Abstract
Recently, genetically targeted cancer therapies have been a topic of great interest. Synthetic lethality provides a new approach for the treatment of mutated genes that were previously considered unable to be targeted in traditional genotype-targeted treatments. The increasing researches and applications in the clinical setting made synthetic lethality a promising anticancer treatment option. However, the current understandings on different conditions of synthetic lethality have not been systematically assessed and the application of synthetic lethality in clinical practice still faces many challenges. Here, we propose a novel and systematic classification of synthetic lethality divided into gene level, pathway level, organelle level, and conditional synthetic lethality, according to the degree of specificity into its biological mechanism. Multiple preclinical findings of synthetic lethality in recent years will be reviewed and classified under these different categories. Moreover, synthetic lethality targeted drugs in clinical practice will be briefly discussed. Finally, we will explore the essential implications of this classification as well as its prospects in eliminating existing challenges and the future directions of synthetic lethality.
Subject terms: Cancer therapy, Cancer genetics, Oncogenes, Cancer metabolism
Introduction
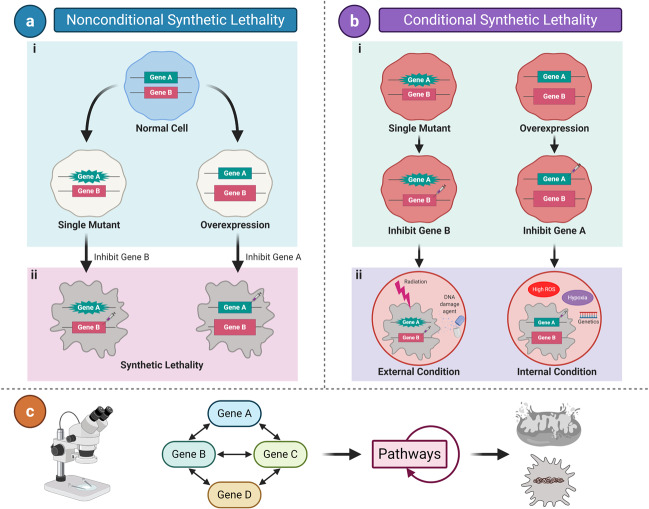
Synthetic lethality (SL) initially originates from studies on fruit flies1,2 and yeast3–5 models. The original concept of SL is based on the simultaneous occurrence of abnormalities in the expression of two or more separate genes, including mutation, overexpression, or gene inhibition, which leads to cell death; whereas abnormality in only one of the genes does not affect cell viability (Fig. 1a).6–8 Tumor cells are the result of mutated or overexpressed genes in otherwise normal cells.9 Hence, inhibitors that target synthetic lethal partners of mutated or overexpressed genes in tumor cells can kill cancers without affecting the survival of normal cells.
Fig. 1.
Synthetic lethality classification. Synthetic lethality is divided into two major categories, nonconditional synthetic lethality and conditional synthetic lethality. a Nonconditional synthetic lethality. (i) Single mutation/overexpression of either gene A or B alone is viable in tumor cells. (ii) Inhibition of gene B or A in cells with a mutation/overexpression of gene A or B results in synthetic lethality. b Conditional synthetic lethality. (ii) Several synthetic lethal interactions may be dependent on certain intrinsic conditions, such as genetic background, hypoxia, high ROS, etc., or extrinsic conditions, such as DNA-damaging agents and radiation. (i) Without these conditions, tumor cells with mutation/overexpression of both gene A and B could still survive. [c] Nonconditional synthetic lethality was further classified into gene level, pathway level, and organelle level according to the degree of studies into its mechanism in the review. Star shape of genes represents mutations; large rectangle represents genetic overexpression; syringe represents inhibitors; viable cells are depicted as ovals; and non-viable cells are depicted in random shapes
With the advancement of tumor research, cancer is now widely recognized as a disease of the genome. Various underlying tumor features, such as genome instability, give rise to the genetic diversity that accelerates their acquisition and inflammation.10 Therefore, targeting oncogenic driving genes, tumor-suppressor genes, and the underlying mechanisms is an applicable direction for cancer therapy.11 The development of genome sequencing and the analysis of thousands of human tumors led to the discovery of the first generation of genetically targeted cancer therapies.12–14 As a result, multiple personalized or precise genotype-targeted cancer treatments have been adopted and shown promising results in cancer patients that failed to respond to standard therapies.7,15,16 For instance, several studies have demonstrated that imatinib, a KIT inhibitor that is effective in treating patients with KIT-mutant gastrointestinal stromal tumors, had approximately 50% response rates and an extended median progression-free survival of 1.5 years.17–20 Imatinib also targets the BCR-ABL fusion tyrosine kinase for patients with chronic myelogenous leukemia.21–24 There are multiple studies that exhibit successful clinical outcomes,11 such as trastuzumab that target encoding HER2 in breast cancer,25 erlotinib, or osimertinib for EGFR mutations in non-small-cell lung cancer (NSCLC), as well as crizotinib for ALK-positive lung cancer, and others.26–30
Although numerous small-molecule and antibody-based drugs for oncogenes or tumor-suppressor genes have proven to be effective for several tumors with certain gene mutations,31 not all oncogenes or tumor-suppressor genes could be targeted and resistance is common,7 In such cases, identifying and exploiting a second or several other functional genes that interact with the primary oncogene or tumor-suppressor gene provides an alternative method for cancer treatment. Therefore, SL is increasingly being explored recently, in an effort to identify new anticancer therapeutic targets through large-scale SL screening in model organisms and human cell lines such as NSCLC (NCI-H1355, NCI-H1299, NCI-H1155), hepatocellular carcinoma (HCC1954, HCC1937, HCC1806), and breast cancer (MDA-MB-468, MDA-MB-436, MDA-MB-415) via clustered regularly interspaced short palindromic repeats (CRISPR),32 tumor genomic sequence database, RNA interference (RNAi) technology,33,34 etc. The most remarkable finding in SL is the hypersensitivity of BRCA1/2-mutant tumor cells to poly-(ADP-ribose) polymerase (PARP) inhibitors.35–37 Several PARP inhibitors (PARPi) were approved by the FDA for the treatment of breast cancer and ovarian cancer in clinical practice.6,38 Furthermore, there have been various findings regarding classical oncogenic driving genes or tumor-suppressor genes, such as TP53, KRAS, MYC, etc.,39 which will be discussed in detail later.
As our understanding of the complexity of cancer-cell signaling networks continues to grow, increasing numbers of targets are being identified as potential synthetically lethal candidates. Many researchers have defined several classes of SL into “synthetic dosage lethality (SDL)”,7,40,41 “collateral SL”,42,43 and others due to its complexity, which is different from the original concept of SL. However, these expanding concepts are scattered and there is no comprehensive classification for various SL that have been discovered. Moreover, the application of SL in clinical practice still faces many challenges. This review will describe a novel and integrated classification of the SL in different situations for a deeper systematic understanding. Multiple SL studies in recent years will be reviewed in different synthetic lethal categories. Furthermore, SL targeted drugs in clinical practice will be briefly discussed. Lastly, the development and limitations of SL as well as the inspiration of this classification for advancements in cancer research will be summarized.
Synthetic lethality classification
This novel classification categorizes SL into various groups that comprehensively summarize different conditions involved in SL reported in recent years that are beyond the original concept. SL is generally divided into two major categories, nonconditional/original SL and conditional SL (Fig. 1a, b). Nonconditional SL is further classified into gene level, functional pathway level, and organelle level, according to the degree of specificity into its biological mechanism (Fig. 1c), while numerous achievements of SL in recent years are reviewed under different synthetic lethal categories we propose accordingly. The existing concepts of SL, namely synthetic dosage lethality, collateral SL, and metabolic SL, are discussed and grouped into this novel classification correspondingly.
Synthetic lethality in genetics
Gene level SL corresponds with the original concept of SL mentioned above (Fig. 1a), in which the interaction between genes forms the basis for SL. The identification of genes with synthetic lethal effects allows researchers to further study the mechanisms at deeper levels. Thus, this category encompasses most of the studies on SL and is the cornerstone of SL at the other levels we mention below. Herein, we focus on describing the examples of oncogenes and tumor-suppressor genes related to SL at the gene level. The significant preclinical findings of synthetic lethal interactions among genes are listed in detail in Table 1.
Table 1.
Representative synthetic lethal interactions among genes in preclinical studies
| Gene | Chromosome | Cellular process and mechanism | SL partners | Cancer type | Reference |
|---|---|---|---|---|---|
| PARP1 (mutant) | 1q41.42 | Regulate cell proliferation and differentiation; repair DNA single- and double-strand breaks. | BRCA1/2 | Breast, ovarian, pancreatic and liver cancer; leukemia | 6,50,132,138 |
| RAD51 | Ovarian cancer; HCC | 139,140 | |||
| ATG5 | Ovarian cancer | 141 | |||
| CDK5 | Cervical and breast cancer | 142,143 | |||
| TP53 (mutant) | 17p13.1 | Major tumor suppressor; regulate the cell cycle, senescence, and apoptosis. | ATM | Glioma | 54 |
| ATR | CLL; osteosarcoma, colon and breast cancer | 55,56 | |||
| WEE1 | HNSCC | 57 | |||
| CHK1 | NSCLC, B-ALL | 58,144 | |||
| BCL-2 | AML | 59 | |||
| SLC711 | NSCLC; renal, esophagus, cervical and gastric cancer | 145 | |||
| mTOR | Pancreatic adenocarcinoma; lung and breast cancer | 102 | |||
| AURKA | Liver cancer | 146 | |||
| PIP4KB | Breast Cancer | 147 | |||
| KRAS (mutant) | 12p12.1 | Transcriptional activator that regulates endothelial cells endothelin-1 gene expression. | CDC6 | Colon cancer | 63 |
| GATA2 | Colon cancer; NSCLC | 63,64 | |||
| SLC25A22 | Colorectal cancer | 65 | |||
| PLK1 and ROCK | Lung and pancreatic cancer | 66 | |||
| CD274 | Colon and lung cancer | 67 | |||
| MYC (mutant) | 8q24.21 | Regulate cell cycle progression, transcription, and apoptosis. | 4EBP1 | Hematological cancer | 68 |
| SAE1/2 | Breast cancer | 69 | |||
| AURKB | T-ALL | 70 | |||
| PIM1 | Breast cancer | 71 | |||
| CDK9 | HCC | 72 | |||
| ARID1A (mutant) | 1p36.11 | Target SWI/SNF complexes, which regulate chromatin remodeling. SWI/SNF complexes are involved in controlling the cell cycle, DNA replication, and repairing DNA damage. | ARID1B | Ovarian cancer | 79 |
| EZH2 | Ovarian cancer | 148 | |||
| PARP1 | Breast and colon cancer | 149 | |||
| MAD2 (overexpress) | 4q27 | A component of the mitotic spindle assembly checkpoint that prevents the onset of anaphase until all chromosomes are properly aligned at the metaphase plate. | PP2A | Lung and liver cancer; malignant lymphoma | 73 |
| CKS1B (overexpress) | 1q21 | Codes for a conserved regulatory subunit of cyclin-CDK complexes that function at multiple stages of cell cycle progression | PLK1 | Breast cancer | 41 |
| TDP1 (overexpress) | 14q32.11 | Encode the protein that repairs stalled topoisomerase I-DNA complexes and repair of free-radical mediated DNA double-strand breaks. | HDAC1/2 | Fibrosarcoma; rhabdomyosarcoma | 74 |
| RPD3 |
SL between homologous recombination-related gene BRCA1/2 and PARP is a classical and fundamental example. Several research groups first reported in 2005 that dysfunctional BRCA1 or BRCA2 cells are significantly more sensitive to PARP inhibitors than cells that have normal BRCA function.35,36 PARP1 is a DNA repair protein that regulates cell proliferation and differentiation by repairing DNA single-strand break (SSB) and double-strand breaks (DSB). The inhibition of PARP1 leads to deleterious mutation accumulation, resulting in the apoptosis of BRCA1/BRCA2-deficient cells.44 Subsequently, the specific mechanisms of SL between these two genes were further investigated and will be discussed at the pathway level later. After PARP inhibitors demonstrated its feasibility, acceptable safety, and considerable efficacy,45 the development of PARP inhibitors increased rapidly and several drugs were evaluated for their use on a wide range of solid tumors and hematologic cancers in clinical trials (Table 2).46–51
Table 2.
Recent clinical trials potentially related to synthetic lethal interactions
| Gene | Targeted SL partners | Agent | Intervention | Cancer type | Phase and ClinicalTrials.gov Identifier |
|---|---|---|---|---|---|
| BRCA1/2 | PARP | Olaparib | Olaparib | Breast and ovarian cancer | IV, NCT04330040 |
| Olaparib + Paclitaxel + Durvalumab | Advanced gastric cancer | II, NCT03579784 | |||
| Olaparib + Abiraterone | Prostate cancer | III, NCT03732820 | |||
| Olaparib + Durvalumab | Bladder cancer | II, NCT03534492 | |||
| Olaparib + Temozolomide | Colorectal cancer | II, NCT04166435 | |||
| Niraparib | Niraparib | Pancreatic cancer | II, NCT03601923 | ||
| Niraparib + Osimertinib | Lung cancer | I, NCT03891615 | |||
| Niraparib + Dostarlimab | Ovarian cancer | III, NCT03602859 | |||
| Niraparib + MGD013 | Gastric and Gastroesophageal junction cancer | I, NCT04178460 | |||
| Niraparib + Dostarlimab | Cervix cancer | II, NCT04068753 | |||
| Rucaparib | Rucaparib | Endometrial cancer | II, NCT03617679 | ||
| Rucaparib + Nivolumab | Biliary cract cancer | II, NCT03639935 | |||
| Rucaparib + Radiotherapy | Breast cancer | I, NCT03542175 | |||
| Rucaparib + Copanlisib | Prostate cancer | I, NCT04253262 | |||
| Rucacparib + Enzalutamide + Abiraterone | Prostate cancer | I, NCT04179396 | |||
| Talazoparib | Talazoparib | Leukemia | I, NCT03974217 | ||
| Talazoparib + Avelumab | Breast cancer | I, NCT03964532 | |||
| Talazoparib + Radiotherapy | Gynecologic cancer | I, NCT03968406 | |||
| Talazoparib + ASTX727 | Breast cancer | I, NCT04134884 | |||
| Talazoparib + Avelumab | Lung cancer | II, NCT04173507 | |||
| Talazoparib + Axitinib | Kidney cancer | I/II, NCT04337970 | |||
| Talazoparib + Atezolizumab | Lung cancer | II, NCT04334941 | |||
| Talazoparib + Gedatolisib | Breast cancer | II, NCT03911973 | |||
| TP53 | ATR | Berzosertib (M6620) | Berzosertib + Radiotherapy | Lung and breast cancer | I, NCT02589522/ I, NCT04052555 |
| Berzosertib + Topotecan (Hydrochloride) | Lung cancer | I/II, NCT02487095/ II, NCT03896503 | |||
| Berzosertib + Carboplatin + Docetaxel | Prostate cancer | II, NCT03517969 | |||
| AZD6738 | AZD6738 + Radiotherapy | Advanced solid tumors | I, NCT02223923 | ||
| AZD6738+ Olaparib | Gynecologic cancer | II, NCT04065269 | |||
| AZD6738 + Olaparib + Durvalumab | Breast cancer | II, NCT03740893 | |||
| AZD6738 + Acalabrutinib | CLL | I/II, NCT03328273 | |||
| AZD6738+ Durvalumab | Biliary tract cancer | II, NCT04298008 | |||
| BAY1895344 | BAY1895344 | Advanced solid tumors | I, NCT03188965 | ||
| BAY1895344 + Pembrolizumab | Advanced solid tumors | I, NCT04095273 | |||
| BAY1895344 + Niraparib | Ovarian cancer | I, NCT04267939 | |||
| M4344 | M4344 + Niraparib | Ovarian cancer | I, NCT04149145 | ||
| M4344 + Carboplatin | Advanced solid tumors | I NCT02278250 | |||
| WEE1 | Adavosertib (AZD1775) | Adavosertib | Advanced solid tumors | I, NCT01748825; II, NCT03253679 / NCT03284385 | |
| Adavosertib + Gemcitabine + Cisplatin + Carboplatin | Advanced solid tumors | I, NCT00648648 | |||
| Adavosertib + Olaparib | Ovarian, primary peritoneal, and fallopian tube cancer | II, NCT03579316 | |||
| Advanced solid tumors | II, NCT02576444 | ||||
| Adavosertib + Olaparib + AZD6738 | Breast cancer | II, NCT03330847 | |||
| Adavosertib + Irinotecan | Advanced solid tumors | I/II, NCT02095132 | |||
| Adavosertib + Cisplatin + Radiotherapy | Cervical, vaginal, and uterine cancer | I, NCT03345784 | |||
| Adavosertib + Temozolomide + Radiotherapy | Glioblastoma | I, NCT01849146 | |||
| CHK1 | SRA737 | SRA737 | Advanced solid tumors | I/II, NCT02797964 | |
| SRA737 + Gemcitabine + Cisplatin | Advanced solid tumors | I/II, NCT02797977 | |||
| Prexasertib (LY2606368) | Prexasertib | Advanced solid tumors | I, NCT01115790 | ||
| Lung cancer | II, NCT02735980 | ||||
| Breast, ovarian, and prostate cancer | II, NCT02203513 | ||||
| mTOR | Temsirolimus | Temsirolimus | Endometrial carcinoma | II, NCT02093598 | |
| Metformin | Metformin + Carboplatin + Paclitaxel | Epithelial ovarian cancer | II, NCT02312661 | ||
| KRAS | PLK1 | CYC140 | CYC140 | Myelodysplastic syndromes, AML, ALL, CML, CLL | I, NCT03884829 |
| BI 2536 | BI 2536 | Pancreatic neoplasms | II, NCT00710710 | ||
| BI 6727 | BI 6727 | Neoplasms | I, NCT01145885 | ||
| NMS-1286937 | NMS-1286937 | Advanced or metastatic solid tumors | I, NCT01014429 | ||
| GSK461364 | GSK461364 | Non-Hodgkins lymphoma | I, NCT00536835 | ||
| Onvansertib (PCM-075) | Onvansertib + Cytarabine+ Decitabine | AML | I/II, NCT03303339 | ||
| CD274/PD-L1 | Sotorasib (AMG 510) | Sotorasib + MEK inhibitor; Sotorasib + PD1 inhibitor; Sotorasib + SHP2 allosteric inhibitor; Sotorasib + Pan-ErbB tyrosine kinase inhibitor; Sotorasib + PD-L1 inhibitor; Sotorasib + EGFR inhibitor + Chemotherapy | Advanced solid tumors | I, NCT04185883 | |
| Pembrolizumab | Pembrolizumab + Docetaxel + Ramucirumab | NSCLC | II, NCT04340882 | ||
| Pembrolizumab + Trametinib | I/II, NCT03225664; I, NCT03299088 | ||||
| Durvalumab | Durvalumab + Carboplatin + Pemetrexed | Lung cancer | II, NCT04470674 | ||
| Avelumab | Avelumab + Binimetinib + Talazoparib | Pancreatic cancer | II, NCT03637491 | ||
| MYC | 4EBP1 | AZD2014 | AZD2014 | Prostate cancer | I, NCT02064608 |
| CC-115 | CC-115 | Glioblastoma multiforme, squamous cell carcinoma of head and neck, prostate cancer, Ewing’s osteosarcoma, and CLL | I, NCT01353625 | ||
| Everolimus | Everolimus + Nelarabine + Cyclophosphamide + Etoposide | Lymphoblastic leukemia and lymphoblastic lymphoma | I, NCT03328104 | ||
| AURKB | GSK1070916A | GSK1070916A | Adult solid tumor | I, NCT01118611 | |
| CDK9 | AZD4573 | AZD4573 | Relapsed or refractory hematological malignancies and Richter’s syndrome | I, NCT03263637 | |
| TP-1287 | TP-1287 | Advanced solid tumors | I, NCT03604783 | ||
| P276-00 | P276-00 | Melanoma | II, NCT00835419 |
The tumor-suppressor gene TP53, a predominant target in SL research, is the most frequently mutated gene in cancers.52 Identifying the synthetic lethal partners of p53 is a feasible method in clinical practice. A previous study conducted by Wang and Simon used gene-expression profiling to select multiple candidates for synthetically lethal gene targets of p53.53 A series of kinase-encoding genes were found to be potential targets of p53-deficient tumors for new drug therapy, including polo-like kinase 1 (PLK1), cyclin-dependent kinase 16 (CDK16), receptor-like tyrosine kinase (RYK), aurora kinase A (AURKA), etc. Recently, increasing studies reported new synthetic lethal partners of p53 such as ATM, ATR, WEE1, CHK1, etc.,54–58 in various types of cancers (listed in Table 1). Furthermore, Pan et al. revealed a different synthetic lethal therapy between p53 and B-cell lymphoma 2 (BCL-2) by activating p53 through MDM2 silencing and inhibiting BCL-2, which accelerates the apoptosis process in acute myeloid leukemia (AML) cells.59 This indicates that a combination of targeting mutated genes with their synthetic lethal partners may improve the synthetic lethal effects in more cancers than only inhibiting partner genes as in previous studies.
SL can also be applied to target oncogenesis drivers such as KRAS and MYC. Recent studies have identified synthetic lethal partners for oncogene KRAS, which was not considered “druggable” by traditional chemotherapy,60 by using large-scale RNAi screening. An earlier study suggested that serine-threonine kinase 33 (STK33) was indispensable for the viability of KRAS-driven tumors; however, this result is considered controversial according to a later study. Previous studies have proved this synthetic lethal effect in colon cancer (DLD-1, HCT-116, SW-480), pancreatic cancer (PANC-1), lung cancer (A549), and other cell lines.61,62 However, some of these cell lines were not verified in the later study. Therefore, the discrepancy of the cell lines and cancer types used in the two studies may be the cause for the opposite result, which suggests that more researches are needed to confirm the application of synthetic lethal effects in various cell lines of the same cancer and in different cancer types. Through RNAi assay, Steckel et al. conducted a series of studies to illuminate the synthetic lethal interactions that DNA replication regulator CDC6 and transcription factor GATA2 have with KRAS.63,64 Downstream regulatory pathways of GATA2 were further studied in NSCLC, which are described in the next section—synthetic lethal pathways. Moreover, SLC25A22 has been identified as a synthetic lethal gene in colorectal cancer cells with KRAS mutations.65 The inhibition of PLK1 and RhoA/Rho kinase (ROCK) has a synergistic effect in KRAS-mutant cancers,66 which is a more complex condition of SL in genetics involving these three genes. Recently, CD274 (encoding PD-L1) blockade has been proved to be a promising KRAS-mutant adenocarcinoma treatment option.67 MYC-targeted therapies, similar to KRAS-targeted therapies, have proven to be a challenge to explore. Recent studies suggest candidate genes that are synthetically lethal in MYC-driven cancers. These include eukaryotic translation initiation factor 4E (eIF4E) binding protein 1 (4EBP1), SUMO-activating enzyme subunit 1/2 (SAE1/2), Aurora-B kinase (AURKB), PIM1, and Cyclin-dependent kinase 9 (CDK9).68–72
Furthermore, mitotic arrest deficiency 2 (MAD2) shares synthetic lethal interaction with PP2A, in which PP2A inhibition in MAD2 overexpressing tumor cells results in SL in several tumors, including lung cancer, liver cancer, and malignant lymphoma.73 Likewise, PLK1 inhibition in CKS1B overexpressed tumor cells leads to breast cancer-cell death, and the inhibition of histone deacetylases (HDACs) or histone deacetylase RPD3 in TDP1 overexpressed cells may kill fibrosarcoma and rhabdomyosarcoma cells.41,74 Several studies referred to these interactions as SDL, an expanding concept of SL, in which the overexpression of one gene combined with the loss of function in another gene that results in cell death, and therefore could be used to target cancer cells with overexpressed, undruggable oncogenes.75 Intriguingly, SDL is without doubt subordinate to the basic concept of SL mentioned above (Fig. 1a) and can be classified into the category of SL in genetics.
Molecular and cellular level in synthetic lethal pathway
Various pathways are crucial for survival in both normal and cancer cells. Proteins are synthesized as a product of multiple gene expression and form the basis for these functional pathways. Ku et al.76 found that the established synthetic lethal effects at the pathway level are more reproducible than those previously reported at the gene level through analysis. Consequently, after identifying genes that have synthetic lethal interactions in tumors, the mechanisms involved in these vital pathways, which these genes participate in, are then studied further by researchers. Pathway mechanisms are primarily studied at the protein level. This section will discuss the classification of the findings of SL at the pathway level.
Single synthetic lethal pathway
Functionally related genes are translated into proteins in sequence to form a pathway that performs essential functions within a cell. In many cases, several components of these pathways are complexes formed by the collaborative expression of multiple genes. Abnormality in two or more genes that constitute the same protein complex on a pathway may lead to cell death. Fang77 also reported that the synthetic lethal effect in essential multiprotein complex subunits that is a component of a single linear essential pathway is a condition of SL. Therefore, this synthetic lethal effect mainly focuses on a pathway (Fig. 2a).
Fig. 2.
Synthetic lethal pathway: single pathway. a Single pathway concept. (i) A pathway performs an essential survival function to maintain cell survival; Protein complex A2 formed by the joint expression of multiple genes (S1, S2, S3, etc.) is an essential factor of this pathway. (ii) Abnormality (mutation, overexpression, or inhibited) of two or more genes in the complex leads the cell death, while only one mutated gene of the complex is viable. b Examples of the SWI/SNF complex. (i) Mutation of the ARID1A subunit of the SWI/SNF complex may turn normal cells into cancers like ovarian cancer and tumor cells still survive. (ii) Inhibition of ARID1B, another subunit of the SWI/SNF complex, will cause the complex collapse and synthetic lethality. Star shape of genes represents a mutation; syringe represents inhibitors; viable cells are depicted as ovals; and inviable cells are depicted as random shapes
The switch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex is assembled by its subunit proteins that some genes, such as SMARCA2/4, SMARCB1, ARID1A/B, and ACTL6A, encode. SWI/SNF complex mainly participates in DNA replication and repair.6,78,79 The gene that encodes AT-rich interactive domain 1 A (ARID1A), a member of the SWI/SNF complex, is frequently mutated across a variety of human cancers. Helming et al. identified that ARID1B knockdown in ARID1A-mutant ovarian cells leads to dissociation of the core catalytic ATPase subunit SMARCA4 (or BRG1) and reduced combination of other subunits in the SWI/SNF complex (Fig. 2b). It was observed that the proliferation of tumors was inhibited.79 Based on this finding, SL between ARID1A and ARID1B could possibly be expanded and applied to more ARID1-mutant tumor cells such as hepatocellular carcinoma (HCC) and colon cancer.80 The other two subunits SMARCA2 and SMARCA4 that make up the SWI/SNF complex share a similar relationship, as SMARCA2 is essential for the survival of tumor cells that possess function mutations in SMARCA4.81,82
Another example of synthetic lethal effect via a pathway was recently reported in pancreatic ductal adenocarcinoma (PDAC). Somatic mutations in SMAD4 are often associated with PDAC.83 Dey et al. studied metabolic gene malic enzyme 2 (ME2) at the SMAD4 site and its paralogous isoform ME3 in PDAC. ME2 and ME3 are both oxidative decarboxylases that are expressed in the same metabolic pathways to catalyze the conversion of malic acid to pyruvate, in which the loss of ME3 causes ME2-mutated PDAC cell death.43
Interestingly, the examples mentioned above are paralogous genes and take part in collateral SL. In collateral SL, mutation or deletion of the gene that encodes a subunit in a specific complex often causes the collateral or passenger gene to become vulnerable. The subunit translated by this collateral gene is a component of the same complex. Further inhibition of the collateral gene causes this complex to collapse and results in the collateral SL of cells.6,84 Collateral SL is consistent with single pathway SL (Fig. 2), thus it can be grouped into this category of SL.
Dual synthetic lethal pathway
This type of SL involves two or more genes and two pathways. Specifically, two pathways perform the same survival function to maintain the cell alive, and abnormality of two or more genes that are key regulatory points in two pathways will cause synthetic lethal interactions in tumors, while the abnormality of genes in only one pathway maintains survival (Fig. 3a).
Fig. 3.
Synthetic lethal pathway: dual pathways. a Dual pathway concept. (i) Pathways 1 and 2 perform the same function to maintain cell survival. (ii) Abnormality (mutation, overexpression, or inhibited) of two or more genes in only one pathway keeps the cell viability. On the contrary, two or more genes on two pathways in abnormal conditions would cause synthetic lethal interactions. b Examples of HR and NHEJ pathways. (i) When DSBs occur in normal cells, BRCA1 is normally expressed and is recruited to sites of breaks, which interacts with 53BP1 to inhibit 53BP1 on the CTIP/MRN complex that promotes end processing to allow HR-mediated repair in S and G2 phases. Whereas in the G0/G1 phase, BRCA1 is silent and 53BP1 is recruited to DSBs to restrain CTIP/MRN activity, which inhibits HR and promotes the classic-NHEJ pathway. (ii) In BRCA-mutated tumors, BRCA1 is not present in S/G2-phase and 53BP1 inhibits CTIP/MRN function, leading to impaired end processing of the breaks, suppression of HR, and promotion of the alternative-NHEJ pathway. In this condition, tumors could still rely on the alternative-NHEJ pathway to repair DSBs and survive. (iii) Use of PARP (a functional gene in the NHEJ pathway) inhibitors will cause synthetic lethality in BRCA-mutated cancers. Star shape of genes represents a mutation; syringe represents inhibitors; viable cells are depicted as ovals; and inviable cells are depicted as random shapes
The synthetic lethal interactions between two major pathways of DNA DSBs repair, homologous recombination (HR) pathway and non-homologous end-joining (NHEJ) pathway,85,86 belong to this category. When DBSs occur in normal cells, BRCA1 is activated in response to DNA damage and recruited at breakpoints to inhibit 53BP1 on the end processing promoting complex C-terminal binding protein interacting protein (CTIP)/ Mre11-Rad50-Nbs1 (MRN), thus allowing HR-mediated repair during S and G2 phases of the cell cycle.87,88 In contrast, BRCA1 is silent in G0 and G1 phases, and 53BP1 is recruited to DSBs to inhibit CTIP/MRN activity, thus inhibiting HR and promoting classic (c)-NHEJ pathway (Fig. 3bi).89–91 In BRCA-mutated tumors, BRCA1 is not present in either S or G2-phases and 53BP1 remains free to inhibit CTIP/MRN function, leading to impaired end processing of breaks, suppression of HR, and promotion of alternative (Alt)-NHEJ pathway.90,91 In this scenario, tumors can rely on the (Alt)-NHEJ pathway to repair DSBs and survive (Fig. 3bii). However, PARP (a functional gene in the NHEJ pathway)85 inhibitors causes SL in BRCA-mutated cancers (Fig. 3biii).36 Further research of the HR pathway revealed that the microhomology-mediated end-joining (MMEJ) pathway also have synthetic lethal interactions with it. Ceccaldi et al. revealed that knockdown of DNA polymerase θ (Polθ also known as POLQ) belonging to MMEJ pathway in HR-deficient epithelial ovarian cancers enhances cell death.92
In addition, p53 and MAPKAP kinase-2 (MK2) also has a synthetic lethal effect between two pathways.39 CDK2 function is inhibited by p21, a downstream target of p53, which interacts with cyclin A and cyclin E to facilitate normal cell cycle entry through participation in the formation of the circ-Foxo3-p21-CDK2 ternary complex. This pathway is regulated by p53 that activates cell cycle checkpoints by inducing cell cycle arrest, thus providing time for DNA damage repair.38,93,94 In contrast, p53-deficient tumors are specifically dependent on the p38/MK2 pathway to prolong G2/M and G1/S checkpoints in response to DNA damage.95 Therefore, MK2 inhibition to block the p38/MK2 pathway could produce a synthetic lethal effect after DNA damage in p53-mutated NSCLC and glioblastoma cells.96,97 To sum up, the synthetic lethal interactions between two pathways that could regulate the same essential survival function of cells belong in this classification.
Multiple synthetic lethal pathway
Aside from SL involving just one or even two pathways as mentioned above, more intricate synthetic lethal interactions involving multiple pathways were identified through further study. Researchers have found that several tumors depend more on some pathways to survive than normal cells and considerable “cross-talk” exists among these pathways at the same time.39 The network of these pathways maintains vital functions in tumors. The co-suppression of these pathways leads to SL while blocking a single pathway or several but not all pathways do not.64 This type of SL involving multi-pathways is described in Fig. 4a.
Fig. 4.
Synthetic lethal pathway: multiple pathways. a Concept of connected multiple pathways. Some pathways form a network and perform their functions to maintain cell survival. Whereas the presence of abnormal (mutation, overexpression, or inhibited) genes in every pathway leads to cell death. However, cells could still survive with abnormal genes in several but not all pathways. b Example: Survival of KRAS oncogene-driven NSCLC depends on the GATA2 transcriptional network. In KRAS-mutated NSCLC, three GATA2 downstream pathways (proteasome pathway, Rho-signaling cascade, and NF-κB signaling pathway) and related cross-talk are essential for the viability of tumors. Combined utilization of Bortezomib (inhibit proteasome and NF-κB) and Fasudil (inhibit Rho-signaling cascade) leads the tumors to death, whereas a single drug could not kill the tumors. Star shape of genes represents a mutation; syringe represents inhibitors; solid black arrows indicate directions of regulation; and dashed lines indicate cross-talk among pathways
One such example is the synthetic lethal effect of the GATA2 transcriptional network in KRAS-mutated NSCLC. Steckel et al. initially demonstrated that transcription factor GATA2 is requisite for KRAS oncogene-dependent cancer cells through RNAi assay.63 Because there is no clinical targeted drug for GATA2, the downstream regulatory pathways of GATA2 were studied. Proteasome pathway, Rho-signaling cascade, NF-κB (nuclear factor kappa light-chain enhancer of activated B cells) signaling pathway, and related cross-talk were proven by Downward and coworkers to be essential for KRAS-mutated NSCLC viability. Each independent member of this three-pathway network is not necessary for mutant NSCLC survival, thus inhibiting only one or two of the pathways does not lead to tumor death. However, the combined suppression of all three pathways will result in cell death (Fig. 4b).64 This synthetic lethal effect does not work on normal lung cells or non-KRAS-mutated NSCLC. Therefore, we assume that this type of SL could be a future direction for further research. Identifying more complex synthetic lethal networks may provide more targets for anticancer therapy.
Organelles-targeted synthetic lethality
Recently, many researchers have explored SL targeting organelles,98–102 a more macro approach compared to synthetic lethal interactions in genes or functional pathways. This type of SL focuses on affecting or utilizing the major functions of organelles to cause tumor cell death. Currently, various experiments regarding SL are targeting mitochondrial function, which belongs to the category of “metabolic SL”, as referred to by some scientists.98,103,104 Herein, we will mainly describe specific examples of mitochondria-targeting SL, whose mutated metabolic enzymes cause cancers. In addition, other organelles-targeted SL will also be discussed.
Succinate dehydrogenase (SDH), also known as mitochondrial respiratory complex II,105 is regarded as one of the most probable mitochondria-linked synthetic lethal targets. SDH is not only an essential mitochondrial enzyme in the tricarboxylic acid (TCA) cycle, it is also a key player in tumorigenesis. Previous studies demonstrated that SDH is inactive in SDH mutated tumors. This damages mitochondrial respiratory function through the shortening of the TCA cycle and abnormal accumulation of succinate.105,106 Although the metabolic adaptations that allow tumor cells to survive in SDH deficiency are not completely understood, recent studies have illustrated several important characteristics of SDH-deficient tumor cells. These experiments have demonstrated that SDHB-deficient tumor cells use more extracellular pyruvate than normal cells due to their insufficient biosynthetic capacity to meet the demands of this amino acid. These cells produce oxaloacetate, a fundamental factor in maintaining the aspartate level, and also transfer glucose-derived carbons for aspartate biosynthesis, which is critical for cell growth,105 through the preferential use of pyruvate carboxylase (PC).106,107 Furthermore, Cardaci et al. proved that PC inhibition not only reduced the proliferation of SDH-deficient tumor cells in vitro but also weakened the capability of these cells to form tumors in vivo.106 Therefore, PC shares a synthetic lethal interaction with SDH, whereby PC inhibition disturbs the TCA cycle (Fig. 5).
Fig. 5.
Organelles-targeted synthetic lethality—mitochondria. In SDH or FH mutant cancers, the main metabolic and signaling pathways involved in the metabolic reprogramming of SDH and FH related to mitochondria are presented above. Use of PC inhibitors in SDH-deficient tumor cells or inhibition of HMOX1 in FH-mutated cancers will disturb the TCA cycle, resulting in synthetic lethality. Solid black arrows indicate single step metabolic reactions; dashed black lines indicate indirect transcriptional cascades; star shape of genes represents a mutation; and syringe represents inhibitors. ACO (aconitase); CS (citrate synthase); FH (fumarate hydratase); HMOX1 (heme oxygenase 1); IDH (isocitrate dehydrogenase); KEAP1 (Kelch-like ECH-associated protein 1); MDH (malate dehydrogenase); NRF2 (nuclear factor erythroid 2-related factor); 2-OG (2-oxoglutarate); OGDH (oxoglutarate dehydrogenase); PC (pyruvate carboxylase); PDH (pyruvate dehydrogenase); SCS (succinyl-CoA synthetase); SDH (succinate dehydrogenase)
Additionally, mutations of mitochondrial enzymes such as fumarate hydratase (FH) and isocitrate dehydrogenases (IDH) are also involved in SL. FH is an enzyme of the TCA cycle that catalyzes the hydration of fumarate to malate.108 The inhibition of Heme Oxygenase 1 (HMOX1) results in a significant reduction in the growth of fumarate hydratase 1 deficient tumor cells but has little or no effect on normal cells.99 Thus, there is a synthetic lethal effect between HMOX1 and FH1 that targets mitochondrial function (Fig. 5). A study conducted by Chan et al. revealed that BCL-2 inhibitor ABT-199 combined with IDH mutations have considerable effects on AML treatment, it is yet another example of SL through mitochondrial metabolism.100 Except for the TCA cycle, glycolysis is another indispensable process and feasible target related to the mitochondria. For example, natural product englerin A (EA) was proved to activate protein kinase C-θ (PKCθ), which induces an insulin-resistant. Moreover, EA simultaneously activates the transcription factor heat shock factor 1 (HSF1), an inducer of glucose dependence. Therefore, through promoting glucose addiction and simultaneously limiting the tumor cells uptake of glucose, EA influences glycolysis and metabolism of mitochondria to have a synthetically lethal effect on highly glycolytic tumors.109
Aside from SL that targets mitochondrial functions, recent research concentrates on SL targeting other organelles. Zhao et al. found that the combination of SU11274 (MET inhibitor) and gefitinib (EGFR inhibitor) could synergistically influence the function of ribosomes to reduce the proliferation of triple-negative breast cancer (TNBC) by reducing the level of ribosomal protein S6 (RPS6).101 In addition, Cordani et al. demonstrated autophagy activation through the formation of autophagic vesicles, in which their fusion with lysosomes by mTOR inhibitor can repress p53- deficient lung, breast, and pancreas cancer-cell growth.102
Conditional synthetic lethality
SL is known to be context-dependent. This context dependence refers to synthetic lethal partner genes of oncogenes and tumor-suppressor genes under the original concept of SL.11 However, in addition to the abnormities of synthetic lethal genes, the heterogeneity of tumor cells, its microenvironment, and external disturbances can affect genetic interactions, resulting in condition-dependent genetic interactions.110,111 Therefore, several synthetic lethal effects (at the gene, functional pathway, and organelle level) mentioned previously will be weaker or unachievable in the absence of particular conditions. This complex phenomenon was called context-specific or contextual SL in earlier studies,6,39 and conditional SL in recent studies.7,112 Conditional SL is a special synthetic lethal effect on tumor cells that also depends on internal or external circumstances (specific genetic backgrounds, hypoxia, high ROS, use of DNA-damaging agents, etc.) (Fig. 1b). Conditional SL could account for the variation in synthetic lethal effects observed in different tumor cells or different cell lines in the same cancer type. When resistance to synthetic lethal tumor-targeting drugs occurs, conditional SL could provide insight on how to solve this problem. In summary, conditional SL is one step further from nonconditional/original SL and will hold great prospects for treating tumors of various complex conditions in the future. We will provide specific examples of conditional SL in this section.
Different genetic properties can suppress synthetic lethal interactions, resulting in therapeutic resistance. As mentioned above, the utilization of PARP inhibitors could lead BRCA mutant tumor cells to SL by destroying the two main DSBs repair pathways, HR and NHEJ.36,85 However, the loss of 53BP1 can inhibit synthetic lethal therapy using PARP inhibitors on BRCA1/2-mutated breast cancer.113–115 In 53BP1+/+ cells, Bouwman et al. proved that the loss of 53BP1 in BRCA1/2-mutated cancers may lead to therapeutic resistance to PARP inhibitors or platinum agents.115 In addition, ATR, ATM, and RAD51 also correlate with PARP inhibitor resistance in BRCA1/2-mutated cancers.116–119 Although uncertainty remains in the underlying mechanism of HR pathway restoration in BRCA1/2-mutated cells with loss of 53BP1, ATR, ATM, or RAD51 after the use of PARP inhibitors, it is evident that these genes are indispensable internal conditions that the synthetic lethal effect of BRCA and PARP requires. Similarly, microsatellite instability (MSI), which results from deficient DNA mismatch repair (MMR), is also a special and essential genetic background for SL in several cancers. Recent studies revealed that targeting WRN helicase has a synthetic lethal effect on the viability of microsatellite instability-high (MSI-H) but not microsatellite stable (MSS) colorectal and endometrial cancer-cell lines.120,121
Other internal conditions also play important roles in conditional SL. A recent study found that acute and chronic hypoxia in the cellular microenvironment may decrease HR protein expression and its function, which sensitizes cells to PARP inhibition.122 This finding can be applied in the treatment of BRCA1/2-mutated tumor cells that are resistant to PARP inhibitors. Furthermore, the use of PARP inhibitors can be extended to tumor cells without BRCA mutations because most solid tumors contain hypoxic cells.122 Similarly, through further study of conditional SL, other internal conditions such as proteotoxic stress and metabolic stress may be proven to increase the range of synthetic lethal interactions.123
In addition to internal conditions, the effect of external factors on tumors, such as radiation and chemotherapy drugs, also have been studied. When exposed to DNA-damaging agents and ionizing radiation, the dependence of tumor cells on PARP, involved in the repair of DNA damage, was enhanced.124 Thus, tumor cells with mutant genes that are synthetic lethal partners of PARP will be more sensitive to PARP inhibitors under those specific conditions. For instance, Bailey et al. identified that cohesin component STAG2 has a synthetic lethal effect with PARP in glioblastoma. Meanwhile, STAG2-mutated glioblastoma cells are more easily destroyed by PARP inhibitors when using temozolomide, a DNA-damaging drug.125
Synthetic lethality targeted drugs in clinical stages
With increasing preclinical studies in the field of SL, SL-targeted drugs in clinical practice have been developed. After Ashworth and Helleday35,36 demonstrated the synthetic lethal interactions of PARP inhibitors in BRCA1/2-deficient tumors in 2005, several inhibitors based on SL for targeted cancer therapy have been applied in clinical practice.
Patients with both BRCA1 and BRCA2 mutations will usually suffer from a lifelong risk of breast and ovarian cancer.126 Initially, PARP inhibitors were applied as a combination therapy of low-dose rucaparib and full-dose temozolomide, a DNA alkylating agent, in clinical trials.127 In phase 1 olaparib clinical trial, which involved patients with BRCA1/2 mutations, 63% of the patients who received olaparib exhibited clinical benefit with minimal side effects than those of conventional chemotherapy regimens.45 Subsequently, phase 2 and 3 clinical trials, which included patients with BRCA1/2-mutated ovarian, breast, prostate and pancreatic cancers, demonstrated the clinical benefit offered by Olaparib.128–132 Based on these clinical trials, the FDA first approved olaparib for the treatment of advanced-stage, BRCA1/2-mutant ovarian cancers in 2014. Subsequently, olaparib was approved for patients with advanced-stage, recurrent ovarian cancer who are in CR or PR after platinum-based chemotherapy and metastatic HER2-negative, BRCA1/2-mutant breast cancer previously treated with chemotherapy in 2017 and 2018, respectively.38 The clinical trials and progress of FDA approval of other PARP inhibitors are presented in Table 2 and Fig. 6. In addition to PARP inhibitors, drugs targeting potential synthetic lethal partners of oncogenes or tumor-suppressor genes such as TP53, KRAS, MYC also have been tested in clinical practice (Table 2).
Fig. 6.

Timeline: landmark discoveries and advances of synthetic lethality in cancer. Several key events of synthetic lethality development. Several expanded concepts of SL beyond the original have been constantly proposed. In contrast, after Ashworth and Helleday demonstrated synthetic lethality of PARP inhibitors in BRCA1/2-deficient tumors, numerous studies on SL in cancer has been significantly increased and several inhibitors, especially PARPi based on SL, has been applied in clinical practice
Conclusion and future perspectives
Since the concept of SL was first proposed, the number of studies on this topic has significantly increased, and many expanded concepts of SL have been proposed. Furthermore, many synthetic lethal drugs, especially several PARP inhibitors approved by the FDA, have achieved major clinical breakthroughs (Fig. 6). In this review, we propose a new and systematic classification of SL that includes those expanded concepts of SL, such as synthetic dosage lethality, collateral SL, metabolic SL, etc. According to the specificity of research on its mechanism, various cases of SL were successively divided into gene-level SL, pathway-level SL, organelle-level SL, and conditional SL—from a more superficial to a deeper level. The classification we proposed above, along with the many recent findings of SL reviewed of each type, provide a more comprehensive understanding and many implications for the future perspectives on SL studies.
Firstly, the identification of SL interactions between genes is the primary step in SL at the gene level. It is imperative to identify more synthetic lethal effects among different genes in multiple cancers to develop a synthetic lethal gene database. In addition, verifying gene synthetic lethal effects in various cell lines of the same cancer and in different cancer types are also important. It could explain the controversies in previous studies, such as the synthetic lethal effect between STK33 and KRAS mentioned above, and provide the basis for further study on more complex conditions using the theory of conditional SL.61,62 Synthetic lethal screening technologies including drug screens,133 RNAi screens,33 bioinformatics screens,34 CRISPR screens,32 and combination of these methods,11 provide the possibility for geneticists to achieve this. At the gene level, many previous studies have focused on identifying a synthetic lethal relationship between two genes. The example of synthetic lethal effects among three genes (PLK1, ROCK, and KRAS) as mentioned above66 indicates that synthetic lethal interactions among multiple genes can be developed in the future. Similarly, previous studies also concentrated on searching for synthetic lethal partners of oncogenes while recent studies tend to target tumor-suppressor genes like p53. Identifying synthetic lethal partners of tumor-suppressor genes may have more potential to be explored. Besides, although targeting the commonly mutated and “undruggable” oncogenes and tumor-suppressor genes are invalid, from the example of activating p53 and inhibiting BCL-2 to kill AML,59 targeting these genes in combination with their synthetic lethal partners may significantly amplify the lethal effects on tumors compared to only inhibiting partner genes as in previous studies, which could expand the application of synthetic lethal effects in targeted cancer therapy.
Secondly, for researchers and clinicians working on targeted therapies for tumors, SL by pathway also provides many future perspectives. After identifying synthetic lethal partner genes, at protein or pathway level, they can further study those that are expressed unusually in specific cancer, while referring to the three synthetic lethal pathway conditions. Since SL has not been applied to the majority of cancers, the mechanism of the synthetic lethal pathway reported could be applied to less studied cancers that have the same mutated oncogenes or tumor-suppressor genes, such as nerve cancers, skin cancers, bone tumors, and biliary tract cancers. Furthermore, synthetic lethal effect in multiple pathways is an important direction in the future. The identification of difficult synthetic lethal effects can lead to further studies regarding pathway networks related to those difficult targets. As previously mentioned, Downward et al. revealed that the GATA2 downstream network (three pathways) has the same effect as GATA2 on KRAS-mutated NSCLC survival.64 Thus, there is a higher chance of finding points of the target for clinical inhibitors in each pathway. Furthermore, inhibitors approved by the FDA were used to block the three-pathway network, consequently killing KRAS-mutated NSCLC efficiently. Through this, the research achievements of SL could be better applied in clinical practice. In addition, organelle-targeted SL, the synthetic lethal effect that destroys mitochondria, ribosomes, lysosomes, and other organelles, is also a more macro direction of future research.
Lastly, conditional SL can provide explanations and future directions for the limitations of SL in anticancer therapeutic targets. The biggest challenge SL faces in clinical practice are drug resistance.11,88 In addition, synthetic lethal interactions that work in one cancer are sometimes ineffective in another. According to conditional SL, synthetic lethal effects on cancers require specific internal and external settings, which helps to explain these problems. Thus, exploring the specific circumstances required by the same cancers in different conditions or different cancer types is of great importance to solve drug resistance and expand the application of SL, which may be addressable using a multi-faceted testing framework.76
For preclinical studies, identifying different microenvironments and genetic backgrounds of cancer cells, which have different sensitivities to the same synthetic lethal effect, may reveal more drug resistance mechanisms.134 For clinical practice, a combination of external conditions including traditional chemotherapy drugs, immunotherapy, or radiation therapy coupled with SL-based drugs, holds great prospects to solve the issue regarding the resistance to synthetic lethal effect.11,67 In addition, due to the fact that high ROS is regarded as an internal condition to promote SL7 and several types of nanoparticles could generate ROS,135 the combination of synthetic lethal cancer therapy with nanotechnology could reduce drug resistance.136 However, Hocsak et al. demonstrated that PARP inhibitors suppress mitochondrial ROS production and decrease ROS-induced apoptosis in oxidative stress, thereby protecting the mitochondrial membrane potential via MKP-1 and ATF4 dependent pathway in A-549, T24/83, and WRL-68 human cell lines.137 Therefore, the role of ROS in conditional SL requires further studying and discussion. In summary, a great deal is yet to be understood and significant amounts of research to be done for tumor-targeted therapy still remains.
In conclusion, although SL is merely a simple genetic concept, its impact on cancer research has been increasing. This novel classification of SL along with the multiple findings of SL reviewed in each type mentioned above not only provides a systematic understanding of this field but also gives research more basis for reference and inspiration for future directions. We firmly believe that SL deserves further study and application in the field of cancer therapy in the future.
Acknowledgements
The authors would like to thank the authors of the primary studies. The figures in this article were created using Adobe Illustrator, Adobe Photoshop, BioRender, and Microsoft PowerPoint.
Funding
This work was supported by the Opening Fund of Engineering Research Center of Cognitive Healthcare of Zhejiang Province (No.2018KFJJ09), Scientific Research Fund of Zhejiang Provincial Education Department (Y201941406), National Natural Science Foundation of China (Nos. 81827804 and 81800540), Zhejiang Clinical Research Center of Minimally Invasive Diagnosis and Treatment of Abdominal Diseases (No. 2018E50003), Science and Technology Innovative Activity Plan and New Seedling Talent Plan for College Students in Zhejiang Province (No. 2020R401217)
Author contributions
S.L., W.T., S.J., X.C., and M.C. wrote this paper. J.C., J.H., B.Z., D.M., X.C., and M.C. reviewed this paper. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Consent for publication
All authors give consent for the publication of the manuscript in “Signal Transduction and Targeted Therapy”.
Footnotes
These authors contributed equally: Shijie Li, Win Topatana, Sarun Juengpanich
Contributor Information
Xiujun Cai, Email: srrsh_cxj@zju.edu.cn.
Mingyu Chen, Email: mychen@zju.edu.cn.
References
- 1.Dobzhansky T. Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. Genetics. 1946;31:269–290. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucchesi JC. Synthetic lethality and semi-lethality among functionally related mutants of Drosophila melanfgaster. Genetics. 1968;59:37–44. doi: 10.1093/genetics/59.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser CA, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-U. [DOI] [PubMed] [Google Scholar]
- 4.Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 5.Bender A, Pringle JR. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell Biol. 1991;11:1295–1305. doi: 10.1128/MCB.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018;15:564–576. doi: 10.1038/s41571-018-0055-6. [DOI] [PubMed] [Google Scholar]
- 7.O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017;18:613–623. doi: 10.1038/nrg.2017.47. [DOI] [PubMed] [Google Scholar]
- 8.Chen M, Cai X. Synthetic lethality is a novel and potential paradigm for precision medicine in advanced hepatocellular carcinoma. Liver Cancer. 2020;9:225–226. doi: 10.1159/000503927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Huang A, Garraway LA, Ashworth A, Weber B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 2020;19:23–38. doi: 10.1038/s41573-019-0046-z. [DOI] [PubMed] [Google Scholar]
- 12.Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell. 2017;168:584–599. doi: 10.1016/j.cell.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoadley, K. A. et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell173, 291–304 (2018). [DOI] [PMC free article] [PubMed]
- 14.Tate JG, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stockley TL, et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016;8:109. doi: 10.1186/s13073-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanton C, et al. Consensus on precision medicine for metastatic cancers: a report from the MAP conference. Ann. Oncol. 2016;27:1443–1448. doi: 10.1093/annonc/mdw192. [DOI] [PubMed] [Google Scholar]
- 17.Demetri GD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 18.Blanke CD, et al. Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J. Clin. Oncol. 2008;26:620–625. doi: 10.1200/JCO.2007.13.4403. [DOI] [PubMed] [Google Scholar]
- 19.Blanke CD, et al. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the kit receptor tyrosine kinase: S0033. J. Clin. Oncol. 2008;26:626–632. doi: 10.1200/JCO.2007.13.4452. [DOI] [PubMed] [Google Scholar]
- 20.Verweij J, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet. 2004;364:1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 21.Roy L, et al. Survival advantage from imatinib compared with the combination interferon-alpha plus cytarabine in chronic-phase chronic myelogenous leukemia: historical comparison between two phase 3 trials. Blood. 2006;108:1478–1484. doi: 10.1182/blood-2006-02-001495. [DOI] [PubMed] [Google Scholar]
- 22.Kantarjian H, et al. Improved survival in chronic myeloid leukemia since the introduction of imatinib therapy: a single-institution historical experience. Blood. 2012;119:1981–1987. doi: 10.1182/blood-2011-08-358135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 24.Hochhaus A, et al. Long-term outcomes of imatinib treatment for chronic myeloid leukemia. N. Engl. J. Med. 2017;376:917–927. doi: 10.1056/NEJMoa1609324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 26.Zhou C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 27.Rosell R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 28.Soria J-C, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 29.First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer. N. Engl. J. Med. 2015;373:1582. doi: 10.1056/NEJMx150034. [DOI] [PubMed] [Google Scholar]
- 30.Crizotinib versus Chemotherapy in Advanced ALK-Positive Lung Cancer. N. Engl. J. Med. 2015;373:1582. doi: 10.1056/NEJMx150036. [DOI] [PubMed] [Google Scholar]
- 31.Pagliarini R, Shao W, Sellers WR. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep. 2015;16:280–296. doi: 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behan FM, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 33.Setten RL, Rossi JJ, Han S-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 34.Carbajosa S, et al. Polo-like kinase 1 inhibition as a therapeutic approach to selectively target BRCA1-deficient cancer cells by synthetic lethality induction. Clin. Cancer Res. 2019;25:4049–4062. doi: 10.1158/1078-0432.CCR-18-3516. [DOI] [PubMed] [Google Scholar]
- 35.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 36.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 37.D’Andrea AD. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair. 2018;71:172–176. doi: 10.1016/j.dnarep.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Pilié, P. G., Tang, C., Mills, G. B. & Yap, T. A. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev.Clin. Oncol.16, 81–104 (2019). [DOI] [PMC free article] [PubMed]
- 39.McLornan DP, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. N. Engl. J. Med. 2014;371:1725–1735. doi: 10.1056/NEJMra1407390. [DOI] [PubMed] [Google Scholar]
- 40.Yan H, Gibson S, Tye BK. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes Dev. 1991;5:944–957. doi: 10.1101/gad.5.6.944. [DOI] [PubMed] [Google Scholar]
- 41.Reid RJD, et al. A synthetic dosage lethal genetic interaction between CKS1B and PLK1 Is conserved in yeast and human cancer cells. Genetics. 2016;204:807–819. doi: 10.1534/genetics.116.190231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller FL, Aquilanti EA, DePinho RA. Collateral lethality: a new therapeutic strategy in oncology. Trends Cancer. 2015;1:161–173. doi: 10.1016/j.trecan.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dey P, et al. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature. 2017;542:119–123. doi: 10.1038/nature21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gogola, E. et al. Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell33, 1078–1093 (2018). [DOI] [PubMed]
- 45.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 46.Clarke, N. W. et al. PROPEL: A randomized, phase III trial evaluating the efficacy and safety of olaparib combined abiraterone first-line therapy in patients with metastatic castration-resistant prostate cancer (mCRPC). J. Clin. Oncol.37, TPS340–TPS340 (2019).
- 47.Rodriguez-Moreno, J. F. et al. Impact of the combination of durvalumab (MEDI4736) plus olaparib (AZD2281) administered prior to surgery in the molecular profile of resectable urothelial bladder Cancer: NEODURVARIB Trial. J. Clin. Oncol.37, TPS503–TPS503 (2019).
- 48.Hardy-Bessard, A.-C. et al. ENGOT-OV44/first study: a randomized, double-blind, adaptive, phase III study of platinum-based therapy with dostarlimab (TSR-042) + niraparib versus standard-of-care (SOC) platinum-based therapy as first-line treatment of stage 3/4 non-mucinous epithelial ovarian cancer (OC). J. Clin. Oncol.37, TPS5600–TPS5600 (2019).
- 49.Sahai V, Tran NH, Griffith KA, Zalupski M. A Multicent. phase II trial rucaparib combination nivolumab Maint. Ther. patients Adv. biliary Trac. cancer. 2019;37:TPS4153–TPS4153. [Google Scholar]
- 50.Vallurupalli M, et al. Talazoparib Treatment Preferentially Depletes Cohesin-Mutant Clones in New In Vivo Models of Cohesin-Mutant Myeloid Diseases. Blood. 2019;134:560–560. doi: 10.1182/blood-2019-130308. [DOI] [Google Scholar]
- 51.Heeke, A. L. et al. Abstract OT2-03-04: a trial of induction Talazoparib followed by a combination of Talazoparib and Avelumab in advanced breast cancer: The TALAVE study. J. Cancer Res.80, OT2-03-04-OT02-03-04 (2020).
- 52.Bykov, V. J. N., Eriksson, S. E., Bianchi, J. & Wiman, K. G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer18, 89–102 (2018). [DOI] [PubMed]
- 53.Wang X, Simon R. Identification of potential synthetic lethal genes to p53 using a computational biology approach. BMC Med. Genomics. 2013;6:30. doi: 10.1186/1755-8794-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durant ST, et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci. Adv. 2018;4:eaat1719. doi: 10.1126/sciadv.aat1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwok M, et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood. 2016;127:582–595. doi: 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- 56.Middleton, F. K., Pollard, J. R. & Curtin, N. J. The Impact of p53 Dysfunction in ATR Inhibitor cytotoxicity and chemo- and radiosensitisation. Cancers(Basel)10, 275 (2018). [DOI] [PMC free article] [PubMed]
- 57.Diab A, et al. Multiple defects sensitize p53-deficient head and neck cancer cells to the WEE1 Kinase Inhibition. Mol. Cancer Res. 2019;17:1115–1128. doi: 10.1158/1541-7786.MCR-18-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tirrò E, et al. Chk1 inhibition restores inotuzumab ozogamicin citotoxicity in CD22-positive cells expressing mutant p53. Front Oncol. 2019;9:57. doi: 10.3389/fonc.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan, R. et al. Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell32, 748–760 (2017). [DOI] [PMC free article] [PubMed]
- 60.Mahadevan, D. et al. Aurora A Inhibitor (MLN8237) plus Vincristine plus Rituximab is synthetic lethal and a potential curative therapy in aggressive B-cell non-hodgkin lymphoma. Clin. Cancer Res.21, 5181 (2015). [DOI] [PubMed]
- 61.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 62.Luo T, et al. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proc. Natl Acad. Sci. USA. 2012;109:2860–2865. doi: 10.1073/pnas.1120589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steckel M, et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 2012;22:1227–1245. doi: 10.1038/cr.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar MS, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 65.Wong, C. C. et al. SLC25A22 promotes proliferation and survival of colorectal cancer cells with KRAS mutations and xenograft tumor progression in mice via intracellular synthesis of aspartate. Gastroenterology151, 945–960 (2016). [DOI] [PubMed]
- 66.Wang J, et al. Suppression of KRas-mutant cancer through the combined inhibition of KRAS with PLK1 and ROCK. Nat. Commun. 2016;7:11363. doi: 10.1038/ncomms11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coelho, M. A. et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity47, 1083–1099 (2017). [DOI] [PMC free article] [PubMed]
- 68.Pourdehnad M, et al. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc. Natl Acad. Sci. USA. 2013;110:11988–11993. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kessler JD, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang, J. et al. Direct phosphorylation and stabilization of MYC by Aurora B kinase promote T-cell leukemogenesis. Cancer Cell37, 200–215 (2020). [DOI] [PMC free article] [PubMed]
- 71.Horiuchi D, et al. PIM1 kinase inhibition as a targeted therapy against triple-negative breast tumors with elevated MYC expression. Nat. Med. 2016;22:1321–1329. doi: 10.1038/nm.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang C-H, et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28:1800–1814. doi: 10.1101/gad.244368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bian Y, et al. Synthetic genetic array screen identifies PP2A as a therapeutic target in Mad2-overexpressing tumors. Proc. Natl Acad. Sci. USA. 2014;111:1628–1633. doi: 10.1073/pnas.1315588111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duffy S, et al. Overexpression screens identify conserved dosage chromosome instability genes in yeast and human cancer. Proc. Natl Acad. Sci. USA. 2016;113:9967–9976. doi: 10.1073/pnas.1611839113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kroll, E. S., Hyland, K. M., Hieter, P. & Li, J. J. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics143, 95–102 (1996). [DOI] [PMC free article] [PubMed]
- 76.Ku AA, et al. Integration of multiple biological contexts reveals principles of synthetic lethality that affect reproducibility. Nat. Commun. 2020;11:2375. doi: 10.1038/s41467-020-16078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fang B. Development of synthetic lethality anticancer therapeutics. J. Med. Chem. 2014;57:7859–7873. doi: 10.1021/jm500415t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Orvis T, et al. BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization. Cancer Res. 2014;74:6486–6498. doi: 10.1158/0008-5472.CAN-14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Helming KC, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat. Med. 2014;20:251–254. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zucman-Rossi, J., Villanueva, A., Nault, J.-C. & Llovet, J. M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology149, 1226–1239 (2015). [DOI] [PubMed]
- 81.Hoffman GR, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl Acad. Sci. USA. 2014;111:3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karnezis AN, et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J. Pathol. 2016;238:389–400. doi: 10.1002/path.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.IThe Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell32, 185–203 (2017). [DOI] [PMC free article] [PubMed]
- 84.Ryan, C. J., Kennedy, S., Bajrami, I., Matallanas, D. & Lord, C. J. A compendium of co-regulated protein complexes in breast cancer reveals collateral loss events. Cell Syst5, 399-409 (2017). [DOI] [PMC free article] [PubMed]
- 85.Chang HHY, Pannunzio NR, Adachi N, Lieber MR, Non-homologous DNA. end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Isono M, et al. BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep. 2017;18:520–532. doi: 10.1016/j.celrep.2016.12.042. [DOI] [PubMed] [Google Scholar]
- 88.Li H, et al. PARP inhibitor resistance: the underlying mechanisms and clinical implications. Mol. Cancer. 2020;19:107. doi: 10.1186/s12943-020-01227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown JS, O’Carrigan B, Jackson SP, Yap TA, Targeting DNA. Repair in cancer: beyond PARP inhibitors. Cancer Disco. 2017;7:20–37. doi: 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aly A, Ganesan S. BRCA1, PARP, and 53BP1: conditional synthetic lethality and synthetic viability. J. Mol. Cell Biol. 2011;3:66–74. doi: 10.1093/jmcb/mjq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ceccaldi R, et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Du WW, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.AbuHammad S, et al. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc. Natl Acad. Sci. USA. 2019;116:17990–18000. doi: 10.1073/pnas.1901323116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cannell IG, et al. A pleiotropic RNA-binding protein controls distinct cell cycle checkpoints to drive resistance of p53-defective tumors to chemotherapy. Cancer Cell. 2015;28:623–637. doi: 10.1016/j.ccell.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morandell S, et al. A reversible gene-targeting strategy identifies synthetic lethal interactions between MK2 and p53 in the DNA damage response in vivo. Cell Rep. 2013;5:868–877. doi: 10.1016/j.celrep.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Phoa, A. F. et al. MK2 inhibition induces p53-dependent senescence in glioblastoma cells. Cancers (Basel)12, 654 (2020). [DOI] [PMC free article] [PubMed]
- 98.Genini D, et al. Mitochondrial dysfunction induced by a SH2 domain-targeting STAT3 inhibitor leads to metabolic synthetic lethality in cancer cells. Proc. Natl Acad. Sci. USA. 2017;114:E4924–E4933. doi: 10.1073/pnas.1615730114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frezza C, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 100.Chan SM, et al. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat. Med. 2015;21:178–184. doi: 10.1038/nm.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yi YW, et al. Dual inhibition of EGFR and MET induces synthetic lethality in triple-negative breast cancer cells through downregulation of ribosomal protein S6. Int J. Oncol. 2015;47:122–132. doi: 10.3892/ijo.2015.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cordani M, et al. Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol. Oncol. 2016;10:1008–1029. doi: 10.1016/j.molonc.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Villanueva MT. Anticancer therapy: metabolic synthetic lethality. Nat. Rev. Drug Disco. 2018;17:543. doi: 10.1038/nrd.2018.123. [DOI] [PubMed] [Google Scholar]
- 104.Megchelenbrink W, Katzir R, Lu X, Ruppin E, Notebaart RA. Synthetic dosage lethality in the human metabolic network is highly predictive of tumor growth and cancer patient survival. Proc. Natl Acad. Sci. USA. 2015;112:12217–12222. doi: 10.1073/pnas.1508573112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zecchini V, Frezza C. Metabolic synthetic lethality in cancer therapy. Biochim. Biophys. Acta Bioenerg. 2017;1858:723–731. doi: 10.1016/j.bbabio.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 106.Cardaci S, et al. Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nat. Cell Biol. 2015;17:1317–1326. doi: 10.1038/ncb3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lussey-Lepoutre C, et al. Loss of succinate dehydrogenase activity results in dependency on pyruvate carboxylation for cellular anabolism. Nat. Commun. 2015;6:8784. doi: 10.1038/ncomms9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schmidt C, Sciacovelli M, Frezza C. Fumarate hydratase in cancer: a multifaceted tumour suppressor. Semin Cell Dev. Biol. 2020;98:15–25. doi: 10.1016/j.semcdb.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sourbier C, et al. Englerin A stimulates PKCθ to inhibit insulin signaling and to simultaneously activate HSF1: pharmacologically induced synthetic lethality. Cancer Cell. 2013;23:228–237. doi: 10.1016/j.ccr.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Filteau M, et al. Evolutionary rescue by compensatory mutations is constrained by genomic and environmental backgrounds. Mol. Syst. Biol. 2015;11:832. doi: 10.15252/msb.20156444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chari S, Dworkin I. The conditional nature of genetic interactions: the consequences of wild-type backgrounds on mutational interactions in a genome-wide modifier screen. PLoS Genet. 2013;9:e1003661. doi: 10.1371/journal.pgen.1003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bukhari AB, et al. Inhibiting Wee1 and ATR kinases produces tumor-selective synthetic lethality and suppresses metastasis. J. Clin. Invest. 2019;129:1329–1344. doi: 10.1172/JCI122622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jaspers JE, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 2013;3:68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu Y, et al. RAD51 mediates resistance of cancer stem cells to PARP inhibition in triple-negative breast cancer. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2017;23:514–522. doi: 10.1158/1078-0432.CCR-15-1348. [DOI] [PubMed] [Google Scholar]
- 117.Schmitt A, et al. Deficiency is associated with sensitivity to PARP1- and ATR inhibitors in lung adenocarcinoma. Cancer Res. 2017;77:3040–3056. doi: 10.1158/0008-5472.CAN-16-3398. [DOI] [PubMed] [Google Scholar]
- 118.Yazinski SA, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31:318–332. doi: 10.1101/gad.290957.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hurley RM, et al. 53BP1 as a potential predictor of response in PARP inhibitor-treated homologous recombination-deficient ovarian cancer. Gynecol. Oncol. 2019;153:127–134. doi: 10.1016/j.ygyno.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lieb, S. et al. Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells. Elife8, e43333 (2019). [DOI] [PMC free article] [PubMed]
- 121.Chan EM, et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature. 2019;568:551–556. doi: 10.1038/s41586-019-1102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chan N, et al. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res. 2010;70:8045–8054. doi: 10.1158/0008-5472.CAN-10-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nagel R, Semenova EA, Berns A. Drugging the addict: non-oncogene addiction as a target for cancer therapy. EMBO Rep. 2016;17:1516–1531. doi: 10.15252/embr.201643030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bailey ML, et al. Glioblastoma cells containing mutations in the cohesin component STAG2 are sensitive to PARP inhibition. Mol. Cancer Therapeutics. 2014;13:724–732. doi: 10.1158/1535-7163.MCT-13-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Elezaby M, et al. Mutation carriers: breast and ovarian cancer screening guidelines and imaging considerations. Radiology. 2019;291:554–569. doi: 10.1148/radiol.2019181814. [DOI] [PubMed] [Google Scholar]
- 127.Plummer R, et al. A phase II study of the potent PARP inhibitor, Rucaparib (PF-01367338, AG014699), with temozolomide in patients with metastatic melanoma demonstrating evidence of chemopotentiation. Cancer Chemother. Pharm. 2013;71:1191–1199. doi: 10.1007/s00280-013-2113-1. [DOI] [PubMed] [Google Scholar]
- 128.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 129.Moore K, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 130.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 131.Mateo J, et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:162–174. doi: 10.1016/S1470-2045(19)30684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Golan T, et al. Maintenance olaparib for germline -mutated metastatic pancreatic cancer. N. Engl. J. Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu L, et al. Synthetic lethality-based identification of targets for anticancer drugs in the human signaling network. Sci. Rep. 2018;8:8440. doi: 10.1038/s41598-018-26783-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 135.Lucky SS, Soo KC, Zhang Y. Nanoparticles in photodynamic therapy. Chem. Rev. 2015;115:1990–2042. doi: 10.1021/cr5004198. [DOI] [PubMed] [Google Scholar]
- 136.Li Z, Di C, Li S, Yang X, Nie G. Smart Nanotherapeutic Targeting of Tumor Vasculature. Acc. Chem. Res. 2019;52:2703–2712. doi: 10.1021/acs.accounts.9b00283. [DOI] [PubMed] [Google Scholar]
- 137.Hocsak E, et al. PARP inhibition protects mitochondria and reduces ROS production via PARP-1-ATF4-MKP-1-MAPK retrograde pathway. Free Radic. Biol. Med. 2017;108:770–784. doi: 10.1016/j.freeradbiomed.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 138.Lin J, et al. Alterations in DNA damage repair genes in primary liver cancer. Clin. Cancer Res. 2019;25:4701–4711. doi: 10.1158/1078-0432.CCR-19-0127. [DOI] [PubMed] [Google Scholar]
- 139.Kaplan, A. R. et al. Cediranib suppresses homology-directed DNA repair through down-regulation of BRCA1/2 and RAD51. Sci. Transl. Med.11, eaav4508 (2019). [DOI] [PMC free article] [PubMed]
- 140.Chen C-C, et al. Corylin increases the sensitivity of hepatocellular carcinoma cells to chemotherapy through long noncoding RNA RAD51-AS1-mediated inhibition of DNA repair. Cell Death Dis. 2018;9:543. doi: 10.1038/s41419-018-0575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Santiago-O’Farrill JM, et al. Poly(adenosine diphosphate ribose) polymerase inhibitors induce autophagy-mediated drug resistance in ovarian cancer cells, xenografts, and patient-derived xenograft models. Cancer. 2020;126:894–907. doi: 10.1002/cncr.32600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bolin C, et al. The impact of cyclin-dependent kinase 5 depletion on poly(ADP-ribose) polymerase activity and responses to radiation. Cell Mol. Life Sci. 2012;69:951–962. doi: 10.1007/s00018-011-0811-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Turner NC, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008;27:1368–1377. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bartucci M, et al. Therapeutic targeting of Chk1 in NSCLC stem cells during chemotherapy. Cell Death Differ. 2012;19:768–778. doi: 10.1038/cdd.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu DS, et al. Inhibiting the system x/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat. Commun. 2017;8:14844. doi: 10.1038/ncomms14844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dauch D, et al. A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat. Med. 2016;22:744–753. doi: 10.1038/nm.4107. [DOI] [PubMed] [Google Scholar]
- 147.Emerling BM, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 2013;155:844–857. doi: 10.1016/j.cell.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bitler BG, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat. Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shen J, et al. ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 2015;5:752–767. doi: 10.1158/2159-8290.CD-14-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]