Abstract
The Anniston Community Health Survey (ACHS-I) was initially conducted from 2005–2007 to assess polychlorinated biphenyl (PCB) exposures in Anniston, Alabama residents. In 2014, a follow-up study (ACHS-II) was conducted to measure the same PCBs as in ACHS-I and additional compounds e.g., polychlorinated dibenzo-p-dioxins (PCDDs), polychlorinated dibenzofurans (PCDFs), and dioxin-like non-ortho (cPCBs) substituted PCBs. In this epigenome-wide association study (EWAS), we examined the associations between PCDD, PCDF, and PCB exposures and DNA methylation. Whole blood DNA methylation was measured using Illumina EPIC arrays (n=292). We modeled lipid-adjusted toxic equivalencies (TEQs) for: ΣDioxins (sum of 28 PCDDs, PCDFs, cPCBs, and mPCBs), PCDDs, PCDFs, cPCBs, and mPCBs using robust multivariable linear regression adjusting for age, race, sex, smoking, bisulfite conversion batch, and estimated percentages of six blood cell types. Among all exposures we identified 10 genome-wide (Bonferroni p≤6.74E-08) and 116 FDR (p≤5.00E-02) significant associations representing 10 and 113 unique CpGs, respectively. Of the 10 genome-wide associations, seven (70%) occurred in the PCDDs and four (40%) of these associations had an absolute differential methylation ≥1.00%, based on the methylation difference between the highest and lowest exposure quartiles. Most of the associations (six, 60%) represented hypomethylation changes. Of the 10 unique CpGs, eight (80%) were in genes shown to be associated with dioxins and/or PCBs based on data from the 2019 Comparative Toxicogenomics Database. In this study, we have identified a set of CpGs in blood DNA that may be particularly susceptible to dioxin, furan, and dioxin-like PCB exposures.
Keywords: Polychlorinated biphenyls (PCBs); mono-ortho PCBs; non-ortho PCBs persistent environmental pollutant (POP); 2,3,7,8-Tetrachlorodibenzodioxin (TCDD/dioxin); Polychlorinated dibenzo-p-dioxins (PCDDs); Polychlorinated dibenzofurans (PCDFs); DNA methylation; Anniston Community Health Survey (ACHS)
1. Introduction
The major US producer of polychlorinated biphenyls (PCBs) operated in Anniston, Alabama from 1929 until 1971. Due to poor environmental controls at the facility, releases of dioxins, furans, and PCBs over time caused wide-spread environmental contamination in the Anniston community. Because these compounds are highly lipophilic and have long half-lives, they bioaccumulate in the food chain and persist in human tissues. The Anniston Community Health Survey (ACHS-I) was established in 2005 to assess the health effects of PCB exposure in the general population of the community (Birnbaum and Staskal-Wikoff, 2010; Pavuk et al., 2014a) and in 2014, a follow-up study was conducted (ACHS-II) (Birnbaum et al., 2016). Prior ACHS-I research has found links between PCB exposures and hypertension (Goncharov et al., 2010), blood pressure (Goncharov et al., 2011), race (Pavuk et al., 2014b), liver disease (Clair et al., 2018), metabolic syndrome (Rosenbaum et al., 2017), serum lipid levels (Aminov et al., 2014), diabetes (Silverstone et al., 2012), and leukocyte telomere length (Callahan et al., 2017).
A number of studies have found associations between altered DNA methylation and dioxin, furan, and PCB exposures in several populations, including Faroe Islanders (Leung et al., 2018), Dutch (van den Dungen et al., 2017), Taiwanese (Su et al., 2019), Greenlandic Inuits (Rusiecki et al., 2008), Koreans (Kim et al., 2010), and Japanese (Kobayashi et al., 2017). We have recently reported on 28 genome-wide significant associations between serum PCB concentrations in ACHS-I and whole blood DNA methylation (Pittman et al. 2019). We carried out a look-up analysis of the 369 FDR significant ACHS-I associations in the ACHS II methylation dataset reported in the present project and determined that the most significantly altered ACHS-I CpG, cg00475490 in the PRSS23 gene, was still significantly associated with the group, summed tri/tetra-ortho substituted, nondioxin-like PCBs (p=1.33E-04) in ACHS-II.
The current ACHS-II study focuses on dioxins and dioxin-like compounds and their toxicities have generally been attributed to sustained aryl hydrocarbon receptor (AHR) activation as measured in model systems, and this property is used in human risk assessment (Theobald and Peterson 1994). Ligand-mediated AHR activation results in upregulation of genes in the AHR pathway, including cytochrome P450s (e.g. CYP1A1 and CYP1B1) and the aryl hydrocarbon receptor repressor (AHRR), a negative regulator of AHR (Hahn et al., 2009). DNA methylation levels at the CpG cg05575921 in AHRR are strongly associated with adult and prenatal tobacco smoke exposure (Joehanes et al., 2016; Joubert et al., 2012; Reynolds et al., 2017; Su et al., 2016; Wan et al., 2018), and this could potentially be related to tobacco smoke polyaromatic hydrocarbons (PAHs), which are also AHR ligands. We recently explored the hypothesis that PCB exposures in ACHS might be associated with whole blood DNA methylation levels in AHRR or other AHR pathway genes, but found no relationship (Pittman et al., 2019). Dioxins are considered stronger ligands for AHR (Theobald and Peterson 1994), so in this work we also tested if dioxin and dioxin-like exposures might lead to methylation changes in AHRR cg05575921. Because dioxin exposures have been linked with altered immune system effects, we used the Houseman model (Houseman et al. 2012; Houseman et al. 2016) to test if exposure was associated with changes in methylation-based estimated cell-type percentages in whole blood. However, the primary objective was to carry out an epigenome-wide association study (EWAS) assessing if exposures to AHR ligands such as PCDDs, PCDFs, cPCBs, and mPCBs were associated with altered whole blood DNA methylation at CpGs across the genome.
2. Methods
2.1. Study population
ACHS-II was conducted in 2014 as a follow-up study to ACHS-I (2005–2007). Both study designs have been previously reported (Birnbaum et al., 2016; Pavuk et al., 2014a). In ACHS-I, two-stage stratified random sampling was used to select households and adults within the household of Anniston, Alabama. We contacted over 1,800 households; individuals living in west Anniston (the area closest to the PCB manufacturing facility) were over-sampled (two-thirds of eligible participants). 1,100 participants were interviewed and 778 volunteered to have their blood samples taken. PCB measurements and covariate data were available for 765 participants. For ACHS-II, 438 of 582 surviving participants from ACHS-I were successfully contacted and 359 eligible individuals were enrolled. While all ACHS-II subjects were recruited from the ACHS-I cohort, due to the lack of available DNA samples, not all ACHS-II subjects overlapped with our previous study (Pittman et al., 2019). We successfully measured DNA methylation in a subset (n=299) of the ACHS-II cohort from frozen (−70°C) whole blood samples. Seven individuals were excluded from analyses as they lacked measures for dioxins, furans and non-ortho substituted PCBs. The Institutional Review Boards at the Centers for Disease Control and the University of Alabama at Birmingham provided clearance for human subjects research. Table 1 contains the ACHS-II study demographics for individuals analyzed here.
Table 1.
Characteristics of the Anniston Community Health Survey, phase II.
| Characteristic | African-Americans (n=150) | Whites (n=142) | Total (n=292) |
|---|---|---|---|
| Mean ± SE | |||
| Agea | 60.9 ± 0.9 | 64.3 ± 1.2 | 62.6 ± 0.8 |
| Body Mass Index (BMI) | 32.3 ± 0.6 | 30.8 ± 0.7 n (%) | 31.6 ± 0.5 |
| n (%) | |||
| Female | 115 (76.7) | 102 (71.8) | 217 (74.3) |
| Age Group (years)b | |||
| <40 | 6 (4.0) | 10 (7.0) | 16 (5.5) |
| 40–59 | 63 (42.0) | 41 (28.9) | 104 (35.6) |
| ≥60 | 81 (54.0) | 91 (64.1) | 172 (58.9) |
| BMI Class (kg/m2) | |||
| <25 | 28 (18.7) | 35 (24.6) | 63 (21.6) |
| 25–29 | 38 (25.3) | 43 (30.3) | 81 (27.8) |
| ≥30 | 84 (56.0) | 64 (45.1) | 148 (50.7) |
| Current smoker | 36 (24.0) | 30 (21.1) | 66 (22.6) |
| Residence in west Annistonc | 140 (93.3) | 113 (79.6) | 253 (86.6) |
| Occupational PCB exposure | 32 (21.3) | 40 (28.2) | 72 (24.7) |
p=2.91E-02 for African-Americans compared to whites using Welch’s two-sided t-test.
p=4.83E-02 for African-Americans compared to whites using using chi-square test.
p=4.46E-04 for African-Americans compared to whites using chi-square test.
2.2. Serum dioxins, furans, and dioxin-like PCBs and lipid measurements
The CDC’s National Center for Environmental Health laboratory measured serum level of 28 dioxins, furans, and dioxin-like PCBs (Pavuk et al., 2014a). We measured 1) seven PCDDs (2378-TCDD, 12378-PeCDD, 123678-HxCDD, 123478-HxCDD, 123789-HxCDD, 1234678-HpCDD, and 12346789-OCDD); 2) ten PCDFs (2378-TCDF, 12378-PeCDF, 23478-PeCDF, 123678-HxCDF, 123789-HxCDF, 123478-HxCDF, 234678-HxCDF, 1234678-HpCDF, 1234789-HpCDF, and 12346789-OCDF); 3) three cPCBs (PCB81, 126, and 169); and 4) eight mPCBs (PCB105, 114, 118, 123, 156, 157, 167, and 189). We assigned exposure values below the limit of detection using the limit of detection divided by square root of 2 (Hornung and Reed, 1990). Lipid-substituted toxic equivalencies (TEQs) were calculated using the World Health Organization’s 2005 toxic equivalency factors list (TEFs; the compound’s potency relative to 2378-TCDD) (Van den Berg et al., 2006). TEQ calculation involved multiplying the lipid-substituted exposure value by the TEF for that compound (Supplemental Table 1). Lipid-substituted TEQs were grouped into five categories: 1) ΣDioxins (the sum of all 28 compounds); 2) PCDDs (2378-TCDD, 12378-PeCDD, 123678-HxCDD, 123478-HxCDD, 123789-HxCDD, 1234678-HpCDD, and 12346789-OCDD); 3) PCDFs (23478-PeCDF, 123678-HxCDF, 123478-HxCDF, 234678-HxCDF, and 1234678-HpCDF); 4) cPCBs (PCB126 and 169); and 5) mPCBs (PCBs 105, 114, 118, 156, 157, 167, and 189) (Table 2). The following compounds from the PCDF and PCB groups were excluded because ≥60% of participants had levels below the limit of detection: 2378-TCDF, 12378-PeCDF, 123789-HxCDF, 1234789-HpCDF, 12346789-OCDF, PCB81, and PCB123. It should be noted that all of the compounds examined in this study were unique to ACHS-II, with the exception of the eight mPCBs. However, in this current study we have used mPCB TEQ values, making these analyses distinct from our previous report (Pittman et al. 2019).
Table 2.
Dioxins and dioxin-like compounds measured in ACHS-II.
| ΣDioxins | PCDDs | PCDFsa | cPCBsa | mPCBsa | |
|---|---|---|---|---|---|
| 2378-TCDD | PCB81 | 2378-TCDD | 23478-PeCDF | PCB126 | PCB105 |
| 12378-PeCDD | PCB105 | 12378-PeCDD | 123678-HxCDF | PCB169 | PCB114 |
| 123678-HxCDD | PCB114 | 123678-HxCDD | 123478-HxCD | PCB118 | |
| 123478-HxCDD | PCB118 | 123478-HxCDD | 234678-HxCDF | PCB156 | |
| 123789-HxCDD | PCB123 | 123789-HxCDD | 1234678-HpCDF | PCB157 | |
| 1234678-HpCDD | PCB126 | 1234678-HpCDD | PCB167 | ||
| 12346789-OCDD | PCB156 | 12346789-OCDD | PCB189 | ||
| 2378-TCDF | PCB157 | ||||
| 12378-PeCDF | PCB167 | ||||
| 23478-PeCDF | PCB169 | ||||
| 123678-HxCDF | PCB189 | ||||
| 123789-HxCDF | |||||
| 123478-HxCDF | |||||
| 234678-HxCDF | |||||
| 1234678-HpCDF | |||||
| 1234789-HpCDF | |||||
| 12346789-OCDF | |||||
Values (lipid-substituted, pg/g) below the limit of detection were imputed by dividing the limit of detection for the assay by the square root of 2.
The following compounds from the PCDFs and PCB groups were excluded because ≥60% of participants had levels below the limit of detection: 2378-TCDF, 12378-PeCDF, 123789-HxCDF, 1234789-HpCDF, 12346789-OCDF, PCB81, and PCB123. The ΣDioxins included all 28 compounds.
We examined if wet-weight-substituted TEQs would differ substantially from using lipid-substituted TEQs. For the ΣDioxins TEQs, there was a strong correlation between the two exposure measurements, r2=0.91, p=6.95E-155 (Supplemental Figure 1). Among the other exposure groups there was also good concordance between the two TEQ measures: PCDDs r2=0.87, p=1.63E-129; PCDFs r2=0.86, p=9.34E-128; cPCBs r2=0.98, p=8.21E-235; and mPCBs, r2=0.96, p=1.03E-209 (Supplemental Figure 2). Based on these data and to be consistent with previous analyses, we used the lipid-substituted TEQs.
Serum lipid levels for total cholesterol, HDL cholesterol, LDL cholesterol, and triglycerides were measured by the University of Washington (Williams et al., 2013), and total lipids were calculated using the Bernert et al. method (Bernert et al., 2007).
2.3. DNA methylation measurement
We isolated genomic DNA from whole blood samples (300 μL per extraction) robotically using the Agencourt Genfind v2 Solid Phase Reversible Immobilization (SPRI) paramagnetic bead-based technology (Beckman Coulter). Prior to bisulfite-conversion, DNA concentrations were measured using a QUBIT dsDNA BR assay kit (Invitrogen). The National Cancer Institute’s Cancer Genomics Research Laboratory performed DNA bisulfite conversion using the EZ-96 DNA Methylation MagPrep kit (Zymo Research). Samples were then run on Illumina EPIC methylation arrays (Illumina).
2.4. Statistical analysis
Bioconductor’s ChAMP package v2.12.0 (Aryee et al., 2014; Fortin et al., 2017; Morris et al., 2014) was used to normalize methylation data. Methylation data exclusions included: 1) any samples which failed array QC standards; 2) all CpG probes on the X and Y chromosomes; 3) probes containing a SNP with a minor allele frequency ≥1% at the CpG site; and 4) probes failing QC standards. We also removed an additional 43,254 probes reported to hybridize to one or more non-target sites in the genome (Pidsley et al., 2016). There were 741,471 CpG probes remaining after exclusions.
Our analysis approach is shown in Figure 1. In brief, after running methylation arrays, associations between exposures and methylation were analyzed using robust multivariable linear regression (M-estimation) with the rlm function in Modern Applied Statistics with S (MASS v.7.3–51.1) (Ripley, 2002). We log10-transformed TEQ exposure measurements, consistent with previous analyses. All models were adjusted for age, race, sex, smoking status (current/never), bisulfite-conversion batch, and estimated white blood cell percentages. We estimated six white blood cell types: CD4 and CD8 T-cells; B-cells; monocytes; natural killer cells; and granulocytes, using the Houseman method (Houseman et al. 2012) (Houseman et al., 2016) based on the Reinius reference panel (Reinius et al., 2012). We also tested if exposure was associated with estimated cell-type percentages in whole blood. We tested other covariates e.g., body mass index and alcohol use; however, no other covariates substantially changed the estimates, contributed to the amount of variance explained, or improved model fit. We used chi-square tests and two-sided t-tests to examine demographic differences between African-Americans and whites.
Figure 1.
Overview of data analyses strategy. Robust linear regression models were adjusted for age, race, sex, smoking status, and percentages for six different white blood cell types. Five different exposure groupings were used in regression: the sum of all 28 compounds (ΣDioxins), polychlorinated dibenzodioxins (PCDDs), polychlorinated dibenzofurans (PCDFs), non-ortho substituted PCBs (cPCBs), and mono-ortho substituted PCBs (mPCBs) Exposure measures were lipid-substituted toxic equivalencies (TEQs).
For CpGs significant at Bonferroni p≤6.74E-08, we calculated differential methylation (ΔM) between the highest and lowest exposure quartiles and identified Benjamini-Hochberg false discovery rate (FDR) significant CpGs at p≤5.00E-02. For testing CpGs with an absolute ΔM≥1.00% across exposure quartiles, we used ANCOVA (adjusting for age, race and sex) and pair-wise two-sided t-tests. We evaluated potential effect measure modification by race and sex for CpGs at p≤6.74E-08 and with an absolute ΔM≥1.00%. Effect modification was assessed by including an interaction term in our regression models. Interaction terms were considered significant at p≤5.00E-02. We identified differential methylated regions (DMRs) using Bioconductor’s DMRcate v.1.18.0 (Peters et al., 2015), comparing the highest versus lowest tertiles for each exposure. We calculated FDR p-values for identified DMRs. Statistical analyses were conducted in R (R Development Core Team, 2018), SAS v9.4, and JMP 13.0.0 (SAS Institute Inc.).
2.5. Functional analysis and Comparative Toxicogenomics Database
Enrichr functional analysis was used to identify functional enrichment of molecular signatures of the group of genes containing FDR significant CpGs among biological processes (BP) or disease (OMIM) (Chen et al., 2013; Kuleshov et al., 2016). In addition, the PCDD-associated FDR-significant CpGs were entered into the eForge web tool hosted at https://eforge.altiusinstitute.org/ and run against the cell type specific epigenomic database (Breeze et al. 2019) and we used the gometh function in the missMethyl package (Phipson et al. 2016) to test enrichment of gene sets defined in Gene Ontology (GO) (Harris et al. 2004) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (Kanehisa and Sato 2020). We characterized gene associations with dioxin and dioxin-like compounds using curated chemical– gene association data from the 2019 Comparative Toxicogenomics Database (CTD) (Davis et al., 2019), MDI Biological Laboratory, Salisbury Cove, Maine, and NC State University, Raleigh, North Carolina. World Wide Web (URL: http://ctdbase.org) [August 2019].
3. Results
3.1. Demographics
Table 1 provides the demographics for subjects with available DNA methylation and exposure data (n=292). The cohort was primarily female (74.3%) and had a higher mean BMI (31.6 ± 0.5) relative to that reported (26.0 ± 0.1) in the National Health and Nutrition Examination Survey 2015–2016 (CDC National Center for Health Statistics, 2018). White study participants were older than African-American participants (64.3 vs 60.9 years). African-American participants were more likely to live in west Anniston (93% vs 80%), the section of Anniston where the PCB production facility was located.
3.2. Serum dioxin levels by age, race, and sex
Dioxin levels in ACHS-II were previously reported to be higher than the general population. Table 3 shows the distribution of our five TEQs groups (ΣDioxins, PCDDs, PCDFs, cPCBs, and mPCBs) by age, race, and sex. TEQs were modeled on the log10-scale.
Table 3.
Arithmetic means of exposure groups TEQs by age, race, and sex for ACHS-II (n=292).
| Exposure groupa | Age group (years) | Arithmetic mean ± SEM | p-valueb | Race | Arithmetic mean ± SEM | p-valueb | Sex | Arithmetic mean ±SEM | p-valuec |
|---|---|---|---|---|---|---|---|---|---|
| ΣDioxins | <40 | 4.92 ± 1.70 | 1.98E-30 | African-American | 32.04 ± 2.34 | 1.00E-12 | Female | 29.53 ± 1.79 | 4.30E-09 |
| 40–59 | 17.20 ± 1.40 | White | 19.70 ± 1.42 | Male | 15.94 ± 1.52 | ||||
| ≥60 | 33.35 ± 2.08 | ||||||||
| PCDDs | <40 | 3.38 ± 1.21 | 2.89E-37 | African-American | 12.54 ± 0.85 | 6.17E-04 | Female | 12.64 ± 0.63 | 1.27E-06 |
| 40–59 | 8.58 ± 0.75 | White | 10.60 ± 0.52 | Male | 8.60 ± 0.66 | ||||
| ≥60 | 14.19 ± 0.65 | ||||||||
| PCDFs | <40 | 1.08 ± 0.73 | 7.13E-24 | African-American | 3.14 ± 0.25 | 5.34E-03 | Female | 3.04 ± 0.17 | 7.13E-04 |
| 40–59 | 2.14 ± 0.15 | White | 2.53 ± 0.11 | Male | 2.26 ± 0.21 | ||||
| ≥60 | 3.43 ± 0.15 | ||||||||
| cPCBs | <40 | 0.21 ± 0.07 | 3.83E-35 | African-American | 12.01 ± 1.37 | 3.02E-17 | Female | 10.27 ± 1.07 | 1.74E-08 |
| 40–59 | 4.55 ± 0.61 | White | 4.77 ± 0.78 | Male | 3.35 ± 0.55 | ||||
| ≥60 | 11.64 ± 1.30 | ||||||||
| mPCBs | <40 | 0.18 ± 0.06 | 6.87E-37 | African-American | 4.18 ± 0.36 | 8.02E-25 | Female | 1.60 ± 0.24 | 2.63E-08 |
| 40–59 | 1.78 ± 0.21 | White | 1.72 ± 0.22 | Male | 3.46 ± 0.29 | ||||
| ≥60 | 3.96 ± 0.34 | ||||||||
Exposure groups: ΣDioxins = sum of 28 dioxin and dioxin-like compounds, PCDDs = polychlorinated dibenzodioxins (n=7), PCDFs = polychlorinated dibenzofurans (n= 5), cPCBs = non-ortho subsituted PCBs (n=2), mPCBs = mono-ortho substituted PCBs (n=7).
TEQs were log10 transformed in a linear regression model adjusted for race and sex. Age was modeled as a continuous variable.
TEQs were log10 transformed in a linear regression model adjusted for age and sex.
TEQs were log10 transformed in a linear regression model adjusted for age and race.
3.3. Correlation among the dioxin, furan and PCBs exposure groups
We examined the correlation among exposure TEQs among the four dioxin exposure groups (PCDDs, PCDFs, cPCBs, and mPCBs). Supplemental File 1, Table S2 shows the coefficient of determination (r2) for each of the comparisons. TEQs were modeled on the log10-scale and adjusted for age, race, and sex. Correlations between the PCDDs and PCDFs and the PCB groups (cPCBs and mPCBs) were relatively weak (r2 range, 0.50–0.58). However, PCDDs and PCDFs had a strong correlation (r2=0.80) as did the cPCBs and mPCBs (r2=0.83). While the summed TEQs for these structurally related groups were relatively strongly correlated, the differences might be reflected in different biological effects, thus we have assessed all of their associations with DNA methylation independently.
3.4. Smoking and methylation
Numerous studies have demonstrated a strong association between tobacco smoke exposure and methylation at the AHRR CpG cg05575921 (Joehanes et al., 2016; Joubert et al., 2012; Monick et al., 2012; Reynolds et al., 2015; Su et al., 2016; Wan et al., 2018). Using robust linear regression, we tested whether this association held for the ACHS-II cohort and found a strong association between current smoking and AHRR cg05575921 methylation levels (p=9.63E-207). AHRR differential methylation between smokers and nonsmokers was 24.1% (t-test, p=2.73E-24). Among nonsmokers, we found no association between ΣDioxins and AHRR cg05575921 (robust linear regression, p=1.71E-01).
3.5. Dioxins, furans, and dioxin-like PCBs, cell-type estimation and DNA methylation
We tested if exposures were associated with methylation-based estimates of cell-type percentages in whole blood and these results are provided in Supplementary Excel File 1, Table S3. We observed nominally significant associations between estimated CD8 T cell percentages and PCDDs (p = 0.029) and PCDFs (p = 0.041). However, as is standard practice in EWAS stud all methylation analyses are adjusted for estimated cell-type percentages to account for any cell type shifts. We observed 10 genome-wide significant CpGs (representing 10 unique genes) associated with the dioxin exposure groups (Table 4). Four (40%) of these associations had ΔM≥1.00% (based on the methylation difference between the highest and lowest exposure quartiles). Seven of the 10 (70%) associations were most significant in the PCDD exposure group. There were 116 CpG/exposure FDR significant associations, representing 113 unique CpGs (Supplemental Excel File 1 Table S4). Figure 2 illustrates the distribution of these CpGs across the dioxin exposure groups, with the majority of CpGs (n=104, 92%) significant exclusively in the PCDD exposure group. There were no FDR significant associations for the mPCB exposure group.
Table 4.
Top ACHS-II Dioxin-associated CpGs, selected by Bonferroni p≤6.74E-08.
| ProbeID | Gene Symbol(s) | Coordinate | Exposurea | % Differential Methylation (Mean ± SEM)b | p-valuec | Exposure Regression Coefficientc | Dioxin/Furan/PCB Associationd |
|---|---|---|---|---|---|---|---|
| cg00999904 | ALLC | chr2:3704751 | PCDDs | −4.64 ± 0.21 | 1.05E-09 | −1.8964 | TCDD |
| cg11354991 | BBS4, HIGD2B | chr15:72978685 | PCDDs | 0.09 ± 0.01 | 6.04E-09 | 0.8172 | TCDD, PCB52, PCB180 |
| cg03510117 | CYCS | chr7:25164991 | PCDDs | 0.37 ± 0.01 | 3.01E-08 | 0.6423 | TCDD, PCB153 |
| cg06169091 | IGSF21 | chr1:18678286 | PCDDs | −2.49 ± 0.16 | 3.08E-08 | −0.5569 | |
| cg21401636 | KIF5Ce | chr2:149629217 | PCDDs | −3.73 ± 0.30 | 1.88E-08 | −0.6405 | TCDD |
| cg03459668 | RAB11FIP3 | chr16:545622 | PCDDs | −2.86 ± 0.14 | 2.26E-08 | −0.4876 | TCDD, PCB126 |
| cg07303330 | TAAR2 | chr6:132945515 | PCDDs | −0.88 ± 0.06 | 7.85E-09 | −0.9439 | |
| cg26787894 | CD83 | chr6:14117621 | PCDFs | 0.29 ± 0.02 | 3.39E-08 | 0.4943 | TCDD |
| cg06242879 | ERBB3, PA2G4 | chr12:56497745 | PCDFs | 0.36 ± 0.04 | 6.13E-09 | 0.3756 | TCDD, PCB77 |
| cg23269663 | BCL3 | chr19:45250480 | cPCBs | −0.51 ± 0.03 | 5.15E-08 | −0.2829 | TCDD |
CpGs are listed in order of exposure category and gene symbol. Individual exposures with ≥60% participants below the limit of detection
were excluded from the exposure groups with the exception of the ΣDioxins.
Exposure groups: ΣDioxins = sum of 28 dioxin and dioxin-like compounds, PCDDs = polychlorinated dibenzodioxins (n=7), PCDFs = polychlorinated dibenzofurans (n= 5), cPCBs = non-ortho subsituted PCBs (n=2), mPCBs = mono-ortho substituted PCBs (n=7).
Differential methylation was calculated by subtracting methylation in lowest exposure quartile from the highest.
Robust linear regression models were adjusted for age, race, sex, smoking status, bisulfite-conversion batch, and estimated percentages of CD4+ and CD8+ T-cells, CD19+ B-cells, monocytes, granulocytes, and natural killer cells.
Genes associated with dioxin, furan, or PCB exposure based on the 2019 Comparative Toxicogenomics Database.
Effect modification by race was detected. See Results 3.6 and Supplemental Table 3 for details.
Figure 2.
Venn diagram of 113 significantly differentially methylated CpGs (FDR p≤5.00E-02) in ACHS-II for four exposure groups: the sum of all 28 compounds (ΣDioxins), polychlorinated dibenzodioxins (PCDDs), polychlorinated dibenzofurans (PCDFs), and non-ortho substituted PCBs (cPCBs). There were no FDR significant CpGs for the mono-ortho substituted PCBs. Exposure measures were lipid-substituted toxic equivalencies (TEQs).
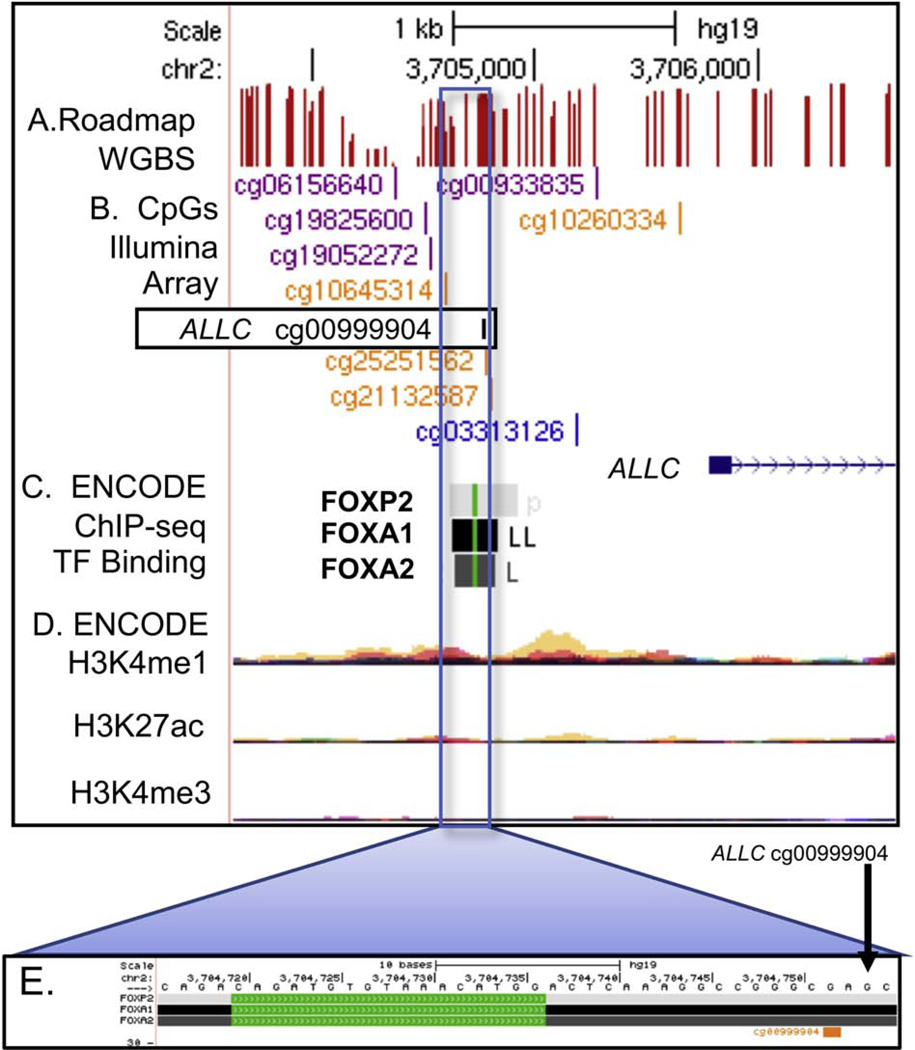
The CpG with the largest absolute ΔM was associated with the allantoicase gene (ALLC, Figure 3). The ALLC CpG cg00999904 had differential methylation of −4.64 ± 0.21 (p=1.05E-09) across low and high quartiles of the PCDD exposure group. ALLC cg00999904 is located ~1.3 kb upstream of the ALLC first exon and is 15 bp downstream of three forkhead-box (FOX) group of transcription factor binding sites (FOXA1, FOXA2, FOXP2) determined by the ENCODE project (Wang et al., 2012).
Figure 3.
ALLC genome browser view showing the altered CpG (boxed) in relation to transcription factor binding sites in the ALLC promoter region. Tracks listed from the top of browser: A. CpG sites as detected by whole genome bisulfite sequencing (Roadmap WGBS, methylation level indicated by height of red bar); B. CpGs on Illumina 450K array, reference cg numbers listed; C. ENCODE project identified transcription factor binding as detected by ChIP-seq, green line indicates position of the TF binding motif sequence; D. ENCODE project enhancer related histone modification measurements layered by color to show multiple cell types (red=GM12878 B cell, tan=H1-hESC stem cell); E. Magnification of genome region containing TFBS.
3.6. Effect modification by race and sex
We examined potential effect modification by race or sex for the four Bonferroni significant CpGs with an absolute ΔM ≥1.00%. We ran multivariable robust linear regression models with an interaction term (sex*exposure or race*exposure). We observed effect modification by race for the PCDD vs cg21401636 (KIF5C) association based on an interaction term with p=2.10E-03. The ΔM for African-Americans was −7.02 ± 0.36 (p=4.33E-03) and for whites −5.02 ± 0.26 (p=1.01E-02) (Supplemental Excel File 1 Table S5).
3.7. DMR analysis
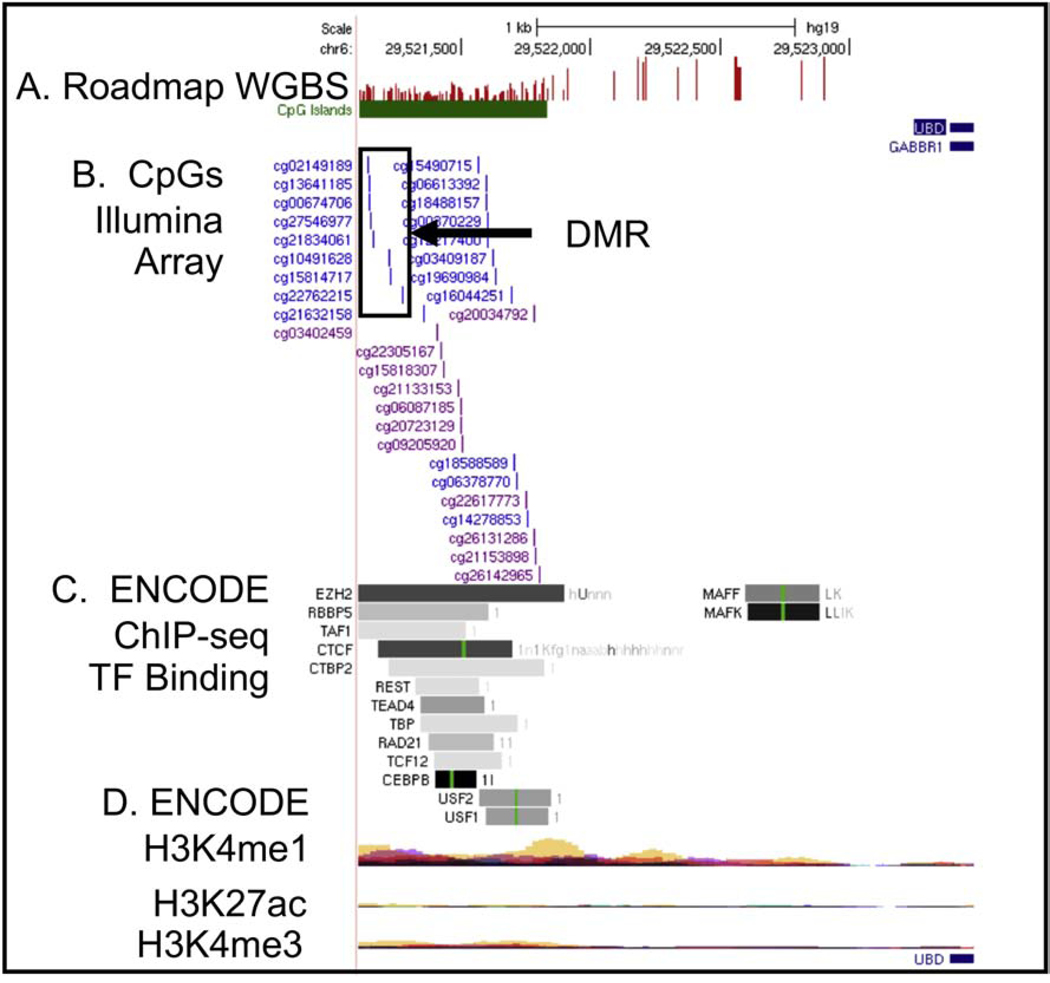
We searched for DMRs, groups of CpGs within a genomic region displaying similar directional changes, that were associated with dioxin and dioxin-like exposures, using DMRcate and exposure group tertiles. We identified one DMR/exposure association in the ΣDioxins data at p=2.75E-13 (Figure 4). This DMR was just beyond the 3-prime end of ubiquitin D (UBD) and gamma-aminobutyric acid type B receptor subunit 1 (GABBR1) at chr6:29521138–29521272 (containing cg02149189, cg13641185, cg00674706, cg27546977, cg21834061, cg10491628, cg15814717, and cg22762215) and was in a CpG island. Also, ENCODE transcription factor binding ChIPseq data indicates this region is potentially transcriptionally active and ENCODE histone modification data from multiple cell types (GM12878, H1-hESC) suggests this region is a potentially repressed enhancer. These DMR CpGs did not overlap with any CpGs in our FDR p≤5.00E-02 list (Supplemental Excel File 1, Table S4).
Figure 4.
Differentially methylated region (DMR, boxed CpGs) in CpG island near UBD and GABBR1 genes. Tracks listed from the top of browser: A. CpG sites as detected by whole genome bisulfite sequencing (Roadmap WGBS, methylation level indicated by height of red bar); B. CpGs on Illumina 450K array, reference cg numbers listed; C. ENCODE project identified transcription factor binding as detected by ChIP-seq, green line indicates position of the TF binding motif sequence; D. ENCODE project enhancer related histone modification measurements layered by color to show multiple cell types (red=GM12878 B cell, tan=H1-hESC stem cell).
3.8. Quartile analysis
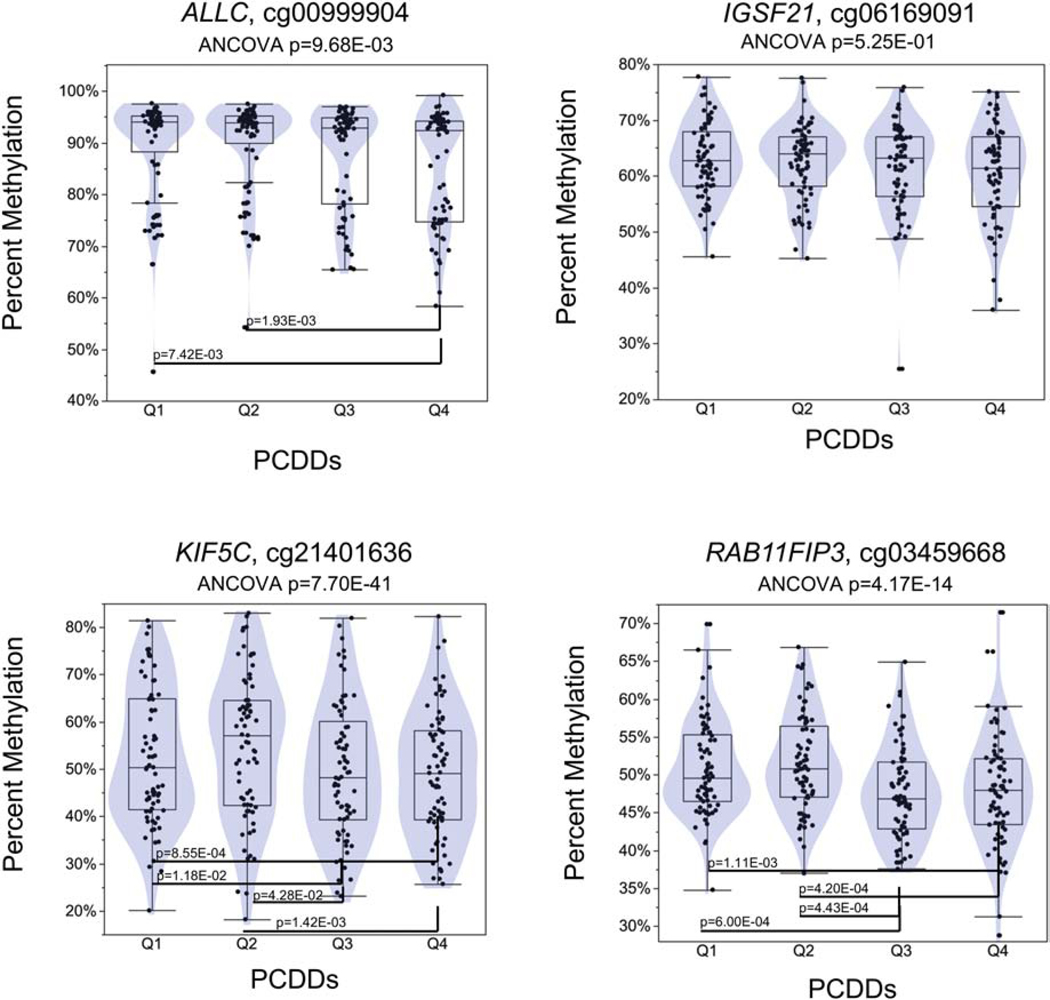
We examined methylation across quartiles for the four CpGs significant at Bonferroni p≤6.70E-08 and that had an absolute ΔM ≥1.00% (Figure 5) using ANCOVA adjusting for race, age and sex. These CpGs were cg00999904 in ALCC; cg06169091 in immunoglobin superfamily member 21 (IGSF21); cg21401636 in kinesin family member 5C (KIF5C); and cg03459668 in RAB11 family interacting protein 3 (RAB11FIP3). For ALCC cg00999904, the methylation in the fourth quartile of PCDDs was significantly different than the first and second quartiles, p=7.42E-03 and p=1.93E-03 respectively (ANCOVA p=9.68E-03). For IGSF21 cg06169091 there were no significant differences among the quartiles (ANCOVA p=5.25E-01). Fourth quartile methylation in the PCDDs was significantly higher than the first and second quartiles for KIF5C cg21401636, p=8.55E-04 and p=1.42E-03 respectively (ANCOVA p=7.70E-41). And for RAB11FIP3 cg03459668, there were significant differences among all quartiles (ANCOVA p=4.17E-14), except between the first vs. second quartile and the third vs. fourth quartile.
Figure 5.
Dioxin and dioxin-like compounds quartile distributions for CpGs with ≥1.0% absolute differential methylation. ANCOVA was adjusted for age, race, and sex. Pair-wise Student’s t-test p-values between quartiles are presented.
3.9. Functional analysis and Comparative Toxicogenomics Database associations
For the genes with FDR significant CpGs, we used Enrichr functional analysis as well as eForge, missMethyl (GO) and missMethyl (KEGG) (Supplemental Excel File 2). Among the Enrichr, KEGG and GO analyses we observed nominal significance for numerous signatures but all failed to reach FDR significance. The eForge analysis identified that altered CpGs were marginally overrepresented in regions with the repressive histone modification H3K27me3 at FDR q= 0.28 in CD8+ T cells, a cell type that displays nominal-dioxin associated changes in estimated percentage. Using the same gene list, we also used the 2019 Comparative Toxicogenomics Database (CTD) to determine if these genes had reports of interactions with dioxins or dioxin-like exposure in scientific literature. For genome-wide significant CpGs, 8 of the 10 (80%) unique genes (Table 4) and 70 (62%) of the 113 FDR significant unique genes (Supplemental Excel File 1,Table S4) were listed as having interactions with dioxins or dioxin-like exposures.
4.0. Look up analysis
We searched the literature for array-based methylation data and associations with dioxins, furans and dioxin-like PCBs. We identified three studies with 450k methylation array data (Leung et al., 2018; Su et al., 2019; van den Dungen et al., 2017). These studies examined the associations between whole blood DNA methylation and PCBs 105 and 118; 23478-PeCDF, 123478-HxCDF, PCBs 153 and 156; and PCBs 28, 52, 105, 118, 138, 153, 156, 170, 180, and 187, respectively (Supplemental File 1 Table S6). Of the 254 unique CpGs identified among the three studies, there was no overlap among the studies. There was also no overlap between these studies’ results and our FDR significant CpGs. Among all CpG look-ups, we observed nominal significance (p<5.0E-02) for 12/254 among PCDF associations and 21/254 mPCB associations; however, none of these reached the calculated Bonferroni-adjusted p-value (p≤2.16E-04) level of significance. We had no data for 22 CpGs probes that failed our quality control screening.
Discussion
In this current study we have examined the associations between dioxin, furan, and dioxin-like PCB exposures and whole blood DNA methylation in the second phase of the Anniston Community Health survey. Among all exposures we identified 10 (Bonferroni p≤6.74E-08) and 116 (FDR p≤5.00E-02) significant associations representing 10 and 113 unique CpGs, respectively. The PCDDs exposure group had the most Bonferroni and FDR significant CpG associations (seven and 107, respectively), and the majority were in genes shown to be associated with dioxins and/or PCBs, based on data from the 2019 CTD. Of the CTD-based dioxin-associated genes, the most significant CpG was upstream of allantoicase (ALLC), a gene in the uric acid degradation pathway. Based on a RNA-seq study, ALLC is expressed primarily in the testis, but also has expression in the adrenal gland, adipose tissue, gall bladder, heart, kidney, liver, and ovaries. ALLC expression in CD34+ hematopoietic stem and progenitor cells was reported to be increased with TCDD exposure (CTD database). However, it is unclear what the function of ALLC is, and it is not considered to be part of the AHR-regulated gene pathway.
Many of the identified CpGs were in gene regulatory regions where numerous transcription factors bind (Figure 4); however, it is not known if changes in methylation in these regions affect transcription factor binding or gene expression. Among FDR-significant CpGs, the B cell lymphoma 3 gene, a proto-oncogene, (BCL3, cg23269663), is of interest due to its connection to NF-kappaB regulation (Keutgens et al., 2010) and lymphoma (Viatour et al., 2004). A CpG (cg26686732) in the FOXA2 gene was in the top 25 FDR-significant CpGs, and it should be noted that several significant CpGs (cg00999904, cg25251562, cg19052272, and cg19825600) in the ALLC gene are near three FOX family transcription factor binding sites (FOXP2, FOXA1, and FOXA2). FOXA2 is a hepatocyte transcription factor, which is of interest because a study in ACHS-I found an association between PCB exposure and toxicant-associated steatohepatitis (Clair et al., 2018).
In our previous study of DNA methylation in ACHS-I, we tested the hypothesis that DNA methylation in AHRR, a gene in the AHR pathway, might be affected by exposure to PCBs, but observed no association. We tested the same hypothesis regarding dioxin-like exposures (stronger AHR ligands) in the present study. Smoking had an association with AHRR cg05575921 methylation levels; however, in nonsmokers we detected no association between dioxin or dioxin-like exposures and changes in AHRR methylation.
In dioxins and furans, the two phenyl groups are in a permanent coplanar orientation, making these compounds strong AHR ligands. In non-ortho and mono-ortho substituted PCBs, the phenyl groups are not in a fixed coplanar configuration, but the phenyl groups can assume this configuration. When PCBs are in this coplanar configuration, they become dioxin-like (i.e. become AHR ligands) (Ericksson, 2001). Based on our original hypothesis, we expected to observe the most significant changes in methylation among the most dioxin-like exposures group i.e., the PCDDs. Indeed, we found the most genome-wide significant (p≤6.74E-08) CpG/exposure associations in the PCDDs, and many of these genes are linked to dioxin-like exposures in model systems (Table 3). However, none of the genes in this group (or FDR list, Supplemental Excel File 1 Table S4) were part of the canonical AHR pathway (e.g. ARNT, AHRR, CYP1A1, CYP1B1). These results suggest blood DNA methylation alterations associated with dioxin and dioxin-like exposures are not directly related to AHR pathway activation.
Other studies have reported associations between dioxins and dioxin-like exposures and altered DNA methylation; however, most of these studies examined global methylation or methylation changes in specific candidate genes. Only a few of these studies had measured TCDD and other dioxins or dibenzofuran congeners or non-ortho PCBs. In the Arctic Monitoring and Assessment Program, it was observed in a cohort of Greenlandic Inuits (n=70) that higher serum levels of 14 PCBs (28, 52, 99, 101, 105, 118, 128, 138, 153, 156, 170, 180, 183, 187) were related to global hypomethylation (Rusiecki et al., 2008); however, only three of these PCBs (105, 118, and 156) have dioxin-like properties. Two studies of TCDD (n=516) (Pilsner et al., 2018) and PCB153 (n=315) (Consales et al., 2016) exposures found an association with sperm DNA hypomethylation. In a cohort of cancer-free Koreans (n=368), researchers reported a non-monotonic association between promoter hypomethylation of MGMT and PCBs 105, 138, and 153 (Park et al., 2015). Two compounds (PCB126 and OCDD) were linked to global DNA hypermethylation (Lind et al., 2013) in a cross-sectional cohort of elderly Swedish men and women (n=524).
Other studies have found potential sex-specific effects. Results from a sub-cohort of the Hokkaido Birth Cohort Study on Environment and Children’s Health (n=169) showed an association between PCBs 170, 178, 180, and 182 and cord blood H19 and LINE1 hypermethylation (Kobayashi et al., 2017). The results also suggested these associations were stronger in females compared to males. A study in a prospective birth cohort of Faroe Islanders with high levels of PCB exposure at birth (n=72) using the 450K array reported an association between PCB105 and sex-specific methylation of a number of CpGs (Leung et al., 2018). Another study in Taiwan used the 450K array and identified 20 CpGs associated with 23478-PeCDF, 123478-HxCDF, PCB153, PCB156, PCB170, and PCB180 exposures in the 2nd generation (n=60) of a cohort whose mothers had been exposed to these compounds due to contaminated cooking oil (Su et al., 2019). However, 13 of the CpG associations reported were in genes that have been associated with smoking exposure in studies by our laboratory (Su et al., 2016) and numerous others (Joehanes et al., 2016; Joubert et al., 2012). The authors of the Taiwanese study reported that their findings could have been confounded by smoke exposure status of the mothers and/or offspring. In in a cohort of Dutch men (n=80), serum levels of dioxin-like PCBs (105, 118, and 156) and the sum of several indicator PCBs (28, 52, 138, 153, 170, 180 and 187) were significantly associated with DMR hypermethylation, with an average 7.4% differential methylation between the subjects with highest and lowest exposure (van den Dungen et al., 2017). From these three array-based studies, we obtained the CpG identifier for the loci associated with dioxin and dioxin-like compounds and compared them to our results. It is of interest to note that the reported CpGs from these studies had no overlap among them; nor was there any overlap with our FDR significant CpGs. The dioxin-like compounds concentrations in these other cohorts were generally lower than in the Anniston population and this may explain the lack of overlap between our findings and these other studies.
Another finding of interest relates to the correlation among PCDDs, PCDFs, cPCBs, and mPCBs. While structurally similar compounds, PCDDs and PCDFs, and the cPCBs and mPCBs were strongly correlated, the correlations among the PCDDs and PCDFs vs the cPCBs and mPCBs were relatively weak. These results are consistent with previously published ACHS-II results (Yang et al., 2018) and suggests that the source of PCDDs/PCDFs exposure may differ from that for cPCBs/mPCBs. Interestingly, significant methylation effects were observed almost exclusively with exposure to the dioxin group (PCDDs).
Our previous analysis of mono-ortho, di-ortho, and tri/tetra-ortho substituted PCB exposures in ACHS-I identified 369 CpG/exposure associations that were significant at FDR p≤5.00E-02 but only one remained significant in a lookup analysis in ACHS-II (Pittman et al., 2019). We compared the ACHS-I FDR results with our current analysis of dioxin-like compounds in ACHS-II. Again, there was no overlap between our previous PCB results and our genome-wide or FDR significant associations with current dioxin and dioxin-like compounds in this report. This finding is not necessarily conflicting. The most significant associations (80%) in our previous study occurred only with tri/tetra-ortho PCBs, which have no dioxin-like properties. Non-dioxin-like PCBs may perturb DNA methylation through a different mechanism than dioxin-like compounds. The CpGs identified in the present study may represent CpG loci that are most sensitive to dioxin-like exposures given the predominant CpG/exposure associations (92%) were found in the PCDDs, the most dioxin-like compounds. Among our most significant CpGs, we tested for sex and race specific effects and observed effect modification by race for cg21401636 in KIF5C. This finding may reflect the significantly (p=6.17E-04) higher PCDD exposure in African-Americans versus whites.
The primary strengths of this current study were the number of dioxin-like compounds measured (n=28) and the use of the Illumina 850K methylation array (the first study we are aware of to examine these exposures using the 850K array). A limitation of our study was the relatively small sample size in ACHS-II (n=292); however, compared to the three referenced 450K assay studies, our sample size is far larger, making our results a significant contribution to the scientific literature. Another limitation was the use of whole blood DNA for methylation analysis. As has been pointed out, cell-type specific effects might be poorly detected in a mixture of cell types because the differential methylation may occur in a relatively rare cell type population or might be in opposite directions in different cell types(Su et al. 2016). In addition, a general issue with the use of genome-wide arrays is the need for multiple-testing correction; therefore, we may have missed true CpG/exposure associations that did not reach FDR significance.
4. Conclusions
This study contributes to the understanding of the effects of dioxin and dioxin-like exposures on whole blood DNA methylation in a human population study. This is the first report to examine dioxin-like exposures and alterations in DNA methylation using the Illumina 850K methylation array. We identified one highly significant DMR and 10 genome-wide significant CpG/exposure associations with 40% having differential methylation ≥1.00%, based on the methylation difference between the highest and lowest exposure quartiles. We observed effect-measure modification by race for cg21401636 (KIF5C). Most of the FDR significant CpGs (113 unique CpGs) we found were associated with PCDD exposure, the most dioxin-like of the exposures we measured. These CpGs and the DMR identified may represent a group of CpGs in blood cell types that are particularly susceptible to dioxin exposure. Our current results did not overlap with three previous 450K methylation array studies; however, all three of those studies had very small sample sizes (60–72 individuals), and only one of those measured dioxins, dibenzofurans and non-ortho PCBs (Su et al., 2019).
In our identification of biological pathway associations, a CpG in FOXA2 (a hepatocyte transcription factor) was significant at 8.87E-07. Since a previous ACHS-I study found an association between PCB exposure and liver disease (Clair et al. 2018), a future goal will be to explore whether changes in DNA methylation modify this exposure/disease association.
Supplementary Material
Highlights.
Anniston, Alabama has high levels of environmental dioxin-like compounds.
Using 850k arrays, we found 10 significant exposure/DNA CpG methylation associations.
Seven methylation associations were with polychlorinated dibenzo-p-dioxins.
Four methylation associations had an absolute differential methylation ≥1.00%.
Eight associations were in genes reported to interact with dioxin exposures.
Acknowledgements
We recognize Dr. Stephen T. Mennemeyer of University of Alabama Birmingham for his study contributions. We express gratitude to the study participants in the ACHS-I and ACHS-II cohorts. We thank Andreas Sjödin and Richard Jones, National Center for Environmental Health CDC, for the expert analyses of the dioxin-like compounds and PCBs. We want to acknowledge Dr. Santica Marcovina, Northwest Lipid Metabolism and Diabetes Research Laboratories, University of Washington, for the analyses of lipids and glycemic parameters. We thank the Cancer Genomics Research Laboratory at the National Cancer Institute (Rockville, MD) for methylation array analysis. We acknowledge Kevin E. Gerrish and Joetta Hitchcock-Smith, NIEHS Molecular Genomics Core Laboratory for ACHS-II DNA extractions (RTP, NC). We also thank Suzanne N. Martos (NIEHS, RTP, NC) for suggestions and feedback regarding data analysis and presentation.
Funding Details
The baseline study (ACHS-I) was conducted using a grant from ATSDR to Jacksonville State University, #5U50TS473215. The follow up study (ACHS II) was funded by the National Cancer Institute through interagency agreements with the Centers for Disease Control and Prevention (CDC) (IAA#: 11-AT1-001-00; IAA#: 12-AT-12-ANNISTON) and by ATSDR. Data collection was supported via contract from ATSDR to the University of Alabama at Birmingham (UAB) (CDC Contract No. 200-2011-40834). The contents of this publication are solely the responsibility of the authors and do not necessarily represent ATSDR/CDC official views. This research was also funded in part by the Intramural Research Program of the National Institute of Environmental Health Sciences-National Institutes of Health project Z01-ES100475.
Footnotes
Disclosure Statement
J.R. Olson served as an expert witness for the plaintiffs in legal actions regarding the residents of Anniston, Alabama being exposed to PCBs. The other authors declare that they have no competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aminov Z, Haase R, Olson JR, Pavuk M, Carpenter DO, Anniston Environmental Health Research, C., 2014. Racial differences in levels of serum lipids and effects of exposure to persistent organic pollutants on lipid levels in residents of Anniston, Alabama. Env. Int 73, 216–223. 10.1016/j.envint.2014.07.022 [DOI] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA, 2014. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369. 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernert JT, Turner WE, Patterson DG, Needham LL, 2007. Calculation of serum “total lipid” concentrations for the adjustment of persistent organohalogen toxicant measurements in human samples. Chemosphere 68, 824–831. [DOI] [PubMed] [Google Scholar]
- Birnbaum LS, Dutton ND, Cusack C, Mennemeyer ST, Pavuk M, 2016. Anniston community health survey: Follow-up and dioxin analyses (ACHS-II)--methods. Env. Sci Pollut Res Int 23, 2014–2021. 10.1007/s11356-015-4684-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum LS, Staskal-Wikoff DS, 2010. 5th International PCB Workshop--summary and implications. Env. Int 36, 814–818. 10.1016/j.envint.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze CE, Reynolds AP, van Dongen J, Dunham I, Lazar J, Neph S, et al. 2019. Eforge v2.0: Updated analysis of cell type-specific signal in epigenomic data. Bioinformatics 35:4767–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan CL, Pavuk M, Birnbaum LS, Ren X, Olson JR, Bonner MR, 2017. Serum polychlorinated biphenyls and leukocyte telomere length in a highly-exposed population: The Anniston Community Health Survey. Env. Int 108, 212–220. 10.1016/j.envint.2017.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC National Center for Health Statistics, 2018. National Health and Nutrition Examination Survey: Analytic Guidelines, 2011–2014 and 2015–2016. Natl. Cent. Heal. Stat. [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clair HB, Pinkston CM, Rai SN, Pavuk M, Dutton ND, Brock GN, Prough RA, Falkner KC, McClain CJ, Cave MC, 2018. Liver Disease in a Residential Cohort With Elevated Polychlorinated Biphenyl Exposures. Toxicol Sci 164, 39–49. 10.1093/toxsci/kfy076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consales C, Toft G, Leter G, Bonde JP, Uccelli R, Pacchierotti F, Eleuteri P, Jonsson BA, Giwercman A, Pedersen HS, Strucinski P, Goralczyk K, Zviezdai V, Spano M, 2016. Exposure to persistent organic pollutants and sperm DNA methylation changes in Arctic and European populations. Env. Mol Mutagen 57, 200–209. 10.1002/em.21994 [DOI] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ, 2019. The Comparative Toxicogenomics Database: update 2019. Nucleic Acids Res 47, D948–d954. 10.1093/nar/gky868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericksson MD, 2001. PCBs : Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky, Lexington, Ky. [Google Scholar]
- Fortin JP, Triche TJ Jr., Hansen KD, 2017. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 33, 558–560. 10.1093/bioinformatics/btw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncharov A, Bloom M, Pavuk M, Birman I, Carpenter DO, 2010. Blood pressure and hypertension in relation to levels of serum polychlorinated biphenyls in residents of Anniston, Alabama. J Hypertens 28, 2053–2060. 10.1097/HJH.0b013e32833c5f3e [DOI] [PubMed] [Google Scholar]
- Goncharov A, Pavuk M, Foushee HR, Carpenter DO, Anniston Environmental Health Reseach, C., 2011. Blood pressure in relation to concentrations of PCB congeners and chlorinated pesticides. Env. Heal. Perspect 119, 319–325. 10.1289/ehp.1002830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn ME, Allan LL, Sherr DH, 2009. Regulation of constitutive and inducible AHR signaling: Complex interactions involving the AHR repressor. Biochem. Pharmacol. 77, 485–497. 10.1016/j.bcp.2008.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. 2004. The gene ontology (go) database and informatics resource. Nucleic Acids Res 32:D258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung RW, Reed LD, 1990. Estimation of Average Concentration in the Presence of Nondetectable Values. Appl. Occup. Environ. Hyg. 5, 46–51. 10.1080/1047322X.1990.10389587 [DOI] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Kile ML, Christiani DC, Ince TA, Kelsey KT, Marsit CJ, 2016. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics 17, 259 10.1186/s12859-016-1140-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, Guan W, Xu T, Elks CE, Aslibekyan S, Moreno-Macias H, Smith JA, Brody JA, Dhingra R, Yousefi P, Pankow JS, Kunze S, Shah SH, McRae AF, Lohman K, Sha J, Absher DM, Ferrucci L, Zhao W, Demerath EW, Bressler J, Grove ML, Huan T, Liu C, Mendelson MM, Yao C, Kiel DP, Peters A, Wang-Sattler R, Visscher PM, Wray NR, Starr JM, Ding J, Rodriguez CJ, Wareham NJ, Irvin MR, Zhi D, Barrdahl M, Vineis P, Ambatipudi S, Uitterlinden AG, Hofman A, Schwartz J, Colicino E, Hou L, Vokonas PS, Hernandez DG, Singleton AB, Bandinelli S, Turner ST, Ware EB, Smith AK, Klengel T, Binder EB, Psaty BM, Taylor KD, Gharib SA, Swenson BR, Liang L, DeMeo DL, O’Connor GT, Herceg Z, Ressler KJ, Conneely KN, Sotoodehnia N, Kardia SL, Melzer D, Baccarelli AA, van Meurs JB, Romieu I, Arnett DK, Ong KK, Liu Y, Waldenberger M, Deary IJ, Fornage M, Levy D, London SJ, 2016. Epigenetic Signatures of Cigarette Smoking. Circ Cardiovasc Genet 9, 436–447. 10.1161/CIRCGENETICS.116.001506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA, Ueland PM, Wu MC, Nystad W, Bell DA, Peddada SD, London SJ, 2012. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Env. Heal. Perspect 120, 1425–1431. 10.1289/ehp.1205412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y. 2020. Kegg mapper for inferring cellular functions from protein sequences. Protein Sci 29:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keutgens A, Shostak K, Close P, Zhang X, Hennuy B, Aussems M, Chapelle JP, Viatour P, Gothot A, Fillet M, Chariot A, 2010. The repressing function of the oncoprotein BCL-3 requires CtBP, while its polyubiquitination and degradation involve the E3 ligase TBLR1. Mol Cell Biol 30, 4006–4021. 10.1128/MCB.01600-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, Chang YS, Jacobs DR, Steffes M, Lee DH, 2010. Association of Low-Dose Exposure to Persistent Organic Pollutants with Global DNA Hypomethylation in Healthy Koreans. Environ. Health Perspect. 118, 370–374. 10.1289/ehp.0901131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Sata F, Miyashita C, Miura R, Azumi K, Kobayashi S, Goudarzi H, Araki A, Ishizuka M, Todaka T, Kajiwara J, Hori T, Kishi R, 2017. Gender-specific association of exposure to non-dioxin-like polychlorinated biphenyls during pregnancy with methylation levels of H19 and long interspersed nuclear element-1 in cord blood in the Hokkaido study. Toxicology 390, 135–145. 10.1016/j.tox.2017.08.010 [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–7. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung YK, Ouyang B, Niu L, Xie C, Ying J, Medvedovic M, Chen A, Weihe P, Valvi D, Grandjean P, Ho SM, 2018. Identification of sex-specific DNA methylation changes driven by specific chemicals in cord blood in a Faroese birth cohort. Epigenetics 13, 290–300. 10.1080/15592294.2018.1445901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind L, Penell J, Luttropp K, Nordfors L, Syvanen AC, Axelsson T, Salihovic S, van Bavel B, Fall T, Ingelsson E, Lind PM, 2013. Global DNA hypermethylation is associated with high serum levels of persistent organic pollutants in an elderly population. Environ. Int. 59, 456–461. 10.1016/j.envint.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Monick MM, Beach SR, Plume J, Sears R, Gerrard M, Brody GH, Philibert RA, 2012. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet B Neuropsychiatr Genet 159B, 141–151. 10.1002/ajmg.b.32021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S, 2014. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics 30, 428–430. 10.1093/bioinformatics/btt684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Kim KS, Lee YM, Kim MJ, Jacobs DR Jr., Porta M, Kim DS, Lee DH, 2015. Persistent organic pollutants and promoter hypermethylation of the O(6)-methylguanine-DNA methyltransferase gene. Biomarkers 20, 136–142. 10.3109/1354750X.2014.1002806 [DOI] [PubMed] [Google Scholar]
- Pavuk M, Olson JR, Sjodin A, Wolff P, Turner WE, Shelton C, Dutton ND, Bartell S, Anniston Environmental Health Research, C., 2014a. Serum concentrations of polychlorinated biphenyls (PCBs) in participants of the Anniston Community Health Survey. Sci Total Env. 473–474, 286–297. 10.1016/j.scitotenv.2013.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavuk M, Olson JR, Wattigney WA, Dutton ND, Sjodin A, Shelton C, Turner WE, Bartell SM, Anniston Environmental Health Research, C., 2014b. Predictors of serum polychlorinated biphenyl concentrations in Anniston residents. Sci Total Env. 496, 624–634. 10.1016/j.scitotenv.2014.06.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, R VL, Clark SJ, Molloy PL, 2015. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8, 6 10.1186/1756-8935-8-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Maksimovic J, Oshlack A. 2016. Missmethyl: An r package for analyzing data from illumina’s humanmethylation450 platform. Bioinformatics 32:286–288. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, Van Djik S, Muhlhausler B, Stirzaker C, Clark SJ, 2016. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 17, 208 10.1186/s13059-016-1066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Shershebnev A, Medvedeva YA, Suvorov A, Wu H, Goltsov A, Loukianov E, Andreeva T, Gusev F, Manakhov A, Smigulina L, Logacheva M, Shtratnikova V, Kuznetsova I, Speranskiy-Podobed P, Burns JS, Williams PL, Korrick S, Lee MM, Rogaev E, Hauser R, Sergeyev O, 2018. Peripubertal serum dioxin concentrations and subsequent sperm methylome profiles of young Russian adults. Reprod Toxicol 78, 40–49. 10.1016/j.reprotox.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman GS, Wang X, Campbell MR, Coulter SJ, Olson JR, Pavuk M, Birnbaum LS, Bell DA, 2019. Polychlorinated biphenyl exposure and DNA methylation in the Anniston Community Health Survey. Epigenetics. 10.1080/15592294.2019.1666654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, Soderhall C, Scheynius A, Kere J, 2012. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 7, e41361. 10.1371/journal.pone.0041361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LM, Lohman K, Pittman GS, Barr RG, Chi GC, Kaufman J, Wan M, Bell DA, Blaha MJ, Rodriguez CJ, Liu Y, 2017. Tobacco exposure-related alterations in DNA methylation and gene expression in human monocytes: the Multi-Ethnic Study of Atherosclerosis (MESA). Epigenetics 12 10.1080/15592294.2017.1403692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LM, Wan M, Ding J, Taylor JR, Lohman K, Su D, Bennett BD, Porter DK, Gimple R, Pittman GS, Wang X, Howard TD, Siscovick D, Psaty BM, Shea S, Burke GL, Jacobs DR, Rich SS, Hixson JE, Stein JH, Stunnenberg H, Barr RG, Kaufman JD, Post WS, Hoeschele I, Herrington DM, Bell DA, Liu Y, 2015. DNA Methylation of the Aryl Hydrocarbon Receptor Repressor Associations with Cigarette Smoking and Subclinical Atherosclerosis. Circ. Cardiovasc. Genet. 8 10.1161/CIRCGENETICS.115.001097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley WNV and B D, 2002. Modern Applied Statistics with S, Fourth. ed Springer. [Google Scholar]
- Rosenbaum PF, Weinstock RS, Silverstone AE, Sjodin A, Pavuk M, 2017. Metabolic syndrome is associated with exposure to organochlorine pesticides in Anniston, AL, United States. Env. Int 108, 11–21. 10.1016/j.envint.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC, 2008. Global DNA Hypomethylation Is Associated with High Serum-Persistent Organic Pollutants in Greenlandic Inuit. Environ. Health Perspect. 116, 1547–1552. 10.1289/ehp.11338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AE, Rosenbaum PF, Weinstock RS, Bartell SM, Foushee HR, Shelton C, Pavuk M, 2012. Polychlorinated biphenyl (PCB) exposure and diabetes: results from the Anniston Community Health Survey. Env. Heal. Perspect 120, 727–732. 10.1289/ehp.1104247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Wang X, Campbell MR, Porter DK, Pittman GS, Bennett BD, Wan M, Englert NA, Crowl CL, Gimple RN, Adamski KN, Huang Z, Murphy SK, Bell DA, 2016. Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. PLoS One 11 10.1371/journal.pone.0166486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su KY, Li MC, Lee NW, Ho BC, Cheng CL, Chuang YC, Yu SL, Guo YL, 2019. Perinatal polychlorinated biphenyls and polychlorinated dibenzofurans exposure are associated with DNA methylation changes lasting to early adulthood: Findings from Yucheng second generation. Environ. Res. 170, 481–486. 10.1016/j.envres.2019.01.001 [DOI] [PubMed] [Google Scholar]
- Theobald HM, Peterson RE. 1994. Developmental and reproductive toxicity of dioxins and other ah receptor agonists In: Dioxins and health, (Schecter A, ed). Boston, MA:Springer US, 309–346. [Google Scholar]
- Van den Berg M, Birnbaum LS, Denison M, De Vito M, Farland W, Feeley M, Fiedler H, Hakansson H, Hanberg A, Haws L, Rose M, Safe S, Schrenk D, Tohyama C, Tritscher A, Tuomisto J, Tysklind M, Walker N, Peterson RE, 2006. The 2005 World Health Organization reevaluation of human and mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol. Sci. 10.1093/toxsci/kfl055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Dungen MW, Murk AJ, Kampman E, Steegenga WT, Kok DE, 2017. Association between DNA methylation profiles in leukocytes and serum levels of persistent organic pollutants in Dutch men. Env. Epigenet 3, dvx001. 10.1093/eep/dvx001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P, Dejardin E, Warnier M, Lair F, Claudio E, Bureau F, Marine JC, Merville MP, Maurer U, Green D, Piette J, Siebenlist U, Bours V, Chariot A, 2004. GSK3mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol Cell 16, 35–45. 10.1016/j.molcel.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Wan M, Bennett BD, Pittman GS, Campbell MR, Reynolds LM, Porter DK, Crowl CL, Wang X, Su D, Englert NA, Thompson IJ, Liu Y, Bell DA, 2018. Identification of smoking-associated differentially methylated regions using reduced representation bisulfite sequencing and cell type-specific enhancer activation and gene expression. Environ. Health Perspect. 126 10.1289/EHP2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, Rando OJ, Birney E, Myers RM, Noble WS, Snyder M, Weng Z, 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22, 1798–1812. 10.1101/gr.139105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PT, Zhao XQ, Marcovina SM, Brown BG, Krauss RM, 2013. Levels of cholesterol in small LDL particles predict atherosclerosis progression and incident CHD in the HDL-Atherosclerosis Treatment Study (HATS). PLoS One 8, e56782. 10.1371/journal.pone.0056782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, Pavuk M, Sjodin A, Lewin M, Jones R, Olson J, Birnbaum L, 2018. Exposure of dioxin-like chemicals in participants of the Anniston community health survey follow-up. Sci Total Env. 637–638, 881–891. 10.1016/j.scitotenv.2018.05.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze CE, Reynolds AP, van Dongen J, Dunham I, Lazar J, Neph S, et al. 2019. Eforge v2.0: Updated analysis of cell type-specific signal in epigenomic data. Bioinformatics 35:47674769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clair HB, Pinkston CM, Rai SN, Pavuk M, Dutton ND, Brock GN, et al. 2018. Liver disease in a residential cohort with elevated polychlorinated biphenyl exposures. Toxicol Sci 164:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. 2004. The gene ontology (go) database and informatics resource. Nucleic Acids Res 32:D258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Kile ML, Christiani DC, Ince TA, Kelsey KT, Marsit CJ. 2016. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics 17:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y. 2020. Kegg mapper for inferring cellular functions from protein sequences. Protein Sci 29:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Maksimovic J, Oshlack A. 2016. Missmethyl: An r package for analyzing data from illumina’s humanmethylation450 platform. Bioinformatics 32:286–288. [DOI] [PubMed] [Google Scholar]
- Pittman GS, Wang X, Campbell MR, Coulter SJ, Olson JR, Pavuk M, et al. 2019. Polychlorinated biphenyl exposure and DNA methylation in the anniston community health survey. Epigenetics 15:337–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Wang XT, Campbell MR, Porter DK, Pittman GS, Bennett BD, et al. 2016. Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. Plos One 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald HM, Peterson RE. 1994. Developmental and reproductive toxicity of dioxins and other ah receptor agonists In: Dioxins and health, (Schecter A, ed). Boston, MA:Springer US, 309–346. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.