Abstract
Abstract
Ovarian cancer is the eighth most commonly occurring cancer in women. Clinically, the limitation of conventional screening and monitoring approaches inhibits high throughput analysis of the tumor molecular markers toward prediction of treatment response. Recently, analysis of liquid biopsies including circulating tumor DNA (ctDNA) open new way toward cancer diagnosis and treatment in a personalized manner in various types of solid tumors. In the case of ovarian carcinoma, growing pre-clinical and clinical studies underscored promising application of ctDNA in diagnosis, prognosis, and prediction of treatment response. In this review, we accumulate and highlight recent molecular findings of ctDNA analysis and its associations with treatment response and patient outcome. Additionally, we discussed the potential application of ctDNA in the personalized treatment of ovarian carcinoma.
Graphical abstract
ctDNA-monitoring usage during the ovarian cancer treatments procedures.
Keywords: Ovarian cancer, Circulating tumor DNA, Prognosis
Introduction
Ovarian cancer was reported with the highest mortality rate (almost 50% of new cases are annually reported by the American cancer society) among the gynecologic malignancies [1]. Most of ovarian cancer patients are diagnosed at the advanced stages, at which the tumors have disseminated. Depending on the stages of the disease, the treatment approach may consist of surgery, chemo, radiation, hormone or targeted therapy toward shrinkage and the elimination of the primary tumor and also suppression of the metastatic progression [2]. Clinically, response to standard of the treatment regimens is vary among the patients due to the complexity of disease, particularly cellular and molecular heterogeneities of tumor. Therefore, the prediction of the treatment efficacy at early stages of therapy can enhance the accuracy of the patient’s selection toward the administration of appropriate treatment regimens, and particularly chemotherapy [2]. Although implementation of liquid biopsy approaches improved the patient outcome in various types of cancers including lung and breast in a personalized manner, in the case of ovarian cancer, utilizing an appropriate approach to identify the proper treatment for the specific patient is the matter of debate [3]. Accordingly, to tackle this problem, more studies must be performed to identify the biomarkers responsible for patient response to chemotherapies such as CA125 (Cancer Antigen 125) level [4] or human epididymis protein 4 (HE4) [5]. However, the accuracy and effectiveness of these biomarkers on the prediction of the chemotherapy response differ among the patients with various epidemic and clinical features [6]. Genomic profiling of tissue biopsy provides a snapshot of the dynamic behavior of tumor information and uncovers the genomic characterization of the tumor at the time of diagnosis [7]. Also, these challenges are particularly obvious in the patients who are resistant to therapy or in the patient’s follow up [8]. In recent years, identification and characterization of cancer-derived components such as circulating tumor cells (CTC), exosomes, and circulating tumor DNA (ctDNA) known as liquid biopsy [8, 9], opened a new way in the patients’ stratification and personalized treatment [7]. Of these, the detection of ctDNA tumor-specific mutations show a great promise in the patient’s selection and precision medicine, and besides, it can be suggested as a prognostic factor for the prediction of treatment response across several tumor types including lung, breast, colorectal, and melanoma cancers [9]. The previous review articles have reported the technological aspects of both detection and isolation of CTC alongside ctDNA as a diagnostic marker in ovarian cancer [10, 11]. While the present review, aim to consider mainly the clinical application of ctDNA in treatment and management of ovarian cancer patients. Moreover, the current review presents studies that compare genetic and molecular changes in ctDNA with tissues in ovarian cancer patients. The potential application of ctDNA as a prognostic factor for the prediction of the patient outcome in ovarian cancer also will be discussed.
The current challenge in the treatment of ovarian cancer
Understanding the molecular mechanisms of ovarian cancer, as a heterogeneous disease and underlying treatment resistance, can lead to discovery of some new therapeutic agents [12]. The standard of care treatment approaches for advance ovarian cancer is relaying on a primary cytoreductive surgery. Accordingly, it can be followed using an adjuvant therapy based on various chemotherapy-accompanied with the combination therapies regimens, which can improve response and the patients overall survival (OS) [13]. Unfortunately, chemotherapy resistance at the cancer advanced-stage is an important clinical challenge [14]. To date, several mechanisms of drug resistance have been explored, including inactivation of the p53 pathway [15], genome wide mutations [16], the enhanced expression of anti-apoptotic genes [17], epigenetic changes [18], dysfunctionality of DNA repair pathways [19], diminished drug accumulation [20], and the elevated drug inactivation [21]. In this regard, all these mechanisms lead to genomic instability, which allows cancer cells to adapt and survive against chemotherapy [22]. Beside these mechanisms, a particular role of cancer stem cells (CSCs) clones are defined within tumor microenvironment (TME) [23] and tumor-associated mesenchymal stem cells [24]. Also it has been observed that, CSCs have an appositive association with platinum, carboplatin, and paclitaxel resistance at the advanced-stage of ovarian cancer. In addition, the TME features including immune cell infiltration, angiogenesis, and hypoxia have been implicated in the platinum chemoresistance [25]. Furthermore, the investigation of molecular ovarian cancer tissue signature has paved the new way for biomarker discovery to assist the clinicians in making better treatment decisions. In this regard, a large number of clinical and preclinical studies suggested potential of tissue-based transcriptomics and proteomics biomarker information for the chemo-resistance prediction [26]. Meanwhile, pre-mediated cellular mechanisms such as clinical phenotypes, chaperones, metabolic proteins, transcription regulators, transporters, and cytoskeletal proteins are up-regulated in the patients with chemo-resistance. Although tissue biopsy is a gold standard to assess the pathological feature of disease, a recently liquid biopsy presented various advantages over this conventional approach [27]. The analysis and role of blood-based biomarker for evaluating the patient response to chemotherapy are undeniable [13]. Also, the plasma-derived ctDNA is the most commonly candidate in clinical practice due to its abundance and overcome on the isolation challenges [28]. The use of ctDNA in monitoring the patients with cancer prevents the risks associated with the repeated tissue biopsies [29].
The current prognostic biomarkers in the prediction of treatment response in ovarian cancer
Lack of an appropriate approach in chemotherapy response prediction in ovarian cancer is likely leading to poor patient’s survival [30]. CA125, which is expressed by epithelial ovarian tumors and other tissues of mullerian origin, was the first ovarian cancer biomarker described by Rober Bast et al. Accordingly, increase in the serum level of CA125 is observed in diverse malignancies, menstruation or pregnancy, and benign gynecological conditions [31]. The measurement of serum CA125, as a vital biomarker in clinical practice, was applied for screening high risk women, and also to predict clinical course and response to chemotherapy. In fact, dynamic changes in serum CA125 levels as a chemo responsiveness predictor can also be used to predict the response to the first-line and the second-line chemotherapy. However, the clinical value of CA125 is uncertain due to its limitations. For instance; it has been evidenced that, CA125 level is not elevated in 50% of the stage I women and in 30% of more advanced ones [32].
HE4 (Human Epididymis Protein 4) is overexpressed in ovarian cancer. Also, the combination of CA-125 and HE4 have the highest sensitivity and specificity in patients sera and also helps in the prediction of malignancy [33]. In addition, it is important to consider its potential false-positive results with CA 125 [34].
Ova1 measures five proteins named as CA-125, transthyretin, apolipoprotein A1, beta-2 microglobulin, and transferrin with the FDA approval [35]. Vascular Endothelial Growth Factor (VEGF) is a glycosylated angiogenesis mediator, which is independently associated with a shorter OS and disease-free survival. Notably, the combination of VEGF with CA-125 and HE4 increased the diagnostic sensitivity up to 84% at the stage I [36].
Kallikreins (KLKs) with 15 family members are responsible in cell growth, angiogenesis, invasion, and metastasis [37]. The decreased incidences of high “false negative” rates were also found in the HE4 and CA-125 positive patients [38].
Osteopontin (OPN) is a secreted extracellular matrix glycoprotein, which is involved in wound healing, the immune response, inflammation, tumorigenesis, bone remodeling, and apoptosis inhibition [39].
Mesothelin is a cell surface glycoprotein that are important in tumor metastasis, cancer cell survival, proliferation, and drug resistance [40]. Also, Mclntosh et al. detected the increased level of serum mesothelin in 60% of ovarian cancer patients with 98% specificity. So, a combination of mesothelin and CA-125 was suggested [41]. Additionally, Obulhasim et al. found that, mesothelin is expressed in 100% of serous cystadenocarcinoma as well as serous borderline ovarian tumor [42].
In addition to these biomarkers, few prognostic biomarkers were introduced as valuable indicators including macrophage colony-stimulating factor (M-CSF), bikunin, EphA2, Transthyretin (TTR), Transferrin receptor 1, B7-H4, Prostasin, and soluble EGF receptor [43–51] (Table 1).
Table 1.
List of known biomarkers in prediction of ovarian cancer treatment response
| Biomarkers | utility | Weakness |
|---|---|---|
| CA125 | Can be assessed in epithelial, endometrial and clear cell types in patients with clinical stage I- IV [52]. |
• Cannot be elevated in some ovarian cancer patients. • Can be elevated in healthy premenopausal women during menses, in pregnancy, in nonmalignant gynecologic diseases, such as ovarian cysts, endometriosis, adenomyosis, and uterine leiomyomas, in several nonmalignant nongynecological diseases, such as peritoneal, pleural, and musculoskeletal inflammatory disorders as well as pelvic inflammatory disease, liver, and renal as well as cardiac disease and in most types of advanced adenocarcinomas, including breast, colorectal, pancreas, lung, endometrium, and cervix as false positive. • Is not expressed in pure mucinous tumors [53]. |
| HE4 | Can be assessed in epithelial ovarian adenocarcinomas high [54]. |
• Can be elevated in endometrioid and clear cell histology [55]. • Cannot be detected in epithelial/ nonepithelial ovarian cancer, including sex cord stromal tumors and germ cell tumors [54] • Overexpressed in gastric cancer, pancreatic cancer as well as occasionally in colon and hepatocellular cancer [56, 57]. |
| Ova1 | Ova1 score ≥ 5 in premenopausal women and ≤ 5 postmenopausal ones were detected, and was considered with higher risk of malignancy [58]. | • Ova1 demonstrated 92.5% sensitivity, but lower specificity of 42.8% [58] |
| VEGF | VEGF level was independently associated with shorter disease-free survival and overall survival [59]. |
• Can be compared with traditional biomarkers, such as CA125 and HE4 moderately [60]. • It must be combined with CA-125 and HE4 to increase the diagnostic sensitivity up to 84% in stage I [36]. • Can be elevated in various cancers, including colorectal, [61], lung [62], gastric [63], endometrial [64] and breast cancer [65]. |
| Kallikreins | Level more than 4.4 mg/L indicated poor prognosis in patients [66]. |
• Exhibit low sensitivity in the early detection of ovarian cancer. • It must be combined with CA-125 for higher specificity and sensitivity [67]. |
| Osteopontin | Has a sensitivity of 83.3% in the detection of ovarian cancer [68]. |
• Its specificity is low. • It must be combined with CA-125 for higher sensitivity [69]. |
| Mesothelin | Elevate in patients with ovarian cancer compared with normal healthy [70]. | • Is not useful markers for early detection [71]. |
| M-CSF | Elevated levels of M-CSF1 in serum and ascites are associated with a poor prognosis [72]. Serum M-CSF appears to improve the diagnostic reliability of serum CA 125 alone [73]. | • This biomarker expressed also in other cancers [74]. |
| Bikunin |
Mediates suppression of tumor cell invasion and metastasis. Low expression is associated with late-stage disease. Low response to chemotherapy, and reduced survival time [44]. |
• Bikunin is present predominantly in amniotic fluid and urine of healthy individuals [75]. |
| EphA2 | Overexpression is associated with poor prognosis [45]. | • EphA2 is overexpressed in many human cancers [76, 77]. |
| Transthyretin | Efficient serum marker for the diagnosis [47]. |
• Plasma levels, affected by acute and chronic diseases. • Its usage must be considerate [78]. |
| Transferrin receptor 1 | Overexpression in high-grade tumor tissues [79]. | • Overexpressed in several cancers [80–82]. |
| B7-H4 | Over expression can be used as a tumor marker with negative prognostic effect for epithelial cell ovarian cancer potential immunotherapeutic target [83]. | • B7-H4 is highly expressed in various human tumors, including breast, ovarian, lung, pancreatic, gastric and urothelial cell carcinoma [84, 85]. |
| Prostasin | Overexpress in ovarian cancer patients at levels significantly higher than normal controls [50]. | • Many human cancers show unusual expression of prostasin like urinary bladder, uterine, prostate, gastric and ovarian cancers [86, 87]. |
| EGF receptor | Is associated with less favorable disease outcomes [88]. | • Little or no difference to survival, either as maintenance treatment after first-line chemotherapy or in association with chemotherapy in recurrent cancer [89]. |
Abbreviations: CA125 cancer antigen 125, HE4 human epididymis secretory protein 4, VEGF vascular endothelial growth factor, M-CSF macrophage colony stimulating factor, EphA2 ephrin type-A receptor 2, B7-H4 a molecule of B7 family
Circulating tumor DNA (ctDNA)
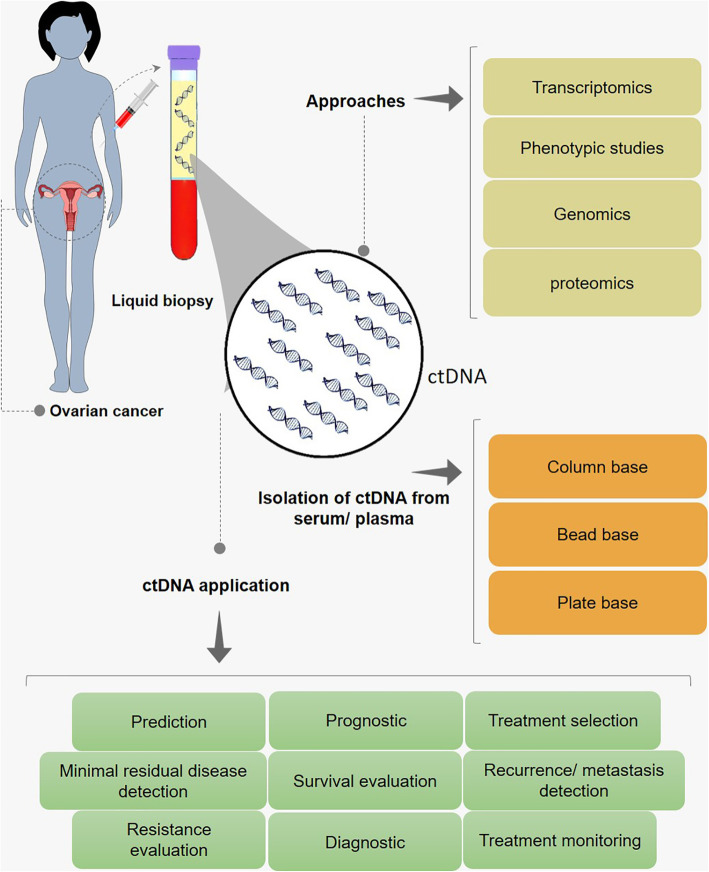
Liquid biopsy is a minimally invasive approach, which can be applied for the detection of molecular biomarkers from body fluids with no need for the costly or invasive procedures [90]. Accordingly, it is considered as a simple and non-invasive alternative to surgical biopsies, which enables discovering a wide range of information on a disease or a tumor through a simple blood sample. Notably, new dedicated methods allow us screening and monitoring cancer cell through circulating tumor cells (CTCs) and extracellular mirco-vesicles (including exosomes) containing small-RNA, mRNA, and ctDNA [91]. Tumor-associated genetic alterations can elucidate the molecular stratification of tumors toward the identification and selection of an appropriate targeted treatment. Historically, the presence of ctDNA in the blood of the patients with cancer was firstly recognized in the 1970s [92]. The elevated level of ctDNA is observed at the advanced stage of the disease progression, partly due to the reduced DNase activity [93]. Besides, ctDNA can reflect some specific genomic alterations of the tumor origin including mutations, methylation, and copy number variations (CNVs) [94] and preserving the genetic characteristics of the original tissue [95] (Fig. 1). The ctDNA-based mutation detection was also reported in 82% of the metastatic patients and 52% of the patients with localized disease [96]. Importantly, 95% concordance in mutational status was found between ctDNA and matched tumor tissue, which suggest that, the analysis of ctDNA, as a non-invasive approach, not only allows the tumor characterization and resolves the complexities of conventional tissue biopsy, but it also acts as an indicator for monitoring the treatment response in a given therapy [97]. A reliable biomarker can ensure that, which patients are more likely to relapse after receiving the adjuvant therapy [98]. Also, usage of ctDNA monitoring for a successful treatment over the resistance was reported in several clinical trials [99, 100]. Moreover, an invasive conventional tumor biopsy cannot be performed, while the frequent imaging can increase the risk of radiation exposure. Thus, the minimally invasive tests can be frequently repeated in a time series framework, which provide constant updates of tumor genetic composition and mutations, suggesting the best course of treatment at an appropriate time [101].
Fig. 1.
ctDNA isolation and application in the ovarian cancer patient
Tumor tissue-based mutations versus ctDNA mutations in ovarian cancer
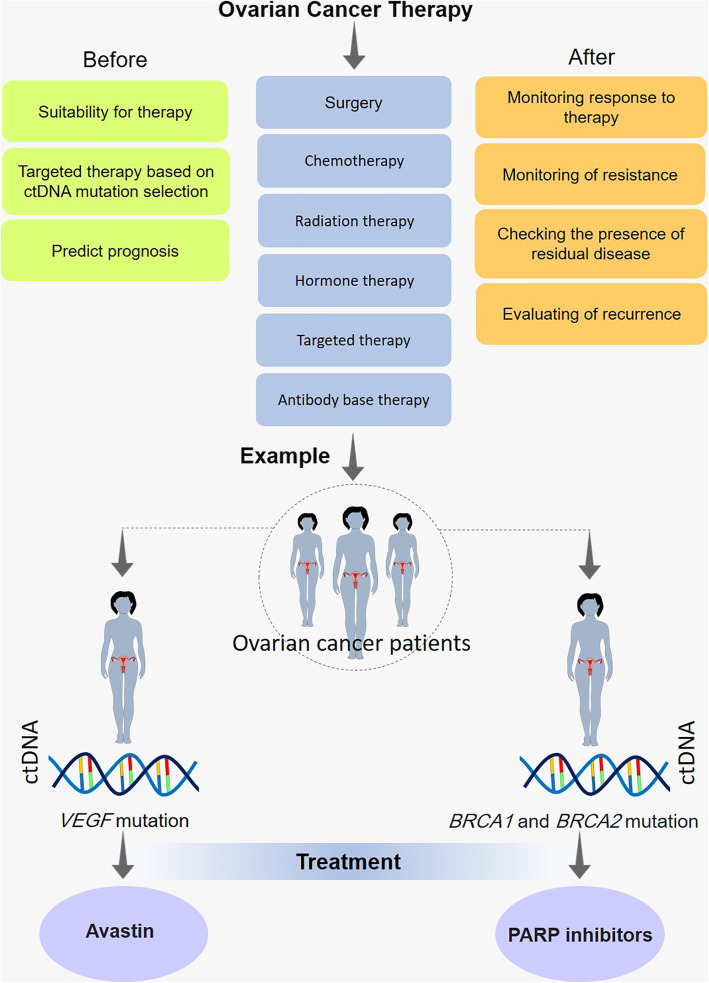
Over the last few years, genome-wide analysis revealed numerous alterations in ovarian cancer genomes including the inactivation of mutations in tumor suppressor genes such as TP53, BRCA1, BRCA2, PTEN, and RB1; and in a SWI/SNF chromatin remodeling gene, ARID1A [102, 103]. Other studies have detected the activation of mutations in the oncogenes KRAS, PIK3CA, BRAF, and ERBB2 [102, 104–106]. In fact, identifying the common gene mutation in blood rather than tissue, can be helpful in determining the patients whom benefit from therapy using the existing molecular targeting drugs. In this regard, Morikawa et al., [107] using the droplet digital PCR (ddPCR) detected PIK3CA-H1047R and KRASG12D mutations in tumor tissue and also matched ctDNA of 33 patients with ovarian clear cell carcinoma and then monitored their response to therapy. Accordingly, they highlighted the detection of mutations in ctDNA as a powerful tool for the diagnosis of ovarian clear cell carcinoma and for predicting its recurrence. In addition to this, Ogasawara et al. [108] assessed the PIK3CA and KRAS mutations in tumor and ctDNA of 304 patients with ovarian cancer. In addition, they indicated that, the detection rates of PIK3CA and/or KRAS ctDNA mutations were associated with the advanced stage; however, they were not related to the histologic subtype or residual tumor status. ctDNA detection was also associated with the shorter progress free survival (PFS) and the increased risk of recurrence independently [108]. In a pre-clinical study, the somatic mutation status of the TP53 was evaluated in both patient-derived tumor specimens and corresponding ctDNA, which resulted into the detection of similar hotspot mutation in TP53 in both sources of biopsy [109]. Moreover, Yong-Man Kim et al. [110] assessed the TP53 mutations across 103 tumor tissues from 61 patients with a high grade ovarian cancer and also confirmed TP53 mutations in 41 patients. They concluded that, detection of TP53 mutation in ctDNA is a potential tumor-specific biomarker for the treatment response monitoring [110]. Clinically, a copy number abnormality was reported in 20% of the patients with ovarian cancer [111] including claudin 4 (CLDN4), RAS oncogene family (RAB25), and ATP binding cassette subfamily F member 2 (ABCF2) [112, 113]. Hong No et al. [114] reported the lack of positive association between mutation in these genes at ctDNA level with the disease-free survival (DFS) and OS [114]. Moreover, BRCA1 and BRCA2 play pivotal roles in DNA repair and germline mutations [115]. Additionally, these BRCA1/2 reversion mutations could be detected by ctDNA sequencing analysis in the patients who received platinum and/or PARP inhibitors [116]. Rebecca et al. [117] compared the genetic variants of a panel of 50 genes between tumor and ctDNA among 20 patients diagnosed with the high-grade ovarian carcinoma during neoadjuvant chemotherapy (NACT). Notably, 38 genetic variants out of six genes (TP53, KIT, KDR, KRAS, PIK3CA, and PTEN) were identified in tumors pre-NACT, while 59 variants out of 19 genes were detected in the ctDNA. In this study, targeted NGS determined the increased level of ctDNA variants with a minimal overlap between ctDNA and tumor DNA. Most of the mutations found in ctDNA were not present in tumor resulting from the amplifying of ctDNA. Besides, the heterogeneity in the tumor can be detected in ctDNA, in contrast with tumor tissue [117].
The potential application of ctDNA to the management of treatment response in ovarian cancer
To date, FDA has not approved any ctDNA-related test in ovarian cancer. In this regard, a clinical ctDNA workflow was recently designed in the management of the high-grade ovarian cancer, to investigate the clinically actionable alterations of 500 cancer-related genes, which was performed in 12 patients. In seven patients, a good concordance of mutations and copy number alterations in ctDNA and tumor samples (NF1, RAD51C, PTEN, BRCA2, STAG2, FANCA, CDKN1B, ERBB2, ERBB4, and MAP 2 K1), and also alterations associated with the clinically available drugs (PI3K/mTOR inhibitor, PARP inhibitor, CDK2/4 inhibitor, CDK4/6 inhibitor, HER2 inhibitor, trastuzumab, ERBB inhibitor, lapatinib, and EGFR inhibitor) were detected. One chemo resistant patient therapy has changed based on the detection of ERBB2 amplification and ctDNA-guided decision. These results can be considered as a proof of using ctDNA concept to guide the clinical decisions during the cycles of chemotherapy in ovarian cancers [118]. In addition, Noguchi et al. [119] compared the variant allele frequency (VAF) of the measured ctDNA mutations during neoadjuvant chemotherapy in 10 plasma samples. In 5 out of 6 NAC-sensitive cases, the VAF of non-synonymous somatic mutations (TP53, KCAN5, and GJA8) decreased following NAC. Also, in two out of the four NAC-resistant cases, the VAF increased in the non-synonymous somatic mutations (KRAS, TRPS, and TP53). The rate of TP53 mutation was significantly higher in the resistant cases compared with the sensitive cases. In addition, the blood tumor mutation burden significantly decreased after the treatment in the sensitive cases. These findings showed that, gene mutation can be profiled and then monitored using ctDNA in ovarian cancer patients during treatment. The Table 2 summarizes the studies that used ctDNA analysis to monitor treatment response in ovarian carcinoma. Lately, a multi-center prospective study demonstrated that, detectable ctDNA following treatment is associated with a subsequent recurrence in ovarian cancer (trial, NCT03691012). In this study, serial blood and tumor samples were collected from 100-stage I-IV debulked ovarian cancer patients under the platinum-based treatment. Each patient was followed for more than a 6- to 8-month period for ctDNA (mutation) and CA125 analysis. After the completion of 6 cycles of chemotherapy, analysis of plasma ctDNA has been shown to exhibit the tumor-related alteration. Regarding the treatment response monitoring in ovarian cancer, an ongoing prospective multicenter trial (NCT03302884) was established to assess ctDNA value for ovarian cancer recurrence after the front-line treatment of chemotherapy to profile the significant gene modifications before the clinical diagnosis of disease relapse. In addition, the assessment of the minimal residual disease through plasma ctDNA in the ovarian cancer patients is currently underway the prospective study (trial, NCT03614689). In this study, match tumor DNA and longitudinal plasma sample was collected from 100 ovarian cancer patients before, during, and after the treatment. The correlation between the clonal status of mutations and therapy response, whether ctDNA detection would be used to predict the ovarian cancer recurrence risk before and after treatment, is an important point that was considered in this study. The immune checkpoint blockade (ICB) recently provides clinical benefits to a subset of patients with ovarian cancer. Bratman et al. [120] have started a prospective phase II clinical trial to assess ctDNA in five distinct cohorts of patients including high grade serous ovarian cancer patients with advanced solid tumors treated with pembrolizumab (NCT02644369). At baseline, 316 serial plasma samples at every three cycles from 94 patients were obtained. These findings indicated that serial ctDNA analysis could serve as a general monitoring strategy for patients treated with ICB and correlates with their survival. The Table 3 summarizes the clinical trial studies that used ctDNA analysis in ovarian cancer management.
Table 2.
Studies of ctDNA in ovarian cancer patients related to treatment response monitoring
| References | year | No of patients | Identified Abnormalities | Methodology |
|---|---|---|---|---|
| Gifford et al. [121] | 2004 | 138 | hMLH1 methylation | Microsatellite PCR |
| Swisher et al. [122] | 2005 | 137 | p53 mutation | DNA sequencing |
| Kamat et al. [123] | 2006 | – | Level | RT-PCR |
| Capizzi et al. [124] | 2008 | 22 | Level | RT-PCR |
| Kamat et al. [125] | 2010 | 164 | Beta-globin | RT-PCR |
| Wimberger et al. [126] | 2011 | 62 | Fluorimetry | Fluorescence |
| Forshew et al. [127] | 2012 | 38 | TP53. Other markers include PTEN, BRAF, KRAS, EGFR, PIK3CA | TAm-Seq, dPCR |
| Murtaza et al. [128] | 2013 | 3 | RB1, ZEB2, BUB1, CES4A, MTOR, PARP8 | NGS, qPCR |
| Choudhuri et al. [129] | 2014 | 100 | Level | RT-PCR |
| Martignetti et al. [130] | 2014 | 1 | FGFR2 fusion transcript | RT-PCR |
| Pereira et al. [131] | 2015 | 22 | TP53, PTEN, PIK3CA, MET, KRAS, FBXW7, BRAF | WES, ddPCR, TGS |
| Cohen et al. [132] | 2016 | 32 | CNV | WES |
| Harris et al. [133] | 2016 | 10 | Aberrant chromosomal junctions | RT-PCR |
| Piskorz et al. [134] | 2016 | 18 | TP53 mutation | NGS |
| Parkinson et al. [135] | 2016 | 40 | TP53 mutation | Digital PCR |
| Vanderstichele [136] | 2017 | 57 | CNV | WGS |
| Phallen et al. [137] | 2017 | 42 | 55 gene panel including TP53, KIT, ALK, APC, ERBB4 etc | NGS (TEC-Seq) and ddPCR |
| Flanagan et al. [138] | 2017 | 247 | Methylation at CpG sites | NGS |
| Widschwendter et al. [139] | 2017 | 151 | Regions linked to COL23A1, C2CD4D and WNT6 | TUC-BS & RRBS |
| Ratajska et al. [140] | 2017 | 121 | BRCA1/2 mutations | NGS |
| Christie et al. [141] | 2017 | 30 | BRCA reversion mutation | NGS |
| Weigelt et al. [116] | 2017 | 19 | BRCA reversion mutation | NGS |
| Giannopoulou et al. [142] | 2018 | 50 | ESR1 | RT-MSP |
| Du et al. [143] | 2018 | 21 | CNV and mutant genes including TP53, BRCA1, NOTCH2, DNMT3A etc | NGS |
| Morikawa et al. [107] | 2018 | 29 | KRAS and PIK3CA | ddPCR |
| Nakabayashi et al. [144] | 2018 | 36 | CNV | WGS |
| Park et al. [109] | 2018 | 4 | TP53 | ddPCR |
| Arend et al. [117] | 2018 | 14 | 50 gene panel | NGS |
| Lin et al. [145] | 2019 | 97 | BRCA reversion mutation, TP53 | NGS |
| Kim et al. [110] | 2019 | 102 | TP53 mutant allele | Sanger sequencing/Digital PCR |
| Oikkonen et al. [118] | 2019 | 12 | ERBB2 amplification | NGS |
| Iwahashi et al. [146] | 2019 | 4 | TP53, APC, BRCA1 and KRAS | CAPP-seq |
| Noguchi et al. [119] | 2020 | 10 | gene mutation profiles and blood tumor mutation burden | CAPP-seq |
| Han et al. [147] | 2020 | 10 | 88 genes panel | NGS |
| Alves et al. [148] | 2020 | 11 | Level | qPCR |
Abbreviations: NGS Next-generation sequencing, RT-PCR Reverse transcription polymerase chain reaction, dPCR droplet Polymerase chain reaction, qPCR Allele-specific quantitative PCR, RT-MSP Real-Time methylation specific PCR, CNV Copy number variation, WGS Whole genome sequencing, WES Whole exome sequencing, ddPCR Droplet digital PCR, TGS Targeted gene sequences, TAm-RSeq Targeted amplicon re-sequencing, RRBS Reduced representation bisulphite sequencing, TUC-BS Targeted ultra-high coverage bisulphite sequencing, CAPP-seq Cancer Personalized Profiling by deep Sequencing
Table 3.
Clinical trial studies related to ctDNA in ovarian cancer patients
| Clinical trial title | Participants | Date | Interventions | Recruitment Status | ClinicalTrials.gov Identifier |
|---|---|---|---|---|---|
| Plasma ctDNA detection in diagnosis of epithelial ovarian cancer. (ctDNA_EOC) | 43 | October 19, 2017 | Diagnostic Test: methylation markers screening | Completed | NCT03155451 |
| Study of circulating tumoral DNA in ovarian cancer. | 25 | January 23, 2017 | Blood sampling | Completed | NCT01350908 |
| Circulating tumor DNA guiding (Olaparib) Lynparza® treatment in ovarian cancer. | 160 | October 18, 2018 |
• Drug: Olaparib • Drug: carboplatin + gemcitabine or carboplatin + paclitaxel or carboplatin + liposomal doxorubicin or liposomal doxorubicin 4-weekly or topotecan or paclitaxel weekly |
Recruiting | NCT02822157 |
| Assessment of the minimal residual disease in ovarian cancer from circulating tumor DNA and immune repertoire. | 100 | August 3, 2018 | – | Recruiting | NCT03614689 |
| Circulating tumor DNA as a marker of residual disease & response to adjuvant chemotherapy in stage I-IV ovarian cancer. | 100 | October 1, 2018 | Diagnostic Test: Circulating tumour DNA testing | Recruiting | NCT03691012 |
| Circulating tumor DNA as an early marker of recurrence and treatment efficacy in ovarian carcinoma (CIDOC). | 150 | September 26, 2019 | biological sampling | Recruiting | NCT03302884 |
| Study of the effects of pembrolizumab in patients with advanced solid tumors | 94 | March 21, 2016 | • Biological: Pembrolizumab | Active, not recruiting | NCT02644369 |
Conclusion and future perspective
In summary, ctDNA detection before treatment facilitates early detection and leads to appropriate treatment decision-making based on patient stratification. Monitoring of the residual disease helps in prevention of recurrence of the tumor. During the course of treatment, regular monitoring of ctDNA can elucidate drug resistance acquired from genetic alterations, which are always present but not detectable by conventional approaches. Therefore, genomic-based drug response prediction can open new horizons in oncology to enable better cancer patient’s management. In addition, a considerable number of clinical trials, mentioned in Table 3, highlight the strong and novel roles of ctDNA in ovarian cancer management guidelines. Further efforts are required in the future for standardization of analysis platforms and incorporation of liquid biopsies as a companion biomarker in large-scale therapeutic trials.
Acknowledgments
We are so grateful from our colleagues in Iran University of Medical Sciences and Royan Stem Cell Technology Company who provided insight and expertise that greatly assisted the research.
Ethical approval and consent to participate
This manuscript is a review paper. So, “Not applicable”.
Authors’ contributions
HAE and MS conceived the concept for the manuscript. MS wrote the initial draft of the manuscript. All authors edited the manuscript. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
This manuscript is a review paper. So, “Not applicable”.
Consent for publication
The manuscript does not contain data from any individual person. So, “Not applicable”.
Competing interests
All the authors declare that they have no conflicts of interest to this work.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mehdi Totonchi, Email: m.totonchi@royaninstitute.org.
Zahra Madjd, Email: Zahra.madjd@yahoo.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Yang R, Niepel M, Mitchison TK, Sorger PK. Dissecting variability in responses to cancer chemotherapy through systems pharmacology. Clin Pharmacol Ther. 2010;88(1):34–38. doi: 10.1038/clpt.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang L, Ni J, Zhu Y, Pang B, Graham P, Zhang H, et al. Liquid biopsy in ovarian cancer: recent advances in circulating extracellular vesicle detection for early diagnosis and monitoring progression. Theranostics. 2019;9(14):4130–4140. doi: 10.7150/thno.34692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zorn KK, Tian C, McGuire WP, Hoskins WJ, Markman M, Muggia FM, et al. The prognostic value of pretreatment CA 125 in patients with advanced ovarian carcinoma: a gynecologic oncology group study. Cancer. 2009;115(5):1028–1035. doi: 10.1002/cncr.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang X, Ye X, Dong L, Cheng H, Cheng Y, Zhu L, et al. Human epididymis protein 4 (HE4) as a serum tumor biomarker in patients with ovarian carcinoma. Int J Gynecol Cancer. 2011;21(5):852–858. doi: 10.1097/IGC.0b013e31821a3726. [DOI] [PubMed] [Google Scholar]
- 6.Zargari A, Du Y, Heidari M, Thai TC, Gunderson CC, Moore K, et al. Prediction of chemotherapy response in ovarian cancer patients using a new clustered quantitative image marker. Phys Med Biol. 2018;63(15):155020. doi: 10.1088/1361-6560/aad3ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 8.Perakis S, Speicher MR. Emerging concepts in liquid biopsies. BMC Med. 2017;15(1):75. doi: 10.1186/s12916-017-0840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmirotta R, Lovero D, Cafforio P, Felici C, Mannavola F, Pelle E, et al. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol. 2018;10:1758835918794630. doi: 10.1177/1758835918794630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng X, Zhang L, Chen Y, Qing C. Circulating cell-free DNA and circulating tumor cells, the “liquid biopsies” in ovarian cancer. J Ovarian Res. 2017;10(1):75. doi: 10.1186/s13048-017-0369-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giannopoulou L, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: recent advances on circulating tumor cells and circulating tumor DNA. Clin Chem Lab Med. 2018;56(2):186–197. doi: 10.1515/cclm-2017-0019. [DOI] [PubMed] [Google Scholar]
- 12.Darcy KM, Birrer MJ. Translational research in the gynecologic oncology group: evaluation of ovarian cancer markers, profiles, and novel therapies. Gynecol Oncol. 2010;117(3):429–439. doi: 10.1016/j.ygyno.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pokhriyal R, Hariprasad R, Kumar L, Hariprasad G. Chemotherapy resistance in advanced ovarian cancer patients. Biomark Cancer. 2019;11:1179299X19860815. doi: 10.1177/1179299X19860815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooke SL, Brenton JD. Evolution of platinum resistance in high-grade serous ovarian cancer. Lancet Oncol. 2011;12(12):1169–1174. doi: 10.1016/S1470-2045(11)70123-1. [DOI] [PubMed] [Google Scholar]
- 15.Yang-Hartwich Y, Soteras MG, Lin ZP, Holmberg J, Sumi N, Craveiro V, et al. p53 protein aggregation promotes platinum resistance in ovarian cancer. Oncogene. 2015;34(27):3605–3616. doi: 10.1038/onc.2014.296. [DOI] [PubMed] [Google Scholar]
- 16.Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521(7553):489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 17.Eliopoulos AG, Kerr DJ, Herod J, Hodgkins L, Krajewski S, Reed JC, et al. The control of apoptosis and drug resistance in ovarian cancer: influence of p53 and Bcl-2. Oncogene. 1995;11(7):1217–1228. [PubMed] [Google Scholar]
- 18.Chang PY, Liao YP, Wang HC, Chen YC, Huang RL, Wang YC, et al. An epigenetic signature of adhesion molecules predicts poor prognosis of ovarian cancer patients. Oncotarget. 2017;8(32):53432–53449. doi: 10.18632/oncotarget.18515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barakat BM, Wang QE, Han C, Milum K, Yin DT, Zhao Q, et al. Overexpression of DDB2 enhances the sensitivity of human ovarian cancer cells to cisplatin by augmenting cellular apoptosis. Int J Cancer. 2010;127(4):977–988. doi: 10.1002/ijc.25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyashita H, Nitta Y, Mori S, Kanzaki A, Nakayama K, Terada K, et al. Expression of copper-transporting P-type adenosine triphosphatase (ATP7B) as a chemoresistance marker in human oral squamous cell carcinoma treated with cisplatin. Oral Oncol. 2003;39(2):157–162. doi: 10.1016/S1368-8375(02)00038-6. [DOI] [PubMed] [Google Scholar]
- 21.Kasahara K, Fujiwara Y, Nishio K, Ohmori T, Sugimoto Y, Komiya K, et al. Metallothionein content correlates with the sensitivity of human small cell lung cancer cell lines to cisplatin. Cancer Res. 1991;51(12):3237–3242. [PubMed] [Google Scholar]
- 22.Yao Y, Dai W. Genomic instability and cancer. J Carcinog Mutagen. 2014;5. 10.4172/2157-2518.1000165. [DOI] [PMC free article] [PubMed]
- 23.Steg AD, Bevis KS, Katre AA, Ziebarth A, Dobbin ZC, Alvarez RD, et al. Stem cell pathways contribute to clinical chemoresistance in ovarian cancer. Clin Cancer Res. 2012;18(3):869–881. doi: 10.1158/1078-0432.CCR-11-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chebouti I, Kasimir-Bauer S, Buderath P, Wimberger P, Hauch S, Kimmig R, et al. EMT-like circulating tumor cells in ovarian cancer patients are enriched by platinum-based chemotherapy. Oncotarget. 2017;8(30):48820–48831. doi: 10.18632/oncotarget.16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senthebane DA, Rowe A, Thomford NE, Shipanga H, Munro D, Mazeedi M, et al. The role of tumor microenvironment in chemoresistance: to survive, keep your enemies closer. Int J Mol Sci. 2017;18(7). 10.3390/ijms18071586. [DOI] [PMC free article] [PubMed]
- 26.Schwartz GW, Petrovic J, Zhou Y, Faryabi RB. Differential integration of transcriptome and proteome identifies pan-cancer prognostic biomarkers. Front Genet. 2018;9:205. doi: 10.3389/fgene.2018.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung AH, Chow C, To KF. Latest development of liquid biopsy. J Thorac Dis. 2018;10(Suppl 14):S1645–S1S51. doi: 10.21037/jtd.2018.04.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castro-Giner F, Gkountela S, Donato C, Alborelli I, Quagliata L, Ng CKY, et al. Cancer diagnosis using a liquid biopsy: challenges and expectations. Diagnostics (Basel). 2018;8(2). 10.3390/diagnostics8020031. [DOI] [PMC free article] [PubMed]
- 29.Alix-Panabieres C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6(5):479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 30.Lloyd KL, Cree IA, Savage RS. Prediction of resistance to chemotherapy in ovarian cancer: a systematic review. BMC Cancer. 2015;15:117. doi: 10.1186/s12885-015-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oikonomopoulou K, Li L, Zheng Y, Simon I, Wolfert RL, Valik D, et al. Prediction of ovarian cancer prognosis and response to chemotherapy by a serum-based multiparametric biomarker panel. Br J Cancer. 2008;99(7):1103–1113. doi: 10.1038/sj.bjc.6604630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta D, Lis CG. Role of CA125 in predicting ovarian cancer survival - a review of the epidemiological literature. J Ovarian Res. 2009;2:13. doi: 10.1186/1757-2215-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore RG, Brown AK, Miller MC, Skates S, Allard WJ, Verch T, et al. The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol Oncol. 2008;108(2):402–408. doi: 10.1016/j.ygyno.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 34.Escudero JM, Auge JM, Filella X, Torne A, Pahisa J, Molina R. Comparison of serum human epididymis protein 4 with cancer antigen 125 as a tumor marker in patients with malignant and nonmalignant diseases. Clin Chem. 2011;57(11):1534–1544. doi: 10.1373/clinchem.2010.157073. [DOI] [PubMed] [Google Scholar]
- 35.Nolen BM, Lokshin AE. Biomarker testing for ovarian cancer: clinical utility of multiplex assays. Mol Diagn Ther. 2013;17(3):139–146. doi: 10.1007/s40291-013-0027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawicki S, Bedkowska GE, Gacuta-Szumarska E, Szmitkowski M. The plasma concentration of VEGF, HE4 and CA125 as a new biomarkers panel in different stages and sub-types of epithelial ovarian tumors. J Ovarian Res. 2013;6(1):45. doi: 10.1186/1757-2215-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong W, Liu Y, Seidl C, Diamandis EP, Kiechle M, Drecoll E, et al. Quantitative assessment and clinical relevance of kallikrein-related peptidase 5 mRNA expression in advanced high-grade serous ovarian cancer. BMC Cancer. 2019;19(1):696. doi: 10.1186/s12885-019-5901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamir A, Jag U, Sarojini S, Schindewolf C, Tanaka T, Gharbaran R, et al. Kallikrein family proteases KLK6 and KLK7 are potential early detection and diagnostic biomarkers for serous and papillary serous ovarian cancer subtypes. J Ovarian Res. 2014;7:109. doi: 10.1186/s13048-014-0109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu ZD, Wei TT, Yang M, Ma N, Tang QQ, Qin BD, et al. Diagnostic value of osteopontin in ovarian cancer: a meta-analysis and systematic review. PLoS One. 2015;10(5):e0126444. doi: 10.1371/journal.pone.0126444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Z, Qian M, Ho M. The role of mesothelin in tumor progression and targeted therapy. Anti Cancer Agents Med Chem. 2013;13(2):276–280. doi: 10.2174/1871520611313020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McIntosh MW, Drescher C, Karlan B, Scholler N, Urban N, Hellstrom KE, et al. Combining CA 125 and SMR serum markers for diagnosis and early detection of ovarian carcinoma. Gynecol Oncol. 2004;95(1):9–15. doi: 10.1016/j.ygyno.2004.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obulhasim G, Fujii H, Matsumoto T, Yasen M, Abe M, Matsuoka S, et al. Mesothelin gene expression and promoter methylation/hypomethylation in gynecological tumors. Eur J Gynaecol Oncol. 2010;31(1):63–71. [PubMed] [Google Scholar]
- 43.Bedkowska GE, Lawicki S, Gacuta E, Pawlowski P, Szmitkowski M. M-CSF in a new biomarker panel with HE4 and CA 125 in the diagnostics of epithelial ovarian cancer patients. J Ovarian Res. 2015;8:27. doi: 10.1186/s13048-015-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuzaki H, Kobayashi H, Yagyu T, Wakahara K, Kondo T, Kurita N, et al. Plasma bikunin as a favorable prognostic factor in ovarian cancer. J Clin Oncol. 2005;23(7):1463–1472. doi: 10.1200/JCO.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Lu C, Shahzad MM, Wang H, Landen CN, Kim SW, Allen J, et al. EphA2 overexpression promotes ovarian cancer growth. Cancer Biol Ther. 2008;7(7):1098–1103. doi: 10.4161/cbt.7.7.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hariprasad G, Hariprasad R, Kumar L, Srinivasan A, Kola S, Kaushik A. Apolipoprotein A1 as a potential biomarker in the ascitic fluid for the differentiation of advanced ovarian cancers. Biomarkers. 2013;18(6):532–541. doi: 10.3109/1354750X.2013.822561. [DOI] [PubMed] [Google Scholar]
- 47.Zheng X, Chen S, Li L, Liu X, Liu X, Dai S, et al. Evaluation of HE4 and TTR for diagnosis of ovarian cancer: comparison with CA-125. J Gynecol Obstet Hum Reprod. 2018;47(6):227–230. doi: 10.1016/j.jogoh.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Nosov V, Su F, Amneus M, Birrer M, Robins T, Kotlerman J, et al. Validation of serum biomarkers for detection of early-stage ovarian cancer. Am J Obstet Gynecol. 2009;200(6):639 e1–639 e5. doi: 10.1016/j.ajog.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 49.Liang L, Jiang Y, Chen JS, Niu N, Piao J, Ning J, et al. B7-H4 expression in ovarian serous carcinoma: a study of 306 cases. Hum Pathol. 2016;57:1–6. doi: 10.1016/j.humpath.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mok SC, Chao J, Skates S, Wong K, Yiu GK, Muto MG, et al. Prostasin, a potential serum marker for ovarian cancer: identification through microarray technology. J Natl Cancer Inst. 2001;93(19):1458–1464. doi: 10.1093/jnci/93.19.1458. [DOI] [PubMed] [Google Scholar]
- 51.Baron AT, Lafky JM, Boardman CH, Cora EM, Buenafe MC, Liu D, et al. Soluble epidermal growth factor receptor: a biomarker of epithelial ovarian cancer. Cancer Treat Res. 2009;149:189–202. doi: 10.1007/978-0-387-98094-2_9. [DOI] [PubMed] [Google Scholar]
- 52.Duffy MJ, Bonfrer JM, Kulpa J, Rustin GJ, Soletormos G, Torre GC, et al. CA125 in ovarian cancer: European group on tumor markers guidelines for clinical use. Int J Gynecol Cancer. 2005;15(5):679–691. doi: 10.1111/j.1525-1438.2005.00130.x. [DOI] [PubMed] [Google Scholar]
- 53.Soletormos G, Duffy MJ, Othman Abu Hassan S, Verheijen RH, Tholander B, Bast RC, Jr, et al. Clinical use of cancer biomarkers in epithelial ovarian cancer: updated guidelines from the European group on tumor markers. Int J Gynecol Cancer. 2016;26(1):43–51. doi: 10.1097/igc.0000000000000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galgano MT, Hampton GM, Frierson HF., Jr Comprehensive analysis of HE4 expression in normal and malignant human tissues. Mod Pathol. 2006;19(6):847–853. doi: 10.1038/modpathol.3800612. [DOI] [PubMed] [Google Scholar]
- 55.Georgakopoulos P, Mehmood S, Akalin A, Shroyer KR. Immunohistochemical localization of HE4 in benign, borderline, and malignant lesions of the ovary. Int J Gynecol Pathol. 2012;31(6):517–523. doi: 10.1097/PGP.0b013e31824fe269. [DOI] [PubMed] [Google Scholar]
- 56.Nozaki K, Ogawa M, Williams JA, Lafleur BJ, Ng V, Drapkin RI, et al. A molecular signature of gastric metaplasia arising in response to acute parietal cell loss. Gastroenterology. 2008;134(2):511–522. doi: 10.1053/j.gastro.2007.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Neal RL, Nam KT, LaFleur BJ, Barlow B, Nozaki K, Lee HJ, et al. Human epididymis protein 4 is up-regulated in gastric and pancreatic adenocarcinomas. Hum Pathol. 2013;44(5):734–742. doi: 10.1016/j.humpath.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller CY. Doctor, should I get this new ovarian cancer test-OVA1? Obstet Gynecol. 2010;116(2 Pt 1):246–247. doi: 10.1097/AOG.0b013e3181e934ba. [DOI] [PubMed] [Google Scholar]
- 59.Harlozinska A, Sedlaczek P, Kulpa J, Grybos M, Wojcik E, Van Dalen A, et al. Vascular endothelial growth factor (VEGF) concentration in sera and tumor effusions from patients with ovarian carcinoma. Anticancer Res. 2004;24(2C):1149–1157. [PubMed] [Google Scholar]
- 60.Zhen S, Bian LH, Chang LL, Gao X. Comparison of serum human epididymis protein 4 and carbohydrate antigen 125 as markers in ovarian cancer: a meta-analysis. Mol Clin Oncol. 2014;2(4):559–566. doi: 10.3892/mco.2014.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kushlinskii NE, Gershtein ES, Nikolaev AA, Delektorskaya VV, Korotkova EA, Dvorova EK, et al. Insulin-like growth factors (IGF), IGF-binding proteins (IGFBP), and vascular endothelial growth factor (VEGF) in blood serum of patients with colorectal cancer. Bull Exp Biol Med. 2014;156(5):684–688. doi: 10.1007/s10517-014-2425-0. [DOI] [PubMed] [Google Scholar]
- 62.Fu ZZ, Sun XD, Li P, Zhang Z, Li GZ, Gu T, et al. Relationship between serum VEGF level and radiosensitivity of patients with nonsmall cell lung cancer among asians: a meta-analysis. DNA Cell Biol. 2014;33(7):426–437. doi: 10.1089/dna.2013.2249. [DOI] [PubMed] [Google Scholar]
- 63.Wu J, Liu X, Wang Y. Predictive value of preoperative serum CCL2, CCL18, and VEGF for the patients with gastric cancer. BMC Clin Pathol. 2013;13:15. doi: 10.1186/1472-6890-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobrzycka B, Terlikowski SJ, Kowalczuk O, Kulikowski M, Niklinski J. Serum levels of VEGF and VEGF-C in patients with endometrial cancer. Eur Cytokine Netw. 2011;22(1):45–51. doi: 10.1684/ecn.2011.0276. [DOI] [PubMed] [Google Scholar]
- 65.Coskun U, Gunel N, Toruner FB, Sancak B, Onuk E, Bayram O, et al. Serum leptin, prolactin and vascular endothelial growth factor (VEGF) levels in patients with breast cancer. Neoplasma. 2003;50(1):41–46. [PubMed] [Google Scholar]
- 66.El Sherbini MA, Sallam MM, Shaban EA, El-Shalakany AH. Diagnostic value of serum kallikrein-related peptidases 6 and 10 versus CA125 in ovarian cancer. Int J Gynecol Cancer. 2011;21(4):625–632. doi: 10.1097/IGC.0b013e31821283c3. [DOI] [PubMed] [Google Scholar]
- 67.Diamandis EP, Scorilas A, Fracchioli S, Van Gramberen M, De Bruijn H, Henrik A, et al. Human kallikrein 6 (hK6): a new potential serum biomarker for diagnosis and prognosis of ovarian carcinoma. J Clin Oncol. 2003;21(6):1035–1043. doi: 10.1200/JCO.2003.02.022. [DOI] [PubMed] [Google Scholar]
- 68.Nakae M, Iwamoto I, Fujino T, Maehata Y, Togami S, Yoshinaga M, et al. Preoperative plasma osteopontin level as a biomarker complementary to carbohydrate antigen 125 in predicting ovarian cancer. J Obstet Gynaecol Res. 2006;32(3):309–314. doi: 10.1111/j.1447-0756.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 69.Mor G, Visintin I, Lai Y, Zhao H, Schwartz P, Rutherford T, et al. Serum protein markers for early detection of ovarian cancer. Proc Natl Acad Sci U S A. 2005;102(21):7677–7682. doi: 10.1073/pnas.0502178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hassan R, Remaley AT, Sampson ML, Zhang J, Cox DD, Pingpank J, et al. Detection and quantitation of serum mesothelin, a tumor marker for patients with mesothelioma and ovarian cancer. Clin Cancer Res. 2006;12(2):447–453. doi: 10.1158/1078-0432.CCR-05-1477. [DOI] [PubMed] [Google Scholar]
- 71.Blyuss O, Gentry-Maharaj A, Fourkala EO, Ryan A, Zaikin A, Menon U, et al. Serial patterns of ovarian cancer biomarkers in a prediagnosis longitudinal dataset. Biomed Res Int. 2015;2015:681416. doi: 10.1155/2015/681416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chambers SK, Kacinski BM, Ivins CM, Carcangiu ML. Overexpression of epithelial macrophage colony-stimulating factor (CSF-1) and CSF-1 receptor: a poor prognostic factor in epithelial ovarian cancer, contrasted with a protective effect of stromal CSF-1. Clin Cancer Res. 1997;3(6):999–1007. [PubMed] [Google Scholar]
- 73.Gadducci A, Cosio S, Zanca G, Genazzani AR. Evolving role of serum biomarkers in the management of ovarian cancer. Womens Health (Lond) 2006;2(1):141–158. doi: 10.2217/17455057.2.1.141. [DOI] [PubMed] [Google Scholar]
- 74.Sidorkiewicz I, Zbucka-Kretowska M, Zareba K, Lubowicka E, Zajkowska M, Szmitkowski M, et al. Plasma levels of M-CSF and VEGF in laboratory diagnostics and differentiation of selected histological types of cervical cancers. BMC Cancer. 2019;19(1):398. doi: 10.1186/s12885-019-5558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fries E, Blom AM. Bikunin--not just a plasma proteinase inhibitor. Int J Biochem Cell Biol. 2000;32(2):125–137. doi: 10.1016/s1357-2725(99)00125-9. [DOI] [PubMed] [Google Scholar]
- 76.Martini G, Cardone C, Vitiello PP, Belli V, Napolitano S, Troiani T, et al. EPHA2 is a predictive biomarker of resistance and a potential therapeutic target for improving antiepidermal growth factor receptor therapy in colorectal cancer. Mol Cancer Ther. 2019;18(4):845–855. doi: 10.1158/1535-7163.MCT-18-0539. [DOI] [PubMed] [Google Scholar]
- 77.Tan YC, Srivastava S, Won BM, Kanteti R, Arif Q, Husain AN, et al. EPHA2 mutations with oncogenic characteristics in squamous cell lung cancer and malignant pleural mesothelioma. Oncogenesis. 2019;8(9):49. doi: 10.1038/s41389-019-0159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lasztity N, Biro L, Nemeth E, Pap A, Antal M. Protein status in pancreatitis--transthyretin is a sensitive biomarker of malnutrition in acute and chronic pancreatitis. Clin Chem Lab Med. 2002;40(12):1320–1324. doi: 10.1515/CCLM.2002.227. [DOI] [PubMed] [Google Scholar]
- 79.Basuli D, Tesfay L, Deng Z, Paul B, Yamamoto Y, Ning G, et al. Iron addiction: a novel therapeutic target in ovarian cancer. Oncogene. 2017;36(29):4089–4099. doi: 10.1038/onc.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosager AM, Sorensen MD, Dahlrot RH, Hansen S, Schonberg DL, Rich JN, et al. Transferrin receptor-1 and ferritin heavy and light chains in astrocytic brain tumors: expression and prognostic value. PLoS One. 2017;12(8):e0182954. doi: 10.1371/journal.pone.0182954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rychtarcikova Z, Lettlova S, Tomkova V, Korenkova V, Langerova L, Simonova E, et al. Tumor-initiating cells of breast and prostate origin show alterations in the expression of genes related to iron metabolism. Oncotarget. 2017;8(4):6376–6398. doi: 10.18632/oncotarget.14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Okazaki F, Matsunaga N, Okazaki H, Azuma H, Hamamura K, Tsuruta A, et al. Circadian clock in a mouse colon tumor regulates intracellular iron levels to promote tumor progression. J Biol Chem. 2016;291(13):7017–7028. doi: 10.1074/jbc.M115.713412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu M, Zhang B, Zhang M, Liu Y, Yin FL, Liu X, et al. Clinical relevance of expression of B7-H1 and B7-H4 in ovarian cancer. Oncol Lett. 2016;11(4):2815–2819. doi: 10.3892/ol.2016.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong Q, Ma X. B7-H4 expression is associated with tumor progression and prognosis in patients with osteosarcoma. Biomed Res Int. 2015;2015:156432. doi: 10.1155/2015/156432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salceda S, Tang T, Kmet M, Munteanu A, Ghosh M, Macina R, et al. The immunomodulatory protein B7-H4 is overexpressed in breast and ovarian cancers and promotes epithelial cell transformation. Exp Cell Res. 2005;306(1):128–141. doi: 10.1016/j.yexcr.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 86.Selzer-Plon J, Bornholdt J, Friis S, Bisgaard HC, Lothe IM, Tveit KM, et al. Expression of prostasin and its inhibitors during colorectal cancer carcinogenesis. BMC Cancer. 2009;9:201. doi: 10.1186/1471-2407-9-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen LM, Wang C, Chen M, Marcello MR, Chao J, Chao L, et al. Prostasin attenuates inducible nitric oxide synthase expression in lipopolysaccharide-induced urinary bladder inflammation. Am J Physiol Renal Physiol. 2006;291(3):F567–F577. doi: 10.1152/ajprenal.00047.2006. [DOI] [PubMed] [Google Scholar]
- 88.Zeineldin R, Muller CY, Stack MS, Hudson LG. Targeting the EGF receptor for ovarian cancer therapy. J Oncol. 2010;2010:414676. doi: 10.1155/2010/414676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morrison J, Thoma C, Goodall RJ, Lyons TJ, Gaitskell K, Wiggans AJ, et al. Epidermal growth factor receptor blockers for the treatment of ovarian cancer. Cochrane Database Syst Rev. 2018;10:CD007927. doi: 10.1002/14651858.CD007927.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xue VW, Wong CSC, Cho WCS. Early detection and monitoring of cancer in liquid biopsy: advances and challenges. Expert Rev Mol Diagn. 2019;19(4):273–276. doi: 10.1080/14737159.2019.1583104. [DOI] [PubMed] [Google Scholar]
- 91.Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn. 2015;17(3):209–224. doi: 10.1016/j.jmoldx.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–650. [PubMed] [Google Scholar]
- 93.Cherepanova AV, Tamkovich SN, Bryzgunova OE, Vlassov VV, Laktionov PP. Deoxyribonuclease activity and circulating DNA concentration in blood plasma of patients with prostate tumors. Ann N Y Acad Sci. 2008;1137:218–221. doi: 10.1196/annals.1448.016. [DOI] [PubMed] [Google Scholar]
- 94.Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res. 2015;21(20):4586–4596. doi: 10.1158/1078-0432.CCR-15-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campos CDM, Jackson JM, Witek MA, Soper SA. Molecular profiling of liquid biopsy samples for precision medicine. Cancer J. 2018;24(2):93–103. doi: 10.1097/PPO.0000000000000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kumar M, Choudhury Y, Ghosh SK, Mondal R. Application and optimization of minimally invasive cell-free DNA techniques in oncogenomics. Tumour Biol. 2018;40(2):1010428318760342. doi: 10.1177/1010428318760342. [DOI] [PubMed] [Google Scholar]
- 98.Popper HH. Commentary on tumor heterogeneity. Transl Lung Cancer Res. 2016;5(4):433–435. doi: 10.21037/tlcr.2016.08.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reece M, Saluja H, Hollington P, Karapetis CS, Vatandoust S, Young GP, et al. The use of circulating tumor DNA to monitor and predict response to treatment in colorectal cancer. Front Genet. 2019;10:1118. doi: 10.3389/fgene.2019.01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Osumi H, Shinozaki E, Yamaguchi K, Zembutsu H. Early change in circulating tumor DNA as a potential predictor of response to chemotherapy in patients with metastatic colorectal cancer. Sci Rep. 2019;9(1):17358. doi: 10.1038/s41598-019-53711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 102.Kanchi KL, Johnson KJ, Lu C, McLellan MD, Leiserson MD, Wendl MC, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun. 2014;5:3156. doi: 10.1038/ncomms4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Testa U, Petrucci E, Pasquini L, Castelli G, Pelosi E. Ovarian cancers: genetic abnormalities, tumor heterogeneity and progression, clonal evolution and cancer stem cells. Medicines (Basel). 2018;5(1). 10.3390/medicines5010016. [DOI] [PMC free article] [PubMed]
- 104.Kalamanathan S, Bates V, Lord R, Green JA. The mutational profile of sporadic epithelial ovarian carcinoma. Anticancer Res. 2011;31(8):2661–2668. [PubMed] [Google Scholar]
- 105.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cancer Genome Atlas Research N Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morikawa A, Hayashi T, Shimizu N, Kobayashi M, Taniue K, Takahashi A, et al. PIK3CA and KRAS mutations in cell free circulating DNA are useful markers for monitoring ovarian clear cell carcinoma. Oncotarget. 2018;9(20):15266–15274. doi: 10.18632/oncotarget.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.A. Ogasawara, Hihara, T,.Yabuno A, S. Shimoyokkaichi, D. Shintani, Y. Ikeda, K. Tai, K. Fujiwara, K. Watanabe, K. Hasegawa. Evaluation of circulating tumor DNA in patients with ovarian cancer harboring somatic PIK3CA or KRAS mutations. ESMO 2017 CONGRESS. [DOI] [PMC free article] [PubMed]
- 109.Park YR, Kim YM, Lee SW, Lee HY, Lee GE, Lee JE, et al. Optimization to detect TP53 mutations in circulating cell-free tumor DNA from patients with serous epithelial ovarian cancer. Obstet Gynecol Sci. 2018;61(3):328–336. doi: 10.5468/ogs.2018.61.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim YM, Lee SW, Lee YJ, Lee HY, Lee JE, Choi EK. Prospective study of the efficacy and utility of TP53 mutations in circulating tumor DNA as a non-invasive biomarker of treatment response monitoring in patients with high-grade serous ovarian carcinoma. J Gynecol Oncol. 2019;30(3):e32. doi: 10.3802/jgo.2019.30.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sonoda G, Palazzo J, du Manoir S, Godwin AK, Feder M, Yakushiji M, et al. Comparative genomic hybridization detects frequent overrepresentation of chromosomal material from 3q26, 8q24, and 20q13 in human ovarian carcinomas. Genes Chromosomes Cancer. 1997;20(4):320–328. doi: 10.1002/(SICI)1098-2264(199712)20:4<320::AID-GCC2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 112.Odicino F, Pecorelli S, Zigliani L, Creasman WT. History of the FIGO cancer staging system. Int J Gynaecol Obstet. 2008;101(2):205–210. doi: 10.1016/j.ijgo.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 113.Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10(11):1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- 114.No JH, Kim K, Park KH, Kim YB. Cell-free DNA level as a prognostic biomarker for epithelial ovarian cancer. Anticancer Res. 2012;32(8):3467–3471. [PubMed] [Google Scholar]
- 115.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12(1):68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weigelt B, Comino-Mendez I, de Bruijn I, Tian L, Meisel JL, Garcia-Murillas I, et al. Diverse BRCA1 and BRCA2 reversion mutations in circulating cell-free DNA of therapy-resistant breast or ovarian cancer. Clin Cancer Res. 2017;23(21):6708–6720. doi: 10.1158/1078-0432.CCR-17-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Arend RC, Londono AI, Montgomery AM, Smith HJ, Dobbin ZC, Katre AA, et al. Molecular response to Neoadjuvant chemotherapy in high-grade serous ovarian carcinoma. Mol Cancer Res. 2018;16(5):813–824. doi: 10.1158/1541-7786.MCR-17-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oikkonen J, Zhang K, Salminen L, Schulman I, Lavikka K, Andersson N, et al. Prospective longitudinal ctDNA workflow reveals clinically actionable alterations in ovarian cancer. JCO Precision Oncol. 2019;3:1–12. doi: 10.1200/PO.18.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Noguchi T, Sakai K, Iwahashi N, Matsuda K, Matsukawa H, Yahata T, et al. Changes in the gene mutation profiles of circulating tumor DNA detected using CAPP-Seq in neoadjuvant chemotherapy-treated advanced ovarian cancer. Oncol Lett. 2020;19(4):2713–2720. doi: 10.3892/ol.2020.11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nature Cancer. 2020. 10.1038/s43018-020-0096-5. [DOI] [PubMed]
- 121.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10(13):4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 122.Swisher EM, Wollan M, Mahtani SM, Willner JB, Garcia R, Goff BA, et al. Tumor-specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am J Obstet Gynecol. 2005;193(3 Pt 1):662–667. doi: 10.1016/j.ajog.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 123.Kamat AA, Bischoff FZ, Dang D, Baldwin MF, Han LY, Lin YG, et al. Circulating cell-free DNA: a novel biomarker for response to therapy in ovarian carcinoma. Cancer Biol Ther. 2006;5(10):1369–1374. doi: 10.4161/cbt.5.10.3240. [DOI] [PubMed] [Google Scholar]
- 124.Capizzi E, Gabusi E, Grigioni AD, De Iaco P, Rosati M, Zamagni C, et al. Quantification of free plasma DNA before and after chemotherapy in patients with advanced epithelial ovarian cancer. Diagn Mol Pathol. 2008;17(1):34–38. doi: 10.1097/PDM.0b013e3181359e1f. [DOI] [PubMed] [Google Scholar]
- 125.Kamat AA, Baldwin M, Urbauer D, Dang D, Han LY, Godwin A, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer. 2010;116(8):1918–1925. doi: 10.1002/cncr.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wimberger P, Roth C, Pantel K, Kasimir-Bauer S, Kimmig R, Schwarzenbach H. Impact of platinum-based chemotherapy on circulating nucleic acid levels, protease activities in blood and disseminated tumor cells in bone marrow of ovarian cancer patients. Int J Cancer. 2011;128(11):2572–2580. doi: 10.1002/ijc.25602. [DOI] [PubMed] [Google Scholar]
- 127.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 128.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 129.Choudhuri S, Sharma C, Banerjee A, Kumar S, Kumar L, Singh N. A repertoire of biomarkers helps in detection and assessment of therapeutic response in epithelial ovarian cancer. Mol Cell Biochem. 2014;386(1–2):259–269. doi: 10.1007/s11010-013-1863-8. [DOI] [PubMed] [Google Scholar]
- 130.Martignetti JA, Camacho-Vanegas O, Priedigkeit N, Camacho C, Pereira E, Lin L, et al. Personalized ovarian cancer disease surveillance and detection of candidate therapeutic drug target in circulating tumor DNA. Neoplasia. 2014;16(1):97–103. doi: 10.1593/neo.131900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pereira E, Camacho-Vanegas O, Anand S, Sebra R, Catalina Camacho S, Garnar-Wortzel L, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One. 2015;10(12):e0145754. doi: 10.1371/journal.pone.0145754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cohen PA, Flowers N, Tong S, Hannan N, Pertile MD, Hui L. Abnormal plasma DNA profiles in early ovarian cancer using a non-invasive prenatal testing platform: implications for cancer screening. BMC Med. 2016;14(1):126. doi: 10.1186/s12916-016-0667-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Harris FR, Kovtun IV, Smadbeck J, Multinu F, Jatoi A, Kosari F, et al. Quantification of somatic chromosomal rearrangements in circulating cell-free DNA from ovarian cancers. Sci Rep. 2016;6:29831. doi: 10.1038/srep29831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Piskorz A, Lin KK, Morris JA, Mann E, Oza AM, Coleman RL, et al. Feasibility of monitoring response to the PARP inhibitor rucaparib with targeted deep sequencing of circulating tumor DNA (ctDNA) in women with high-grade serous carcinoma on the ARIEL2 trial. J Clin Oncol. 2016;34(15_suppl):5549. doi: 10.1200/JCO.2016.34.15_suppl.5549. [DOI] [Google Scholar]
- 135.Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016;13(12):e1002198. doi: 10.1371/journal.pmed.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vanderstichele A, Busschaert P, Smeets D, Landolfo C, Van Nieuwenhuysen E, Leunen K, et al. Chromosomal instability in cell-free DNA as a highly specific biomarker for detection of ovarian cancer in women with adnexal masses. Clin Cancer Res. 2017;23(9):2223–2231. doi: 10.1158/1078-0432.CCR-16-1078. [DOI] [PubMed] [Google Scholar]
- 137.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403). 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed]
- 138.Flanagan JM, Wilson A, Koo C, Masrour N, Gallon J, Loomis E, et al. Platinum-based chemotherapy induces methylation changes in blood DNA associated with overall survival in patients with ovarian cancer. Clin Cancer Res. 2017;23(9):2213–2222. doi: 10.1158/1078-0432.ccr-16-1754. [DOI] [PubMed] [Google Scholar]
- 139.Widschwendter M, Zikan M, Wahl B, Lempiainen H, Paprotka T, Evans I, et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 2017;9(1):116. doi: 10.1186/s13073-017-0500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ratajska M, Koczkowska M, Zuk M, Gorczynski A, Kuzniacka A, Stukan M, et al. Detection of BRCA1/2 mutations in circulating tumor DNA from patients with ovarian cancer. Oncotarget. 2017;8(60):101325–101332. doi: 10.18632/oncotarget.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Christie EL, Fereday S, Doig K, Pattnaik S, Dawson SJ, Bowtell DDL. Reversion of BRCA1/2 Germline mutations detected in circulating tumor DNA from patients with high-grade serous ovarian cancer. J Clin Oncol. 2017;35(12):1274–1280. doi: 10.1200/JCO.2016.70.4627. [DOI] [PubMed] [Google Scholar]
- 142.Giannopoulou L, Mastoraki S, Buderath P, Strati A, Pavlakis K, Kasimir-Bauer S, et al. ESR1 methylation in primary tumors and paired circulating tumor DNA of patients with high-grade serous ovarian cancer. Gynecol Oncol. 2018;150(2):355–360. doi: 10.1016/j.ygyno.2018.05.026. [DOI] [PubMed] [Google Scholar]
- 143.Du ZH, Bi FF, Wang L, Yang Q. Next-generation sequencing unravels extensive genetic alteration in recurrent ovarian cancer and unique genetic changes in drug-resistant recurrent ovarian cancer. Mol Genet Genomic Med. 2018. 10.1002/mgg3.414. [DOI] [PMC free article] [PubMed]
- 144.Nakabayashi M, Kawashima A, Yasuhara R, Hayakawa Y, Miyamoto S, Iizuka C, et al. Massively parallel sequencing of cell-free DNA in plasma for detecting gynaecological tumour-associated copy number alteration. Sci Rep. 2018;8(1):11205. doi: 10.1038/s41598-018-29381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lin KK, Harrell MI, Oza AM, Oaknin A, Ray-Coquard I, Tinker AV, et al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2019;9(2):210–219. doi: 10.1158/2159-8290.CD-18-0715. [DOI] [PubMed] [Google Scholar]
- 146.Iwahashi N, Sakai K, Noguchi T, Yahata T, Matsukawa H, Toujima S, et al. Liquid biopsy-based comprehensive gene mutation profiling for gynecological cancer using CAncer personalized profiling by deep sequencing. Sci Rep. 2019;9(1):10426. doi: 10.1038/s41598-019-47030-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Han MR, Lee SH, Park JY, Hong H, Ho JY, Hur SY, et al. Clinical implications of circulating tumor DNA from ascites and serial plasma in ovarian cancer. Cancer Res Treat. 2020;52(3):779–788. doi: 10.4143/crt.2019.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Alves MC, Fonseca FLA, Yamada A, Barros L, Lopes A, Silva L, et al. Increased circulating tumor DNA as a noninvasive biomarker of early treatment response in patients with metastatic ovarian carcinoma: a pilot study. Tumour Biol. 2020;42(5):1010428320919198. doi: 10.1177/1010428320919198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This manuscript is a review paper. So, “Not applicable”.