Abstract
韦荣球菌是口腔生物膜的早期定植菌之一,高丰度分布于口腔微生态中。目前在口腔中已检出7种韦荣球菌,其在不同部位、不同患者口腔中存在分布差异。近年来研究发现,韦荣球菌与口腔疾病的关系密切,韦荣球菌有助于变异链球菌的黏附且能分解链球菌代谢产生的乳酸,是被公认的致龋因素之一;韦荣球菌为牙龈卟啉单胞菌提供黏附位点,并可通过促进免疫反应参与牙周病的发生发展;韦荣球菌脂多糖的致病性及代谢产生H2S也与牙髓及根尖周病、口臭等相关。韦荣球菌与疾病的相关性已有研究进行阐释,与相关致病菌的作用机制也有一定进展,但其影响口腔疾病发生发展的分子机制仍不明确,本文就韦荣球菌与龋病、牙周病等口腔感染性疾病的研究进展作一综述。
Keywords: 韦荣球菌, 龋病, 牙周病, 口腔疾病
Abstract
Veillonella species, known as the early colonizer of oral biofilm, are prevalent in oral microbiota. Seven Veillonella species have been isolated from oral cavity. Their distribution varies not only with different people but also with different sites in the oral cavity. Oral Veillonella are associated with oral diseases. They contribute to the adhesion of Streptococcus mutans and consume the lactate generated by streptococci. Veillonella species play an important role in the occurrence and development of periodontal diseases by providing adhesion sites for Porphyromonas gingivalis and boosting immune responses. The production of lipopolysaccharide and H2S is related to other oral diseases, such as pulpitis, periapical periodontitis, and halitosis. Several studies have been conducted on the relationship between Veillonella and oral diseases and the interaction between Veillonella and other pathological microorganisms, but limited knowledge is available at the molecular level. This article reviews the research progress in the relationship between Veillonella and oral infectious diseases, such as dental caries and periodontal diseases.
Keywords: Veillonella, dental caries, periodontal diseases, oral diseases
1898年,法国微生物学家Veillon和Zuber从人的感染阑尾中首次分离出一种革兰阴性专性厌氧球菌,1933年,Prévot将之命名为“Veillonella parvula”,之后韦荣球菌属下的各韦荣球菌种被科学家们陆续分离得到。韦荣球菌属是口腔微生态的重要成员之一,除了口腔,韦荣球菌也存在于人类上呼吸道、肠道和阴道等位置。有研究[1]显示,健康成人唾液、牙菌斑中韦荣球菌丰度仅次于链球菌。目前报道过的韦荣球菌共有14种,其中有7种可从人口腔中分离得到:小韦荣菌(Veillonella parvula,V. parvula)、殊异韦荣菌(Veillonella dispar,V. dispar)、非典型韦荣菌(Veillonella atypica,V. atypica)、Veillonella rogosae、Veillonella denticariosi、Veillonella tobetsuensis、Veillonella infantium[2]–[5]。由于具有相似的生化特性和表型特征,韦荣球菌种类的鉴别目前主要是基于16S rDNA、dnaK、rpoB和gltA等基因的差异[6]–[7]。在人口腔中,韦荣球菌主要分布于唾液、舌黏膜及颊黏膜、牙龈及龈沟,不同种韦荣球菌的分布存在一定差异。Doel等[8]分析样本后发现了V. atypica的检出率为舌面>唾液>牙菌斑,V. dispar的检出率为舌面>唾液,V. parvula在牙菌斑中检出率最高。V. dispar主要在口腔卫生指标良好或社会经济地位高者口腔内检出[9];V. parvula在儿童中检出最频繁,而严重的早发性孩童期龋齿与V. atypica有关[10];牙周袋中V. parvula的检出率比在牙龈沟中更高,与慢性牙周炎有关[11]。地区、年龄、吸烟、饮食、口腔卫生及患病情况等影响因素也会造成菌种分布的差异[12]–[14]。韦荣球菌不能代谢碳水化合物和多元醇,但能利用短链有机酸,特别是利用乳酸作为能源,转化为酸性较弱的乙酸和丙酸。口腔韦荣球菌因其代谢乳酸可降低pH的特性曾被认为是防龋的益生菌,后多项研究[15]–[16]表明,口腔韦荣球菌与龋病密切相关,可协同变异链球菌(Streptococcus mutans,S. mutans)致龋。除此之外,口腔韦荣球菌与牙髓、根尖周感染以及慢性牙周炎等口腔疾病也有一定关系。因此,韦荣球菌属在口腔微生态中的作用及其与口腔疾病的相关性不容忽视。
1. 韦荣球菌与龋病
龋病是在以细菌为主的多种因素影响下,牙体硬组织发生慢性进行性破坏的一种疾病,链球菌属为常见的致龋微生物之一。链球菌可分解碳水化合物产酸,使牙面局部pH下降造成脱矿而致龋,因此能利用其他细菌产生的乳酸而减少牙脱矿的韦荣球菌与龋病的关系值得探讨。
1.1. 韦荣球菌与龋病相关性的研究
关于韦荣球菌属在龋病与非龋病人群口腔中的分布情况仍存在争议。有关于根面龋、乳牙龋等的研究[10],[17]认为,韦荣球菌属在龋病组中的丰度和检出率高于无龋组,是龋病的优势菌之一;然而有研究[18]–[19]则得出相反结论,也有实验[20]–[21]证明,其丰度无显著差别。Tanner等[21]则发现,V. parvula在严重儿童龋的样本中检出率显著高于无龋组,而V. dispar在无龋组的检出率显著高于严重儿童龋组。综上,近来的研究[22]更倾向于认为,韦荣球菌属在龋病发生发展中有重要作用,其与链球菌属在生物膜形成过程中的形态结构、生长代谢、细菌间共聚集及其他黏附现象、环境因素等方面的相互作用不可忽视。Guggenheim等[23]观察到在生物膜形成过程中,V. dispar发生了形态上的转变,由在整个生物膜中稀疏散布的单个球菌和大多数小球状小菌落,到不存在单细胞且大多数小菌落是细长细胞桥连接而成的大扁圆结构。Kara等[24]发现,S. mutans及V. parvula双菌种生物膜中的细菌会聚集成距离小于1.2 µm的空间排列。在口腔内唾液流动的环境中,链球菌的黏附为韦荣球菌提供了位点,当链球菌不存在,韦荣球菌则不能附着并生长[25]。许多研究[15],[25]–[26]也通过链球菌与韦荣球菌的双菌种生物模型反复验证了,韦荣球菌的存在能促进S. mutans、戈登链球菌(Streptococcus gordonii,S. gordonii)及唾液链球菌(Streptococcus salivarius,S. salivarius)生物膜的形成,而与口腔链球菌(Streptococcus oralis,S. oralis)及血链球菌(Streptococcus sanguinis,S. sanguinis)的生长无关甚至抑制。
1.2. 韦荣球菌与链球菌的相互作用
S. mutans、S. gordonii、韦荣球菌这3种菌种的相互作用涉及多个层面(图1)。S. gordonii可以通过产生H2O2抑制S. mutans的生长,而S. mutans产生细菌素抑制S. gordonii[27],V. parvula参与的生物膜形成则能减少H2O2对S. mutans的抑制[28],Zhou等[29]将V. parvula PK1910的过氧化氢酶基因(catA)整合到V. atypica OK5,证实了catA基因产物能保护具核梭杆菌在S. gordonii产生的H2O2环境下生长的猜想。Liu等[27]通过向S. mutans与S. gordonii双菌种模型中加入V. parvula,S. mutans生长速率和最终生物量的增加证明了V. parvula能平衡两者的竞争。链球菌发酵碳水化合物产生的有机酸可作为韦荣球菌的碳源,V. dispar和V. atypica是口腔中硝酸盐还原细菌的重要组成部分[30],有助于硝酸盐转化成抑制龋洞中产酸细菌生长的还原产物[31]。V. atypica PK1910与相对分子质量为4.5×104的蛋白有关的黏附素、V. atypica OK5的表面蛋白血凝素(hemagglutinin,Hag)1,S. gordonii的表面黏附素Hsa均可介导两者的属间共聚集[32]–[34],而V. tobetsuensis产生的环状二肽会抑制S. gordonii生物膜的形成[35],Egland等[36]和Johnson等[37]则观察到S. gordonii与V. atypica PK1910之间有特异性可扩散信号传导现象,由于转录因子CcpA的诱导作用,V. atypica使S. gordonii的α-淀粉酶amyB基因的淀粉酶产物活性增加,促进糖原水解,提高碳水化合物利用率和乳酸生成量,有助于细菌生长。
图 1. S. mutans、S. gordonii、韦荣球菌的相互作用.
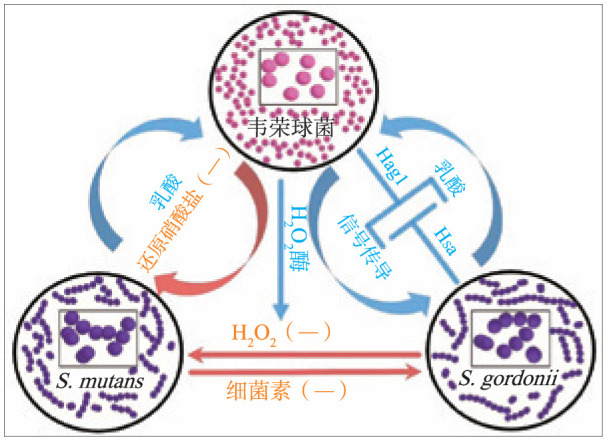
Fig 1 The internaction between S. mutans, S. gordonii and Veillonella
2. 韦荣球菌与牙周病
牙周病是发生在牙的支持组织的疾病,主要临床症状是牙龈出血、牙槽骨吸收、牙周袋形成、牙齿松动,牙菌斑生物膜是牙周病的一个最重要致病因素,牙周病是牙菌斑微生态失衡使牙周致病菌成为优势菌群的结果。较牙周健康人群来说,牙周病患者口腔中韦荣球菌丰度显著增高[38]。
2.1. 韦荣球菌直接参与牙周病的进展
通过调节中性粒细胞[39]等免疫细胞的功能及炎症介质反应[40],微生物可影响牙周炎的发生发展。Hirschfeld等[41]通过研究人外周血中性粒细胞对19种牙周菌斑优势菌的反应发现,牙周炎组织损伤可能是由牙菌斑诱导的细胞外活性氧释放过多引起的,活性氧虽然有助于微生物的杀灭,但其却并不能区分病原体和宿主组织。研究还提示,中性粒细胞在牙周细菌刺激下,会促进细胞外、细胞内和超氧化物释放,且具有物种特异性,与其他微生物相比,V. parvula能刺激更高水平的中性粒细胞胞外杀菌网络和活性氧的释放。Ji等[42]在研究非牙周致病菌与牙周致病菌对牙龈上皮细胞先天免疫应答的影响中发现,作为非牙周致病菌的V. atypica可显著诱导牙龈上皮细胞产生一种在炎症或感染状态下才产生的广谱抗菌肽,即人β防御素-3(human beta defensin-3,HBD-3)。此外,韦荣球菌具有强毒性内毒素,可能通过引起非特性免疫反应参与牙周病的进展[43],其脂多糖(lipopolysaccharide,LPS)也被证明可通过介导前列腺素E2生成而促进骨的吸收[44]。
2.2. 韦荣球菌协同牙龈卟啉单胞菌影响牙周病的发生发展
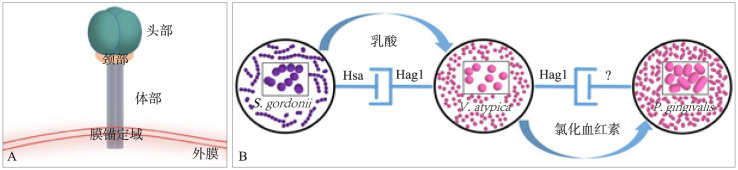
韦荣球菌作为“桥接菌”,附着于早期定植菌后,吸附中、晚期定植菌。牙龈卟啉单胞菌(Porphyromonas gingivalis,P. gingivalis)是公认的牙周致病菌[45],也是晚期定植菌,韦荣球菌与P. gingivalis在形成生物膜过程中的协同作用[46]与二者间的黏附密切相关。一方面,韦荣球菌可为P. gingivalis提供黏附位点。Zhou等[34]研究发现V. atypica表面存在一种蛋白Hag1,可以介导韦荣球菌与P. gingivalis的黏附。Hag1是韦荣球菌第一个被证实的表面蛋白,为目前已知最大的Hag基因。Hag1可以介导韦荣球菌与一些早期定植菌如S. gordonii等的聚集,以及晚期定植菌如P. gingivalis等的黏附[47]。Hag1基因位于单基因操纵子上,编码蛋白为三聚体自转运蛋白,由头、颈、体、膜锚定域4部分组成,头部形成浅口袋形的受体结合域,颈部具有弹性连接头部与体部,体部借锚定域锚定在外膜上(图2A)。Eke等[48]曾得到韦荣球菌与P. gingivalis的黏附受乳糖、半乳糖抑制的结论,由此推断其黏附机制包括糖结合作用,且可能受菌种、菌液的密度标准、加入乳糖的顺序、加入乳糖的量、允许两菌反应的时间长度影响。Zhou等[34]发现,乳糖、半乳糖处理对V. atypica与P. gingivalis的黏附无影响。Park等[49]研究表明,血清型、菌株特异性的黏附在口腔菌群间十分常见,即韦荣球菌、P. gingivalis不同亚型之间的相互黏附均有差异,可分为乳糖敏感性及非乳糖敏感型。S. gordonii通过唾液酸结合性蛋白(sialic acid-binding adhesin,Hsa)与韦荣球菌表面蛋白Hag1结合黏附[33]–[34],P. gingivalis表面与Hag1的结合受体仍未知,而胎球蛋白能够在一定程度上抑制韦荣球菌与P. gingivalis的黏附,证明其黏附机制中包括唾液酸结合作用(图2B)。
图 2. V. atypica与S. gordonii、P. gingivalis的相互作用.
Fig 2 The internaction between V. atypica, S. gordonii and P. gingivalis
A:Hag1基因编码的蛋白结构;B:V. atypica与S. gordonii、P. gingivalis的结合黏附模型,S. gordonii代谢碳水化合物产生的乳酸为V. atypica提供碳源,V. atypica合成的氯化血红素促进P. gingivalis的生长,S. gordonii通过唾液酸结合性蛋白Hsa与V. atypica表面蛋白Hag1结合黏附,而P. gingivalis表面与Hag1的结合受体仍未知。
另一方面,韦荣球菌可以为P. gingivalis提供营养来源。Zhou等[50]在V. atypica的细胞裂解液中检测到氯化血红素及其相关产物,表明存在氯化血红素生物合成途径,他们还证明了韦荣球菌通过提供P. gingivalis生长所必需的氯化血红素促进其生长,P. gingivalis又可使韦荣球菌的氯化血红素合成表达上调。在组成成分不包含氯化血红素的培养基中无法存活的P. gingivalis,加入灭活氯化血红素合成基因的V. atypica后数量可倍增,说明韦荣球菌还可通过其他途径促进P. gingivalis生长,亟待进一步研究。此外,V. parvula对四环素等牙周炎常用抗菌药物具有天生抗性[51],生物膜中韦荣球菌的存在降低其对抗菌素的敏感性。
3. 韦荣球菌与其他口腔疾病
牙髓和根尖周病变中频繁检出韦荣球菌[52],多为V. parvula,韦荣球菌致病性可能与其产生的LPS有关,LPS能够激活机体的补体系统[53],持续释放促炎症因子。对于其引起根尖周病变的免疫潜力方面,有研究发现,V. parvula蛋白和LPS均对巨噬细胞和淋巴细胞具有免疫调节作用,能显著增强人外周血单核细胞迁移活性[54],且V. parvula产生的LPS是人外周血单个核细胞产生细胞因子的弱刺激因子[55]。牙髓及根尖周感染通常为混合感染,韦荣球菌和其他细菌之间的相互作用也不容忽视,V. parvula能够产维生素K2供卟啉单胞菌属和普雷沃菌属利用[56]。此外,V. parvula在与牙密螺旋体的共培养中能够促进其生长[57]。
口臭的主要来源是口腔内细菌代谢产生的H2S等含硫化合物,该过程受到口腔环境因素pH和乳酸的调节。韦荣球菌可通过分解L-半胱氨酸产H2S[58],Washio等[59]的研究发现,口腔中脱落细胞中的主要蛋白质——角蛋白,可以作为L-半胱氨酸在口腔中的来源,而位于细胞质中的多种酶参与了其代谢过程,而关于韦荣球菌产生H2S的具体酶和通路仍不明确;他们还发现,乳酸的存在可以通过激活L-半胱氨酸降解前的过程,如细胞膜上L-半胱氨酸的摄取,来显著提高韦荣球菌静息细胞H2S生成量,并且在pH=6~7时,H2S生成量较高,pH=5时,H2S生成量较低。已有研究[60]证明,由产碱韦荣球菌(Veillonella alcalescens)产生的膜泡具有对谷氨酸和丝氨酸的摄取活性,并伴有乳酸作为电子供体参与电子传递系统。韦荣球菌是否可通过类似的系统影响H2S的产生还有待进一步研究。
4. 韦荣球菌分子机制的研究进展
韦荣球菌曾经由于基因改造、遗传操作的困难,是人类口腔微生物群中最普遍但研究最少的微生物之一。而Liu等[61]通过电穿孔成功将靶向突变引入到V. parvula PK1910的染色体中,第一次验证了韦荣球菌的可转化性。但是转化效率较低,因此,他们又基于韦荣球菌的内源质粒构建了第一个大肠杆菌-韦荣球菌穿梭载体,开发了第一个遗传可转化菌株V. atypica OK5[62],并测定了完整基因组序列[63]。但是在下游基因不产生极性效应的情况下,单交叉诱变系统不能在多基因操纵子中产生突变,因此,他们再次利用V. atypica建立了第一个反向无标记诱变系统[64],这样的遗传转化系统不仅可以用于基因缺失研究,还可以用于插入序列。由于通过电穿孔的等位基因交换突变已经被证实是无效的,Knapp等[65]验证了V. parvula菌株等位基因置换突变的自然转化能力较常见。Zhou等[33]–[34]利用该遗传可转化菌株V. atypica OK5鉴定出Hag1黏附素参与韦荣球菌与链球菌、P. gingivalis和人体颊细胞的分子识别、共聚集过程;并证明了存在促进牙周病原体生长的血红素生物合成途径[50];证实了韦荣球菌的catA基因产物过氧化氢酶能保护具核梭杆菌等其他细菌在H2O2环境的生长[29]。韦荣球菌遗传转化系统的建立为韦荣球菌在口腔生物膜形成中的桥接作用及疾病防治的研究提供新的思路,有利于深入了解人类口腔生物膜的生态学。
5. 总结
综上所述,韦荣球菌与龋病、牙周病、牙髓及根尖周病、口臭等口腔疾病存在相关性,但在分子水平上的具体机制仍不够清楚。龋病方面,早期认为可利用乳酸减少牙脱矿的韦荣球菌有防龋作用,尽管针对龋病患者口腔韦荣球菌的分布情况仍无统一定论,但目前研究更倾向于认可韦荣球菌在致龋过程中发挥重要作用,由于其与链球菌属间的共聚集和黏附、并能促进形成生物膜,以及在代谢方面有协同生长作用,而其与链球菌的具体黏附方式及促进链球菌生物膜形成的机制尚不清楚。牙周病方面,韦荣球菌通过调节免疫反应中的中性粒细胞和释放活性氧、为牙周致病菌提供黏附位点和营养来源参与牙周病的进展,但与P. gingivalis表面的结合位点仍不明确,也需要进一步研究促进P. gingivalis生长的其他代谢途径。口腔疾病是多种微生物、多因素共同作用的结果,目前韦荣球菌与致病菌在某些方面的协同关系已被证实,其在分子水平上影响口腔疾病的具体机制、是否促进疾病发生发展仍需要进一步研究。
Funding Statement
[基金项目] 国家自然科学基金(81700963);四川省科技计划项目(2018JY0561)
Supported by: The National Natural Science Foundation of China (81700963); Sichuan Science and Technology Program (2018JY0561).
Footnotes
利益冲突声明:作者声明本文无利益冲突。
References
- 1.Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries[J] Immunol Lett. 2014;162(2 Pt A):22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byun R, Carlier JP, Jacques NA, et al. Veillonella denticariosi sp. nov., isolated from human carious dentine[J] Int J Syst Evol Microbiol. 2007;57(Pt 12):2844–2848. doi: 10.1099/ijs.0.65096-0. [DOI] [PubMed] [Google Scholar]
- 3.Mashima I, Liao YC, Miyakawa H, et al. Veillonella infantium sp. nov., an anaerobic, Gram-stain-negative coccus isolated from tongue biofilm of a Thai child[J] Int J Syst Evol Microbiol. 2018;68(4):1101–1106. doi: 10.1099/ijsem.0.002632. [DOI] [PubMed] [Google Scholar]
- 4.Arif N, Do T, Byun R, et al. Veillonella rogosae sp. nov., an anaerobic, Gram-negative coccus isolated from dental plaque[J] Int J Syst Evol Microbiol. 2008;58(Pt 3):581–584. doi: 10.1099/ijs.0.65093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mashima I, Kamaguchi A, Miyakawa H, et al. Veillonella tobetsuensis sp. nov., an anaerobic, gram-negative coccus isolated from human tongue biofilms[J] Int J Syst Evol Microbiol. 2013;63(Pt 4):1443–1449. doi: 10.1099/ijs.0.042515-0. [DOI] [PubMed] [Google Scholar]
- 6.Aujoulat F, Bouvet P, Jumas-Bilak E, et al. Veillonella seminalis sp. nov., a novel anaerobic Gram-stain-negative coccus from human clinical samples, and emended description of the genus Veillonella[J] Int J Syst Evol Microbiol. 2014;64(Pt 10):3526–3531. doi: 10.1099/ijs.0.064451-0. [DOI] [PubMed] [Google Scholar]
- 7.Mashima I, Djais AA, Haase EM, et al. Establishment of a species-specific primer pair for detecting Veillonella infantium based on the 70 kDa heat shock protein gene dnaK[J] Anaerobe. 2018;52:79–82. doi: 10.1016/j.anaerobe.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Doel JJ, Benjamin N, Hector MP, et al. Evaluation of bacterial nitrate reduction in the human oral cavity[J] Eur J Oral Sci. 2005;113(1):14–19. doi: 10.1111/j.1600-0722.2004.00184.x. [DOI] [PubMed] [Google Scholar]
- 9.Mashima I, Theodorea CF, Thaweboon B, et al. Identification of Veillonella species in the tongue biofilm by using a novel one-step polymerase chain reaction method[J] PLoS One. 2016;11(6):e0157516. doi: 10.1371/journal.pone.0157516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanasi E, Dewhirst FE, Chalmers NI, et al. Clonal analysis of the microbiota of severe early childhood caries[J] Caries Res. 2010;44(5):485–497. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mashima I, Fujita M, Nakatsuka Y, et al. The distribution and frequency of oral Veillonella spp. associated with chronic periodontitis[J] Int J Curr Microbiol App Sci. 2015;4(3):150–160. [Google Scholar]
- 12.Al-Ahmad A, Roth D, Wolkewitz M, et al. Change in diet and oral hygiene over an 8-week period: effects on oral health and oral biofilm[J] Clin Oral Investig. 2010;14(4):391–396. doi: 10.1007/s00784-009-0318-9. [DOI] [PubMed] [Google Scholar]
- 13.Stahringer SS, Clemente JC, Corley RP, et al. Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood[J] Genome Res. 2012;22(11):2146–2152. doi: 10.1101/gr.140608.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitsui T, Saito M, Harasawa R. Salivary nitrate-nitrite conversion capacity after nitrate ingestion and incidence of Veillonella spp. in elderly individuals[J] J Oral Sci. 2018;60(3):405–410. doi: 10.2334/josnusd.17-0337. [DOI] [PubMed] [Google Scholar]
- 15.Mashima I, Nakazawa F. The influence of oral Veillonella species on biofilms formed by Streptococcus species[J] Anaerobe. 2014;28:54–61. doi: 10.1016/j.anaerobe.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Do T, Sheehy EC, Mulli T, et al. Transcriptomic analysis of three Veillonella spp. present in carious dentine and in the saliva of caries-free individuals[J] Front Cell Infect Microbiol. 2015;5:25. doi: 10.3389/fcimb.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, Chen YX, Xie LZ, et al. Pyrosequencing of plaque microflora in twin children with discordant caries phenotypes[J] PLoS One. 2015;10(11):e0141310. doi: 10.1371/journal.pone.0141310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belstrøm D, Fiehn NE, Nielsen CH, et al. Altered bacterial profiles in saliva from adults with caries lesions: a case-cohort study[J] Caries Res. 2014;48(5):368–375. doi: 10.1159/000357502. [DOI] [PubMed] [Google Scholar]
- 19.Thomas RZ, Zijnge V, Ciçek A, et al. Shifts in the microbial population in relation to in situ caries progression[J] Caries Res. 2012;46(5):427–431. doi: 10.1159/000339482. [DOI] [PubMed] [Google Scholar]
- 20.Jiang S, Gao XL, Jin LJ, et al. Salivary microbiome diversity in caries-free and caries-affected children[J] Int J Mol Sci. 2016;17(12):E1978. doi: 10.3390/ijms17121978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanner AC, Mathney JM, Kent RL, et al. Cultivable anaerobic microbiota of severe early childhood caries[J] J Clin Microbiol. 2011;49(4):1464–1474. doi: 10.1128/JCM.02427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishiro T, Oka K, Kuroki Y, et al. Oral microbiome alterations of healthy volunteers with proton pump inhibitor[J] J Gastroenterol Hepatol. 2018;33(5):1059–1066. doi: 10.1111/jgh.14040. [DOI] [PubMed] [Google Scholar]
- 23.Guggenheim M, Shapiro S, Gmür R, et al. Spatial arrangements and associative behavior of species in an in vitro oral biofilm model[J] Appl Environ Microbiol. 2001;67(3):1343–1350. doi: 10.1128/AEM.67.3.1343-1350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kara D, Luppens SB, van Marle J, et al. Microstructural differences between single-species and dual-species biofilms of Streptococcus mutans and Veillonella parvula, before and after exposure to chlorhexidine[J] FEMS Microbiol Lett. 2007;271(1):90–97. doi: 10.1111/j.1574-6968.2007.00701.x. [DOI] [PubMed] [Google Scholar]
- 25.Chalmers NI, Palmer RJ, Jr, Cisar JO, et al. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque[J] J Bacteriol. 2008;190(24):8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thurnheer T, Belibasakis GN. Streptococcus oralis maintains homeostasis in oral biofilms by antagonizing the cariogenic pathogen Streptococcus mutans[J] Mol Oral Microbiol. 2018;33(3):234–239. doi: 10.1111/omi.12216. [DOI] [PubMed] [Google Scholar]
- 27.Liu JM, Wu CG, Huang IH, et al. Differential response of Streptococcus mutans towards friend and foe in mixed-species cultures[J] Microbiology (Reading, Engl) 2011;157(Pt 9):2433–2444. doi: 10.1099/mic.0.048314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luppens SB, Kara D, Bandounas L, et al. Effect of Veillonella parvula on the antimicrobial resistance and gene expression of Streptococcus mutans grown in a dual-species biofilm[J] Oral Microbiol Immunol. 2008;23(3):183–189. doi: 10.1111/j.1399-302X.2007.00409.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhou P, Li XL, Huang IH, et al. Veillonella catalase protects the growth of Fusobacterium nucleatum in microaerophilic and Streptococcus gordonii-resident environments[J] Appl Environ Microbiol. 2017;83(19):e01079–e01017. doi: 10.1128/AEM.01079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burleigh MC, Liddle L, Monaghan C, et al. Salivary nitrite production is elevated in individuals with a higher abundance of oral nitrate-reducing bacteria[J] Free Radic Biol Med. 2018;120:80–88. doi: 10.1016/j.freeradbiomed.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 31.Doel JJ, Hector MP, Amirtham CV, et al. Protective effect of salivary nitrate and microbial nitrate reductase activity against caries[J] Eur J Oral Sci. 2004;112(5):424–428. doi: 10.1111/j.1600-0722.2004.00153.x. [DOI] [PubMed] [Google Scholar]
- 32.Hughes CV, Andersen RN, Kolenbrander PE. Characterization of Veillonella atypica PK1910 adhesin-mediated coaggregation with oral Streptococcus spp[J] Infect Immun. 1992;60(3):1178–1186. doi: 10.1128/iai.60.3.1178-1186.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou P, Liu JM, Li XL, et al. The sialic acid binding protein, Hsa, in Streptococcus gordonii DL1 also mediates intergeneric coaggregation with Veillonella species[J] PLoS One. 2015;10(11):e0143898. doi: 10.1371/journal.pone.0143898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou P, Liu JM, Merritt J, et al. A YadA-like autotransporter, Hag1 in Veillonella atypica is a multivalent hemagglutinin involved in adherence to oral streptococci, Porphyromonas gingivalis, and human oral buccal cells[J] Mol Oral Microbiol. 2015;30(4):269–279. doi: 10.1111/omi.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mashima I, Miyakawa H, Scannapieco FA, et al. Identification of an early stage biofilm inhibitor from Veillonella tobetsuensis[J] Anaerobe. 2018;52:86–91. doi: 10.1016/j.anaerobe.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Egland PG, Palmer RJ, Jr, Kolenbrander PE. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition[J] Proc Natl Acad Sci USA. 2004;101(48):16917–16922. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson BP, Jensen BJ, Ransom EM, et al. Interspecies signaling between Veillonella atypica and Streptococcus gordonii requires the transcription factor CcpA[J] J Bacteriol. 2009;191(17):5563–5565. doi: 10.1128/JB.01226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei YP, Shi M, Zhen M, et al. Comparison of subgingival and buccal mucosa microbiome in chronic and aggressive periodontitis: a pilot study[J] Front Cell Infect Microbiol. 2019;9:53. doi: 10.3389/fcimb.2019.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts HM, Ling MR, Insall R, et al. Impaired neutrophil directional chemotactic accuracy in chronic periodontitis patients[J] J Clin Periodontol. 2015;42(1):1–11. doi: 10.1111/jcpe.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebersole JL, Steffen MJ, Thomas MV, et al. Smoking-related cotinine levels and host responses in chronic periodontitis[J] J Periodont Res. 2014;49(5):642–651. doi: 10.1111/jre.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirschfeld J, White PC, Milward MR, et al. Modulation of neutrophil extracellular trap and reactive oxygen species release by periodontal bacteria[J] Infect Immun. 2017;85(12):e00297–e00217. doi: 10.1128/IAI.00297-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji S, Kim Y, Min BM, et al. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria[J] J Periodont Res. 2007;42(6):503–510. doi: 10.1111/j.1600-0765.2007.00974.x. [DOI] [PubMed] [Google Scholar]
- 43.Papapanou PN, Neiderud AM, Papadimitriou A, et al. “Checkerboard” assessments of periodontal microbiota and serum antibody responses: a case-control study[J] J Periodontol. 2000;71(6):885–897. doi: 10.1902/jop.2000.71.6.885. [DOI] [PubMed] [Google Scholar]
- 44.Xiao L, Ornatowska M, Zhao GQ, et al. Lipopolysaccharide-induced expression of microsomal prostaglandin E synthase-1 mediates late-phase PGE2 production in bone marrow derived macrophages[J] PLoS One. 2012;7(11):e50244. doi: 10.1371/journal.pone.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue[J] Periodontol 2000. 2010;52(1):68–83. doi: 10.1111/j.1600-0757.2009.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Periasamy S, Kolenbrander PE. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel[J] J Bacteriol. 2010;192(12):2965–2972. doi: 10.1128/JB.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hughes CV, Kolenbrander PE, Andersen RN, et al. Coaggregation properties of human oral Veillonella spp.: relationship to colonization site and oral ecology[J] Appl Environ Microbiol. 1988;54(8):1957–1963. doi: 10.1128/aem.54.8.1957-1963.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eke PI, Rotimi VO, Laughon BE. Coaggregation of black-pigmented Bacteroides species with other oral bacteria[J] J Med Microbiol. 1989;28(1):1–4. doi: 10.1099/00222615-28-1-1. [DOI] [PubMed] [Google Scholar]
- 49.Park J, Shokeen B, Haake SK, et al. Characterization of Fusobacterium nucleatum ATCC 23726 adhesins involved in strain-specific attachment to Porphyromonas gingivalis[J] Int J Oral Sci. 2016;8(3):138–144. [Google Scholar]
- 50.Zhou P, Li XL, Qi FX. Identification and characterization of a haem biosynthesis locus in Veillonella[J] Microbiology. 2016;162(10):1735–1743. doi: 10.1099/mic.0.000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodrigues RM, Gonçalves C, Souto R, et al. Antibiotic resistance profile of the subgingival microbiota following systemic or local tetracycline therapy[J] J Clin Periodontol. 2004;31(6):420–427. doi: 10.1111/j.1600-051X.2004.00493.x. [DOI] [PubMed] [Google Scholar]
- 52.Pourhajibagher M, Raoofian R, Ghorbanzadeh R, et al. An experimental study for rapid detection and quantification of endodontic microbiota following photo-activated disinfection via new multiplex real-time PCR assay[J] Photodiagnosis Photodyn Ther. 2018;21:344–350. doi: 10.1016/j.pdpdt.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Matera G, Liberto MC, Berlinghieri MC, et al. Biological effects of Veillonella parvula and Bacteroides intermedius lipopolysaccharides[J] Microbiologica. 1991;14(4):315–323. [PubMed] [Google Scholar]
- 54.Nagashima Y. Immunobiological activities of Veillonella parvula isolated from infected root canals[J] Kanagawa Shigaku. 1990;25(2):209–220. [PubMed] [Google Scholar]
- 55.Matera G, Muto V, Vinci M, et al. Receptor recognition of and immune intracellular pathways for Veillonella parvula lipopolysaccharide[J] Clin Vaccine Immunol. 2009;16(12):1804–1809. doi: 10.1128/CVI.00310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramotar K, Conly JM, Chubb H, et al. Production of menaquinones by intestinal anaerobes[J] J Infect Dis. 1984;150(2):213–218. doi: 10.1093/infdis/150.2.213. [DOI] [PubMed] [Google Scholar]
- 57.ter Steeg PF, van der Hoeven JS. Growth stimulation of Treponema denticola by periodontal microorganisms[J] Antonie Van Leeuwenhoek. 1990;57(2):63–70. doi: 10.1007/BF00403156. [DOI] [PubMed] [Google Scholar]
- 58.Washio J, Sato T, Koseki T, et al. Hydrogen sulfide-producing bacteria in tongue biofilm and their relationship with oral malodour[J] J Med Microbiol. 2005;54(Pt 9):889–895. doi: 10.1099/jmm.0.46118-0. [DOI] [PubMed] [Google Scholar]
- 59.Washio J, Shimada Y, Yamada M, et al. Effects of pH and lactate on hydrogen sulfide production by oral Veillonella spp[J] Appl Environ Microbiol. 2014;80(14):4184–4188. doi: 10.1128/AEM.00606-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Konings WN, Boonstra J, De Vries W. Amino acid transport in membrane vesicles of obligately anaerobic Veillonella alcalescens[J] J Bacteriol. 1975;122(1):245–249. doi: 10.1128/jb.122.1.245-249.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu JM, Merritt J, Qi FX. Genetic transformation of Veillonella parvula[J] FEMS Microbiol Lett. 2011;322(2):138–144. doi: 10.1111/j.1574-6968.2011.02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu JM, Xie ZJ, Merritt J, et al. Establishment of a tractable genetic transformation system in Veillonella spp[J] Appl Environ Microbiol. 2012;78(9):3488–3491. doi: 10.1128/AEM.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou P, Xie G, Li XL, et al. Complete genome sequence of Veillonella atypica OK5, the first transformable strain in the species[J] Genome Announc. 2017;5(22):e00391–e00317. doi: 10.1128/genomeA.00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou P, Li XL, Qi FX. Establishment of a counter-selectable markerless mutagenesis system in Veillonella atypica[J] J Microbiol Methods. 2015;112:70–72. doi: 10.1016/j.mimet.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knapp S, Brodal C, Peterson J, et al. Natural competence is common among clinical isolates of Veillonella parvula and is useful for genetic manipulation of this key member of the oral microbiome[J] Front Cell Infect Microbiol. 2017;7:139. doi: 10.3389/fcimb.2017.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]