Abstract
IKZF1 belongs to the IKAROS family of transcription factors, and its deletion/mutation frequently affects acute lymphoblastic leukemia. In acute myeloid leukemia, IKZF1 deletion has been demonstrated recurrent, but whether IKZF1 mutation also exists in AML remained largely unknown. Herein, we analyzed the IKZF1 mutation in AML. In our cohort, the frequency of IKZF1 mutation was 2.6% (5/193), and 5 frameshift/nonsense mutations as well as 2 missense mutations were identified in total. Molecularly, IKZF1 mutation was absent in fusion gene-positive AML, but it was demonstrated as the significant concomitant genetic alteration with SF3B1 or bi-allele CEBPA mutation in AML. Clinically, two IKZF1, PTPN11 and SF3B1-mutated AML patients exhibited one aggressive clinical course and showed primary resistant to chemotherapy. Furthermore, we confirmed the recurrent IKZF1 mutation in AML with cBioPortal tool from OHSU, TCGA and TARGET studies. Interestingly, OHSU study also showed that SF3B1 mutation was the significant concomitant genetic alteration with IKZF1 mutation, indicating their strong synergy in leukemogenesis. In conclusion, IKZF1 mutation recurrently affected AML.
Keywords: IKZF1 mutation, Acute myeloid leukemia, Recurrence
IKZF1 belongs to the IKAROS family of transcription factors. It contains four zinc fingers at the N-terminal that directly bind to DNA at the core motif A/GGAAA and additional two zinc fingers at the C-terminal required for forming homo- and hetero-dimerization between different IKZF proteins [1]. DNA binding activity of IKZF1 can be enhanced by its dimerization, so both DNA-binding and dimer-forming defects alter IKZF1 function. IKZF1 deletions and mutations have been reported to affect B-cell precursor ALL and contribute to its poor prognosis [2]. IKZF1 alterations are less studied in AML. Recurrent IKZF1 deletions have been identified in AML [3, 4], but whether IKZF1 mutations affect AML in general remains unknown. Herein, we analyzed IKZF1 mutation in AML.
A total of 193 adult AML patients, who subjected to TES, were retrospectively analyzed in our center (01/05/2018–29/02/2020), while APL was excluded. Among these patients, 100 were male and 93 were female, and the median age was 56 (range 18–82). A total of 169 patients were diagnosed with de novo AML, 10 with refractory/relapsed AML, 6 with MDS/AML, 5 with MLL (5 de novo cases), and 3 with MS/AML (1 de novo case, 2 refractory/relapsed cases). The panel of TES included 236 genes recurrently mutated in hematological malignancies, and TES was displayed by NovaSeq platform (Illumina). The average raw sequencing depth on target per sample was ≥ 1000, and VAF ≥ 1% was considered significant. For TES, 184 samples were collected from BM and 9 samples from PB. In addition, fusion gene screening for common rearrangements in AML was employed.
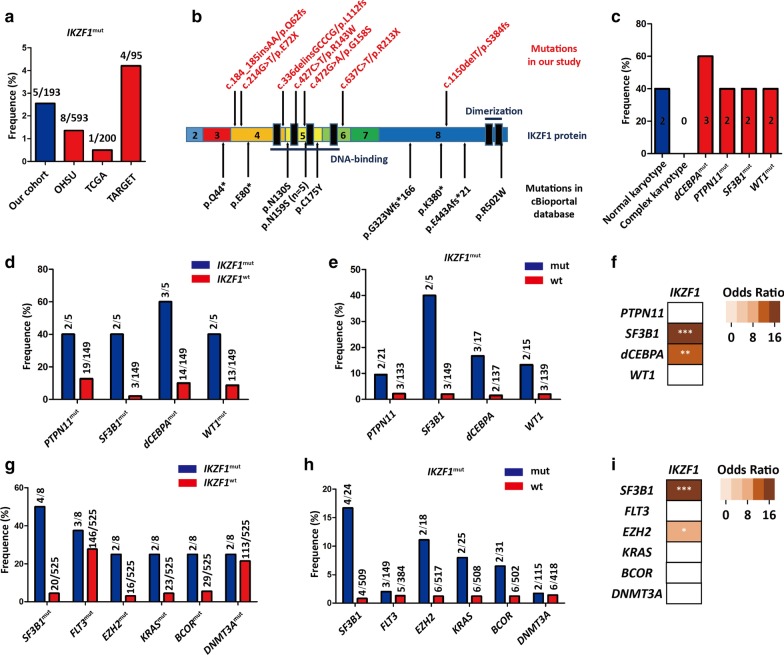
IKZF1 mutation affected 2.6% (5/193) or 1.8% (3/169) of all AML patients or de novo AML patients from our cohort, respectively (Fig. 1a and Table 1). Totally, 7 different types of IKZF1 mutations were found, and 5 were frameshift or nonsense mutations, while 2 were missense mutations (Fig. 1b). Interestingly, IKZF1 mutation was absent in fusion gene-positive AML, while IKZF1 mutation co-occurred with PTPN11, SF3B1, bi-allelic CEBPA or WT1 mutation in our study (Fig. 1c). Their association was further determined by Chi-square test with continuity correction, and OR was calculated. In 154 fusion gene-negative patients, we found that SF3B1 and bi-allelic CEBPA but not PTPN11 or WT1 mutations were the significant concomitant genetic alteration with IKZF1 mutation (P < 0.05; OR > 1) (Fig. 1d–f). In clinic, treatment response was evaluated in 4/5 patients with IKZF1-mutated AML, and CR was achieved in 2 patients. Notably, 2 primary chemotherapy-resistant patients had IKZF1, PTPN11 and SF3B1-mutated AML, so this subtype of AML seemingly exhibited an aggressive clinical course. However, the impact of IKZF1 mutation in AML could not be determined in our study due to limited positive cases and short follow-up duration.
Fig. 1.
IKZF1 mutation in AML. a The frequency of IKZF1 mutation in our AML cohort and literature reports. b The type of IKZF1 mutation identified in our cohort and literature reports. c Genetic lesion in AML with IKZF1 mutation. d The frequency of PTPN11, SF3B1, bi-allele CEBPA and WT1 mutations in IKZF1mut-AML and IKZF1wt-AML from our cohort, respectively. e The frequency of IKZF1 mutation in PTPN11, SF3B1, bi-allele CEBPA, WT1-wild type/-mutated AML from our cohort, respectively. f The statistical significance of associations between IKZF1 and other gene mutations in our study was assessed by Chi-square test with continuity correction. Odds ratio was also calculated to define whether the correlation was positive or negative. g The frequency of SF3B1, FLT3, EZH2, KRAS, BCOR and DNMT3A mutations in IKZF1mut-AML and IKZF1wt-AML from OHSU study, respectively. h The frequency of IKZF1 mutation in SF3B1, FLT3, EZH2, KRAS, BCOR and DNMT3A-wild type/-mutated AML from OHSU study, respectively. i Associations between IKZF1 and other gene mutations in OHSU study were also analyzed as f indicated. NS, P ≥ 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001
Table 1.
Acute myeloid leukemia with IKZF1 mutation in our cohort
| No. | Gender/age | Diagnosis | PB | BM blast | IKZF1 mutation (VAF, mutational site) | Karyotype | Gene fusion | Gene mutation | Response | OS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M/45 | AML-M0 |
WBC 19.1 G/L HB 77 g/l PLT 129 G/L Blast 89% |
90% | 44.51%, Exon4:c.184_185insAA/p.Q62fs*32 | 46,XY,t(3;3)(q13;q27)[10] | ND | PTPN11,SF3B1 | HAA, NR | Dead, 3.5 months |
| 2 | M/61 | AML-M0 |
WBC 5.2 G/L HB 83 g/l PLT 917 G/L |
28% |
38.19%, Exon4:c.214G>T/p.E72X 42.29%, Exon8:c.1150delT/p.S384fs*31 |
47,XY,+3(q21)[20] | ND | BCOR,PTPN11,FLT3, SF3B1 | AZA + IDA, NR | Dead, 3 months |
| 3 | M/24 | AML-M2 |
WBC 6.6 G/L HB 85 g/l PLT 10 G/L Blast 34% |
57% |
1.69%, Exon5:c.427C>T/p.R143W 3.49%, Exon6:c.637C>T/p.R213X |
46,XY,del(8)(q22)[5]/46,XY[5] | ND | dCEBPA,MSH6,DNMT3A,WT1 | IA, CR | Live, 5 months |
| 4 | M/21 | MLL-M2 |
WBC 46.6 G/L HB 76 g/l PLT 23 G/L Blast 75% |
88.5% | 23.04%, Exon5:c.472G>A/p.G158S | 46,XY[20] | ND | CCND3,dCEBPA,GATA2 | VEN + CAG, CR | Live, 4 months |
| 5 | F/52 | AML-M2/ MS (r/r) |
WBC 4.2 G/L HB 98 g/l PLT 84 G/L |
35% | 19.93%, Exon4:c.336delinsGCCCG/ p.L112fs*4 | 46,XX[20] | ND | dCEBPA,CSF3R,CTCF,WT1 | GHAA, NA | Live, 0.5 months |
PB peripheral blood, BM bone marrow, VAF variant allele frequency, OS overall survival, M male, F female, AML acute myeloid leukemia, MLL mixed lineage leukemia, AML/MS acute myeloid leukemia with myeloid sarcoma, R/R relapsed or refractory, WBC white blood cell, HB hemoglobin, PLT platelet, ND not detected, NR no response, CR complete remission, NA not available
In addition to our study, we also used the cBioPortal tool to analyze the frequency of IKZF1 mutation in other three independent studies (OHSU [5], TCGA [6] and TARGET [7]). The frequency was 1.35% (8/593), 0.5% (1/200) and 4.21% (4/95), respectively, while the relatively high frequency in our study was possibly attributed to the criterion of enrollment and the limited cases (Fig. 1a). In total, 13 mutations were found in these studies, but there were no patients with 2 different mutations simultaneously (Additional file 1: Table S1). Of these 13 patients, 6 had frameshift or nonsense mutations and the rest 7 had missense mutations. IKZF1N195S was a hotspot mutation with the frequency of 38.5% (5/13), but it was absent in COSMIC and our study (Fig. 1b). Due to limited positive cases in TCGA and solely pediatric cases in TARGET, we further analyzed the related genetic events of IKZF1 mutations in OHSU and found that SF3B1 and EZH2, but not KRAS, BCOR, FLT3 or DNMT3A mutations were the significant concomitant alteration with IKZF1 mutation (Fig. 1g–i). Remarkably, SF3B1 mutation appeared in both concomitant alteration lists of our study and OHSU, suggesting their strong synergy in leukemogenesis.
Compared to AML, IKZF1 alteration is well studied in ALL. Churchman et al. reported that IKZF1 alteration affected 25% of childhood and 44% of young adult pre-B-cell ALL, especially BCR-ABL1-positive ALL with frequency of over 80%. In ALL, the most common type of alterations in IKZF1 is deletions, whereas IKZF1 mutations accounted only 2.6% of childhood and 3.4% of young adult ALL. The latter were observed in 11.9% of BCR-ABL1-negative and 2.2% of BCR-ABL1-positive ALL cases [8]. The frameshift or nonsense mutations of IKZF1 often occurred at the N-terminal or the region between DNA binding and dimerization domains, while missense mutations affected both domains. Consistently, IKZF1 mutations followed the same pattern in AML. Similarly to IKZF1N159S in AML, IKZF1N159Y is a hotspot mutation in ALL that affects its DNA binding domain. IKZF1N159Y-ALL exhibited one unique transcriptional profile characterized by downregulation of B-cell receptor and JAK-STAT signaling and upregulation of SALL1 [9]. Nevertheless, whether IKZF1N159S-AML could be defined as one independent subtype remains to be investigated.
In conclusion, besides of IKZF1 deletion, IKZF1 mutation is also recurrent in AML.
Supplementary information
Additional file 1: Table S1. The variant allele frequency of IKZF1 mutation in AML from cBioPortal database.
Acknowledgements
Targeted-exome-sequencing was supported by Acornmed Company (Beijing, China). We thanked Bulat Abdrahimov for helping edit our English language.
Abbreviations
- ALL
Acute lymphoblastic leukemia
- AML
Acute myeloid leukemia
- APL
Acute promyelocytic leukemia
- BM
Bone marrow
- CR
Complete remission
- MDS/AML
Myelodysplastic syndrome-transformed AML
- MLL
Mixed lineage leukemia
- MS/AML
Myeloid sarcoma with bone marrow infiltration
- OR
Odds ratio
- PB
Peripheral blood
- TES
Targeted exome sequencing
- VAF
Variant allele frequency
Authors’ contributions
XZ designed the experiments. X-WZ, XL, Y-FL, Y-NZ and J-HW collected and integrated clinical materials. XZ, JJ and W-JY integrated and analyzed all the data. XZ wrote the manuscript. JJ and W-JY revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81800199, 81670124).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
This study was approved by the ethical review committees of the First Affiliated Hospital to Zhejiang University School of Medicine.
Consent for publication
Written informed consent was obtained from this patient.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jie Jin, Email: jiej0503@zju.edu.cn.
Wenjuan Yu, Email: drwjyu1977@zju.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13045-020-00972-5.
References
- 1.Marke R, van Leeuwen FN, Scheijen B. The many faces of IKZF1 in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2018;103:565–574. doi: 10.3324/haematol.2017.185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milosevic JD, Puda A, Malcovati L, Berg T, Hofbauer M, Stukalov A, Klampfl T, Harutyunyan AS, Gisslinger H, Gisslinger B, et al. Clinical significance of genetic aberrations in secondary acute myeloid leukemia. Am J Hematol. 2012;87:1010–1016. doi: 10.1002/ajh.23309. [DOI] [PubMed] [Google Scholar]
- 4.de Rooij JD, Beuling E, van den Heuvel-Eibrink MM, Obulkasim A, Baruchel A, Trka J, Reinhardt D, Sonneveld E, Gibson BE, Pieters R, et al. Recurrent deletions of IKZF1 in pediatric acute myeloid leukemia. Haematologica. 2015;100:1151–1159. doi: 10.3324/haematol.2015.124321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, Long N, Schultz AR, Traer E, Abel M, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ, Jr, Laird PW, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolouri H, Farrar JE, Triche T, Jr, Ries RE, Lim EL, Alonzo TA, Ma Y, Moore R, Mungall AJ, Marra MA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24:103–112. doi: 10.1038/nm.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churchman ML, Low J, Qu C, Paietta EM, Kasper LH, Chang Y, Payne-Turner D, Althoff MJ, Song G, Chen SC, et al. Efficacy of retinoids in IKZF1-mutated BCR-ABL1 acute lymphoblastic leukemia. Cancer Cell. 2015;28:343–356. doi: 10.1016/j.ccell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JF, Dai YT, Lilljebjorn H, Shen SH, Cui BW, Bai L, Liu YF, Qian MX, Kubota Y, Kiyoi H, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci U S A. 2018;115:E11711–E11720. doi: 10.1073/pnas.1814397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The variant allele frequency of IKZF1 mutation in AML from cBioPortal database.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.