Abstract
Methyllysine sites in proteins are recognized by an array of reader domains that mediate protein-protein interactions for controlling cellular processes. Herein, we engineer a chromodomain, an essential methyllysine reader, to carry 4-azido-L-phenylalanine (AzF) via amber suppressor mutagenesis and demonstrate its potential to bind and crosslink methylated proteins in human cells. We further develop a first-of-its kind chromodomain variant bearing two AzF units with enhanced crosslinking potential suitable for profiling the transient methyllysine interactome.
Graphical Abstract

A methyllysine reader is engineered to carry photoactivatable amino acid in mammalian cells to profile interacting partners.
Site- and degree-specific methylations of ε-amino group of lysine residues in histones have emerged as an important regulatory mark for controlling chromatin structure and function.1, 2 While the steady-state pattern of lysine methylation is carefully maintained by reverse-acting lysine methyltransferases (KMTs) and demethylases (KDMs), its biological function is primarily manifested by the reader or effector modules that are often subunits of a multimeric protein complex (Fig. 1A).3, 4 These binding domains recognize methylated lysine through cation-π interactions in a conserved hydrophobic pocket known as the ‘aromatic cage’.5, 6
Fig. 1.
Engineering of methyllysine reader. (A) A lysine methyltransferase (KMT) establishes site-specific methylation pattern that is recognized by an array of reader proteins. A lysine demethylase (KDM) removes the mark for steady-state composition of methyllysine. (B) Engineering of methyllysine readers to introduce a photoactive amino acid for crosslinking with interacting partners. The exact site and nature of the crosslinking are undefined.
The human genome encodes for more than 15 unique methyllysine readers in about 200 proteins.7 These include chromodomains, plant homeodomains (PHD), Tudor domains and ankyrin repeat units, to name a few.6 The reader-mediated recognition of methyllysine is highly dynamic and responsive to intrinsic and extrinsic stimuli leading to context-dependent biological outcomes.8, 9 Furthermore, recent proteomic studies have identified more than 5000 methyllysine sites beyond the canonical histones.1, 10 Many of these non-histone methylated sites could crosstalk synergistically or antagonistically with other modifications and are recognized by the same set of methyllysine effectors for controlling protein stability and localization.11 For example, dimethylation at the evolutionarily conserved lysine residue in DNA ligase 1 (K126 in human LIG1) by G9a and GLP (G9a-like protein) is recognized by the Tandem Tudor domain of UHRF1 for recruitment of UHRF1 to replicating genome to maintain DNA methylation in daughter cells.12
Potential readout of 5000 methyllysine sites by 200 readers could establish an extensive array of protein-protein interactions. Structural studies have detailed the interaction between methyllysine and its readers.12, 13 Due to the transient and weak nature of such associations, it has remained a significant challenge to characterize context-dependent methyllysine interactions mediated by a specific reader. The available methods based on immunoprecipitation suffer in efficiency to enrich weak interactions because the dissociation constants of the majority of such interactions are in the range of tens of micromolar. These techniques oftentimes lack temporal control as the interacting partners are netted in homogenous cellular extracts. To address such limitations, we envisioned developing photo-responsive methyllysine readers to capture weak and transient interacting partners (Fig. 1B). We recently developed photo-crosslinkable bromodomains, a reader module for acetylated lysine, to identify reader-specific acetylome.14, 15 Such an approach for methyllysine readers is yet to be developed.
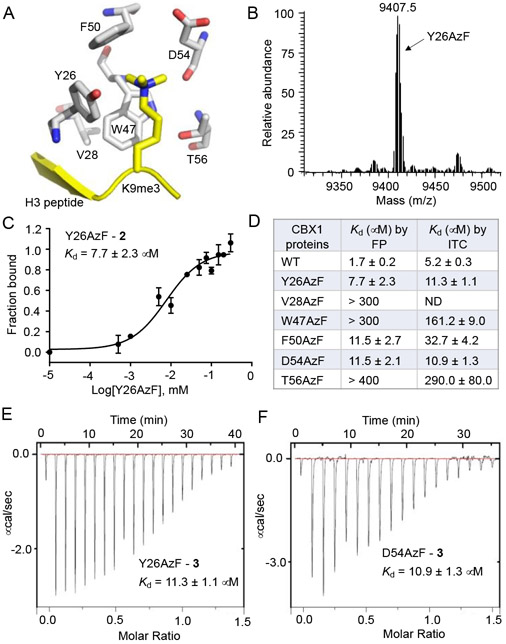
To access photo-crosslinkable methyllysine reader analogues, we focused on chromodomains that have shown to regulate cellular processes by recognizing methylated histone and non-histone proteins.16, 17 We have determined a high-resolution crystal structure (PDB: 6D07) of H3K9me3-bound chromodomain of Chromobox protein 1 (CBX1).18 The structure revealed presence of a canonical hydrophobic pocket binding the Kme3 moiety via cation-π interaction mediated by conserved aromatic residues (Fig. 2A). Earlier structural studies on chromodomains have revealed a unique mode of sequence recognition with the histone tail inserted between two β-strands of the chromodomain forming an antiparallel three-stranded β-sheet that imparts sequence selectivity such as H3K9me3 over H3K27me3 lacks connection.5, 19-21 Given that engineering the amino acids involved in the β-sheet interactions may lead to a change in sequence preference, we specifically focused on residues in the aromatic cage. We surmised that replacing these amino acids, one at a time, with light-sensitive amino acids would lead to photo-crosslinkable CBX1 variants that could be employed to capture the CBX1 interactome.
Fig. 2.
Engineering of CBX1 chromodomain with photo-crosslinkable amino acid AzF 1. (A) Crystal structure showing chromodomain of CBX1 bound with H3K9me3 peptide (PDB: 6D07). The indicated residues are replaced, one at a time, with AzF. (B) ESI LC-MS spectrum of the Y26AzF mutant. (C) Dissociation constant of Y26AzF mutant from peptide 2 as measured by fluorescence anisotropy. (E, F) Dissociation constants of Y26AzF and D54AzF mutants, respectively, as measured by isothermal titration calorimetry.
The selected residues include Y26, V28, W47, F50, D54 and T56. We focused on 4-azido-L-phenylalanine (AzF, 1), an analogue of phenylalanine, as a potential crosslinker because of its high crosslinking efficiency, minimal structural perturbation and availability of suitable methods for site-specific incorporation into proteins (Fig. 1B).22, 23 For this purpose, we employed amber nonsense codon (TAG) mutagenesis using the evolved orthogonal M. jannaschii TyrRS-tRNACUATyr pair developed by Schultz and colleagues.24 Using a bacterial expression system, we successfully obtained all the desired photo-responsive variants (Y26AzF, V28AzF, W47AzF, F50AzF, D54AzF and T56AzF) in high yields (Fig. S1). ESI LC-MS analysis confirmed the integrity of the intact proteins, each carrying one AzF unit (Fig. 2B, S2, Table S1).
To examine the ability of these mutants to bind H3K9me3 peptide, we employed a fluorescence polarization (FP) assay using a tracer peptide 2 carrying tetramethyl rhodamine (TAMRA) at the N-terminus (Fig. S3). In the assay, wild type chromodomain of CBX1 bound to the peptide with a dissociation constant (Kd) of 1.7 ± 0.2 μM, similar to previously reported values.18, 21 While the mutants displayed varied degrees of binding, the affinity was consistently lower than that of their wild type counterpart (Fig. 2D, S4). Among the tested mutants, Y26AzF, F50AzF and D54AzF retained significant binding towards the peptide with Kd values of 7.7 ±2.3 μM, 11.5 ± 2.7 μM and 11.5 ± 2.1 μM, respectively (Fig. 2C, D, S4). Given the structural similarity between tyrosine/ phenylalanine and AzF, Y26AzF and F50AzF mutations encountered minimum steric and electronic perturbations as reflected by their binding efficiency being comparable to the wild type reader. Although the affinity of D54AzF towards the tracer peptide appears to be counterintuitive as this mutation resulted in a loss of electrostatic interactions between the carboxylic group of D54, water molecule in the aromatic cage and the trimethylammonium ion of K9me3, we reasoned that the recovery of binding is likely due to new cation-π interaction between methylated lysine and AzF54.13, 25 Consistently, we have noted that certain methyllysine readers such as PHD fingers carry an aromatic residue in that particular site, which can engage in cation-π interaction with methyllysine.26
In order to obtain accurate values of the binding constants, we measured the thermodynamic parameters of binding using isothermal titration calorimetry (ITC). We synthesized a non-TAMRA peptide 3 with sequence identical to that of 2 (Fig. S3, Table S2). The Kd of wild type chromodomain with H3K9me3 peptide was measured to be 5.3 ±0.4 μM (Fig. 2D, S5). We next determined dissociation constants for the selected mutants which showed moderate to strong binding in the initial FP assay. Y26AzF and D54AzF indeed exhibited strong affinity towards the peptide with Kd of 11.3 ± 1.1 μM and 10.9 ± 1.3 μM, respectively, closely matching with the data obtained through our FP assay (Fig. 2D, E, F). V28AzF, W47AzF and T56AzF mutants all showed significantly weaker binding affinity than the wild type CBX1, consistent with the fluorescence anisotropy-based results (Fig. 2D, S5). Collectively, these results obtained from two independent binding experiments using different techniques demonstrate robust and reliable binding of the engineered CBX1 chromodomains carrying AzF towards methylated histone peptide.
Encouraged by the above results, we decided to explore other possibilities to engineer the CBX1 chromodomain. We noted that trimethyllysine readers carry multiple aromatic residues in the hydrophobic pocket to maximize cation-π interaction. We reasoned that site-specific incorporation of two AzF units in the chromodomain is expected to enhance crosslinking efficiency without compromising the integrity of the aromatic cage (Fig. 3A).27 We introduced amber codon (TAG) to generate Y26TAG/F50TAG and Y26TAG/D54TAG mutants by combining the variants that showed strong binding towards H3K9me3 peptide as noted above. Quite remarkably, we observed efficient readthrough of the nonsense codons by the orthogonal M. jannaschii TyrRS-tRNACUATyr pair to access the combined mutants, albeit in a lower yield than the respective single mutant. We confirmed the double incorporation of AzF in these mutants by ESI LC-MS (Fig. S6). In the FP assay, both Y26AzF/F50AzF and Y26AzF/D54AzF showed moderate binding to the peptide with Kd values of 48.9 ± 8.7 μM and 50.8 ± 5.7 μM, respectively (Fig. 3B, C). These results show that CBX1, and likely other chromodomains, can be engineered to bear multiple photoactive amino acids.
Fig. 3.
Engineering of CBX1 chromodomain with AzF 1. (A) Schematic showing site-specific incorporation of two AzF units in CBX1 chromodomain. (B, C) Dissociation constant of the double mutants from peptide 2 as measured by fluorescence anisotropy.
We proceeded to examine if the selected mutants were capable of crosslinking with the methylated interacting partners upon exposure to ultraviolet (UV) light. A mixture of TAMRA-attached peptide 2 and individual mutants was subjected to photo irradiation and resolved in SDS-PAGE. Successful crosslinking is expected to generate a fluorescently labeled protein band in polyacrylamide gel. Indeed, Y26AzF, F50AzF and Y26AzF/F50AzF mutants underwent crosslinking with 2 only when the engineered reader and peptide complexes were exposed to 365 nm light (Fig. S7). No fluorescent band was observed for the wild type CBX1 in spite of its strong association (Kd ~2 μM) with the peptide, thus demonstrating that crosslinking ability of the mutants must have been acquired through incorporation of AzF in the binding pocket (Fig. S7). We further noted that Y26AzF/F50AzF, although a weak binder, has higher crosslinking efficiency than corresponding single mutants, due to the presence of two AzF groups that enhances the likelihood of crosslinking via either or both of the photoactive moieties (Fig. S7). We next synthesized TAMRA-H3K9mel 4 and TAMRA-H3K9me2 5 peptides and photo-irradiated in presence of the mutants (Fig. S3, S7). We noted a varied degree of crosslinking, consistent with the observation that CBX1 chromodomain is also capable of binding with the mono- and di-methyllysine marks.
To assess the ability of the mutants to crosslink full-length proteins, we prepared semisynthetic histone H3 carrying trimethylated thialysine derivative at position 9 (H3KC9me3) (Fig. 4A, S8).28 This site-specifically methylated H3 protein was incubated with selected mutants, one at a time, followed by UV exposure. Western blot analysis using anti 6xHis antibody (CBX1 chromodomain carries 6xHis tag) confirmed robust crosslinking of Y26AzF with H3 only in the presence UV light as evident from the appearance of higher molecular weight protein bands (Fig. 4A). The D54AzF mutant, although a strong binder, consistently failed to crosslink with H3 as observed with peptide 3 as well, likely because of an orientation of the AzF moiety towards solvent-exposed protein surface. As expected, wild type CBX1 did not undergo crosslinking with the modified H3 (Fig. 4A).
Fig. 4.
Photo-crosslinking of engineered of CBX1 with histone and non-histone proteins. (A) Semi-synthetic H3 carrying trimethylated thialysine. Western blot with anti 6xHis antibody demonstrating successful crosslinking of semisynthetic H3 with Y26AzF mutant. (B) Crosslinking of CBX1 mutants with nucleosomal H3 isolated from HEK293T cells. Western blot with anti 6xHis and H3 antibodies showing successful crosslinking of H3 with selected mutants. (C) Schematic showing expression of full-length CBX1-Y26AzF in HEK293T cells using evolved tRNA-synthetase pair. (D) Western blot with anti HA antibody confirmed the expression of full-length Y26AzF mutant in HEK293T cells. Expression of wild type CBX1 (Mol. wt. 21.4 KDa including affinity tags) does not require AzF. (E) Coomassie gel showing putative interacting partners of CBX1 upon affinity enrichment following photoirradiation of HEK293T cells expressing Y26AzF variant.
We next tested the ability of the mutants to crosslink nucleosomal H3 which is a biologically more relevant interacting partner of reader proteins.29-31 Upon UV irradiation followed by western blot analysis with anti 6xHis antibody, the selected mutants underwent varied degrees of crosslinking with histone (Fig. 4B, S8). To further confirm that the crosslinked species contained H3, we repeated the western blot using anti H3 antibody that recognizes C-terminal of human H3. We indeed observed the protein band at higher molecular weight for the successful mutants (Fig. 4B). It is worth noting that F50AzF which failed to crosslink methylated peptide 2 or the semisynthetic histone, did show noticeable crosslinking with the functionally relevant endogenous H3, likely due to enhanced association between the engineered reader and H3 (Fig. 4B). Crosslinking efficiency of the double mutant (Y26AzF/F50AzF) was comparable or marginally higher than that of the individual mutants in spite of its weaker binding capacity (Kd ~50 μM), further demonstrating the potential benefit of two AzF groups in the hydrophobic pocket (Fig. S8). No crosslinking was noticed in samples not exposed to UV light.
Finally, we examined the expression of CBX1 analogues in cultured human cells and their ability to crosslink cellular proteins upon photoactivation (Fig. 4C). Employing the orthogonal TyrRS-tRNACUATyr pair developed by Sakmar and colleagues,32 full-length CBX1 Y26AzF mutant was successfully expressed in HEK293T cells, only when treated with AzF, as determined by western blot with anti-hemagglutinin (HA) antibody (Fig. 4D). Subsequently, AzF-treated cells were irradiated with 365nm light, lysed and the crosslinked proteins were enriched via affinity tag. Coomassie staining of the pulldown samples revealed distinct protein bands present only in the irradiated cells (Fig. 4E). Collectively, the results demonstrate that CBX1 mutant with photoactive amino acid can be expressed in human cells and that these mutants are capable of covalently binding with putative interacting partners upon UV irradiation.
In this work, we introduced an amino acid carrying arylazide moiety into the aromatic cage of CBX1 chromodomain. We demonstrated that setting up an azide-methyllysine photoreaction deep inside the hydrophobic pocket allows covalent bond formation between the reader and a methylated protein to help capture the interacting dimer. We also developed a first-of-its kind chromodomain variant with two AzF units introduced into the aromatic cage that could undergo robust crosslinking with nucleosomal H3 in spite of its relatively weaker affinity towards methylated peptide. Furthermore, a CBX1 mutant was expressed in human cells to capture in-cellulo binding partners for profiling context-dependent interactome. By reason of the ubiquitous presence of an aromatic cage in methyllysine readers, the current approach will find widespread application to more than 200 such reader modules in humans.
Supplementary Material
Acknowledgments
We thank the University of Pittsburgh, the National Institutes of Health (R01GM123234, R01GM130752) and the National Science Foundation (MCB-1817692) for financial support; Prof. T. Sakmar for the evolved tRNA and synthetase pair; Dr. D. Dey, Dr. M. Waldman, S. Albright for reagents; Dr. D. Chakraborty and members of our laboratory for editing of the manuscript.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/d0cc03814h
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Murn J and Shi Y, Nat. Rev. Mol. Cell Biol 2017, 18, 517–527. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T, Cell, 2007, 128, 693–705. [DOI] [PubMed] [Google Scholar]
- 3.Ruthenburg AJ, Allis CD and Wysocka J, Mol. Cell, 2007, 25, 15–30. [DOI] [PubMed] [Google Scholar]
- 4.Black JC, Van Rechem C and Whetstine JR, Mol. Cell, 2012, 48, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV and Laue ED, Nature, 2002, 416, 103–107. [DOI] [PubMed] [Google Scholar]
- 6.Taverna SD, Li H, Ruthenburg AJ, Allis CD and Patel DJ, Nat. Struct. Mol. Biol 2007, 14, 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yun M, Wu J, Workman JL and Li B, Cell Res. 2011, 21, 564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vermeulen M, et al. Cell, 2007, 131, 58–69. [DOI] [PubMed] [Google Scholar]
- 9.Shi X, et al. T Nature, 2006, 442, 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frye SV, Future Med. Chem 2015, 7, 1831–1833. [DOI] [PubMed] [Google Scholar]
- 11.Biggar KK and Li SS, Nat. Rev. Mol. Cell Biol 2015, 16, 5–17. [DOI] [PubMed] [Google Scholar]
- 12.Ferry L, et al. Mol. Cell, 2017, 67, 550–565.e555. [DOI] [PubMed] [Google Scholar]
- 13.Baril SA, Koenig AL, Krone MW, Albanese KI, He CQ, Lee GY, Houk KN, Waters ML and Brustad EM, J. Am. Chem. Soc 2017, 139, 17253–17256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudhamalla B, Dey D, Breski M, Nguyen T and Islam K, Chem. Sci 2017. 8, 4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner S, Sudhamalla B, Mannes P, Sappa S, Kavoosi S, Dey D, Wang S and Islam K, Chem. Commun 2020, 56, 3641–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB and Misteli T, Science 2003, 299, 721–725. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Galka M, Mori E, Liu X, Lin YF, Wei R, Pittock P, Voss C, Dhami G, Li X, Miyaji M, Lajoie G, Chen B and Li SS, Mol. Cell, 2013, 50, 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arora S, Horne WS and Islam K, J. Am. Chem. Soc 2019, 141, 15466–15470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisert RJ, Kennedy SA and Waters ML, Biochemistry, 2015, 54, 2314–2322. [DOI] [PubMed] [Google Scholar]
- 20.Hard R, Li N, He W, Ross B, Mo GCH, Peng Q, Stein RSL, Komives E, Wang Y, Zhang J and Wang W, Sci. Adv. 2018, 4, eaau1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaustov L, Ouyang H, Amaya M, Lemak A, Nady N, Duan S, Wasney GA, Li Z, Vedadi M, Schapira M, Min J and Arrowsmith CH, J. Biol. Chem. 2011, 286, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pham ND, Parker RB and Kohler JJ, Curr. Opin. Chem. Biol 2013, 17, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TA, Cigler M and Lang K, Angew. Chem. Int. Ed 2018, 57, 14350–14361. [DOI] [PubMed] [Google Scholar]
- 24.Chin JW, Santoro SW, Martin AB, King DS, Wang L and Schultz PG, J. Am. Chem. Soc 002, 124, 9026–9027. [DOI] [PubMed] [Google Scholar]
- 25.Eisert RJ and Waters ML, ChemBioChem 2011, 12, 2786–2790. [DOI] [PubMed] [Google Scholar]
- 26.Patel DJ, Cold Spring Harbor Perspec. Biol 2016, 8, a018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, Gilgenast MJ, Hauc S and Chatterjee A, ACS Chem. Biol 2018. 13, 1137–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikarand GJ Shokat KM, Cell, 2007, 128, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews FH, Gatchalian J, Krajewski K, Strahl BD and Kutateladze TG, ACS Chem. Biol 2016, 11, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H and Allis CD, Nature, 2005, 438, 1116–1122. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z and Patel DJ, J. Biol. Chem 2011, 286, 18363–18368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grunbeck A, Huber T, Sachdev P and Sakmar TP, Biochemistry, 2011, 50, 3411–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.