Figure 5.
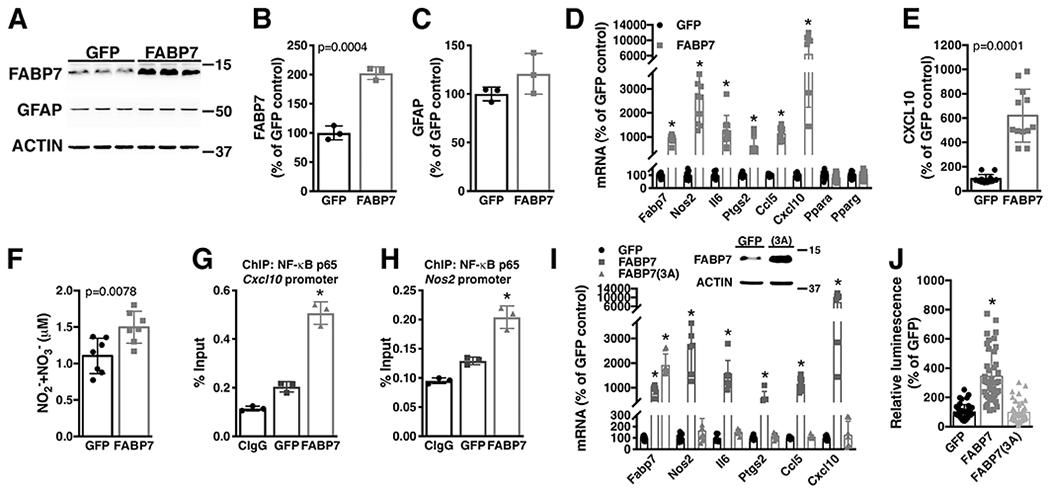
Over-expression of FABP7 confers upon non-transgenic astrocytes a proinflammatory phenotype. A) Primary confluent spinal cord astrocyte cultures obtained from neonatal NonTG mice were transduced with adenovirus expressing GFP or FABP7. 48hs later, FABP7 and GFAP protein levels were determined and corrected by actin levels. B, C) Quantification of the images shown in A. D) Fabp7, Nos2, Il6, Ptgs2, Ccl5, Cxcl10, Ppara and Pparg mRNA levels in astrocytes treated as in A. mRNA levels were determined by real-time PCR and corrected by Actin mRNA levels. Data is expressed as percentage of GFP control cells. E) CXCL10 levels in conditioned media (48hs) from GFP or FABP7 over-expressing NonTG astrocytes. Data is expressed as percentage of GFP control cells. F) Nitrite (NO2−) and nitrate (NO3−) levels in conditioned media (48hs) from GFP or FABP7 over-expressing NonTG astrocytes. G, H) Astrocytes were treated as in A and 48hs later chromatin immunoprecipitation was performed with a pre-immune IgG (CIgG) or an anti-NF-κB p65 subunit antibody. Purified DNA was analyzed by real-time PCR with specific primers flanking an NF-κB binding site in the Cxcl10 (G) or Nos2 (H) promoter. I) Real-time PCR analysis of Fabp7, Nos2, Il6, Ptgs2, Ccl5, Cxcl10 mRNA expression levels 48hs after FABP7 or FABP7(3A) over-expression in neonatal NonTG astrocytes. mRNA levels were corrected by Actin levels and expressed as percentage of GFP control cells. The data points for GFP and FABP7 in panel K were also included in panel F. Panel F includes additional biological replicates performed independently of the experiment described in panel K. J) Relative luminescence produced by firefly luciferase expressed under an NF-κB-driven promoter 48hs after FABP7 or FABP7(3A) over-expression in neonatal NonTG astrocytes. Relative firefly luciferase luminescence was corrected by the amount of Renilla luciferase activity controlled by a constitutive promoter and expressed as percentage of GFP control cells. For all graph panels, data are expressed as mean±S.D. (*p<0.05).
