Villin, encoded by Vil1, is an actin-binding protein expressed by intestinal, hepatobiliary, and renal tubular epithelial cells.1 Transgenic mice in which the Vil1 promoter drives Cre recombinase expression are widely used to inactivate genes in the intestinal epithelium. The 4 most commonly used Vil1Cre lines were generated by independent groups using distinct promoter constructs.2,3 Although these lines have been well-characterized in the intestine, their extraintestinal Cre activity has not been fully defined or compared.
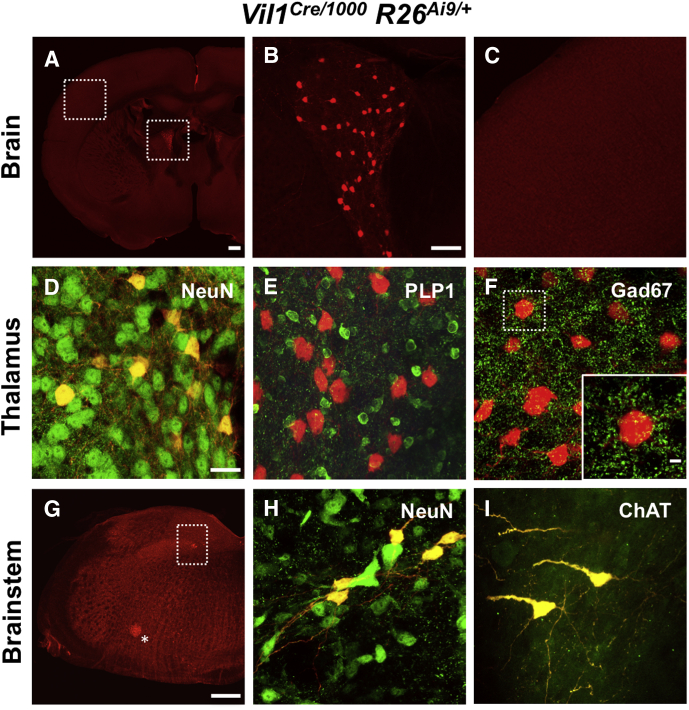
We first studied the Tg(Vil1-cre)997Gum and Tg(Vil1-cre)1000Gum transgenic lines in which a 12.4-kilobase region of the Vil1 promoter drives Cre expression.3 We crossed them with Rosa26-Ai9 reporter mice, in which Cre-mediated excision of a stop cassette leads to expression of fluorescent TdTomato (Supplementary Methods).4 Both the Vil1Cre/997 and Vil1Cre/1000 lines exhibited Cre activity in epithelial cells throughout the small intestine and colon, as previously described. Unexpectedly, Cre activity was also evident in specific central nervous system (CNS) nuclei: the anterodorsal thalamic nucleus (important for spatial navigation) and 2 small groups of cholinergic neurons in the medulla (Figure 1). All TdTomato+ cells in these nuclei co-localized with the neuronal marker NeuN, and none expressed the oligodendrocyte marker PLP1 (Figure 1). Consistent with these observations, Vil1 transcripts were reported in a subset of thalamic excitatory neurons at postnatal day 215 but not in the adult mouse CNS (Allen Mouse Brain Atlas),6 suggesting at least transient expression in these neurons.
Figure 1.
The Vil1 promoter drives Cre recombinase expression in the CNS. (A–C) Coronal section from a Vil1Cre/1000 Rosa26Ai9/+ mouse brain shows TdTomato expression in cells within a periventricular region of the forebrain consistent with the anterodorsal thalamic nucleus (A and B) but not in the cerebral cortex (A and C). (D–F) Immunohistochemical staining of a Vil1Cre/1000 Rosa26Ai9/+ mouse brain shows that all TdTomato+ neurons within the anterodorsal thalamus (AD) co-localize with the pan-neuronal marker NeuN (D) but not the glial marker PLP1 (E). Thirty-eight percent ± 15% of NeuN+ neurons in the AD thalamus expressed TdTomato (mean ± standard deviation, n = 3 animals). TdTomato+ neurons are not immunoreactive for GAD67 but are contacted by many GAD67+ puncta, likely the terminals of GABAergic inhibitory neurons (F). (E) is from a mouse that was also hemizygous for the PLP1eGFP transgene. (G–I) Cross sections of brainstem from Vil1Cre/1000 Rosa26Ai9/+ mice show TdTomato expression in the dorsal and ventral medulla. All TdTomato+ cells co-localized with NeuN and ChAT, the biosynthetic enzyme for acetylcholine (shown in H and I for dorsal medulla and boxed region in G). TdTomato+ neurons in the dorsal medulla localized to the nucleus of the solitary tract (boxed region in G) and those in the ventral medulla to the nucleus ambiguus (asterisk in G). Scale bars for A and G = 400 μm; B and C = 75 μmol/L; D–F, H, and I = 25 μm.
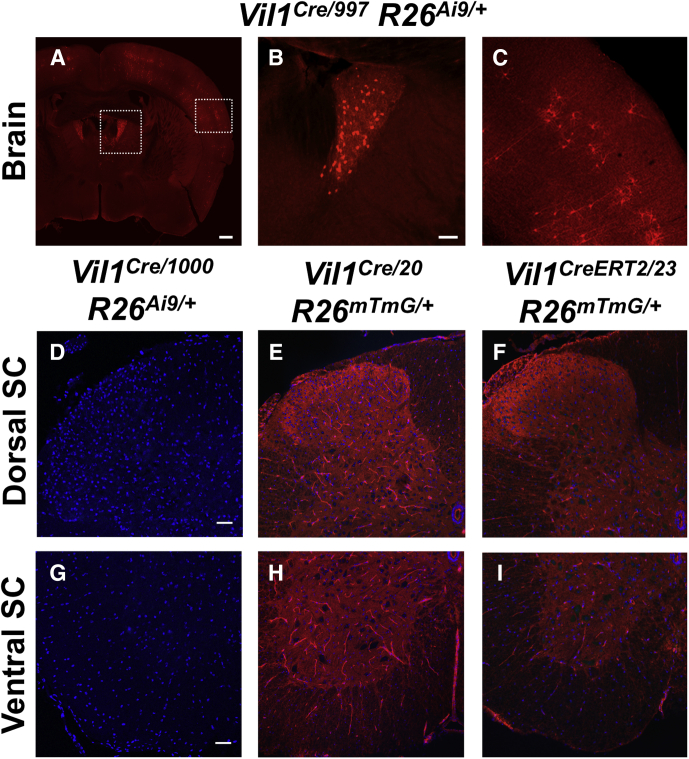
The Vil1Cre/997 line also drove patchy Cre-mediated recombination throughout the cerebral cortex (Supplementary Figure 1). In contrast, Vil1Cre/1000 mice had no detectable TdTomato in cells in the cerebral cortex (Figure 1) or spinal cord (Supplementary Figure 1) or within intrinsic and extrinsic nerve fibers innervating the gut (Supplementary Figure 2). Therefore, we focused further characterization on these mice. Vil1Cre/1000 mice exhibited Cre-dependent reporter expression in small areas of the exocrine pancreas, rare epithelial cells within the glandular stomach (consistent with prior studies7), and some proximal tubules in the kidney (Supplementary Figures 2 and 3, Table 1). Patchy reporter expression was also detected in oviduct epithelium, but not the uterus, ovary, or testis (Supplementary Figures 3 and 4).
Supplementary Figure 1.
Vil1 promoter-driven Cre recombinase expression is present in the brain but undetectable in the spinal cord. (A–C) Coronal sections of brain tissue from an adult Vil1Cre/997 R26Ai9/+ mouse show Cre-dependent TdTomato reporter expression in the anterodorsal thalamic nucleus (A and B) and patchy recombination throughout the cerebral cortex (A and C). Scale bars for A = 400 μm; B and C = 100 μm. (D–I) No Cre-dependent reporter expression was observed in cross sections of dorsal or ventral horn spinal cord (SC) tissue from adult Vil1Cre/1000 R26Ai9/+, Vil1Cre/20 R26mTmG/+, or Vil1CreERT2/23 R26mTmG/+ mice. Scale bars = 50 μm.
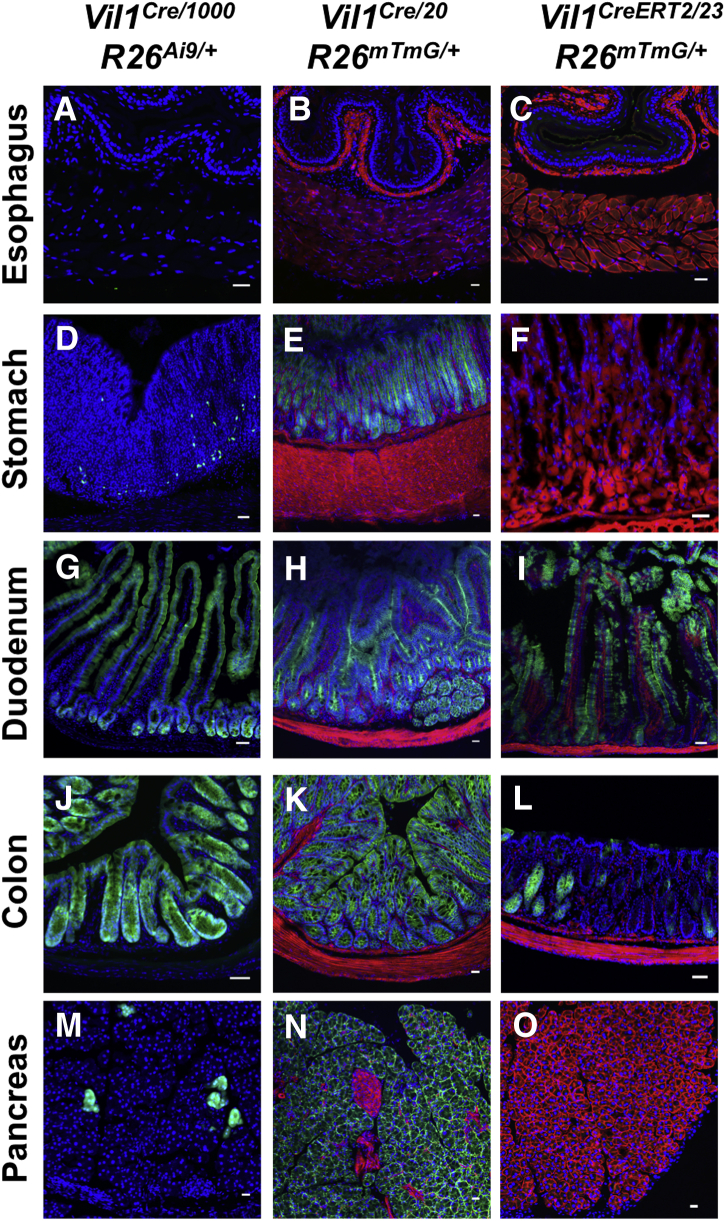
Supplementary Figure 2.
Vil1 promoter-driven Cre recombinase expression in the digestive tract varies between transgenic lines. (A, D, G, J, M) Cross sections of tissue from an adult Vil1Cre/1000 R26Ai9/+ mouse show Cre-dependent TdTomato reporter expression (pseudo-colored green) in rare epithelial cells of the glandular stomach, throughout the epithelia of the small and large intestines, and in isolated exocrine cells of the pancreas. No reporter expression was detected in the esophagus. (B, E, H, K, N) Cross sections of tissue from an adult Vil1Cre/20 R26mTmG/+ mouse show Cre-dependent GFP reporter expression (green) throughout the epithelia of the glandular stomach, small intestine, and colon. Extensive GFP expression was also evident throughout the pancreatic parenchyma, except in blood vessels and islets (which retain TdTomato fluorescence [red] and are morphologically distinct). No GFP was detected in the esophagus. (C, F, I, L, O) Cross sections of tissue from an adult Vil1CreERT2/23 R26mTmG/+ mouse 7 days status post a 5-day course of tamoxifen show Cre-dependent GFP reporter expression (green) in subsets of epithelial cells in the small and large intestines. No GFP was detected in the esophagus, stomach, or pancreas. Scale bars = 25 μm.
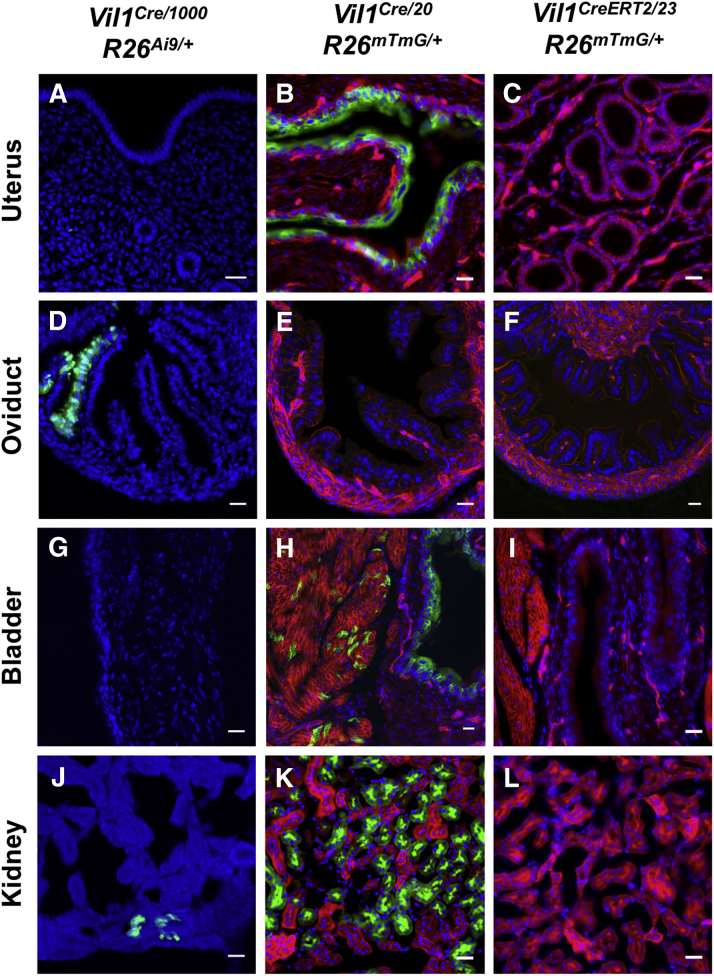
Supplementary Figure 3.
Vil1 promoter-driven Cre recombinase expression in the bladder, kidney, and female reproductive organs varies between transgenic lines. (A–C) Cre-dependent GFP reporter expression was detected throughout the uterine epithelium in some Vil1Cre/20 R26mTmG/+ mice (such as example shown here), but not the other 2 lines examined. (D–F) Cre-dependent TdTomato reporter expression (pseudo-colored green) is seen in patchy areas of the oviduct epithelium in Vil1Cre/1000 R26Ai9 mice, but not the other 2 lines. (G–I) Cre-dependent GFP reporter expression was detected throughout the bladder epithelium in Vil1Cre/20 R26mTmG/+ mice, but not the other 2 lines. (J–L) Cre-dependent reporter expression was seen in scattered cells in the kidneys of Vil1Cre/1000 R26Ai9 mice and was extensive throughout the renal proximal tubular epithelium in Vil1Cre/20 R26mTmG/+ mice. No reporter expression was detected in Vil1CreERT2/23 R26mTmG/+ mouse kidney. Scale bars = 25 μm.
Table 1.
Cre-Dependent Reporter Expression in Tissues From Vil1 Promoter-Driven Transgenic Mouse Lines
| Vil1Cre/1000 | Vil1Cre/20 | Vil1CreERT2/23 | |
|---|---|---|---|
| Central nervous system | |||
| Brain | Rare neurons | – | – |
| Spinal cord | – | – | – |
| Alimentary tract | |||
| Tongue | – | – | – |
| Esophagus | – | – | – |
| Stomach (nonglandular) | – | – | – |
| Stomach (glandular) | Rare | Extensive | – |
| Small intestine | Extensive | Extensive | Patchy |
| Large intestine | Extensive | Extensive | Patchy |
| Pancreas | Scattered | Extensive | – |
| Liver | – | Scattered | – |
| Urogenital | |||
| Kidney | Scattered | Extensive | – |
| Bladder | – | Extensive | – |
| Testis | – | Scattered | Scattered |
| Ovary | – | – | – |
| Oviduct | Scattered | – | – |
| Uterus | – | +/– | – |
| Other | |||
| Heart | – | – | – |
| Lung | – | – | – |
| Spleen | – | Scattered | – |
| Adrenal | – | – | – |
NOTE. – indicates reporter expression was not detected. +/– indicates patchy reporter expression was detected in some, but not all, animals.
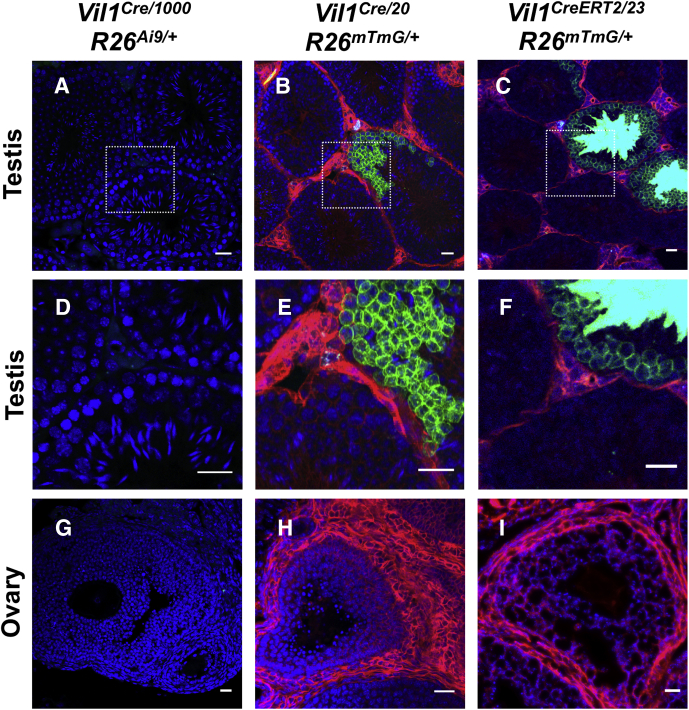
Supplementary Figure 4.
The Vil1 promoter drives Cre recombinase expression in the male, but not female, gonads in some transgenic lines. (A, D, G) Cross sections of tissue from adult Vil1Cre/1000 R26Ai9/+ mice show no evident Cre-dependent TdTomato reporter expression (pseudo-colored green) in the testis or the ovary. (B, E, H) Cross sections of tissue from adult Vil1Cre/20 R26mTmG/+ mice show Cre-dependent GFP reporter expression (green) in subset of seminiferous tubules of the testis but no expression in the ovary. (C, F, I) Cross sections of tissue from adult Vil1CreERT2/23 R26mTmG/+ mice 7 days after tamoxifen show Cre-dependent GFP reporter expression (green) in subset of seminiferous tubules of the testis but none in the ovary. Boxed regions in A–C are shown at higher magnification in D–F. Scale bars = 25 μm.
We next examined Tg(Vil1-cre)20Syr and Tg(Vil1-cre/ERT2)23Syr, transgenic lines in which a 9-kilobase region of the Vil1 promoter drives Cre expression.2 These lines were crossed with Rosa26-mTmG reporter mice in which Cre activity terminates TdTomato expression and activates green fluorescent protein (GFP) expression.8 Vil1Cre/20 mice exhibited Cre activity in epithelia throughout the small and large intestines (Supplementary Figure 2), as previously described. Cre activity was also detected in the exocrine pancreas and renal proximal tubules to a much greater extent than in Vil1Cre/1000 mice (Supplementary Figures 2 and 3). Unlike Vil1Cre/1000 mice, Vil1Cre/20 mice exhibited diffuse Cre activity in epithelia of the glandular stomach and bladder and patchy activity in liver and spleen (Supplementary Figures 2 and 3, Table 1). Cre activity was also evident in the uterine epithelium of some females (Supplementary Figure 3). There was no GFP expression in oviduct or ovary, but germ cell expression was observed within a subset of seminiferous tubules in the testis in all males, raising the possibility of germline recombination (Supplementary Figure 4).
The Vil1CreERT2/23 line is unique among the 4 lines studied because it expresses a Cre recombinase–estrogen receptor fusion protein that requires tamoxifen to trigger nuclear translocation and recombination.2 Vil1CreERT2/23 Rosa26mTmG/+ mice were administered tamoxifen daily for 5 doses and then analyzed 7 days after the last dose. Reporter expression was diffuse in the small intestinal epithelium and patchy in the colon (Supplementary Figure 2), as previously described. In contrast to other lines, Cre activity was not detected in the stomach, pancreas, kidney, or female reproductive tract (Supplementary Figure 2, Supplementary Figure 3, Supplementary Figure 4). However, there was reporter expression in germ cells within some seminiferous tubules (Supplementary Figure 4). Neither this line nor the Vil1Cre/20 line exhibited any Cre activity in the nervous system.
These data show that the 4 most commonly used Vil1Cre mouse lines are nonequivalent and have distinct and unexpected sites of extraintestinal Cre expression. Therefore, the Vil1Cre line to be used for any given experiment should be chosen carefully on the basis of the gene of interest and the potential for “off-target” effects outside the gut. The extensive Cre activity in Vil1Cre/997 mouse brains provides particularly strong rationale for selecting Vil1Cre/1000 or Vil1Cre/20 lines as alternatives for most studies. Conversely, although Cre activity within the exocrine pancreas, glandular stomach, and bladder of Vil1Cre/20 mice might be useful for some experiments, it is an important confounding variable in studies where gene deletion in these organs could impact the interpretation of observations attributed to gene functions in the intestinal epithelium. The Vil1CreERT2/23 line was the most selective for the intestinal epithelium, but like any inducible Cre system, it exhibited relatively less recombination than the constitutively active Cre lines. Importantly, both Vil1CreERT2/23 and Vil1Cre/20 lines exhibited Cre activity in a subset of male germ cells, raising concern that when these lines are used to delete genes, some progeny could have globally null alleles. One limitation of our study is that 2 different Cre-reporter lines were used, although both targeted the Rosa26 locus. Recombination efficiency at different loxP sites and genomic loci can vary.9 Thus, although our findings provide guidance on Cre activity in these 4 Vil1 transgenic lines, it is possible that this activity may manifest differently in the context of other floxed alleles.
Our findings highlight the necessity of rigorously characterizing transgenic lines in all studies to ensure accurate data interpretation and reproducibility. Just as some Cre lines presumed to be selective for the nervous system were later found to be expressed in the germline or gut epithelium,9,10 our data show that Cre lines commonly used to study intestinal epithelial biology can be active in the nervous system and other tissues that might affect gut functions. These observations do not detract from the value of these genetic tools, but they do emphasize the need for careful reagent choice, data interpretation, and methods reporting in all studies using these mice.
Acknowledgments
This work used core facilities supported by the Harvard Digestive Disease Center (NIH P30DK034854) and the Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center (NIH 1U54HD090255). The authors thank L. Feinstein and C. A. Hertz for technical assistance.
CRediT Authorship Contributions
Michael Rutlin, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Lead; Investigation: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Daniella Rastelli (Data curation: Equal; Investigation: Equal; Writing – review & editing: Supporting)
Wei-Ting Kuo, PhD (Conceptualization: Supporting; Data curation: Equal; Investigation: Equal; Writing – review & editing: Supporting)
Jason A. Estep (Investigation: Supporting; Writing – review & editing: Supporting)
Adriell Louis (Investigation: Supporting; Writing – review & editing: Supporting)
Martin M. Riccomagno, PhD (Funding acquisition: Supporting; Investigation: Supporting; Writing – review & editing: Supporting)
Jerrold R. Turner, MD, PhD (Conceptualization: Equal; Formal analysis: Supporting; Funding acquisition: Equal; Writing – review & editing: Equal)
Meenakshi Rao, MD, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Lead; Funding acquisition: Equal; Investigation: Equal; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest This author discloses the following: J. R. Turner is a co-founder of Thelium Therapeutics. The remaining authors disclose no conflicts.
Supplementary Methods
Mouse Lines
Mice were housed in a facility with a 12-hour dark cycle and handled in accordance with protocols approved by the Animal Care and Use Committees of Boston Children’s Hospital and Brigham and Women’s Hospital. Hemizygous Tg(Vil1-cre)1000Gum (JAX 021504; Jackson Laboratory, Bar Harbor, ME) and Tg(Vil1-cre)997Gum (JAX 004586) mice were bred with the homozygous Rosa26-Ai9 reporter mice (JAX 007909). Hemizygous Tg(Vil1-cre)20Syr (JAX 033019) and Tg (Vil1-cre/ERT2)23Syr (JAX 020282) mice, gifts of S. Robine, were bred with homozygous Rosa26-mTmG reporter mice (JAX 007676). Cre-positive male and female progeny from these crosses were studied at 6–10 weeks of age. Vil1Cre/1000 Rosat26Ai9/+ mice that were hemizygous for the PLP1eGFP transgene (JAX 033357) were analyzed for data shown in Figure 1E. Vil1CreERT2/23 Rosat26mTmG/+ mice were administered 1 mg tamoxifen (Cayman Chemical 13258; Ann Arbor, MI) dissolved in 100 μL corn oil by intraperitoneal injection once daily for 5 consecutive days, and tissues were collected 7 days after the last dose of tamoxifen. All reported results were observed and confirmed in a minimum of 3 mice per genotype.
Tissue Isolation and Processing
Mice were euthanized by CO2 inhalation and then underwent intracardiac perfusion of phosphate-buffered saline (PBS) followed by 4% paraformaldehyde in PBS. All tissues were collected and post-fixed with 4% PFA/PBS at 4°C for 16 hours. Brain and brainstem tissues were cut into 100- to 120-μm sections using a Leica VT1000 S vibratome (Wetzlar, Germany). All other tissues were equilibrated in 30% sucrose/PBS overnight, embedded in Tissue-Tek O.C.T. compound (Sakura Finetek 4583; Torrance, CA), and cut into 14-μm frozen sections by using a Leica CM1950 cryostat.
Immunohistochemical Staining and Imaging
Free-floating, 100- to 120-μm brain sections were washed 6 times with PBS with 0.5% Triton X-100 (PBST) for 20 minutes per wash and then incubated with primary antibodies for 24–72 hours with gentle rocking. Sections were washed again 6 times with PBST and then incubated with secondary antibodies with gentle rocking. Samples were then washed 6 times with PBST and mounted with Vectashield (Vector Laboratories H-1200; Burlingame, CA) or Fluoromount Aqueous Mounting media (Sigma-Aldrich, St Louis, MO). Primary and secondary antibodies were diluted in 5% heat inactivated normal goat serum and 20% dimethyl sulfoxide in PBST. For brain sections, visualization of TdTomato and GFP reporter fluorescence was achieved by using rabbit anti-DsRed (1:500; Takara cat. no. 632496; St. Louis, MO) and chicken anti-GFP (1:1000, Aves GFP-1020; Tigard, OR) with goat anti-rabbit 594 (1:500; Invitrogen, Carlsbad, CA) and goat anti-chicken 488 (1:500; Invitrogen) secondary antibodies, respectively. For non-brain tissues, we found no difference in reporter expression in 14-μm frozen sections whether reporter proteins were detected by endogenous fluorescence or by immunohistochemical staining as previously described.1 The data reported in Supplementary Figure 1, Supplementary Figure 2, Supplementary Figure 3, Supplementary Figure 4 and Table 1 for non-brain tissues reflect endogenous reporter fluorescence without any amplification introduced by immunohistochemical staining. Images of brain sections in Figure 1A–C and G and Supplementary Figure 1A–C were acquired by using a Zeiss LSM880 (Oberkochen, Germany) microscope with Fast Airyscan and represent tiled composites of images taken at a single z-plane with 10% overlap. For Figure 1D–F, H, and I, primary antibodies used were mouse anti-NeuN (1:1000, Millipore-Sigma MAB377; Burlington, MA), mouse anti-GAD67 (1:500, Millipore-Sigma MAB5406), and goat anti-ChAT (1:500, Millipore-Sigma AB144P). Confocal images were taken by using a Leica SPE II microscope. For cell counts, the ImageJ cell counter plugin was used.2 All other images are single planar images or maximum intensity projections of z-stacks, as detailed in the figure legends, and were acquired by using either a Zeiss LSM700 or Zeiss LSM880 microscope.
All authors had access to the study data and approved the final manuscript.
References
- 1.Maunoury R. Development. 1992;115:717–728. doi: 10.1242/dev.115.3.717. [DOI] [PubMed] [Google Scholar]
- 2.el Marjou F. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 3.Madison B.B. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 4.Madisen L. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeisel A. Cell. 2018;174:999–1014 e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lein E.S. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 7.Qiao X.T. Gastroenterology. 2007;133:1989–1998. doi: 10.1053/j.gastro.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muzumdar M.D. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 9.Luo L. Neuron. 2020;106:37–65.e5. doi: 10.1016/j.neuron.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter D.V. eNeuro. 2018;5 doi: 10.1523/ENEURO.0077-18.2018. ENEURO.0077-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Rao M., Nelms B.D., Dong L., Salinas-Rios V., Rutlin M., Gershon M.D., Corfas G. Enteric glia express proteolipid protein 1 and are a transcriptionally unique population of glia in the mammalian nervous system. Glia. 2015;63:2040–2057. doi: 10.1002/glia.22876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Vos K. Cell counter plugin. 2001. https://imagej.nih.gov/ij/plugins/cell-counter.html Available at: Accessed March 2020.