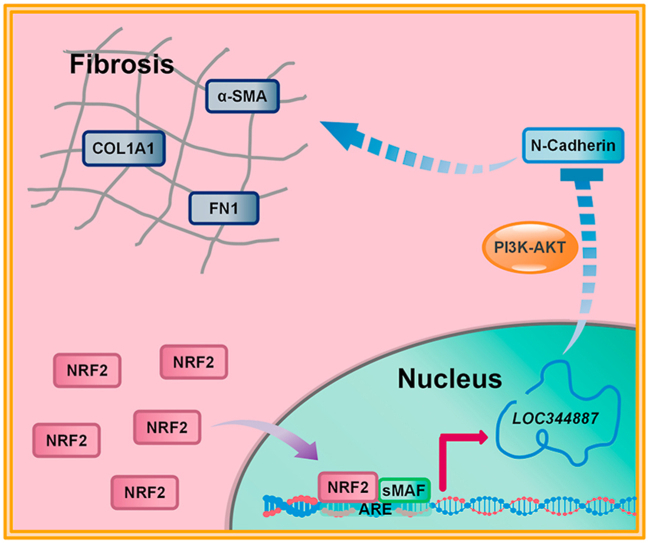
Abstract
Idiopathic pulmonary fibrosis (IPF) is a progressive and irreversible disease characterized by an increase in differentiation of fibroblasts to myofibroblasts and excessive accumulation of extracellular matrix in lung tissue. Pharmacological activation of NRF2 has proved to be a valuable antifibrotic approach, however the detailed mechanisms of how NRF2 mediates antifibrotic function remain unclear. In this study, we found that the antifibrotic function of sulforaphane (SFN), an NRF2 activator, was largely dependent on LOC344887, a long noncoding RNA. Two functional AREs were identified in both the promoter and intron 1 of LOC344887, which defines LOC344887 as a novel anti-fibrotic NRF2 target gene. RNA-seq analysis revealed that LOC344887 controls genes and signaling pathways associated with fibrogenesis. Deletion or downregulation of LOC344887 enhanced expression of CDH2/N-cadherin, as well as a number of other fibrotic genes and blunted the antifibrotic effects of SFN. Furthermore, LOC344887-mediated downregulation of fibrotic genes may involve the PI3K-AKT signaling pathway, as pharmacologic inhibition of PI3K activity blocked the effects of LOC344887 knockdown. Our findings demonstrate that NRF2-mediated LOC344887 upregulation contributes to the antifibrotic potential of SFN by repressing the expression of CDH2 and other fibrotic genes, providing novel insight into how NRF2 controls the regulatory networks of IPF. This study provides a better understanding of the molecular mechanisms of NRF2 activators against pulmonary fibrosis and presents a novel therapeutic axis for prevention and intervention of fibrosis-related diseases.
Keywords: NRF2, LOC344887, CDH2, PI3K-AKT, Extracellular matrix, Pulmonary fibrosis
Graphical abstract
1. Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic lung disease caused by different factors, such as environmental pollutants, drug side effects, and viral or fungal infection [[1], [2], [3]]. In fact, pulmonary fibrosis is also regarded as one of the major thoracic complications in patients with coronavirus disease 2019 (COVID-19) [4,5]. Pulmonary fibrosis is characterized by activation of lung fibroblasts, excessive accumulation of extracellular matrix (ECM) in lung tissues, and progressive lung scarring. In recent years, the incidence of pulmonary fibrosis has increased sharply [2], but effective treatments for fibrotic diseases are limited. The current treatments for pulmonary fibrosis, immunosuppressants (e.g. cyclophosphamide) and corticosteroids (e.g. dexamethasone), are limited in their potential because of severe side effects and low efficacy [6,7]. Nintedanib and pirfenidone, two drugs recently approved by the FDA for the treatment of pulmonary fibrosis, can slow down the progression of lung fibrosis and preserve lung function, however, neither of those drugs are able to reverse fibrosis. Furthermore, these new drugs display severe side effects, causing injury of many organs including the liver and skin [[8], [9], [10], [11]]. Therefore, understanding the molecular events underlying the pathogenesis of IPF, coupled with developing novel and effective means of prevention/intervention that target early stages of fibrogenesis remain an important area of research.
Nuclear factor erythroid 2 (NFE2)-related factor 2 (NRF2) is an important transcription factor that regulates cellular antioxidant and anti-inflammatory responses by controlling expression of its target genes containing antioxidant response elements (AREs) in their regulatory regions. In response to oxidants and electrophiles, NRF2 forms heterodimeric complexes with small musculoaponeurotic fibrosarcoma (sMAF) protein and binds to AREs to transcriptionally activate its target genes and restore cellular homeostasis [[12], [13], [14]]. The role of NRF2 in pulmonary fibrosis has been investigated, and those studies indicate that NRF2 protects against the development of pulmonary fibrosis, indicating NRF2 activators could be used to inhibit fibrogenesis [[15], [16], [17]]. For instance, sulforaphane (SFN), a well-studied NRF2 activator, attenuates bleomycin-induced pulmonary fibrosis by effectively suppressing the epithelial-mesenchymal transition (EMT) process [15], an effect that was also observed when NRF2 was activated by another inducer artemisitene [17]. In addition, trans-4,4′-dihydroxystilbene significantly ameliorates cigarette smoke-induced pulmonary fibrosis via NRF2 activation [18]. Therefore, NRF2 activators hold promising potential to be developed into preventive agents against pulmonary fibrosis. The current mechanistic action of NRF2 activators against pulmonary fibrosis are thought to derive from its mediation of the antioxidant response and inhibition of EMT, which is essential for the overexpression of ECM genes [15,19,20]. However, the detailed molecular mechanisms linking NRF2 activation to the reduction of fibrogenesis are missing and warrant investigation.
Long noncoding RNAs (lncRNAs) have attracted widespread attention in various fields. LncRNAs are non-coding transcripts longer than 200 nucleotides involved in various biological processes, such as cell proliferation and cell fate decisions. More importantly, aberrant IncRNA expression has been associated with developmental disorders and the pathogenesis of many human diseases [21,22]. LncRNAs have also been demonstrated to play a role in pulmonary fibrosis. Knockdown of lncRNA H19 was able to attenuate pulmonary fibrosis in vitro and in vivo, through modulation of the miR-140-TGF-β/Smad 3 signaling pathway [23]. LncRNA ZEB1-AS1 was reported to promote lung fibrosis by enhancing ZEB1-mediated EMT [24]. Another study demonstrated that lncRNA FENDRR also plays a role in mitigating pulmonary fibrosis [25]. Therefore, modulating lncRNAs has been considered as a novel therapeutic strategy for treating pulmonary fibrosis as well as other respiratory diseases. In this study, we report that a lncRNA, NmrA like redox sensor 2, pseudogene (NMRAL2P/LOC344887), is a novel anti-fibrotic NRF2 target gene that is largely responsible for the previously observed antifibrotic function of NRF2. Knockdown or knockout of LOC344887 was associated with enhanced fibrosis, including increased expression of N-cadherin and ECM via the PI3K-AKT pathway.
2. Materials and methods
2.1. Chemicals, reagents, and antibodies
Sulforaphane (S6317), as well as anti-mouse IgG (HRP) (1:3000, A-9044) and anti-rabbit IgG (HRP) (1:3000, A-0545) antibodies were purchased from Sigma-Aldrich. LY294002 (ab120243) was purchased from Abcam. The rabbit normal IgG and antibodies against NRF2 (1:1000, sc-13032), sMAF (1:1000, sc-22831), NQO1 (1:1000, sc-32793), FN1 (1:1000, sc-8422), COL1A1 (1:1000, sc-293,182), α-SMA (1:1000, sc-53142), N-Cadherin (1:500, sc-7939) and GAPDH (1:3000, sc-32233) were purchased from Santa Cruz Biotechnology. The antibody against E-Cadherin (1:1000, 610,181) was purchased from BD Biosciences. Antibodies against AKT (1:2000, 4691S) and phospho-AKT (Ser473) (1:1000, 4060S) were purchased from Cell Signaling Technology. TGF-β (100–21) was purchased from PeproTech.
2.2. Cell culture
Human non-small cell lung cancer cell lines, NCI–H1299 and A549, human lung bronchus epithelial cell line, BEAS-2B, and human lung fibroblast cell line, HFL1, were purchased from American Type Culture Collection (ATCC). A549, H1299 and BEAS-2B cells were grown in DMEM medium supplemented with 10% FBS (Gibco, 26,140–079) and 1% penicillin/streptomycin. HFL1 cells were cultured using DMEM:F12 Medium supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were cultured in a humidified incubator at 37 °C with 5% CO2, and the medium was changed every two days.
2.3. Generation of gene knockout cell lines
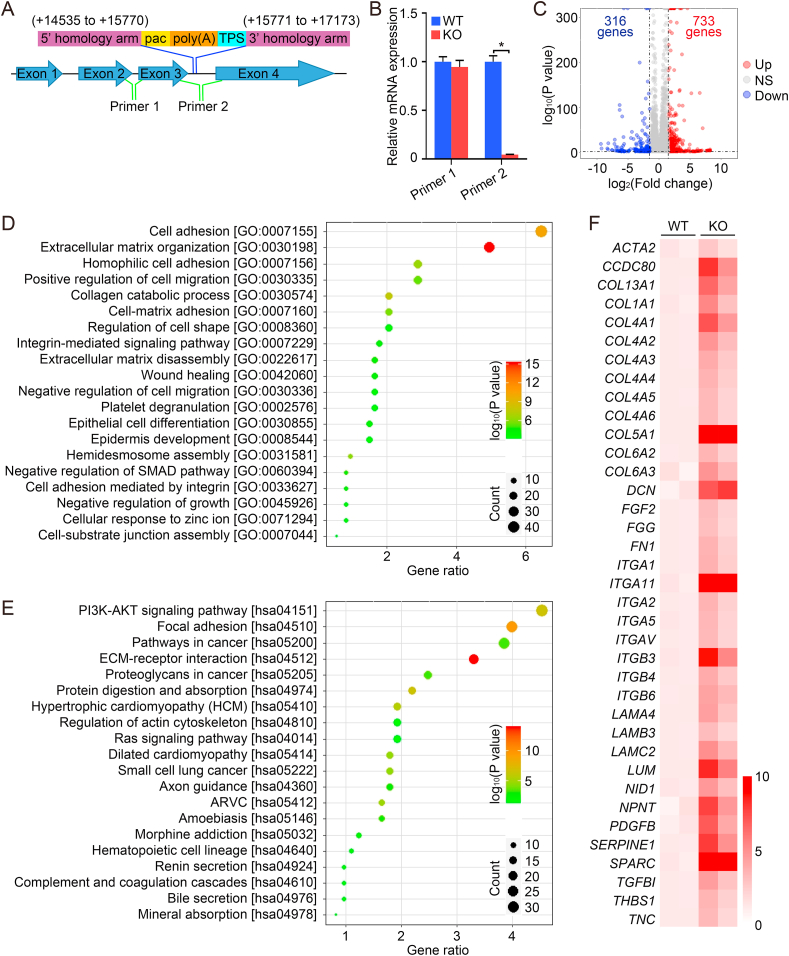
LOC344887 knockout cells were generated using CRISPR-Cas9-mediated homologous recombination as reported previously [26,27]. In brief, one single guide RNA (sgRNA) sequence was used to target the sequence around the insert point, and the sgRNA sequence is as follows: 5′-TCGTCCAGAGCTAGCTGATG-3′. The puromycin N-acetyltransferase (pac) plus a combined poly(A) and a transcriptional pause site (TPS), were inserted into the third intron (between +15,770 and + 15,771, Fig. 2A). The positive cells were selected by puromycin treatment (3 μg/ml, Gibco), and the result was further confirmed by qPCR.
Fig. 2.
LOC344887 knockout enhances the expression of fibrotic genes. (A–B) A schematic diagram of generation of a LOC344887 knockout A549 cell line using CRISPR/Cas9 by insertion of the pac gene (for puromycin resistance), followed by a combined poly(A) and a transcriptional pause sites (A). Two primer pairs, one located upstream (primer pair 1), the other downstream (primer pair 2), of the insertion site (A) were used for qRT-PCR analysis of the mRNA level of LOC344887 (B). (C–F) LOC344887 WT and KO cells were subjected to RNA-seq analysis. A volcano plot showing differentially expressed genes in WT vs. KO cells (C). GO (D) and KEGG pathway enrichment (E) analyses were performed to analyze the enrichment of genes or pathways. Heat map showing the differential expression of fibrotic genes in WT vs. KO cells (F). *: P < 0.05 between two different groups.
NRF2 knockout cells were generated using CRISPR-Cas9 mediated gene editing as per our previous studies [28,29]. A pair of sgRNA sequences (sgRNA-A: 5′-TATTTGACTTCAGTCAGCGA-3′; sgRNA-B: 5′-TAGTTGTAACTGAGCGAAAA-3′) were used to target coding sequences, and successful knockout was confirmed using immunoblot.
2.4. Affymetrix human transcriptome array (HTA) and RNA seq analysis
Total RNAs were extracted using TRIzol (Invitrogen) and further purified using RNeasy MinElute Cleanup Kit (Qiagen). RNA samples were sent to the UCLA Clinical Microarray Core for Affymetix HTA or RNA seq analysis. Briefly, in Affymetix HTA assay, the samples were first fragmented and labeled with GeneChip WT Terminal Labeling Kit (Affymetrix), and the ssDNA was further hybridized onto Gene Chip Human Gene 1.0 ST Arrays (Affymetrix) according to the manufacturer's instructions. Finally the arrays were analyzed using Agilent Microarray System. For RNA seq analysis, the rRNA in each sample was depleted using Ribo-Zero Gold rRNA Removal Kit (Illumina) and further prepared for sequencing using the Ultra Directional RNA library Prep Kit (New England Biolabs). Then, the samples were run on a HiSeq 3000 Illumina Sequencing Platform as 50-bp single-end read runs. Herein, the results were sorted and indexed for quicker access using SAMtools, and the reading number of each gene was annotated with a custom in-house generated script.
2.5. Real-time quantitative reverse transcription PCR (qRT-PCR)
Total mRNA was extracted using TRIzol (Invitrogen), and 2 μg of RNA was used to synthesize cDNA as reported previously [30,31]. Herein, ACTB was used for all of qPCR normalization, and all experiments were performed in triplicate. The primer sequences (5′–3′) are as follows:
LOC344887-Primer 1-F 5′-CGTGAAAGCCTCTGATGGAGA-3′
LOC344887-Primer 1-R 5′-AGACGGCTGCTCCAATATCA-3′
LOC344887-Primer 2-F 5′-GGTTTTGGGGAAAGAAGTCC-3′
LOC344887-Primer 2-R 5′-CGACACATATTGGCTATTTCCTT-3′
COL1A1-F 5′-GAGGGCCAAGACGAAGACATC-3′
COL1A1-R 5′-CAGATCACGTCATCGCACAAC-3′
FN1–F 5′-GAGAATAAGCTGTACCATCGCAA-3′
FN1-R 5′-CGACCACATAGGAAGTCCCAG-3′
ACTA2-F 5′-CTATGAGGGCTATGCCTTGCC-3′
ACTA2-R 5′-GCTCAGCAGTAGTAACGAAGGA-3′
CDH1-F 5′-CGAGAGCTACACGTTCACGG-3′
CDH1-R 5′-GGGTGTCGAGGGAAAAATAGG-3′
CDH2-F 5′-AGCCAACCTTAACTGAGGAGT-3′
CDH2-R 5′-GGCAAGTTGATTGGAGGGATG-3′
NQO1-F 5′-ATGTATGACAAAGGACCCTTCC-3′
NQO1-R 5′-TCCCTTGCAGAGAGTACATGG-3′
ACTB-F 5′-CCCAGAGCAAGAGAGG-3′
ACTB-R 5′-GTCCAGACGCAGGATG-3′
2.6. Luciferase reporter gene assay
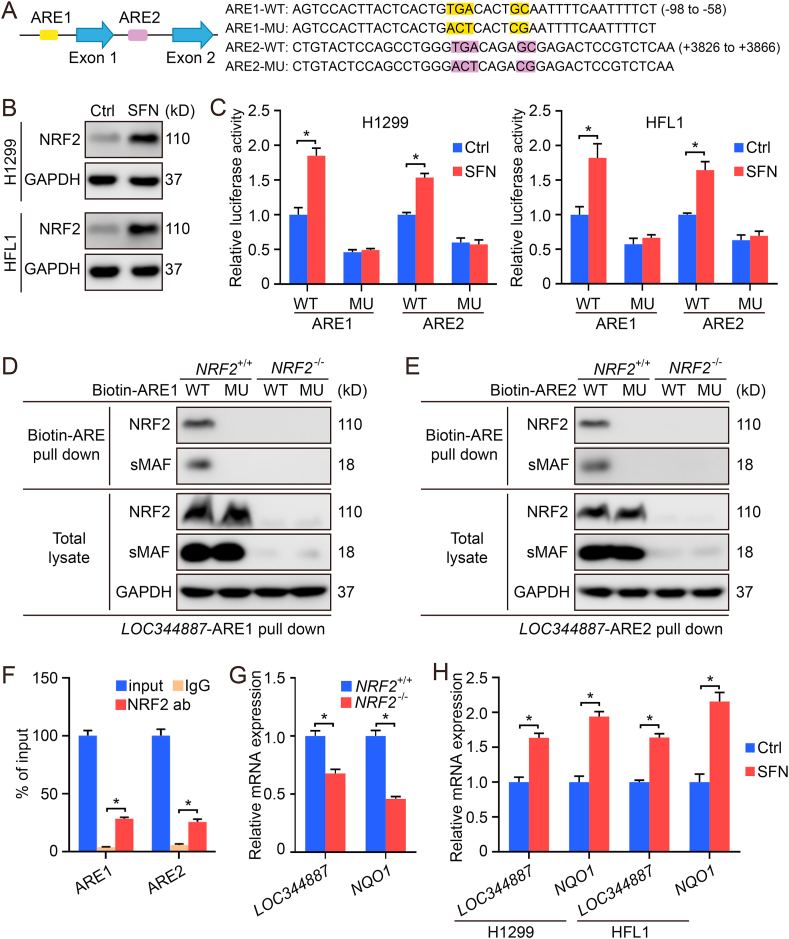
Both wild type and mutated 41-bp putative ARE-containing sequences (Fig. 1A) were cloned into the pGL4.22-luciferase vector, and transfected using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. In addition, luciferase activity was measured using the dual luciferase reporter assay system (Promega). For relative luciferase activity analysis, the value of Firefly-luciferase was normalized to the value of Renilla luciferase as reported previously [28,29].
Fig. 1.
LOC344887 is an NRF2 target gene. (A) The location and sequences of two putative AREs in LOC344887. MU indicates the highlighted nucleotides in the putative ARE were mutated. (B–C) H1299 and HFL1 cells transfected with ARE-firefly luciferase or TK-renilla luciferase vectors for 24 h were left untreated or treated with SFN (5 μM) for 16 h. The cells were harvested for immunoblot (B) and dual luciferase assay (C). (D–E) Biotinylated DNA probes (41bp) containing the LOC344887-ARE1-WT/MU (D) or LOC344887-ARE2-WT/MU (E) were incubated with lysates from NRF2+/+ or NRF2−/− A549 cells. The DNA-bound proteins were pulled down using streptavidin beads and measured by immunoblot analysis (D–E). (F) Chip-qPCR using A549 NRF2+/+ cells. Rabbit IgG was used as a negative control. (G–H) mRNA levels of LOC344887 and NQO1 were compared in NRF2+/+ vs. NRF2−/− cell lines (G), or in untreated vs SFN-treated NRF2+/+ cells. *: P﹤0.05 between two different groups.
2.7. Small interfering RNA (siRNA) transfection
Human LOC344887 siRNA (#5: SI04712344 and #8: SI05178838), human CDH2 siRNA (#5: SI02663927, #6: SI02757335, #10: SI04434612 and #11: SI04434619) and non-targeted control siRNA (SI03650318) were purchased from Qiagen. siRNA constructs were transfected using Hiperfect reagent (Qiagen) as per the manufacturer's protocol. Briefly, 1 × 105 cells/well were seeded into a 12-well plate and transfected with different siRNAs (10 nM) for 72 h. Finally, the knockdown efficiency was evaluated via qPCR or immunoblot.
2.8. Immunoblot analysis
In our study, all of the cell samples were harvested using 1X Sample buffer (100 mM DTT, 2% SDS, 10% glycerol, 0.1% bromophenol blue and 50 mM Tris-HCl pH = 8) and resolved by SDS-PAGE. The samples were further transferred onto a nitrocellulose membrane and blocked with 5% nonfat dried milk diluted in 1X PBS for 1 h. The membrane was then incubated with primary antibody overnight at 4 °C, followed by incubation with secondary antibody for 1 h at room temperature. Finally, the protein bands were visualized using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) on the C600 Imaging System (Azure Biosystems). The results in different groups were quantified using ImageJ 1. x (National Institutes of Health).
2.9. Chromatin immunoprecipitation assay (ChIP)-qPCR
The CHIP-qPCR assay (EZ-CHIPTM, Merck, Germany) was performed as per our previous study [29]. For qPCR detection, 1 μL of purified DNA was amplified using primers specific for the sequence flanking the AREs located in the LOC344887 promoter and first intron. For qPCR, the following primers were used:
LOC344887-ARE1-F 5′-CTCAAGCCTGGGTAACAAGAAC-3′
LOC344887-ARE1-R 5′-AACCCATTCCAATTACCACGCC-3′
LOC344887-ARE2-F 5′-TGAGGCAGGAGAATGGCGTG-3′
LOC344887-ARE2-R 5′-CTCACAAAAGTGAGAACGGTGC-3′
2.10. Biotin-DNA pull-down
Biotin-DNA pull-down was performed as reported previously [28,29]. Briefly, BEAS-2B cells were lysed using RIPA buffer containing 1 mM DTT (Golden Biotech), 1% protease inhibitor cocktail (Sigma-Aldrich) and 1 mM phenylmethylsulfonyl fluoride (PMSF, Thermo Fisher). Then the samples were precleared with streptavidin beads and further incubated with 2 μg of the biotinylated DNA probes overnight. The sequences of the different DNA probes are shown in Fig. 1A. Finally, the DNA–protein complexes were pulled down using streptavidin beads and subjected to immunoblot analysis.
2.11. Statistical analysis
The results a presented as means ± SD. Statistical analysis was performed using SPSS 17.0. Unpaired Student's t-tests were used to compare the means of two groups. One-way ANOVA with Bonferroni's correction was used to analyze the means of three or more groups. P < 0.05 was considered statistically significant.
3. Results
3.1. LOC344887 is an NRF2 target gene
To determine what lncRNAs might be regulated by NRF2 in lung cells, analysis of a Human Transcriptome Array comparing all transcripts in BEAS-2B NRF2+/+ vs. BEAS-2B NRF2−/− cell lines was performed. Interestingly, array analysis indicated a significant reduction in the expression of lncRNA (LOC344887) in NRF2 knockout cells compared to the wild type (data not shown). Computational analysis identified two putative AREs in the promoter region (−82GTGACACTGCA−72) and first intron (+3842GTGACAGAGCG+3852) of LOC344887. To verify the authenticity of these two putative AREs, both wild type (WT) and mutated (MU) ARE-containing 41-bp sequences (Fig. 1A) were cloned into the pGL4.22-luciferase vector. Human H1299 lung adenocarcinoma cells and HFL1 lung fibroblasts were transfected with the different ARE-reporter containing plasmids, and treated with DMSO (vehicle control) or SFN, an NRF2 activator, for 16 h. Immunoblot analysis indicated the induction of NRF2 by SFN (Fig. 1B), and relative luciferase activity was only increased in response to SFN in cells transfected with reporter plasmids containing ARE1/2-WT, but not ARE1/2-MU (Fig. 1C). Furthermore, NRF2-sMAF binding to both ARE1 (Fig. 1D) and ARE2 (Fig. 1E) was confirmed using biotinylated 41-bp ARE-containing DNA probes, as ARE1-WT (Fig. 1D) and ARE2-WT (Fig. 1E) were only able to pull down NRF2 and sMAF in NRF2+/+ cells, but not in NRF2−/− cells (Fig. 1D and E). Endogenous binding of NRF2 to ARE1 and ARE2 of LOC344887 was further confirmed using CHIP-qPCR (Fig. 1F). In addition, the transcript level of LOC344887 was much lower in NRF2−/− cells, compared to NRF2+/+ cells (Fig. 1G), and was enhanced significantly by SFN in both H1299 and HFL1 wild type cell lines (Fig. 1H). Importantly, the expression pattern of LOC344887 following loss of NRF2, or upon SFN treatment was similar to NQO1, a well-established NRF2 target gene (Fig. 1G and H). Collectively, these results demonstrate that there are two functional AREs located in the promoter and intron 1 region of LOC344887, which defines LOC344887 as an NRF2 target gene in lung cells.
3.2. LOC344887 knockout enhances the expression of fibrotic genes
A LOC344887 knockout (KO) A549 cell line was established using the CRISPR/Cas9 technique to insert a DNA sequence encoding the pac gene (for puromycin resistance), followed by a combined poly(A) and a transcriptional pause site (TPS) (Fig. 2A). Two primer pairs, one targeting upstream (Primer 1), and the other downstream (Primer 2), of the pac gene insertion site (Fig. 2A) were used for qRT-PCR analysis. It was confirmed that transcription of LOC344887 downstream of the insertion site (downstream of exon 3) did not occur in LOC344887-KO cells (Fig. 2B). RNA-seq analysis was performed to compare genome-wide mRNA expression changes in LOC344887-WT and LOC344887-KO cells. The results showed that 733 genes were upregulated and 316 genes were downregulated in LOC344887-KO compared with the LOC344887-WT cell line (Fig. 2C). Gene Ontology (GO) (Fig. 2D) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment (Fig. 2E) analyses indicated that deletion of LOC344887 resulted in overexpression of numerous genes involved in fibrogenic processes, including reorganization and production of ECM, ECM-receptor interactions, and dysregulation of the actin cytoskeleton (Fig. 2D and E). The KEGG pathway mapping indicated that the PI3K-AKT signaling pathway might be associated with the enhanced fibrogenic process (Fig. 2E). A heat map of transcripts that were significantly increased in LOC344887 knockout cells indicated enhanced expression of a number of genes involved in the fibrogenic process, such as ACTA2 (encoding alpha smooth muscle actin, a-SMA), COL1A1 (encoding the major component of type I collagen), and FN1 (encoding fibronectin, a high-molecular weight extracellular matrix protein that binds to integrins), among others (Fig. 2F).
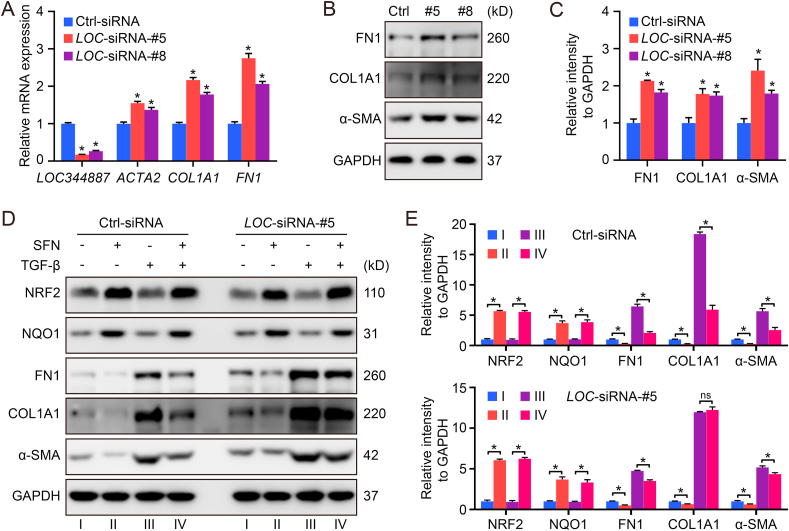
3.3. The antifibrotic function of SFN largely depends on LOC344887
To confirm the function of LOC344887 in controlling pulmonary fibrosis, LOC344887-siRNA was used to knockdown the LOC344887 transcript in HFL1 cells. Consistent with the RNA-seq data shown in Fig. 2, LOC344887 knockdown enhanced the expression of ACTA2/α-SMA, COL1A1, and FN1 at both the mRNA and protein level (Fig. 3A–C). Next, the contribution of LOC344887 on the previously reported therapeutic effects of SFN against pulmonary fibrosis was evaluated. To simulate fibrosis in vitro, the fibrotic activator TGF-β, was used. While TGF-β activated the expression of the fibrotic gene (FN1), it did not affect LOC344887 expression (Figs. S1A–S1B). In addition, SFN induced NRF2 expression and suppressed the expression of ACTA2/α-SMA, COL1A1 and FN1 in both basal and TGF-β-stimulated conditions. However, LOC344887 knockdown significantly limited the effect of SFN on the reduced expression of these proteins in both basal and TGF-β-stimulated conditions (Fig. 3D and E). It is worth mentioning that while the effect of SFN on the expression of these fibrogenic genes was much weaker in LOC344887 knockdown than Ctrl group, the induction of NQO1 by SFN was not affected by LOC344887 knockdown (Fig. 3D and E).
Fig. 3.
The antifibrotic function of SFN largely depends on LOC344887. (A–C) HFL1 cells transfected with siRNA against LOC344887 (10 nM) for 72 h were harvested for qRT-PCR (A), and immunoblot analysis (B, quantification in C). (D–E) HFL1 cells transfected with LOC-siRNA-#5 or Ctrl-siRNA (10 nM) for 72 h were treated with SFN (5 μM) or TGF-β (10 ng/ml) for 16 h. The cells were harvested for immunoblot analysis (D, quantification in E). *P < 0.05 between different groups.
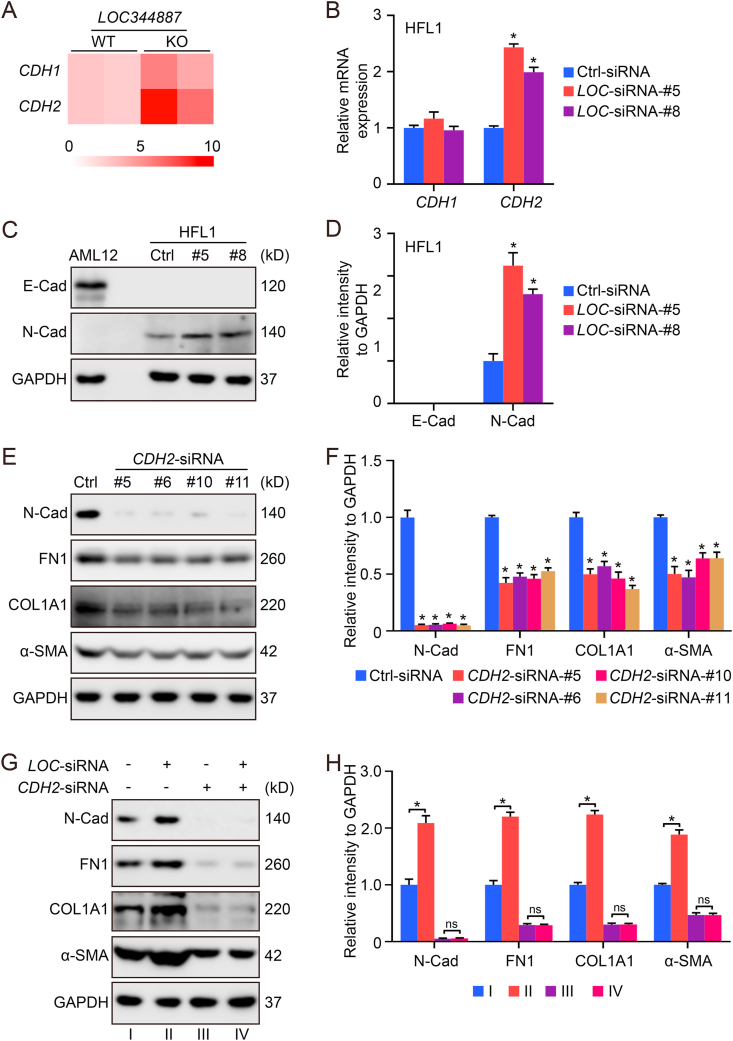
3.4. LOC344887 knockdown increases the expression of CDH2/N-cadherin
Another gene whose expression is upregulated by the knockout of LOC344887 is CDH2, a gene that encodes the transmembrane cell adhesion protein N-Cadherin (N-Cad) (Fig. 4A). SiRNA mediated knockdown of LOC344887 in HFL1 cells confirmed the transcriptional upregulation of CDH2, but not CDH1 (Fig. 4B). The protein level of N-Cad was also enhanced when LOC344887 was knocked down (Fig. 4C and D). Since N-Cad has been reported to be associated with EMT, a process known to contribute to lung fibrosis [32,33], the importance of N-Cad in fibrotic gene expression was tested. Knockdown of CDH2 resulted in a decreased expression of ACTA2/α-SMA, COL1A1, and FN1 (Fig. 4E and F). More importantly, the effect of LOC344887 on the expression of these genes could be abolished by CDH2 knockdown (Fig. 4G and H). Taken together, LOC344887 may modulate the expression of ACTA2/α-SMA, COL1A1, and FN1 through altering CDH2 gene expression.
Fig. 4.
LOC344887 knockdown increases the expression of CDH2/N-Cadherin. (A) A heat map showing the expression of CDH1 and CDH2 in A549 LOC344887-WT and LOC344887-KO cells. (B–D) HFL1 cells transfected with LOC-siRNA-#5/#8 (10 nM) for 72 h were harvested for qRT-PCR (B) and immunoblot analysis (C, quantification in D). Herein, AML12, a liver epithelial cell line, was used as a positive control for E-Cad detection. (E–F) HFL1 cells transfected with CDH2-siRNA-#5/#6/#10/#11 (10 nM) for 72 h were harvested for immunoblot analysis (E, quantification in F). (G–H) HFL1 cells co-transfected with LOC-siRNA-#5 and CDH2-siRNA-#5 (10 nM) for 72 h were harvested for immunoblot analysis (G, quantification in H).
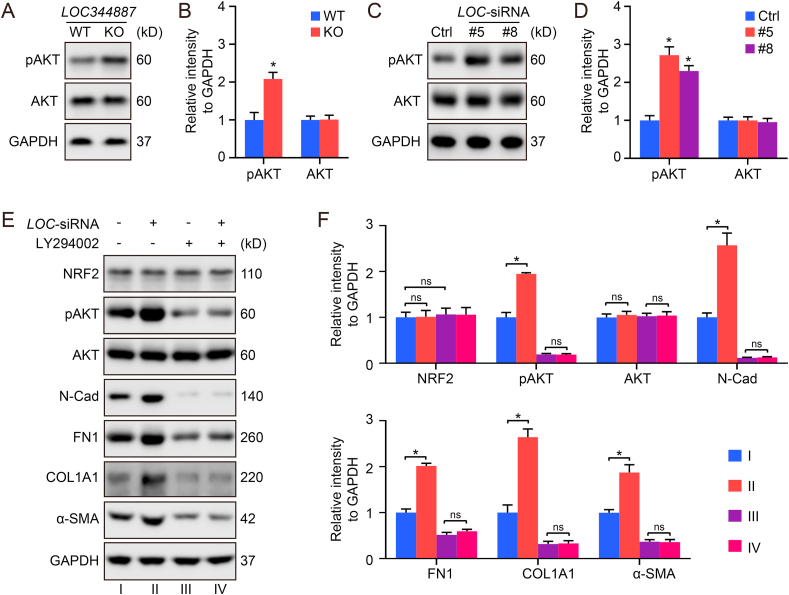
3.5. LOC344887 regulates the expression of CDH2/N-cad via the PI3K-AKT signaling pathway
KEGG pathway enrichment analysis indicated that the pathway most affected by LOC344887 knockout is the PI3K-AKT signaling pathway (Fig. 2E). Indeed, LOC344887 knockout in A549 cells (Fig. 5A and B) and knockdown in HFL1 cells (Fig. 5C and D) increased the level of phospho-AKT (Ser473), without altering the total level of AKT. To further confirm the requirement of PI3K-AKT signaling in mediating the antifibrogenic effects of loss of LOC344887, LY294002, a PI3K inhibitor was used. LY294002 not only decreased the level of phospho-AKT, N-Cad, FN1, COL1A1, and ACTA2/α-SMA, but also completely abolished the effect of LOC344887 in increasing these proteins (Fig. 5E and F). Thus, PI3K-AKT signaling appears to play a key role in the mediating the effect of LOC344887 on the expression of CDH2, FN1, COL1A1, and ACTA2/α-SMA.
Fig. 5.
LOC344887 negatively regulates the expression of CDH2/N-Cad via the PI3K-AKT signaling pathway. (A–B) A549 LOC344887-WT and LOC344887-KO cells were harvested for immunoblot analysis (A, quantification in B). (C–D) HFL1 cells transfected with LOC-siRNA-#5/#8 (10 nM) for 72 h were harvested for immunoblot analysis (C, quantification in D). (E–F) HFL1 cells transfected with LOC-siRNA-#5 (10 nM) and treated with LY294002 (30 μM) for 72 h were harvested for immunoblot analysis (E, quantification in F).
4. Discussion
IPF is a progressive and irreversible lung disease with an average life expectancy of only 3–5 years after diagnosis in untreated patients [34,35]. The current drugs used to treat pulmonary fibrosis slow down the progression of the disease and preserve lung function, but do not reverse or halt its progression. In addition, patients must balance the limited lung preserving effects and toxic side-effects associated with current drug regimes. Therefore, the development of better antifibrotic therapies, especially targeting early stages of pulmonary fibrosis, represents an unmet medical need. Targeting activation of the NRF2 pathway for IPF prevention or intervention has been investigated, and the therapeutic potential of NRF2 activators against pulmonary fibrosis has been demonstrated [[15], [16], [17], [18],20]. Also, the NRF2 activator SFN has recently been used in a phase II clinical trial to treat chronic obstructive pulmonary disease (NCT01335971).
Mechanistically, inhibition of EMT and TGF-β1/Smad signaling has been considered the major mechanism for the antifibrotic function of NRF2 [15,18,20]. In addition, treatment with SFN has been shown to suppress TGF-β1-induced EMT via decreased SNAIL protein levels, with SNAIL knockdown also abolishing the ability of SFN to reduce EMT and fibrosis [36]. These studies clearly indicate that NRF2 negatively regulates fibrogenic gene expression; however, which set of NRF2 targets is primarily responsible for reduced fibrogenesis is unclear. For example, it is unknown if the observed inhibition of EMT, compromised TGF-β1/Smad signaling, and decreased SNAIL expression in response to NRF2 activation is driven by antioxidant or anti-inflammatory NRF2 target genes. Interestingly, our study identified LOC344887 as an NRF2 target gene that was largely responsible for SFN-mediated antifibrotic activity in vitro. LOC344887 inhibited the expression of CDH2/N-cad via PI3K-AKT signaling pathway, which is essential for the progression of fibrosis, i.e. excess production of FN1, COL1A1, and ACTA2/α-SMA. This work provides a novel molecular insight into the antifibrotic function of NRF2 and highlights the importance of a specific NRF2 target gene, LOC344887, in suppressing pulmonary fibrosis.
LncRNAs have emerged as key regulators of many cellular processes, including chromatin remodeling, scaffolding, transcriptional and translational regulation, splicing modulation, and RNA degradation [37]. Proper function of lncRNAs has been demonstrated as a key mechanism controlling cellular homeostasis and is critical in the development of organisms [37]. Dysregulation of lncRNAs has been associated with many human diseases [37]. In this study, we demonstrated that a lncRNA, LOC344887, is an anti-fibrotic NRF2 target gene with two functional AREs (Fig. 1) that plays an important role in mediating fibrosis based on RNA-seq data analysis (Fig. 2). Functional analysis confirmed that LOC344887 negatively regulates fibrogenic gene expression (Fig. 3, Fig. 4). Importantly, the effects of SFN in reducing the expression of FN1, COL1A1, and ACTA2/α-SMA was largely abolished when LOC344887 was knocked down by siRNA (Fig. 3), indicating the significant contribution of this newly-discovered NRF2 target gene in controlling the fibrogenic process. At this point, it remains unclear if the antifibrogenic function of LOC344887 involves the modulation of antioxidant or anti-inflammatory pathways; however, the role of the NRF2-LOC344887 axis in mediating at least some aspects of the fibrotic stress response is clear. Another important finding of this study is that LOC344887 regulates the expression of these fibrotic genes via inactivation of PI3K-AKT signaling (Fig. 5). It is reasonable to assume an indirect effect of LOC344887 on the PI3K-AKT pathways, considering the wide spectrum of roles that lncRNAs play in regulating cellular signaling networks. The detailed molecular events that link LOC344887 upregulation to dephosphorylation of AKT, thus blunting PI3K-AKT signaling, warrant further investigation.
The role of LOC344887 in cancer development has been investigated previously. The expression of LOC344887 is upregulated in hepatocellular carcinoma compared with normal tissue [38]. In other studies, LOC344887 might function as an oncogene in gallbladder cancer and colon cancer, as high expression of LOC344887 is associated with larger tumor size and the downregulation of LOC344887 inhibited cell proliferation in gallbladder cancer cells and colon cancer cells [39,40]. These observations are in line with the “dark side” of NRF2 in cancer, i.e. certain cancer cells have constitutively high NRF2 levels, which increases growth rate and leads to resistance to anti-cancer drug treatment. Clinically, high expression of NRF2 in patient tumors is strongly correlated with a poor prognosis. NRF2 is highly expressed in many cancer types including: lung, head and neck, and bladder, due to somatic mutations in KEAP1 or NRF2 [41]. Furthermore, KEAP1 is mutated in lung cancer as frequently (>30%) as the tumor suppressor gene TP53 [12,42,43]. It would be interesting to dissect the degree of contribution of LOC344887 to the observed NRF2-dependent resistance and high proliferation in these cancer cells.
Recent recognition that a large fraction of the noncoding genome that was previously considered “junk” DNA actually encodes many lncRNAs, coupled with an increasing understanding of the diverse roles of IncRNAs in human disease, has created exciting new therapeutic opportunities [37]. Our study demonstrating that the antifibrotic activity of NRF2 is largely derived from this new NRF2 target gene LOC344887, provided an important new avenue for developing nucleic acid-based drugs to suppress fibrogenesis at early stages. However, studying LOC344887 in vivo has proved to be a challenge because LOC344887 is only found in the human genome, and a similar gene has not yet been identified in mice. If the mouse counterpart of the LOC344887 gene cannot be identified, a humanized mouse model might be desirable to study the in vivo function of LOC344887 in the future. At this point, we do not know if LOC344887 is expressed in a tissue-specific manner. If LOC344887 is also expressed in liver tissues, it would be interesting to investigate if LOC344887 also negatively regulates liver fibrosis. In addition, our study also indicates an effect of LOC344887 on the PI3K-AKT signaling pathway. Therefore, other diseases related with PI3K-AKT signaling, such as ischemia/reperfusion injury [44] and diabetes [45], could also be affected by LOC344887, along with cancer and fibrosis.
5. Conclusion
In this study, we have identified the lncRNA LOC344887 as an NRF2 target gene with two functional AREs in the promoter and the first intron. GO annotation and KEGG pathway enrichment implied an antifibrotic function of LOC344887. Using lung fibroblasts, we demonstrated that the antifibrotic function of NRF2 was largely dependent on LOC344887, since deletion or downregulation of LOC344887 enhanced expression of CDH2/N-cadherin and fibrotic genes and blunted the antifibrotic effects of SFN. Furthermore, the LOC344887-mediated downregulation of CDH2/N-cadherin and fibrotic genes was connected to the PI3K-AKT signaling pathway, as pharmacologic inhibition of PI3K activity blocked the effects of LOC344887 knockdown. Although the detailed mechanism and function of NRF2-LOC344887 signaling in pulmonary fibrosis requires further investigation, our study illustrates the distinct function of this axis in suppressing fibrosis, and opens a new therapeutic avenue for treating human pulmonary fibrosis or other fibrotic diseases.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the following NIH grants: R35 ES031575, P42 ES004940, and R01 DK109555 to DDZ, R01 ES031463 to EC and P01 HL126609, P01 HL134610, R42 HL152888, and R01 HL141387 to JGNG.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101766.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Weng T., Ko J., Masamha C.P., Xia Z., Xiang Y., Chen N.Y. Cleavage factor 25 deregulation contributes to pulmonary fibrosis through alternative polyadenylation. J. Clin. Invest. 2019;129:1984–1999. doi: 10.1172/JCI122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall D.C., Salciccioli J.D., Shea B.S., Akuthota P. Trends in mortality from idiopathic pulmonary fibrosis in the European Union: an observational study of the WHO mortality database from 2001-2013. Eur. Respir. J. 2018:51. doi: 10.1183/13993003.01603-2017. [DOI] [PubMed] [Google Scholar]
- 3.Spinello I.M., Munoz A., Johnson R.H. Pulmonary coccidioidomycosis. Semin. Respir. Crit. Care Med. 2008;29:166–173. doi: 10.1055/s-2008-1063855. [DOI] [PubMed] [Google Scholar]
- 4.Lechowicz K., Drozdzal S., Machaj F., Rosik J., Szostak B., Zegan-Baranska M. COVID-19: the potential treatment of pulmonary fibrosis associated with SARS-CoV-2 infection. J. Clin. Med. 2020;9:1917. doi: 10.3390/jcm9061917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George P.M., Wells A.U., Jenkins R.G. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir Med. 2020;8:807–815. doi: 10.1016/S2213-2600(20)30225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novelli L., Ruggiero R., De Giacomi F., Biffi A., Faverio P., Bilucaglia L. Corticosteroid and cyclophosphamide in acute exacerbation of idiopathic pulmonary fibrosis: a single center experience and literature review. Sarcoidosis Vasc. Diffuse Lung Dis. 2016;33:385–391. [PubMed] [Google Scholar]
- 7.Luppi F., Cerri S., Beghe B., Fabbri L.M., Richeldi L. Corticosteroid and immunomodulatory agents in idiopathic pulmonary fibrosis. Respir. Med. 2004;98:1035–1044. doi: 10.1016/j.rmed.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Li X.H., Xiao T., Yang J.H., Qin Y., Gao J.J., Liu H.J. Parthenolide attenuated bleomycin-induced pulmonary fibrosis via the NF-kappaB/Snail signaling pathway. Respir. Res. 2018;19:111. doi: 10.1186/s12931-018-0806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher T.M., Corte T.J., Fischer A., Kreuter M., Lederer D.J., Molina-Molina M. Pirfenidone in patients with unclassifiable progressive fibrosing interstitial lung disease: a double-blind, randomised, placebo-controlled, phase 2 trial. Lancet Respir Med. 2020;8:147–157. doi: 10.1016/S2213-2600(19)30341-8. [DOI] [PubMed] [Google Scholar]
- 10.Behr J., Bendstrup E., Crestani B., Gunther A., Olschewski H., Skold C.M. Safety and tolerability of acetylcysteine and pirfenidone combination therapy in idiopathic pulmonary fibrosis: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med. 2016;4:445–453. doi: 10.1016/S2213-2600(16)30044-3. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda S., Sekine A., Baba T., Yamakawa H., Morita M., Kitamura H. Hepatotoxicity of nintedanib in patients with idiopathic pulmonary fibrosis: a single-center experience. Respir Investig. 2017;55:51–54. doi: 10.1016/j.resinv.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Canc. Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidlin C.J., Dodson M.B., Madhavan L., Zhang D.D. Redox regulation by NRF2 in aging and disease. Free Radic. Biol. Med. 2019;134:702–707. doi: 10.1016/j.freeradbiomed.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T., Yamamoto M. Molecular basis of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2015;88:93–100. doi: 10.1016/j.freeradbiomed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z., Qu J., Zheng C., Zhang P., Zhou W., Cui W. Nrf2 antioxidant pathway suppresses Numb-mediated epithelial-mesenchymal transition during pulmonary fibrosis. Cell Death Dis. 2018;9:83. doi: 10.1038/s41419-017-0198-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walters D.M., Cho H.Y., Kleeberger S.R. Oxidative stress and antioxidants in the pathogenesis of pulmonary fibrosis: a potential role for Nrf2. Antioxidants Redox Signal. 2008;10:321–332. doi: 10.1089/ars.2007.1901. [DOI] [PubMed] [Google Scholar]
- 17.Chen W., Li S., Li J., Zhou W., Wu S., Xu S. Artemisitene activates the Nrf2-dependent antioxidant response and protects against bleomycin-induced lung injury. Faseb. J. 2016;30:2500–2510. doi: 10.1096/fj.201500109R. [DOI] [PubMed] [Google Scholar]
- 18.Wang T., Dai F., Li G.H., Chen X.M., Li Y.R., Wang S.Q. Trans-4,4'-dihydroxystilbene ameliorates cigarette smoke-induced progression of chronic obstructive pulmonary disease via inhibiting oxidative stress and inflammatory response. Free Radic. Biol. Med. 2020;152:525–539. doi: 10.1016/j.freeradbiomed.2019.11.026. [DOI] [PubMed] [Google Scholar]
- 19.Qu J., Zhang Z., Zhang P., Zheng C., Zhou W., Cui W. Downregulation of HMGB1 is required for the protective role of Nrf2 in EMT-mediated PF. J. Cell. Physiol. 2019;234:8862–8872. doi: 10.1002/jcp.27548. [DOI] [PubMed] [Google Scholar]
- 20.Zeng H., Wang Y., Gu Y., Wang J., Zhang H., Gao H. Polydatin attenuates reactive oxygen species-induced airway remodeling by promoting Nrf2-mediated antioxidant signaling in asthma mouse model. Life Sci. 2019;218:25–30. doi: 10.1016/j.lfs.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz S.U., Grote P., Herrmann B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Cheng Z., Dai L., Jiang T., Jia L., Jing X. Knockdown of long noncoding RNA H19 represses the progress of pulmonary fibrosis through the transforming growth factor beta/smad 3 pathway by regulating MicroRNA 140. Mol. Cell Biol. 2019;39 doi: 10.1128/MCB.00143-19. e00143-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian W., Cai X., Qian Q., Peng W., Yu J., Zhang X. lncRNA ZEB1-AS1 promotes pulmonary fibrosis through ZEB1-mediated epithelial-mesenchymal transition by competitively binding miR-141-3p. Cell Death Dis. 2019;10:129. doi: 10.1038/s41419-019-1339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C., Liang Y., Zeng X., Yang X., Xu D., Gou X. Long noncoding RNA FENDRR exhibits antifibrotic activity in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2020;62:440–453. doi: 10.1165/rcmb.2018-0293OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu P., Dodson M., Fang D., Chapman E., Zhang D.D. NRF2 negatively regulates primary ciliogenesis and hedgehog signaling. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P., Rojo de la Vega M., Sammani S., Mascarenhas J.B., Kerins M., Dodson M. RPA1 binding to NRF2 switches ARE-dependent transcriptional activation to ARE-NRE-dependent repression. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E10352–E10361. doi: 10.1073/pnas.1812125115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P., Tian W., Tao S., Tillotson J., Wijeratne E.M.K., Gunatilaka A.A.L. Non-covalent NRF2 activation confers greater cellular protection than covalent activation. Cell Chem Biol. 2019;26:1427–14235 e5. doi: 10.1016/j.chembiol.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao S., Liu P., Luo G., Rojo de la Vega M., Chen H., Wu T. p97 negatively regulates NRF2 by extracting ubiquitylated NRF2 from the KEAP1-CUL3 E3 complex. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00660-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian X., Anzovino A., Kim S., Suyama K., Yao J., Hulit J. N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene. 2014;33:3411–3421. doi: 10.1038/onc.2013.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotb A.M., Hierholzer A., Kemler R. Replacement of E-cadherin by N-cadherin in the mammary gland leads to fibrocystic changes and tumor formation. Breast Cancer Res. 2011;13:R104. doi: 10.1186/bcr3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolters P.J., Blackwell T.S., Eickelberg O., Loyd J.E., Kaminski N., Jenkins G. Time for a change: is idiopathic pulmonary fibrosis still idiopathic and only fibrotic? Lancet Respir Med. 2018;6:154–160. doi: 10.1016/S2213-2600(18)30007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins B.F., Raghu G. Antifibrotic therapy for fibrotic lung disease beyond idiopathic pulmonary fibrosis. Eur. Respir. Rev. 2019;28 doi: 10.1183/16000617.0022-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou W., Mo X., Cui W., Zhang Z., Li D., Li L. Nrf2 inhibits epithelial-mesenchymal transition by suppressing snail expression during pulmonary fibrosis. Sci. Rep. 2016;6:38646. doi: 10.1038/srep38646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin B., Wang W., Du G., Huang G.Z., Han L.T., Tang Z.Y. Identifying hub genes and dysregulated pathways in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2015;19:592–601. [PubMed] [Google Scholar]
- 39.Wu X.C., Wang S.H., Ou H.H., Zhu B., Zhu Y., Zhang Q. The NmrA-like family domain containing 1 pseudogene Loc344887 is amplified in gallbladder cancer and promotes epithelial-mesenchymal transition. Chem. Biol. Drug Des. 2017;90:456–463. doi: 10.1111/cbdd.12967. [DOI] [PubMed] [Google Scholar]
- 40.Johnson G.S., Li J., Beaver L.M., Dashwood W.M., Sun D., Rajendran P. A functional pseudogene, NMRAL2P, is regulated by Nrf2 and serves as a coactivator of NQO1 in sulforaphane-treated colon cancer cells. Mol. Nutr. Food Res. 2017;61 doi: 10.1002/mnfr.201600769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau A., Villeneuve N.F., Sun Z., Wong P.K., Zhang D.D. Dual roles of Nrf2 in cancer. Pharmacol. Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H., Wang L., Weng X., Chen H., Du Y., Diao C. Inhibition of Brd 4 alleviates renal ischemia/reperfusion injury-induced apoptosis and endoplasmic reticulum stress by blocking FoxO4-mediated oxidative stress. Redox Biol. 2019;24:101195. doi: 10.1016/j.redox.2019.101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J., Yang W., Chen Z., Chen J., Meng Y., Feng B. Long noncoding RNA lncSHGL recruits hnRNPA1 to suppress hepatic gluconeogenesis and lipogenesis. Diabetes. 2018;67:581–593. doi: 10.2337/db17-0799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.