; protocols #02100220-2 and #02550418-3).
Mice
Per1–/–Per2–/– (Zheng et al., 2001) were back crossed to C57BL/6 (Adamovich et al., 2019) and single Per1–/–, Per2–/–, were generated (kindly provided by Gad Asher, WIS). These mice and WT C57Bl/6 controls (ENVIGO RMS, Israel) were maintained under specific-pathogen-free conditions at the Weizmann Institute’s animal facility.
Cell Culture
MC38 mouse colon cancer cells (kindly provided by Lea Eisenbach, WIS), LLC mouse lung carcinoma cells (kindly provided by Zvika Granot, HUJI), E0771 mouse breast cancer cells (kindly provided by Ronen Alon, WIS), and NIH3T3 mouse fibroblasts cell line (ECACC, #93061524) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Biological Industries, 01-052-1A) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 1% pen-strep and 1% L-glutamine (Biological Industries). All cell lines were maintained at 37°C in 5% CO2 and regularly tested for mycoplasma by PCR assay (Biological Industries). For orthotropic and portal vein injections, the cells were suspended at 80–90% confluence by treatment with 0.25% trypsin, 0.02% EDTA and washed with 1x phosphate buffered saline (PBS).
Generation of Per2::Luc Cells by Lentiviral Infection
pLenti6-B4B2 construct expressing Per2-dLuc (Liu et al., 2008) (kindly provided by Gad Asher, WIS) was inserted into MC38, LLC, NIH-3T3, and E0771 cells using lentiviral infection. After infection, the cells were selected by blasticidin-containing complete DMEM for 10 days.
Real-Time Luminescence Monitoring
Cells were seeded into black, clear bottom 24-well plates (Provair) and synchronized with 100 nM Dexamethasone treatment for 20 min. Bioluminescence was measured by a LumiCycle luminometer (Actimetrics) every 12 min for at least 3 days in the presence of luciferin (Promega) in the cell culture medium. For siRNA experiments, 24 h after seeding, NIH3T3 Per2::luc cells were transfected with siRNA for the specified genes (SMART Pool siRNA, Dharmacon). After 24 h cells were synchronized by Dexamethasone and bioluminescence was measured as described above.
Mathematical Analysis of Bioluminescence Data
To evaluate the rhythmicity of Per2::luc cells we created a CircadLib package for the MATLAB R2017b environment using a non-linear regression model. The model includes a constant level and one sinusoid to estimate the curve parameters – amplitude, phase and period. To remove effects of cell proliferation and signal decline the algorithm uses a de-trending function. To remove technical noise, the moving average method was used. This analysis yields two parameters: R2 – indicating how similar the curve is to a sinusoid, and π – indicating the period length.
Quantitative RT–PCR Analysis
Total RNA was extracted from cells using Bio-Tri reagent (Bio-Lab). mRNA was reverse-transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative RT–PCR analysis was performed using Fast SYBR Green Master mix (Applied Biosystems) with the primers listed in Table 1.
TABLE 1.
Primers used in this study.
| Forward | Reverse | |
| HPRT | CATAACCTGGTTCATCATCGC | TCCTCCTCAGACCGCTTTT |
| TBP | CCCTATCACTCCTGCCACACCAGC | GTGCAATGGTCTTTAGGTCA |
| AGTTTACAGCC | ||
| b2m | ACCGGCCTGTATGCTATCCAGAAA | GGTGAATTCAGTGTGA |
| GCCAGGAT | ||
| Per1 | ACCAGCCATTCCGCCTAAC | CGGGGAGCTTCATAACCAGA |
| Per2 | GAAAGCTGTCACCACCATAGAA | AACTCGCACTTCCTTTTCAGG |
Liver Metastatic Colonization via Portal Vein Injection
Sixteen weeks old WT C57Bl/6, Per1–/–, Per2–/–, and Per1–/–Per2–/– mice, n = 9–16 per genotype, were injected with an analgesic agent (Buprenorphine, 0.1 mg/kg SC), and anesthetized using isoflurane 2.5%. To form liver experimental metastases, 20,000 MC38 cells were injected into the hepatic portal vein. To prevent bleeding, light pressure was applied to the injection site with a cotton applicator for 4 min. Then, the muscle and skin were sutured using 6/0 polypropylene monofilament sutures. At day 21 post-injection, mice were sacrificed, and body weights were measured. Livers were harvested and weighed, and macrometastases were quantified by visual inspection. To normalize liver weights, liver/body weight ratios were calculated. Then, livers were formalin-fixed and paraffin-embedded for histological analysis.
Quantification of Liver Metastasis Analysis
Hematoxylin and Eosin (H&E) slides of liver tissues were scanned by a Pannoramic SCAN II scanner, 20×/0.8 objective (3DHISTECH, Budapest, Hungary). Quantification of liver metastases area was done by QuPath (version 0.2.0-m8) (Bankhead et al., 2017) with pixel classification using the simple threshold method, with prefilter Gaussian, smoothing sigma 4 and a threshold of 180.
Total RNA Isolation From Livers of WT and Per2–/– Mice
Per2–/– or WT C57Bl/6 male mice were injected at the age of 16 weeks, under anesthesia, with 20,000 MC38 cells (n = 4 mice per genotype) and PBS (n = 2 control mice per genotype) into the hepatic portal vein as described above. At day 7 post-injection, mice were sacrificed, and body weights were measured. Livers were weighed and dissociated by Bead Ruptor Elite (OMNI International) with metal beads in Micro-tube (Sarstedt #72.694.006). RNA isolation was performed by RNeasy Fibrous Tissue Mini Kit (Qiagen #74704) according to manufacturer’s instructions.
Library Preparation and RNA-Sequencing Analysis
Libraries were prepared using the SENSE mRNA-Seq Library Prep Kit V2 (Lexogen United States) according to the instructions of the manufacturer. Libraries were sequenced on an Illumina NovaSeq 6000 machine, at 10 M reads per liver sample, to provide sufficient reads to pass quality control (quality of reads and mapping quality percentage). Read counts were normalized and tested for differences using DEseq2 (Love et al., 2014). RNA sequencing results are detailed in Supplementary Table 1. Hierarchical clustering was performed using Euclidian distance on differentially expressed genes which were filtered with the following parameters: baseMean > 5, padj < 0.05, and |log fold change| > 1. Analysis was performed by Partek Genomics Suite software. Pathway analysis was performed using Metascape (Zhou et al., 2019). Significant pathways were determined if p < 0.05 (for details see Supplementary Table 2). Liver populations analysis was performed using CIBERSORT (Newman et al., 2015) based on published single-cell RNA-sequencing data (Heng and Painter, 2008; Dobie et al., 2019; Xiong et al., 2019). The gene signatures of liver subpopulations used for this analysis and the results table are detailed in Supplementary Tables 3, 4, respectively. The RNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession number GSE156450.
CD8+ CD4+ Depletion
WT C57Bl/6 and Per1–/–Per2–/– mice, n = 7–8 per genotype, were injected intraperitoneally with anti-mouse antibodies for CD8 (Clone 53–6.7) and CD4 (Clone GK1.5) or IgG2a control (Clone 2A3; 300 μg per injection per mouse for all antibodies; Bio X Cell), 2 days prior to the injection of MC38 cells to the portal vein as described above, and every 5 days to maintain full depletion until the endpoint. 50 μl blood were drawn to validate depletion of CD8+ and CD4+ cells by flow cytometry. At day 21, mice were sacrificed, body weights were measured, livers were fixed with 4% PFA, and macrometastases were quantified by visual inspection.
Orthotropic Injection to the Colon
Per2–/– or WT C57Bl/6 male mice, n = 10–14 per genotype, were injected at the age of 16 weeks, under anesthesia, with 200,000 MC38 cells suspended in 20 μl PBS into the colon wall (submucosal). The injection was performed under endoscopic guidance. Body weights were measured using an analytical balance, the appearance of tumors was monitored by colonoscopy at days 10 and 14. At day 14, mice were sacrificed, colons were harvested, and tumor volumes were measured using a digital caliper and calculated using the formula: V = (W2 × L)/2.
Orthotropic Injection to the Mammary Fat Pad
Per2–/– or WT C57Bl/6 female mice, n = 11–13 per genotype, were injected at the age of 8–11 weeks, under anesthesia, with 200,000 E0771 cells into the upper left mammary fat pad. Body weights were measured, and tumor volumes were measured using a digital caliper and calculated using the formula: V = (W2 × L)/2. Mice were sacrificed at day 32.
Immunohistochemistry of Mouse Tissues
Livers were fixed in 4% Paraformaldehyde (PFA), processed and embedded in paraffin blocks, cut into 4–5 μm sections and immunostained as follows: Formalin-fixed, paraffin-embedded (FFPE) sections were deparaffinized, treated with 0.3% H2O2 and antigen retrieval was performed by microwave with citrate acid buffer (pH 6.0). Slides were blocked with 10% normal horse serum, and anti-SMA antibodies were used (Sigma #F3777). Visualization was achieved with 3, 30-diaminobenzidine (DAB) as a chromogen (#SK4100, Vector Labs Kit, CA, United States)/M.O.M Kit (Vector Labs). Counterstaining was performed with Mayer’s Hematoxylin (MHS-16, Sigma-Aldrich, Rehovot, Israel). Images were taken with a Nikon Eclipse Ci microscope.
Statistical Analysis
Statistical analysis and visualization were performed using R (Version 3.6.0, R Foundation for Statistical Computing, Vienna, Austria) and Prism 8.2.0. (GraphPad, United States). Statistical tests were performed as described in each Figure Legend.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156450.
Ethics Statement
The animal study was reviewed and approved by the Weizmann Institute of Science Institutional Animal Care and Use Committee.
Author Contributions
LS, SM, and CL designed and performed the experiments and analyses and wrote the manuscript. HL performed the experiments. AN wrote code for mathematical analysis of bioluminescence data and assisted with writing the manuscript. RS-S designed and supervised the study and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank G. Asher for fruitful discussions, Per constructs, and Per KO mice, M. Golik for the generation of the Per1–/– and Per2–/– mice, and S. Ben-Moshe (Itzkovitz Lab, WIS) for her assistance with CIBERSORT analysis of the single cell data. We also thank members of the RS-S Lab and R. Aviram and G. Manella from the Asher Lab for valuable input on the manuscript.
Footnotes
Funding. This study was supported by a research grant from the Estate of Aliza Yemini. RS-S was supported by the Israel Science Foundation (Grants Nos. 401/17 and 1384/1), the European Research Council (ERC Grant Agreement 754320), the Israel Cancer Research Fund, the Laura Gurwin Flug Family Fund, the Peter and Patricia Gruber Awards, the Comisaroff Family Trust, the Estate of Annice Anzelewitz, and the Estate of Mordecai M. Roshwal. RS-S is the incumbent of the Ernst and Kaethe Ascher Career Development Chair in Life Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.587697/full#supplementary-material
References
- Adamovich Y., Ladeuix B., Sobel J., Manella G., Neufeld-Cohen A., Assadi M. H., et al. (2019). Oxygen and carbon dioxide rhythms are circadian clock controlled and differentially directed by behavioral signals. Cell Metab. 29 1092.e3–1103.e3. 10.1016/j.cmet.2019.01.007 [DOI] [PubMed] [Google Scholar]
- Bae K., Jin X., Maywood E. S., Hastings M. H., Reppert S. M., Weaver D. R. (2001). Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30 525–536. 10.1016/S0896-6273(01)00302-6 [DOI] [PubMed] [Google Scholar]
- Bankhead P., Loughrey M. B., Fernández J. A., Dombrowski Y., McArt D. G., Dunne P. D., et al. (2017). QuPath: open source software for digital pathology image analysis. Sci. Rep. 7:16878. 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J., Takahashi J. S. (2010). Circadian integration of metabolism and energetics. Science 330 1349–1354. 10.1126/science.1195027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovici N., Guérin M. V., Trautmann A., Donnadieu E. (2019). The remarkable plasticity of macrophages: a chance to fight cancer. Front. Immunol. 10:1563. 10.3389/fimmu.2019.01563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeman V., Williams J. L., Meng Q.-J., Streuli C. H. (2016). Circadian clocks and breast cancer. Breast Cancer Res. 18:89. 10.1186/s13058-016-0743-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borniger J. C., Walker W. H., II, Gaudier-Diaz M. M., Stegman C. J., Zhang N., Hollyfield J. L., et al. (2017). Time-of-day dictates transcriptional inflammatory responses to cytotoxic chemotherapy. Sci. Rep. 7:41220. 10.1038/srep41220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Kakan X., Wang S., Dong W., Jia A., Cai C., et al. (2013). Deletion of clock gene Per2 exacerbates cholestatic liver injury and fibrosis in mice. Exp. Toxicol. Pathol. 65 427–432. 10.1016/j.etp.2011.12.007 [DOI] [PubMed] [Google Scholar]
- Chen S.-T., Choo K.-B., Hou M.-F., Yeh K.-T., Kuo S.-J., Chang J.-G. (2005). Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis 26 1241–1246. 10.1093/carcin/bgi075 [DOI] [PubMed] [Google Scholar]
- Dobie R., Wilson-Kanamori J. R., Henderson B. E. P., Smith J. R., Matchett K. P., Portman J. R., et al. (2019). Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep. 29 1832.e8–1847.e8. 10.1016/j.celrep.2019.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. M., Chiang J. Y. L. (2015). Circadian rhythms in liver metabolism and disease. Acta Pharm. Sin. B 5 113–122. 10.1016/j.apsb.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Pelicano H., Liu J., Huang P., Lee C. C. (2002). The circadian gene period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111 41–50. 10.1016/S0092-8674(02)00961-3 [DOI] [PubMed] [Google Scholar]
- Fu X.-J., Li H.-X., Yang K., Chen D., Tang H. (2016). The important tumor suppressor role of PER1 in regulating the cyclin-CDK-CKI network in SCC15 human oral squamous cell carcinoma cells. Onco. Targets Ther. 9 2237–2245. 10.2147/OTT.S100952 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fuhr L., Abreu M., Carbone A., El-Athman R., Bianchi F., Laukkanen M. O., et al. (2019). The interplay between colon cancer cells and tumour-associated stromal cells impacts the biological clock and enhances malignant phenotypes. Cancers 11:988. 10.3390/cancers11070988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery S., Komatsu N., Baldjyan L., Yu A., Koo D., Koeffler H. P. (2006). The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell 22 375–382. 10.1016/j.molcel.2006.03.038 [DOI] [PubMed] [Google Scholar]
- Hadadi E., Taylor W., Li X., Aslan Y., Villote M., Rivière J., et al. (2019). Chronic circadian disruption modulates breast cancer cell stemness and their immune microenvironment to drive metastasis in mice. bioRxiv 10.1101/777433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Stevens R. G. (2012). Case-control study of shift-work and breast cancer risk in Danish nurses: impact of shift systems. Eur. J. Cancer 48 1722–1729. 10.1016/j.ejca.2011.07.005 [DOI] [PubMed] [Google Scholar]
- Heng T. S. P., Painter M. W. (2008). The immunological genome project: networks of gene expression in immune cells. Nat. Immunol. 9 1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Hou L., Li H., Wang H., Ma D., Liu J., Ma L., et al. (2020). The circadian clock gene PER2 enhances chemotherapeutic efficacy in nasopharyngeal carcinoma when combined with a targeted nanosystem. J. Mater. Chem. B 8 5336–5350. 10.1039/D0TB00595A [DOI] [PubMed] [Google Scholar]
- Jin K., Pandey N. B., Popel A. S. (2017). Crosstalk between stromal components and tumor cells of TNBC via secreted factors enhances tumor growth and metastasis. Oncotarget 8 60210–60222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettner N. M., Katchy C. A., Fu L. (2014). Circadian gene variants in cancer. Ann. Med. 46 208–220. 10.3109/07853890.2014.914808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling S., Beaulieu-Laroche L., Blum I. D., Landgraf D., Welsh D. K., Storch K.-F., et al. (2017). Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol. 15:13. 10.1186/s12915-017-0349-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Wood P. A., Hrushesky W. J. M. (2002). Circadian chemotherapy for gynecological and genitourinary cancers. Chronobiol. Int. 19 237–251. 10.1081/CBI-120002600 [DOI] [PubMed] [Google Scholar]
- Lauriola M., Enuka Y., Zeisel A., D’Uva G., Roth L., Sharon-Sevilla M., et al. (2014). Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nat. Commun. 5:5073. 10.1038/ncomms6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Stone M. L., Porrett P. M., Thomas S. K., Komar C. A., Li J. H., et al. (2019). Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature 567 249–252. 10.1038/s41586-019-1004-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Wei T., Meng H., Luo P., Zhang J. (2019). Role of the dynamic tumor microenvironment in controversies regarding immune checkpoint inhibitors for the treatment of non-small cell lung cancer (NSCLC) with EGFR mutations. Mol. Cancer 18:139. 10.1186/s12943-019-1062-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A. C., Tran H. G., Zhang E. E., Priest A. A., Welsh D. K., Kay S. A. (2008). Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 4:e1000023. 10.1371/journal.pgen.1000023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Gong X., Yang K. (2020). Overexpression of the clock gene Per2 suppresses oral squamous cell carcinoma progression by activating autophagy via the PI3K/AKT/mTOR pathway. J. Cancer 11 3655–3666. 10.7150/jca.42771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Weaver V. M., Werb Z. (2012). The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 196 395–406. 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. M., Liu C. L., Green M. R., Gentles A. J., Feng W., Xu Y., et al. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12 453–457. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. M., Steen C. B., Liu C. L., Gentles A. J., Chaudhuri A. A., Scherer F., et al. (2019). Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37 773–782. 10.1038/s41587-019-0114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K., Billaud M. (2017). Desynchronization of circadian clocks in cancer: a metabolic and epigenetic connection. Front. Endocrinol. 8:136. 10.3389/fendo.2017.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakopoulos T., Bauer M. R., Davidson S. M., Heimann M., Subbaraj L., Bhutkar A., et al. (2016). Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. 24 324–331. 10.1016/j.cmet.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D. F., Joyce J. A. (2013). Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19 1423–1437. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan C., Xu H., Khan S. K., Shen Y., Gitis P. J., Welsh D. K., et al. (2014). Cell type-specific functions of period genes revealed by novel adipocyte and hepatocyte circadian clock models. PLoS Genet. 10:e1004244. 10.1371/journal.pgen.1004244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H., Asher G. (2016). Circadian clock control of liver metabolic functions. Gastroenterology 150 574–580. 10.1053/j.gastro.2015.11.043 [DOI] [PubMed] [Google Scholar]
- Reinke H., Asher G. (2019). Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 20 227–241. 10.1038/s41580-018-0096-9 [DOI] [PubMed] [Google Scholar]
- Relógio A., Thomas P., Medina-Pérez P., Reischl S., Bervoets S., Gloc E., et al. (2014). Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet. 10:e1004338. 10.1371/journal.pgen.1004338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvidis C., Koutsilieris M. (2012). Circadian rhythm disruption in cancer biology. Mol. Med. 18 1249–1260. 10.2119/molmed.2012.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schernhammer E. S., Laden F., Speizer F. E., Willett W. C., Hunter D. J., Kawachi I., et al. (2003). Night-shift work and risk of colorectal cancer in the nurses’ health study. JNCI J. Natl. Cancer Inst. 95 825–828. 10.1093/jnci/95.11.825 [DOI] [PubMed] [Google Scholar]
- Shostak A. (2017). Circadian clock, cell division, and cancer: from molecules to organism. Int. J. Mol. Sci. 18:873. 10.3390/ijms18040873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Chen D., Yang K., Zhao Q., Zhao D., Lv X., et al. (2017). The circadian clock gene PER2 plays an important role in tumor suppression through regulating tumor-associated genes in human oral squamous cell carcinoma. Oncol. Rep. 38 472–480. 10.3892/or.2017.5653 [DOI] [PubMed] [Google Scholar]
- Tahara Y., Shibata S. (2016). Circadian rhythms of liver physiology and disease: experimental and clinical evidence. Nat. Rev. Gastroenterol. Hepatol. 13 217–226. 10.1038/nrgastro.2016.8 [DOI] [PubMed] [Google Scholar]
- Tamiya H., Ogawa S., Ouchi Y., Akishita M. (2016). Rigid cooperation of Per1 and Per2 proteins. Sci. Rep. 6:32769. 10.1038/srep32769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Liu S., Zhang S., Min L., Zhu S. (2020). Cellular and extracellular components in tumor microenvironment and their application in early diagnosis of cancers. Anal. Cell. Pathol. 2020:6283796. 10.1155/2020/6283796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh D. K., Yoo S.-H., Liu A. C., Takahashi J. S., Kay S. A. (2004). Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 14 2289–2295. 10.1016/j.cub.2004.11.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Tao B., Zhang T., Fan Y., Mao R. (2019). Pan-cancer analysis reveals disrupted circadian clock associates with T Cell exhaustion. Front. Immunol. 10:2451. 10.3389/fimmu.2019.02451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Kuang H., Ansari S., Liu T., Gong J., Wang S., et al. (2019). Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 75 644.e5–660.e5. 10.1016/j.molcel.2019.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Takahashi J. S. (2005). Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 393 288–301. 10.1016/S0076-6879(05)93012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wood P. A., Ansell C., Hrushesky W. J. M. (2009). Circadian time-dependent tumor suppressor function of period genes. Integr. Cancer Ther. 8 309–316. 10.1177/1534735409352083 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yuan G., Xie H., Wei T., Zhu D., Cui J., et al. (2019). Circadian clock associates with tumor microenvironment in thoracic cancers. Aging 11 11814–11828. 10.18632/aging.102450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., et al. (2004). PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101 5339–5346. 10.1073/pnas.0308709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Jiang Y.-C., Sun C.-K., Chen Q.-M. (2016). Role of the tumor microenvironment in tumor progression and the clinical applications (Review). Oncol. Rep. 35 2499–2515. 10.3892/or.2016.4660 [DOI] [PubMed] [Google Scholar]
- Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., Hogenesch J. B. (2014). A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 111 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Albrecht U., Kaasik K., Sage M., Lu W., Vaishnav S., et al. (2001). Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105 683–694. 10.1016/S0092-8674(01)00380-4 [DOI] [PubMed] [Google Scholar]
- Zheng B., Larkin D. W., Albrecht U., Sun Z. S., Sage M., Eichele G., et al. (1999). The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400 169–173. 10.1038/22118 [DOI] [PubMed] [Google Scholar]
- Zhou J., Li X., Zhang M., Gong J., Li Q., Shan B., et al. (2020). The aberrant expression of rhythm genes affects the genome instability and regulates the cancer immunity in pan-cancer. Cancer Med. 9 1818–1829. 10.1002/cam4.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A. H., Tanaseichuk O., et al. (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10:1523. 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Wang Q., Hu Y., Wang F. (2019). The circadian gene Per1 plays an important role in radiation-induced apoptosis and DNA damage in glioma. Asian Pac. J. Cancer Prev. 20 2195–2201. 10.31557/apjcp.2019.20.7.2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E., Halpern Z., Elinav E., Brazowski E., Jung S., Varol C. (2011). Utilization of murine colonoscopy for orthotopic implantation of colorectal cancer. PLoS One 6:e28858. 10.1371/journal.pone.0028858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156450.