Summary
Macrophages play an important role in structural cardiac remodeling and the transition to heart failure following myocardial infarction (MI). Previous research has focused on the impact of blood-derived monocytes on cardiac repair. Here we examined the contribution of resident cavity macrophages located in the pericardial space adjacent to the site of injury. We found that disruption of the pericardial cavity accelerated maladaptive post-MI cardiac remodeling. Gata6+ macrophages in mouse pericardial fluid contributed to the reparative immune response. Following experimental MI, these macrophages invaded the epicardium, lost Gata6 expression, but continued to perform anti-fibrotic functions. Loss of this specialized macrophage population enhanced interstitial fibrosis after ischemic injury. Gata6+ macrophages were present in human pericardial fluid, supporting the notion that this reparative function is relevant in human disease. Our findings uncover an immune cardioprotective role for the pericardial tissue compartment and argue for the reevaluation of surgical procedures that remove the pericardium.
Keywords: Pericardial macrophage, myocardial infarction, fibrosis
Graphical Abstract
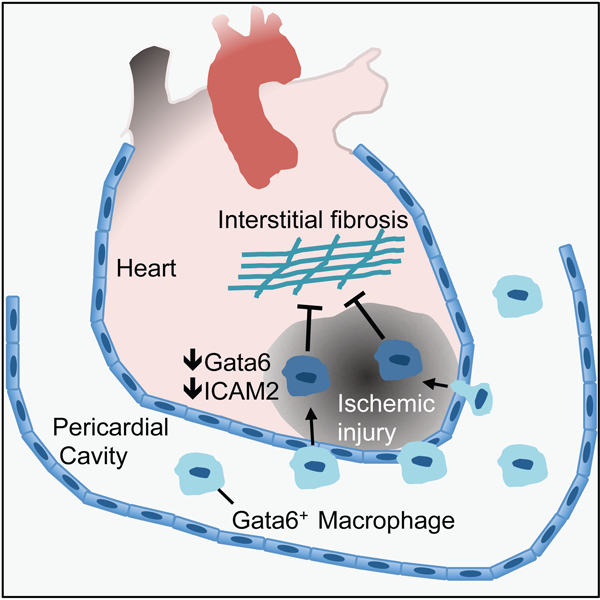
In Brief
Deniset et al. describe the pericardial cavity as an important source of resident macrophages that migrate into the heart following ischemic injury and prevent detrimental repair caused by excessive fibrosis.
Introduction
Macrophages are central players of the inflammatory response in the heart following myocardial infarction (MI). Removal or alteration of these cells in either the pro-inflammatory or repair phases of the immune response has important consequences for functional cardiac recovery (Hilgendorf et al., 2014; Horckmans et al., 2017; Nahrendorf et al., 2007). Most recruited cardiac macrophages following MI are believed to be uniquely monocyte-derived originating from hematopoietic sites (Heidt et al., 2014; Swirski et al., 2009). These cells are thought to be recruited early into the infarcted site, together with neutrophils, and induce a potent inflammatory response that causes tissue injury and eventually a fibrotic scar. Studies also note an ability of resident cardiac macrophages to proliferate locally to contribute to both adaptive healing and/or maladaptive responses in the heart (Dick et al., 2019; Epelman et al., 2014; Sager et al., 2016).
Recent work demonstrated that Gata6+ resident peritoneal cavity macrophages directly migrate to injured liver via the peritoneal cavity - not the vasculature - to promote tissue repair (Wang and Kubes, 2016). The maintenance and phenotype of this peritoneal macrophage population is critically dependent on expression of the transcription factor Gata6, which is driven by local cues (Gautier et al., 2014; Okabe and Medzhitov, 2014; Rosas et al., 2014). The heart is similarly surrounded by a cavity, but whether Gata6+ repair macrophages are also found in the pericardial space and contribute to the immune response following cardiac injury remains undefined. It is also important to note that despite coronary artery disease being one of the most prevalent diseases in the world, the conventional coronary ligation method routinely used to model MI in rodents does not fully mimic the human condition. In rodent models the pericardium is excised or disrupted to gain access to the heart during the surgical procedure, potentially causing the loss of pericardial cells. As such, the direct contribution of the pericardial cavity and its associated immune cells remains largely unexplored.
Here we examined the impact of the pericardial tissue compartment on cardiac functional recovery after MI by using a modified coronary ligation model in mice. We found a population of resident Gata6+ pericardial macrophages that exhibited transcriptional profiles similar to peritoneal and pleural cavity Gata6+ macrophages, but distinct from that of resident cardiac macrophages. GATA6+ macrophages were also identified in human pericardial fluid. Pericardial Gata6+ macrophages were recruited into the heart following cardiac injury, where Gata6 expression and the associated transcriptional signature was lost. Absence of this immune population in the system using Lyz2cre;Gata6fl/fl mice resulted in an increase in adverse cardiac fibrosis following MI. Thus, pericardial Gata6+ macrophages contribute to cardiac repair upon injury, highlighting the potential of the pericardial cavity as a site for immune modulation.
Results
An intact pericardial cavity preserves post-MI cardiac function
We modified the open-chest permanent coronary ligation method to evaluate the influence of the pericardial tissue compartment on cardiac functional recovery after MI. In this modified approach the pericardium is preserved, unopened, and the ligation suture is passed through the parietal pericardium and around the left anterior descending (LAD) coronary artery. This approach maintains an intact pericardium and unlike the conventional model, immune cells that surround the heart remain in their physiological environment (as we demonstrated using lavage). The use of either the conventional (disrupted pericardium) or modified (intact pericardium) ligation approaches (Fig. S1A) resulted in similar infarct sizes (Fig. S1B) which contributed to significant decreases in ejection fraction, an overall indicator of cardiac function, at 4 weeks post-MI compared to baseline (Fig. S1C, Table S1). More in depth assessment of left ventricle function by pressure-volume loop analysis revealed that animals with an intact pericardium had improved left ventricle hemodynamics compared to animals with a disrupted pericardium. The intact pericardium cohort displayed reduced left ventricle stiffness as noted by a lower end diastolic pressure volume relationship (EDPVR) and improved left ventricle relaxation capacity measured by both the isovolumic relaxation constant Tau and dP/dtmin. This was complemented by improvements in left ventricle contractile performance as indicated by superior preload recruitable stroke work (PRSW) and dP/dt max indices in the intact pericardium group as compared to the disrupted pericardium group (Fig. S1D, Table S1). It is worth noting that some parameters including another contractile function index end-systolic pressure volume relationship (ESPVR) were not improved (Fig. S1D, Table S1). Further, animals undergoing sham surgeries revealed that simply disrupting the pericardium without inducing the coronary ligation injury did not result in alteration of these relaxation and contractile parameters (Table S2). Collectively, this supports the concept that an intact pericardial cavity preserves cardiac function following MI.
The pericardial cavity serves as a reservoir for resident Gata6+ macrophages
We next evaluated the potential contribution of resident pericardial immune cells to this protective phenotype. We performed pericardial lavage in naïve mice and identified the Gata6+(MHCII− CD102+) macrophages as the most abundant cell type (34.2%) under steady-state conditions (Fig. 1A). In addition, MHCII+CD11c+(Gata6−CD102−) and MHCII+CD11c− myeloid populations along with different lymphoid populations including T, B2, B1a, and B1b cell subsets were also noted in varying amounts within pericardial immune compartment (Fig. 1A). Further characterization of the myeloid populations confirmed that expression of conventional macrophage markers F4/80, MERTK, and CD64 was limited to Gata6+CD102+ cells, thus Gata6+ pericardial macrophage (GPCM). Similar to populations in pleural and peritoneal cavities, MHCII+ cells in the pericardium expressed CD226 with the CD11c+ population also expressing CD26 but not C5aR (CD88), consistent with a CD11b+ conventional dendritic cell (DC) (Nakano et al., 2015) (Fig. 1A). Deletion of Gata6 in myeloid cells using Lyz2Cre;Gata6fl/fl mice resulted in specific depletion of GPCMs, highlighting the importance of Gata6 for the maintenance of this population within the pericardium (Figure 1B). Gata6+ pericardial macrophages (GPCMs) were not restricted to mice as we also documented these cells in pig and human pericardial fluid, where they are observed to be even more abundant than in rodents (Fig. 1C and 1D). This supports GPCMs as a highly conserved population of resident pericardial cavity immune cells across mammalian species.
Fig. 1-. Gata6+ macrophages are a major component of the pericardial immune cell compartment.
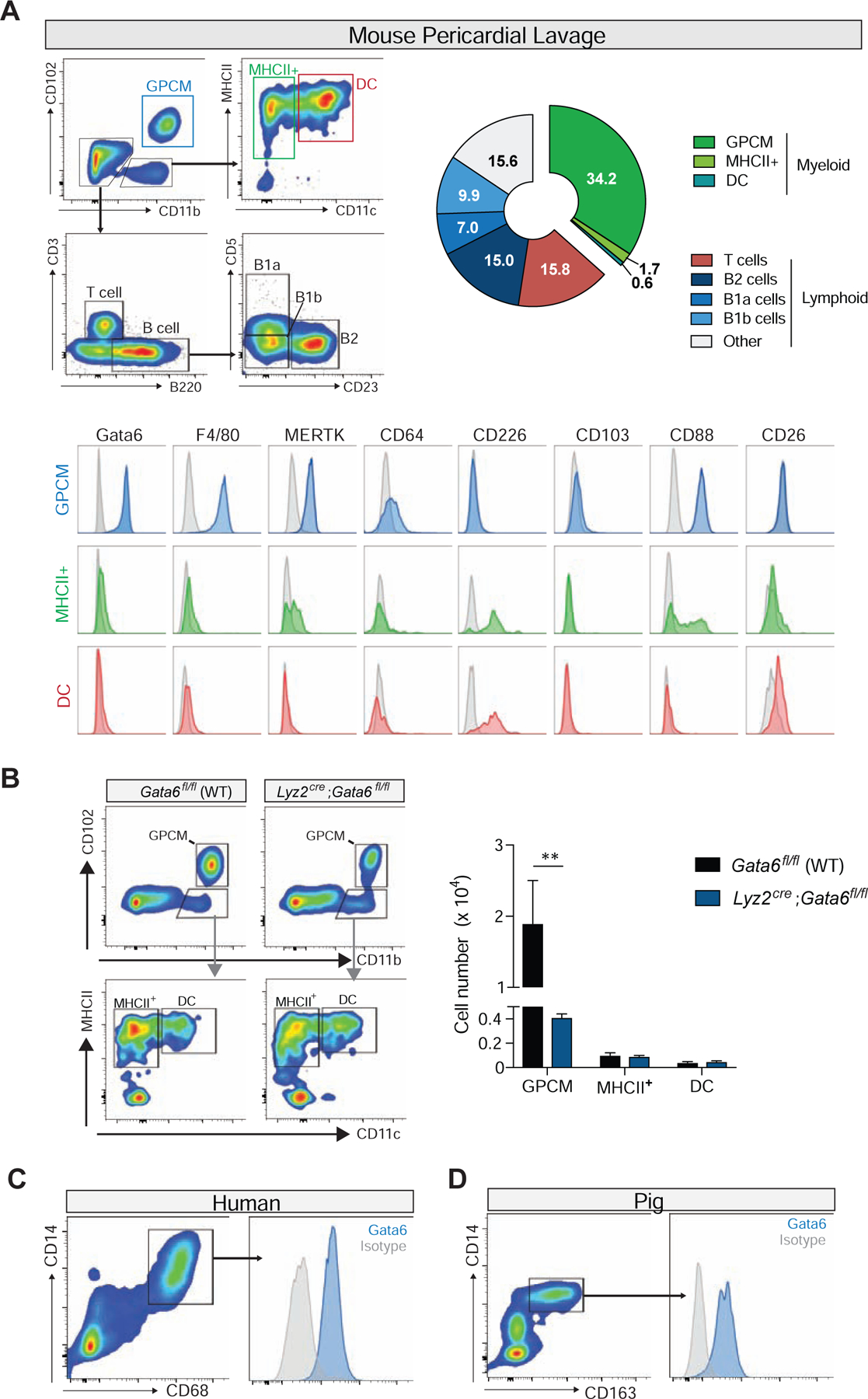
(A) Quantification and representative flow cytometry plots of mouse pericardial immune cells populations. Representative of 2 independent experiments, n= 4. Bottom panels-Representative histograms of macrophages and dendritic cell marker expression for pericardial myeloid cell populations. Representative of 2 independent experiments. (B) Representative flow cytometry plots and quantification of pericardial lavage derived GPCMs, MHCII+ , and DC populations at baseline in Gata6fl/fl (WT) and Lyz2Cre;Gata6fl/fl mice. Representative of n=4 from 2 independent experiments. **=p<0.01. Representative flow plots and Gata6 expression of pericardial cavity macrophages from (C) human pericardial fluid, and (D) pig pericardial fluid. Representative of 5 independent experiments for both human and porcine.
Pericardial Gata6+ macrophages are transcriptionally distinct from cardiac macrophages
Due to their anatomical location and Gata6 expression, we aimed to determine whether GPCMs were unique relative to tissue resident macrophages in the neighbouring heart as well as Gata6 expressing macrophages in other serosal cavities (pleura, peritoneum). Bulk RNAseq of sorted macrophage populations from these different tissue environments was used to evaluate these relationships (Fig. 2A). Principal component analysis and hierarchical clustering revealed that Gata6+ macrophages display more similar RNA expression profiles amongst the different cavities as compared to cardiac macrophages (Fig. 2B, 2C). Further analysis noted differential expression of 4296 (1353 increased vs GPCMs, 2943 decreased vs. GPCMs) genes between cardiac macrophages and GPCMs, 33 (12 increased vs GPCMs, 21 decreased vs. GPCMs) genes between peritoneal macrophages and GPCMs, and none between pleural macrophages and GPCMs (Fig. 2D). Pathway analysis revealed that cardiac macrophages in comparison to pericardial macrophages were enriched for expression of genes related to structural tissue organization and inflammatory mediators (Fig 2E). Conversely, relative to cardiac macrophages, pericardial macrophages displayed enhanced expression of genes associated with protein and nucleic acid metabolism (Fig 2E). Peritoneal macrophages were enriched for the expression of cell adhesion- and angiogenesis-related genes and inhibited for expression of malaria-associated genes, relative to pericardial macrophages (Fig 2D and 2E). All cavity macrophage populations expressed genes associated with hemostasis and importantly markers linked with cavity macrophages (e.g. Gata6, Icam2) (Fig 2D). This highlights that Gata6 is likely an important determinant of macrophage phenotype within all cavities, which distinguishes these populations from tissue resident macrophages in neighboring organs such as the heart.
Fig. 2-. Pericardial macrophages are transcriptionally distinct from cardiac macrophages.
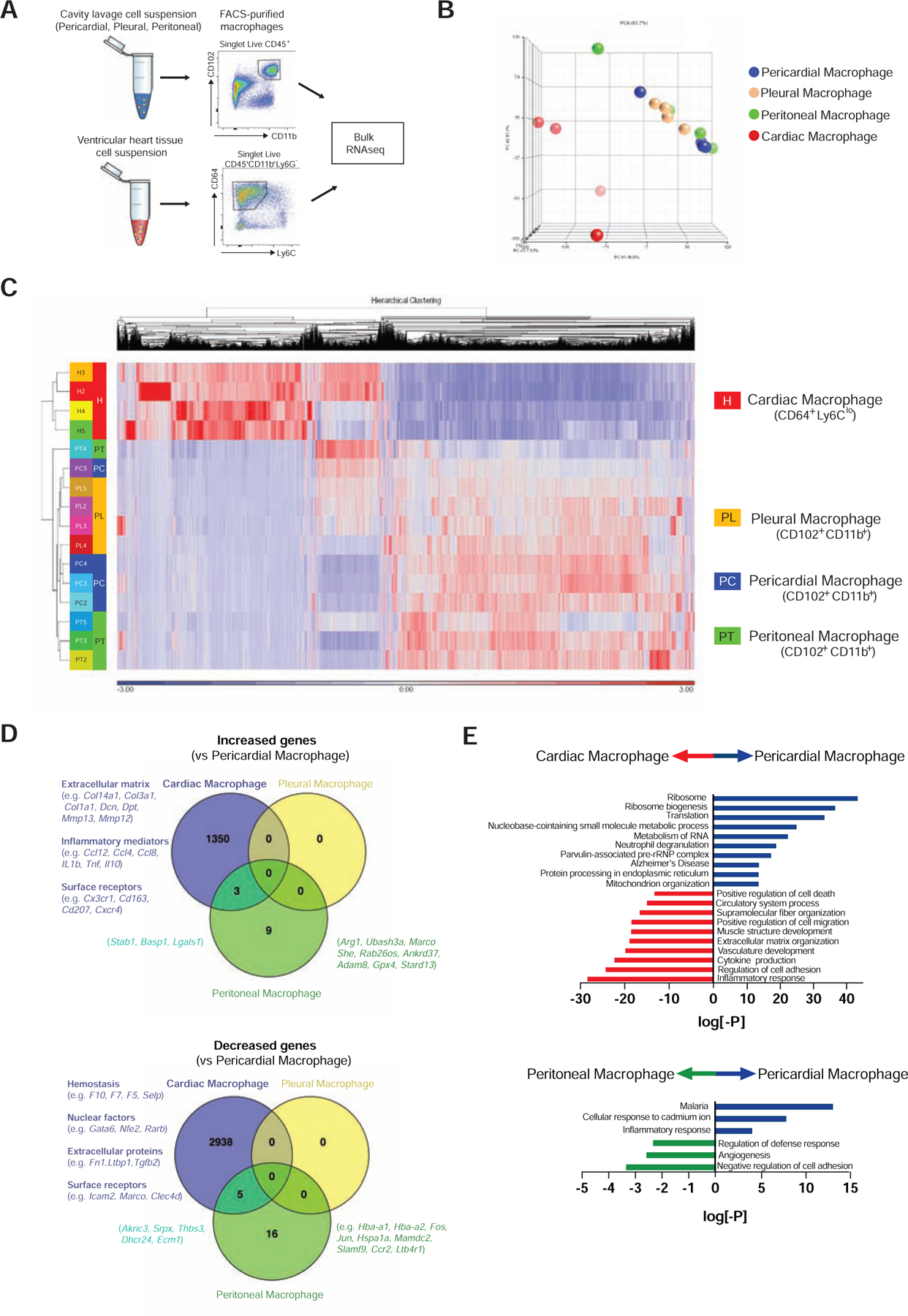
(A) Schematic of bulk RNAseq workflow from sorted macrophage populations. (B) Principle component analysis and (C) hierarchical clustering analysis of RNAseq profiles from cardiac, pericardial, pleural, and peritoneal macrophages. Data generated from 4 independent experiments. PCA plot was generated using all genes in the annotation, whereas the Heat Map shows only those genes where at least one sample has an RPKM > 1 and where the Coefficient of Variation across all 12 samples is > 0.3. (D) Venn diagrams representing increased and decreased mRNA expression of genes (≥2 fold, raw p-value ≤ 0.05, FDR adjusted p-value≤ 0.1) in cardiac (purple), peritoneal (green), and pleural (yellow) macrophages compared to pericardial macrophages. Differentially expressed genes in pericardial macrophages were compared to cardiac macrophages and peritoneal macrophages. Pathway enrichment is expressed as the log[–P].
Gata6+ pericardial macrophage are recruited to the heart following cardiac injury
To understand the fate of these myeloid cells following ischemic injury, pericardial myeloid cells were evaluated after experimental MI using the modified coronary ligation approach. Shortly following coronary ligation, a rapid disappearance of GPCMs in the pericardial lavage fluid was noted at day 3 through day 7 and followed by a slow rebound of this population at 28 days post-MI (Fig. 3). GPCM reduction coincided with a large influx of neutrophils and Ly6Chi monocytes into the pericardium that returned to basal levels by 28 days following injury (Fig. 3), consistent with recruitment kinetics previously described for these cells in the heart proper (Nahrendorf et al., 2007; Yan et al., 2013). The MHCII+ myeloid population displayed a trending increase in cell numbers at 28 days, whereas DC cell numbers were initially reduced before rebounding from day 7 onwards after injury (Fig. 3). We postulated that this specific disappearance of GPCMs during the recruitment phase of the immune response in the heart might be related to their relocation capacity to the heart.
Fig. 3-. Resident pericardial Gata6+ macrophage undergo dynamic changes following MI.
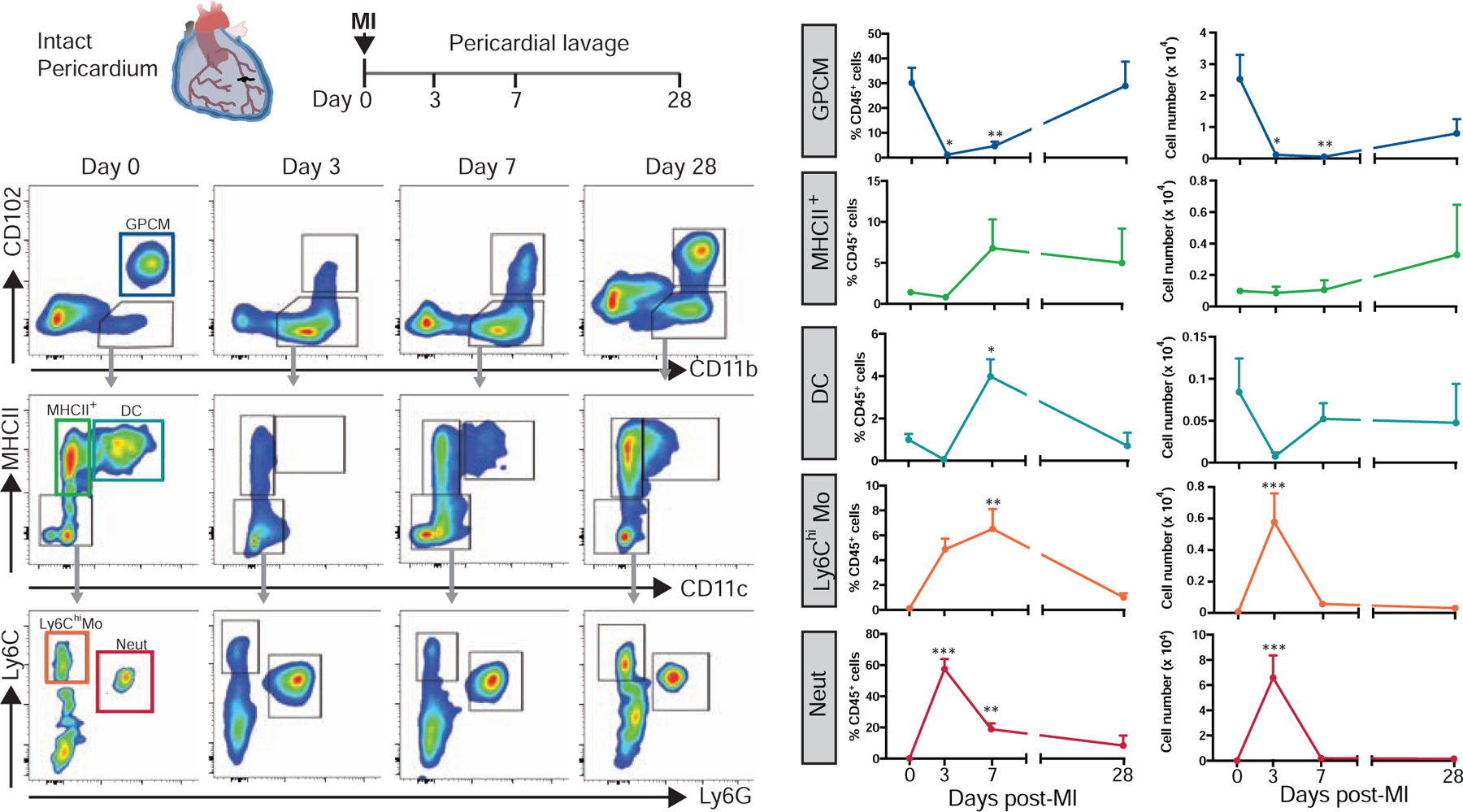
Representative flow cytometry plots and quantification of mouse pericardial GPCMs, MHCII+ myeloid cells, DCs, Ly6Chi monocytes, and neutrophils at baseline and days 3, 7, and 28 post-MI. Data represented as mean ± SEM, n=3–7 from at least 2 independent experiments per timepoint. *=p<0.05, **=p<0.01, ***=p<0.001 vs. baseline, one-way ANOVA.
Gata6 expression can be found in myocardial-derived cells (Freyer et al., 2015). Therefore, to evaluate the capacity of GPCMs to migrate from the pericardial cavity into the myocardium following MI, we generated Gata6H2B-Venus:C57BL/6J bone marrow chimeras (Fig. 4A). This approach resulted in roughly 70–80% reconstitution of GPCMs that were phenotypically identical to be GPCMs in control GataH2B-Venus animals (Venus+Gata6+F4/80hiCD11bhi) (Fig. S2). Venus+ parenchymal cells were not detected in the heart of these chimeric mice (Fig. S2). Whole mount confocal imaging of these bone marrow chimera mice revealed a robust recruitment of Venus+ GPCMs to the mesothelial layer that surrounds the heart at 7 days post-infarct (Fig. 4B). 3D reconstructions of the infarcted, peri-infarct, and remote zones of the left ventricle revealed the presence Venus+ macrophages only in the infarcted region of the cardiac tissue where they penetrated past the epicardial surface and into the heart tissue (Fig. 4C, Video S1). Whether these GPCMs are restricted to this region throughout subsequent cardiac remodeling remains unclear. Flow cytometry analysis of the pericardial cavity and ventricular heart tissue confirmed that Venus+ cells within the heart after injury were macrophages (CD11b+ CD64+) and differed from cardiac macrophages that lacked Venus expression based on their expression of CD102 and Gata6 (Fig. 4D). Time course analysis using a CD11b+CD102+ gating strategy revealed low level binding of GPCM to the heart early which increased over time reaching significance at day 7 post-MI (Fig. S3). Deletion of the chemokine receptor CCR2, which decreases monocyte-derived macrophage recruitment to the heart (Epelman et al., 2014), had no impact on CD102+ macrophage recruitment to the heart suggesting they are likely not dependent on monocyte influx (Fig. S3). Gata6 expression was limited to this CD102+ population, however, in a portion of this population its protein expression was decreased relative to pericardial cavity levels (Fig 4D, Fig. S3). This change in Gata6 protein expression coincided with decreased CD102 protein expression (Fig. 4D), highlighting the fact that GPCMs may be losing their cavity specific phenotype as they move into a new tissue environment.
Fig. 4-. Gata6+ pericardial macrophage are recruited into the heart following cardiac injury.
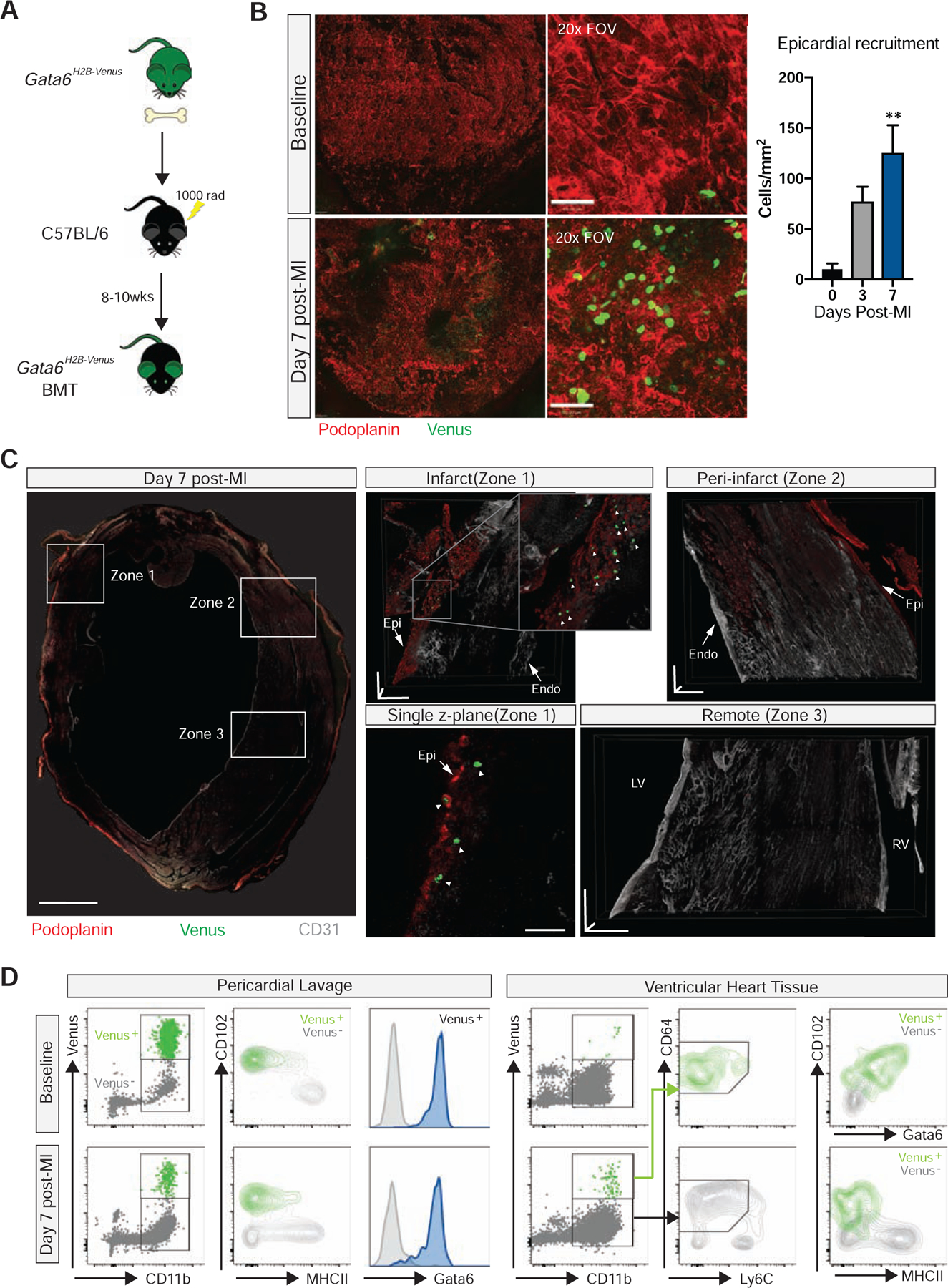
(A) Schematic of Gata6-venus bone marrow chimera generation. (B) Confocal composite stitch images (left panels) and single 20x FOVs (right panels) of epicardial localization and quantification of Venus+ GPCM at baseline and post-MI. Scale bars for stitched images = 200 μm and scale bars for single 20x tiles = 50 μm. Data represented as mean ± SEM, n= 3–4 from 2 independent experiments, **=p<0.01, one-way ANOVA. (C) Confocal composite stitch images and 3D projections of Venus+ GPCM within the infarct (zone 1), peri-infarct (zone 2), and remote (zone 3) areas of heart cross-section at 7 days post-MI. Scale bar= 1mm for global stitch images, scale bars= 150 μm in each plane for individual zone images, scale bar= 50 μm for single z-plane. Representative of n= 4 from 3 independent experiments. White arrowheads indicate GPCMs. Epi- epicardium, Endo- endocardium, LV- left ventricle, RV- right ventricle. (D) Flow cytometry analysis of pericardial cavity and cardiac tissue associated Venus+ GPCM pre- and post-MI. Representative of n=4 from 2 independent experiments. See also Figure S2.
Gata6+ pericardial macrophage alter their phenotype upon entry into the injured heart
To more directly track these cells from the pericardial space into the heart, we labeled GPCMs with fluorescent beads that were delivered locally using the intercostal approach to the pleural space (ICAPS) method (Weber, 2015), which allows delivery of substances into the pleural space without the need to ventilate the mice. Due to pores that exist between the pleural and pericardial cavities in rodents (Nakatani et al., 1988), local pleural injection of beads lead to specific labeling of GPCMs (>95% of bead+ cells) and pleural cavity cells without labeling cardiac macrophages or blood monocytes (Fig. S4). Using this approach (Fig 5A), we observed a dramatic displacement of GPCMs from the pericardial cavity, which was noted by a significant decrease in bead+ myeloid cells (Fig. 5B), into the heart as evidenced by an increase in bead+ cells following MI (Fig. 5C). Consistent with the concept that GPCMs change their phenotype upon entering heart tissue, only a limited number of bead+ GPCMs maintained Gata6 and CD102 protein expression (Fig. 5C). In fact, we noted a 10-fold increase in cell number of GPCMs making their way into the heart using this approach than was predicted by the CD102+ gating scheme (Fig S3). This further suggests that CD102 and Gata6 markers are reduced in the heart following MI, These data indicate that Gata6+ macrophages are recruited to the heart from the pericardial cavity upon myocardial infarction and start to penetrate into the tissue changing their phenotype significantly. However, we cannot fully discount the possibility that cardiac macrophages could take up beads released by dying pericardial macrophages in the infarcted area. While we do not support this contention, since no cardiac macrophages according to our specific markers had any intracellular beads. Free beads were also not seen in the tissues. Regardless, the data clearly demonstrated that the pericardial macrophages are at the very least entering the cardiac tissue.
Figure 5-. Pericardial macrophage down-regulate their cavity phenotypic markers upon entry into the heart.
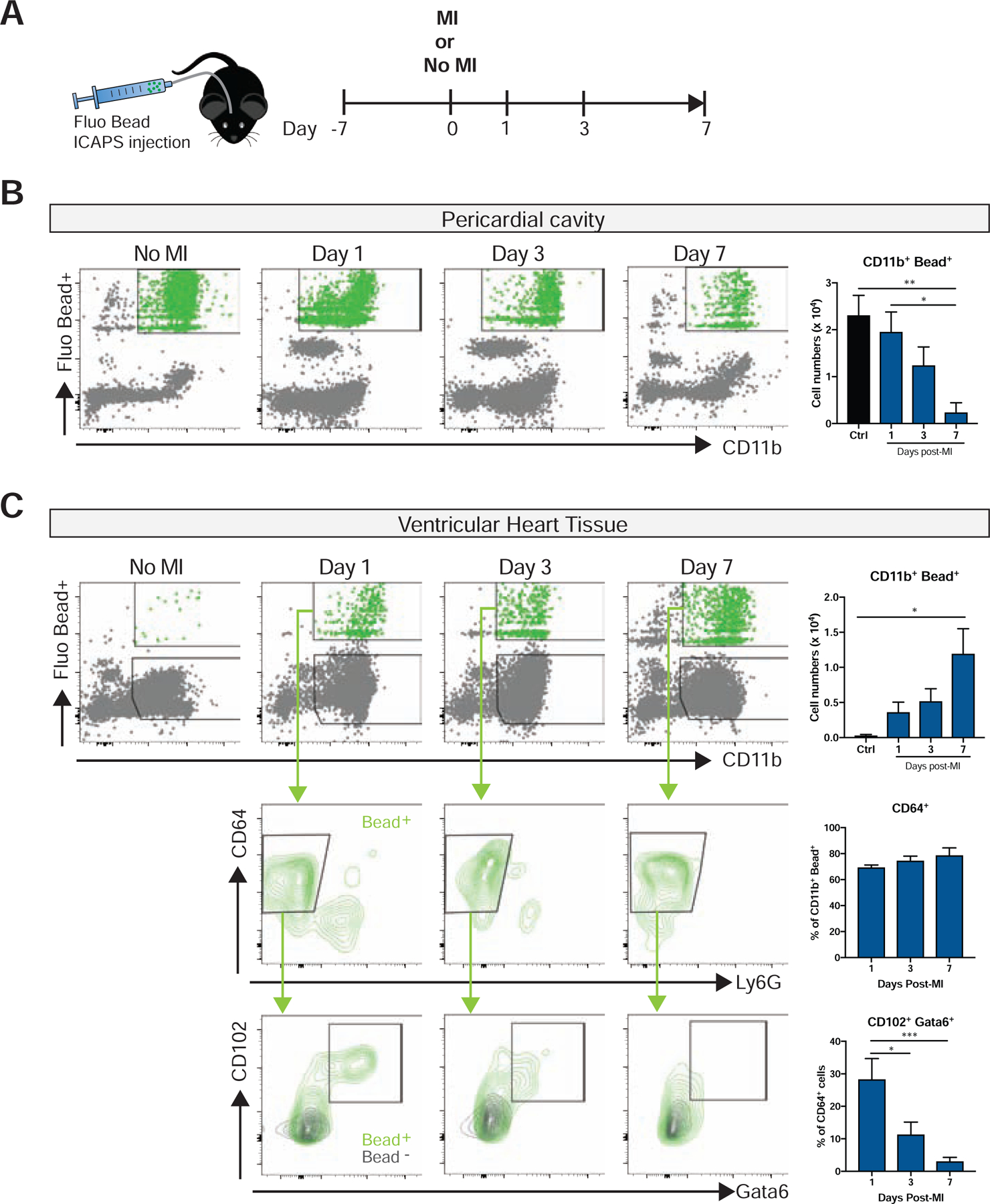
(A) Schematic timeline of ICAPS mediated fluorescent bead delivery combined with or without subsequent coronary ligation procedure. Flow cytometry analysis and quantification of fluorescent bead labeled pericardial myeloid cells in (B) pericardial cavity and (C) cardiac tissue with or without MI. Lower panels depict representative expression of pericardial specific markers (CD102+, Gata6+) in fluorescent bead positive macrophage. Representative of 2–3 independent experiments, n= 4–7. *=p<0.05, **=p<0.01, ***= p<0.001, one-way ANOVA. See also Figure S3 and S4.
Pericardial Gata6+ macrophage prevent adverse cardiac fibrosis
We used Lyz2Cre;Gata6fl/fl mice mice to determine the role of GPCMs on post-MI functional cardiac recovery. As previously noted, the number of GPCMs in the pericardial cavity of these mice was greatly diminished when compared to littermate controls (Fig. 1B). MHCII+ myeloid and DC numbers in the pericardium were unaffected in the Lyz2Cre;Gata6fl/fl mice (Fig. 1B). Neutrophils, Ly6Chi monocytes and Ly6Clo macrophage cell numbers were the same in the heart at baseline and were recruited in equal amounts following MI in Lyz2Cre;Gata6fl/fl mice and littermate controls, suggesting that Lyz2Cre;Gata6fl/fl effects are not related to the recruitment of the other major myeloid cells and consistent with the fact that none of these populations express Gata6 (Fig. S3, S5). Collectively, this supports the view that targeted deletion of Gata6 in the myeloid compartment localized in or around the heart was restricted to GPCMs. Coronary ligation with intact pericardium of Lyz2Cre;Gata6fl/fl mice and control animals (Fig. 6A) resulted in reductions of overall cardiac function (ejection fraction) at 4 weeks post-infarct as compared to baseline levels as assessed by echocardiography (Fig. 6B) and similar infarct sizes (TTC staining, 14.71 ± 4.42% and 12.98 ± 1.05%, p=0.79). Similar to animals with MI in the setting of a disrupted pericardium (Fig. 1C), Lyz2Cre;Gata6fl/fl mice displayed an elevated end-diastolic pressure volume relationship, which is associated with a stiffer left ventricle (Fig. 6C). In addition, a trend towards reduced contractile performance as represented by the preload recruitable stroke work (PRSW) index was also noted in Lyz2Cre;Gata6fl/fl mice as compared to WT littermates (Fig. 6C). Other parameters remained unchanged between groups (Table S3). Diffuse fibrosis in the remote myocardium from the ischemic scar has been noted to contribute to adverse remodeling, increased ventricular stiffness, and reduced contractile capacity (Jellis et al., 2010). Total fibrotic area measured by Sirius red positivity in the entire left ventricle did not differ between groups (Fig. 6D). However, regional evaluation of collagen deposition in the infarct, peri-infarct, and remote areas of the left ventricle myocardium by Sirius red staining and collagen I expression revealed enhanced interstitial fibrosis in the remote myocardium in Lyz2Cre;Gata6fl/fl animals compared to control littermates (Fig. 6E, 6F). Collectively, this observation supports the view that increased collagen deposition in remaining viable tissue in Lyz2Cre;Gata6fl/fl mice contributes to increased left ventricular stiffness as documented in these animals.
Figure 6-. Pericardial Gata6+ macrophage prevent adverse cardiac fibrosis.
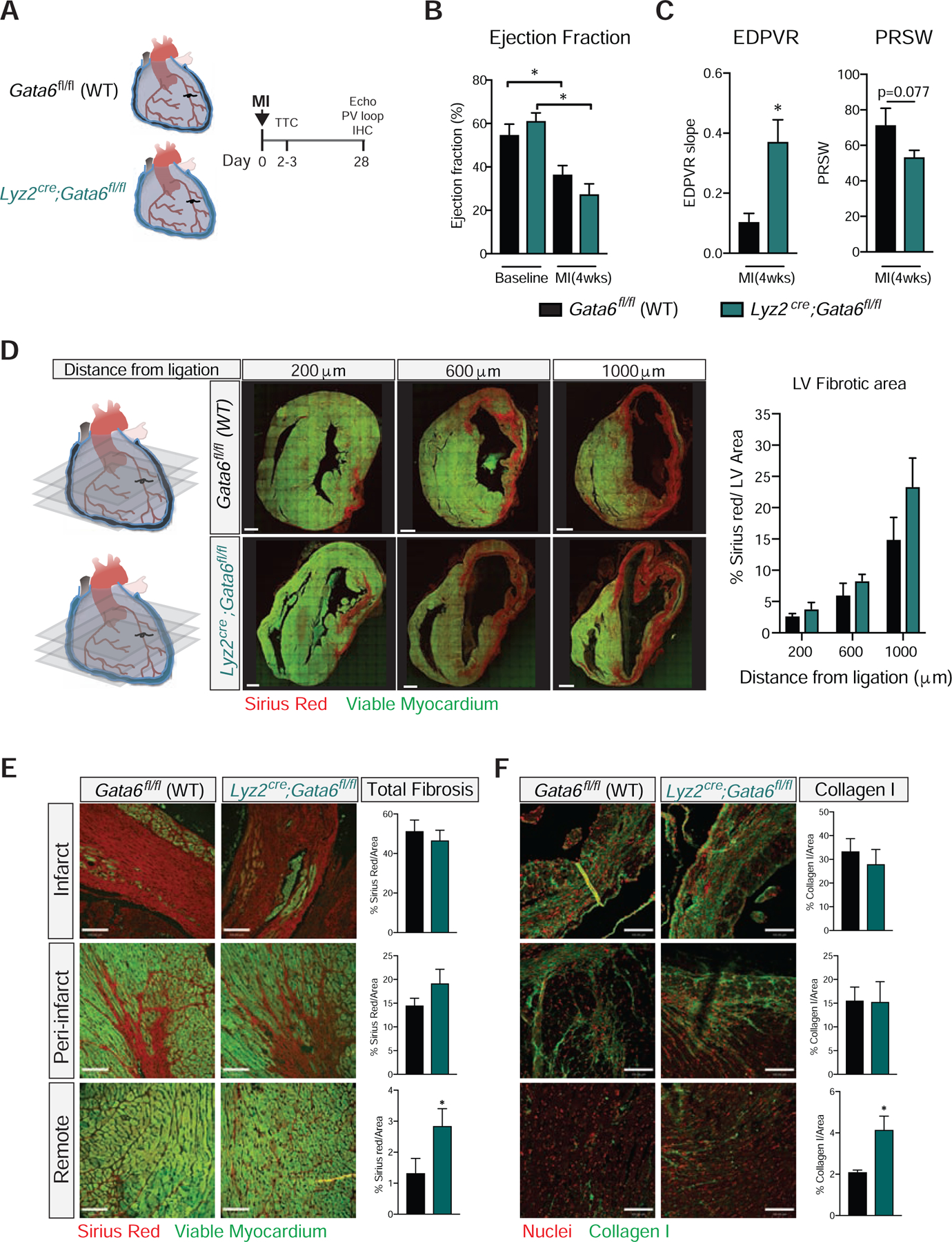
(A) Schematic timeline of coronary ligation procedure with Gata6fl/fl (WT) and Lyz2Cre;Gata6fl/fl mice. LV functional parameters at 28 days post-infarct for Gata6-WT and Gata6-KOmye mice measured by (B) echocardiography (EF) and (C) pressure-volume assessment (EDPVR, PRSW). Data represented as mean ± SEM, n= 7–9 for echocardiography, n= 8–9 for pressure volume loop assessment. *=p<0.05, one-way ANOVA for echocardiography, t-test for pressure volume measurements. (D) Confocal composite stitch images and quantification of picrosirius red staining in cardiac cross-sections at 28 days post-infarct for Gata6fl/fl (WT) and Lyz2Cre;Gata6fl/fl mice. Scale bars = 600 μm, Data represented as mean ± SEM, n=4–5. Representative images and quantification of sirius red (E) and collagen I (F) staining in infarct, peri-infarct, and remote areas of the left ventricle 28 days post-infarct for Gata6fl/fl (WT) and Lyz2Cre;Gata6fl/fl mice. Scale bars= 100 μm. Data represented as mean ± SEM, n=4–5, t-test. See also Figure S5 and Table S3.
Discussion
We found that resident Gata6+ macrophages in pericardial fluid played a crucial role in preventing fibrosis of healthy myocardium and improved functional cardiac recovery after ischemic injury. Our findings highlight the importance of serous pericardial cavity cells in cardiac repair, which conventional experimental methodologies currently overlook. While widely recognized for its lubricating and biomechanical properties, the pericardium and its fluid can contain cytokines, growth factors and miRNAs associated with pro-inflammatory and reparative responses under different pathological conditions (Beltrami et al., 2017; Butts et al., 2017; Fujita et al., 1996; Fujita et al., 1998). Here we found that immune cells, including GPCMs, are also localized within the cavity under steady-state and respond to injury. In addition, pericardial adipose tissue contains lymphoid cell populations that can influence the cardiac immune response (Horckmans et al., 2018). Thus, a combination of these factors likely accounts for the more exaggerated functional phenotype observed in the animals with a disrupted pericardial cavity.
This study contributes to our growing understanding of macrophage heterogeneity in the heart following cardiac injury. Using single cell RNA sequencing, Dick et al. recently describe the presence of 13 different myeloid clusters in the mouse heart following myocardial infarction with 6 unique macrophage populations within the infarct area that are distinct from baseline populations (Dick et al., 2019). Amongst these 6 populations, none of them are enriched for Gata6. It should be noted that the coronary ligation model that was used involved disrupting the outer pericardial layer, thus potentially removing Gata6+ macrophages from the local environment. Secondly as demonstrated here, Gata6 protein expression is decreased in GPCMs upon entry into the heart thus would not be represented in these different macrophage clusters. Functionally we found that with an intact pericardial cavity, GPCMs directly modulated adverse remodeling in the remote area of the heart leading to cardiac stiffness and diastolic dysfunction. In contrast to monocyte-derived macrophages and resident cardiac macrophages, which appear to modulate cardiac fibrosis in their local milieu (Dick et al., 2019; Sager et al., 2016), GPCMs impacted remodeling at a distance. This remote remodeling is a compensatory mechanism due to the increased workload on this portion of the heart and is dependent on characteristics of the infarct (e.g. size, composition) and infarct-derived factors. Relevant to this study, epicardial-derived cells and therapies that target the epicardial side of the infarct can reduce cardiac stiffness through local release of soluble mediators (Mewhort et al., 2017; Zhou et al., 2011). Based on the localization of GPCMs near the epicardial surface of the infarct, it is likely they act through a similar paracrine fashion to mediate functional changes in the heart. GPCMs remaining in the pericardial space may also contribute to this paracrine effect. A limitation of our study is the lack of a specific fate mapping approach. Development of such a tool for Gata6 expressing macrophages will be integral for assessing the localization and effector function of these cells within the pericardial cavity and heart tissue following cardiac injury.
Our findings also have significant clinical implications, as the pericardial cavity is intact during a myocardial infarction in the human context. Further, open-heart procedures often involve the removal of the pericardial fluid and disruption or removal of pericardial tissue without re-closure. In doing so, this may be inadvertently removing beneficial macrophages and their factors. This also emphasizes the poor understanding of this compartment and its impact on the heart. Our study suggests the pericardial cavity and associated immune cells play an active role in cardiac remodeling and function. Specifically, GPCMs that reside locally responded to cardiac injury and protected against maladaptive remodeling. More broadly these data highlight that the pericardial space is a potential immune-modulatory therapeutic avenue for many cardiac-related pathologies and interventions.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Paul Kubes (pkubes@ucalgary.ca). This study did not generate new unique reagents.
Experimental Model and Subject Details
Animals
C57BL/6J mice were purchased from Jackson Laboratories. Gata6H2B-Venus reporter mice (Freyer et al., 2015) were kindly provided by Dr. Hadjantonakis (Memorial Sloan Kettering). Ccr2Rfp/Rfp gene targeted mice (Saederup et al., 2010) were kindly provided by Richard M. Ransohoff (Lerner Research Institute, Cleveland Clinic, Cleveland) and Israel F. Charo (University of California San Francisco, San Francisco) . Gata6 floxed mice (Okabe and Medzhitov, 2014) were kindly provided by Dr. Medzhitov from Yale University and bred in-house with Lyz2cre mice. Lyz2cre;Gata6 fl/fl were subsequently bred with Gata6fl/fl to generate Cre+ and Cre− littermates. 10–14 week old male were used for experiments. All mice were house under specific pathogen-free, double-barrier unit at the University of Calgary. Mice were fed autoclaved rodent feed and water ad libitum. Farm pigs were obtained from Britestone Farming Co. 10–12 week old pigs were used. All protocols used were in accordance with the guidelines drafted by the University of Calgary Animal Care Committee and the Canadian Council on the Use of Laboratory Animals.
Human subjects
Pericardial fluid samples (N=5) were obtained from consenting patients undergoing valve replacement surgery (4- aortic valve replacement (all male, ages 67, 70, 65, and 69), 1-bicuspid aortic valve patient (female, age 42)) at the Foothills Medical Centre (Calgary, Alberta). All pericardial fluid samples were collected in a sterile environment on ice. Experiments involving human tissue usage were approved by the Conjoint Health Research Ethics Board at the University of Calgary and conform to the Declaration of Helsinki.
Method Details
In vivo interventions
Myocardial infarction (MI) was induced by permanent ligation of the left anterior descending (LAD) coronary artery either with (conventional) or without (modified) excising and tearing the outer pericardial tissue layer. For the procedure, mice were anesthetized using isoflurane (2% isoflurane with oxygen as carrier gas), intubated and ventilated using a VentElite Small Animal Ventilator Ventilator (Harvard Apparatus). The chest wall was shaved and cleaned with ethanol and iodine prior to a left thoracotomy in the fourth intercostal space. The left ventricle was landmarked and the LAD was ligated with monofilament 8–0 suture (Ethicon). The chest and skin were closed with a 5–0 Vicryl suture (Ethicon) and air in the thorax was evacuated via a pleural catheter. The same surgeon performed all procedures in a blinded fashion. For bone marrow chimeras generation, 6 week old male C57BL/6J mice were lethally irradiated (2× 525cGY) and subsequently reconstituted with Gata6 H2B-Venus bone marrow cells for 8 weeks.
The intercostal approach of the pleural space (ICAPS) method was carried out as previously noted (Weber, 2015). For the procedure, mice were anesthetized using isoflurane (2% isoflurane with oxygen as carrier gas) and placed on their left side with legs and arms fixed in place with the use of adhesive tape. The right antero-lateral thoracic area was shave and cleaned with ethanol and baxedin prior to performing a 3 cm long incision. A fluorescent bead loaded catheter syringe is then guided into the intercostal space. Once in the pleural space the total volume (50μl, 5μl of fluoresbrite fluorescent microsphere-1μm(Polysciences, Inc.) + 45 μl saline) is slowly injected and the catheter is removed in one motion. Wound closure was subsequently performed using staples and pain medication (buprenorphine, 0.05 mg/kg) was administered subcutaneously.
Cardiac Function assessment
Cardiac function was assessed via both non-invasive echocardiology and invasive pressure volume loop assessment. Functional assessment was performed prior to surgery (baseline) and 28 days post-MI. For PV-loop analysis, mice were anesthetized using isoflurane (4% induction, 2% maintenance), intubated and ventilated with a VentElite Small Animal Ventilator Ventilator (Harvard Apparatus). The neck was shaved, cleaned and excised to expose the right carotid artery. The carotid was occluded distally and a 1F conductance catheter (Millar Instruments) was gently advanced down the carotid artery into the left ventricle chamber. After recording baseline pressure volume measurements, an abdominal occlusion of the vena cava was performed to obtain a family of loops with varying afterload and preloads. After recording was complete, a parallel conductance value was obtained by jugular vein injection of hypertonic saline and blood was withdrawn to calibrate the conductance catheter. Animals were euthanized and tissues collected for subsequent analysis. Data were analyzed using the PV loop analysis module in Labchart (ADI Instruments).
Transthoracic echocardiography was performed under light anesthesia (isofluorane) at baseline and 28 days post myocardial infarction. Images were acquired using a 30 MHz linear transducer and analyzed with Vevo 770 software (Visual Sonics). LV ejection fraction was calculated using end-diastolic (ED) and end-systolic (ES) volumes (LV ejection fraction=(LVED volume–LVES volume)/LVED volume x100%).
Cell isolation, Flow Cytometry, Cell sorting
Mouse pericardial lavage was performed in anesthetised animals by a single injection of 100 μl of sterile saline into the pericardial cavity via PE-10 catheter and subsequent retrieval. Human pericardial fluid was collected during cardiac surgery using a syringe prior to opening the pericardial cavity. Pericardial samples were subsequently centrifuged at 1500 rpm for 5 minutes at 4°C and cell pellets was subsequently processed for flow cytometry. The murine heart ventricular tissue was excised, minced and subsequently digested in 450 U/ml collagenase I(Sigma), 125 U/ml collagenase XI(Sigma), 60 U/ml DNase I(Roche) and 60 U/ml hyaluronidase (Sigma) PBS for 1 hour at 37°C on an orbital shaker. For cell sorting exp eriments enzyme concentrations were doubled and digestion was performed for 30 minutes. Homogenates were initially passed through a 70 μm cell stainer and spun down at 60g for 5 minutes at 4°C to remove cardiac parenchymal cells. Supernatant was collected and passed through a 40 μm cell stainer for a single cell suspension. For remainder of experiments, residual red blood cells were lysed using ACK (Invitrogen). The cells were blocked using anti-mouse CD16/32 antibody (2.4G2 clone, BioXcell) or human FcγR binding inhibitor (eBioscience) for 20–30 minutes. Cells were then stained for 30 minutes with specified markers (Key Resources Table). Appropriate isotype control antibodies were used to confirm positive signals. Non-viable cells were identified using viability dye efluor 780 (eBioscience) or ghost dye™ red 710 (TONBO Biosciences). For intracellular staining the Foxp3 staining kit (Thermo Fisher) was used. Samples were run using BD FACS Canto flow cytometer and analyzed using FlowJo software (Tree Star). Neutrophils were identified as CD11b+ Ly6Ghi Ly6Cint. Classical monocytes were identified as CD11b+ Ly6G−Ly6Chi. Cardiac macrophages were identified as CD11b+ Ly6Clo CD64+ MHCII+/−. Gata6+ pericardial macrophage (GPCM) were identified as CD11bhiCD102+MHCII−Gata6+. Pericardial MHCII+ myeloid and DC populations were identified as CD11b+ CD102− MHCII+ CD11c− and CD11b+CD102− MHCII+CD11c+, respectively. For cell sorting experiments, cardiac macrophages were identified as viable CD45+ CD11b+ CD64+ Ly6Clo and cavity macrophages from the pericardium, pleura, and peritoneum were identified as viable CD45+ CD11b+ CD102+.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AF647 anti- Rabbit IgG | Life Technologies | Cat# A-21244 |
| APC anti-mouse MHCII | Biolegend | Cat# 107614 |
| eFluor660 anti-mouse CD23 | ThermoFisher | Cat# 50–0232-82 |
| APC-Cy7 anti-mouse CD45 | Biolegend | Cat# 103116 |
| APC-Cy7 anti-mouse MHCII | Biolegend | Cat# 107628 |
| BV421 anti-human CD163 | Biolegend | Cat# 333612 |
| BV510 anti-mouse CD45 | Biolegend | Cat# 103138 |
| BV605 anti-mouse CD102 | BD | Cat# 740346 |
| BV786 anti-mouse CD64 | BD | Cat# 741024 |
| eFluor450 anti-mouse CD3 | ThermoFisher | Cat# 48–0031-82 |
| FITC anti-mouse CD26 | Biolegend | Cat# 137806 |
| FITC anti-mouse CD226 | Biolegend | Cat# 128803 |
| FITC anti-mouse CD102 | BD | Cat# 557444 |
| FITC anti-mouse CD5 | Biolegend | Cat# 100605 |
| FITC anti-mouse F4/80 | Biolegend | Cat#123108 |
| FITC anti-mouse CD11c | Biolegend | Cat# 117306 |
| FITC anti- mouse Ly6G | ThermoFisher | Cat# 11–9668-82 |
| FITC anti-human CD14 | Biolegend | Cat# 325603 |
| FITC anti-pig CD14 | Bio-Rad | Cat# MCA12186A |
| FITC Rat IgG2a (Isotype control) | Biolegend | Cat# 400506 |
| FITC Rat IgG2b (Isotype control) | Biolegend | Cat# 400606 |
| Pacific Blue anti-mouse Ly6G | Biolegend | Cat# 127612 |
| PE anti-mouse CD64 | BD | Cat# 558455 |
| PE anti-mouse F4/80 | ThermoFisher | Cat# 12–4801-82 |
| PE anti-mouse MERTK | Biolegend | Cat# 151506 |
| PE anti-mouse CD102 | ThermoFisher | Cat# A15451 |
| PE anti-mouse B220 | BD | Cat# 553090 |
| PE anti-mouse CD103 | BD | Cat# 557495 |
| PE anti-mouse CD88 | Biolegend | Cat# 135806 |
| PE Mouse IgG1 (Isotype control) | ThermoFisher | Cat# 12–4714-81 |
| PE Rat IgG2a (Isotype control) | Biolegend | Cat# 400507 |
| PE Rat IgG2b (Isotype control) | BD | Cat# 553989 |
| PerCP-Cy5.5 anti-mouse Ly6C | Biolegend | Cat# 117328 |
| PerCP-Cy5.5 anti-mouse Ly6C | Biolegend | Cat# 128012 |
| PE-Cy7 anti-mouse CD11b | ThermoFisher | Cat# 25–0112-82 |
| PE-Cy7 anti-human CD68 | Biolegend | Cat# 333816 |
| Unconjugated anti-human FcγR | ThermoFisher | Cat# 14–9161-71 |
| Unconjugated anti-human GATA6 | Cell Signaling | Cat# 5851 |
| Unconjugated rabbit IgG (Isotype control) | Cell Signaling | Cat#3900S |
| Unconjugated anti-mouse CD16/32 | BioXcell | Cat# BE0307 |
| Unconjugated anti-mouse Collagen I | Abcam | Cat# ab21286 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fixable viability dye efluor 780 | ThermoFisher | Cat# 65–0865-14 |
| Ghost Dye red 710 | TONBO Biosciences | Cat# 13–0871-T100 |
| Formalin | HARLECO | Cat# R04586–82 |
| Collagenase I(Sigma), | Sigma | Cat# C0130 |
| Collagenase XI(Sigma) | Sigma | Cat# C7657 |
| DNase IX | Sigma | Cat# D5025–375ICU |
| Hyaluronidase (Sigma) | Sigma | Cat# H4272 |
| EDTA (0.5M, pH 8.0) | Ambion | Cat# AM9261 |
| PFA (16% solution) | EMS | Cat# 15710 |
| Ethyl cinnamate (Sigma). | Sigma | Cat# 112372 |
| Ammonia solution (2.0M in ethanol) | Sigma | Cat# 392685 |
| Sudan Black B (HARLECO) | HARLECO | Cat# 3545–12 |
| BSA (Sigma) | Sigma | Cat# A4503 |
| Triton X-100 (Sigma) | Sigma | Cat# X-100 |
| Syto 9 green fluorescent nucleic acid stain | ThermoFisher | Cat# S34854 |
| Fluorescent mounting media | Dako | Cat# S3023 |
| Tetrozolium chloride (TTC) | Sigma | Cat# T8877 |
| Fluoresbrite® YG Microspheres | Polysciences Inc. | Cat# 17154–10 |
| TRIzol Reagent | ThermoFisher | Cat# 15596026 |
| Critical Commercial Assays | ||
| Foxp3 / transcription factor staining buffer set | ThermoFisher | Cat# 00–5523-00 |
| Retrievagen kit | BD | Cat# 550524 |
| Direct-zol RNA Miniprep | Zymo Research | Cat# R2051 |
| NEBNext® Ultra™ II RNA Library Prep Kit for Illumina® | New England Biolabs | Cat# E7770S |
| NEBNext Multiplex kit | New England Biolabs | Cat# E7335 |
| NEBNext Poly(A) mRNA Magnetic isolation module | New England Biolabs | Cat# E7490 |
| Deposited Data | ||
| Cardiac and cavity MF RNAseq Dataset | GEO | GEO:GSE131724 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57: C57BL/6J | The Jackson Laboratory | JAX:000664 |
| Mouse: Gata6H2B-Venus | Dr. Hadjantonakis (Memorial Sloan Kettering) | Published in (Freyer et al., 2015) |
| Mouse: Ccr2Rfp/Rfp | Dr. Ransohoff (Lerner Research Institute, Cleveland Clinic, Cleveland) and Dr. Charo (University of California San Francisco, San Francisco) | Published in (Saederup et al., 2010) |
| Mouse: Gata6 fl/fl | Dr. Medzhitov from Yale University | Published in (Okabe and Medzhitov, 2014) |
| Mouse:LysMcre: B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | JAX: 004781 |
| Porcine: Farm pigs | Britestone Farming Co. | |
| Software and Algorithms | ||
| FlowJo software | FlowJo | www.flowjo.com |
| Volocity software | Perkin Elmer | |
| GraphPad Prism v6.0 software | GraphPad | www.graphpad.com |
| Labchart software | ADI Instruments | www.adinstruments.com/products/labchart |
| Vevo 770 software | Visual Sonics | www.visualsonics.com/product/imaging-systems |
| LAS X software | Leica | www.leica-microsystems.com/products/microscope-software |
| ImageJ software | NIH | www.imagej.nih.gov/ij/ |
| Partek Genomics Suite | Partek Inc. | www.partek.com |
| Adobe Illustrator CC 2018 | Adobe | www.adobe.com/ca/products/illustrator |
RNAseq Analysis
Pericardial cavity, pleural cavity, peritoneal cavity, and cardiac macrophage samples were collected and pooled from multiples naïve C57 wild-type mice and 1×105 cells from each group were sorted directly into TRIzol and stored at −80°C. A total of 4 biological replicates were prepared for each macrophage group. RNA was isolated using the Direct-zol RNA isolation kit (Zymo Research) following the included protocol. The harvested RNA was prepared for sequencing using the NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs). Sample Concentrations were calculated using PicoGreen and multiplexed samples were mixed in equimolar concentration and sent to the NIAMS Sequencing Core (Bethesda, MD) for sequencing on an Illumina HiSeq 3000. Data were demultiplexed and converted to FastQ using Bcl2fastq 2.17.1.14 (Illumina) and mapped to mm10 with Tophat 2.1.1. RPKM counting and normalization were performed using Partek GS 7.18.0723. Principal Component Analysis and Hierarchical clustering were performed on gene level RPKM data using Partek GS. Differential expression analysis was performed using fold change ≥ 2, with a raw p-value of < 0.05 and a false discovery rate (FDR) adjusted p-value ≤ 0.1 as thresholds. Venn diagrams were generated using Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Metascape (Metascape.org) was employed to identify enriched pathways based on the differentially expressed genes between two groups. Pathway gene sets from Gene Ontology (GO) biological processes (http://www.geneontology.org/), KEGG pathway (https://www.genome.jp/kegg/), Reactome molecular pathways (http://www.reactome.org/), and CORUM complexes database (http://mips.gsf.de/genre/proj/corum/index.html) were used.
Whole-mount imaging and tissue clearing
Prior to sacrifice, mice were anesthetized (10 mg/Kg xylazine hydrochloride and 200 mg/Kg ketamine hydrochloride), intubated and ventilated with a MiniVent ventilator for mice (Model 845, Harvard Apparatus). Thoracotomy was performed and Ef660 conjugated anti-mouse podoplanin antibody (clone, eBioscience) was injected into the pericardial space using a PE10 cathether and allowed to label the pericardial mesothelium for 10–15min.
For whole-mount imaging, the animal was subsequently euthanized, the outer pericardial layer removed and the heart excised and placed in 2% PFA containing PBS solution for 30 minutes. The fixed heart was placed left ventricle down onto a glass coverslip on the inverted microscope stage and kept in place using modeling putty. Image acquisition of the heart was performed using Olympus IX81 inverted microscope, equipped with an Olympus focus drive and a motorized stage (Applied Scientific Instrumentation, Eugene, OR) and fitted with a motorized objective turret equipped with 4x/0.16 UPLANSAPO, 10X/0.40 UPLANSAPO and 20x/0.70 UPLANSAPO objective lenses and coupled to a confocal light path (WaveFx; Quorum Technologies, Guelph, ON) based on a modified Yokogawa CSU-10 head (Yokogawa Electric Corporation, Tokyo, Japan). Laser excitation wavelengths 491-, 561- , and 642nm (Cobolt) were used in a rapid succession together with the appropriate band-pass filters (Semrock). A back-thinned EMCCD 512 × 512 pixel camera was used for fluorescence detection. Volocity software (Perkin Elmer) was used to drive the confocal microscope and for acquisition and analysis of images. Composite stitch images of the heart were acquired combining multiple 10x FOVs and placed together by the Volocity software. Venus+ cells were identified using the “Find object” function within the measurement modality and quantified for the entire area for the heart captured in the stitch.
Heart tissue clearing was performed as previously described (Klingberg et al., 2017). In brief following intrapericardial administration of antibody, mice were perfused using 5mM EDTA containing PBS and 4% PFA containing PBS. Heart was excised, placed in 4% PFA containing PBS for 2 hours followed by sequential dehydration steps (50%, 70%, 99%, 99% EtOH) of 4 hours at 4°C. After dehydration, hearts were place in ethyl cinnamate (Sigma). For imaging, cleared heart were placed LV upwards in an ethyl cinnamate containing vessel and topped with a coverslip. Image acquisition of was performed using a Leica TCS SP8 upright resonant scanning dual white-light laser equipped confocal microscope. Fluorescently labeled cells were excited with with a white-light laser at 488, 594 and 642nm and detected using HyD hybrid internal descanned detectors. Leica software was used to drive the confocal microscope, 3D rendering, acquisition of images, and video generation.
Histological staining
Hearts were fixed in 10% formalin and sent to Calgary Laboratory Services for paraffin embedding, sectioning (6 μm) and picrosirius staining. Sections were prepared at different levels from the ligation site. Unstained sections were used for immunohistochemistry staining of Collagen I. For this purpose, sections were deparafinized, rehydrated and treated with an 50mM ammonia solution (Sigma) in 70% EtOH for 3 hours followed by a 5 minutes incubation 0.1% Sudan Black B (HARLECO) in 70% EtOH to reduce tissue autofluorescence. Antigen retrieval was performed prior to primary antibody incubation as per manufacturer’s instructions (Retrievagen kit, BD). Sections were blocked with 2% rat serum before primary staining with rabbit anti-mouse Collagen I antibody (polyclonal, Abcam) and secondary staining with anti-rabbit fluorophore-conjugated antibody (Life Technologies) in 1% BSA (Sigma) 0.2% Triton-X (Sigma) containing PBS solution. Nuclei were stained using Syto 9 nucleic acid stain (ThermoFisher). Anti-fade fluorescence mounting media (Dako) was added to the section before mounting the slides for imaging. Image acquisition of both picrosirius staining and collagen I were obtained using an inverted spinning disk confocal microscope as described above. Composite stitch images of heart cross-sections were generated using the Volocity software. Quantification of staining was performed using ImageJ software.
Cardiac tissue viability was assessed acutely following MI by TTC staining. Hearts were collected, covered in plastic wrap and chilled at −20°C prior to 1mm sectioning of the ventricular tissue using a brain matrix and sectioning blades. 1mm sections were subsequently placed in individual wells of a 24-well plate and incubated with 1% TTC (Sigma) containing PBS solution while shaking at 37°C for 15 minutes. Sections were washed and fixed in 2% PFA containing PBS. Image acquisition of heart sections was performed using a camera equipped optical microscope (Zeiss). Infarct area for each section as assessed using ImageJ software was determined by lack of deep red coloration (indicative of viable tissue) and tabulated for the entire volume of the ventricular portion of the heart.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are presented as mean ± SEM. Statistical comparisons were performed using GraphPad Prism v6.0 software. Data were compared either by unpaired two- tailed t-test, one-way, or two-way ANOVA. Statistical significance was set at p<0.05. The numbers of independent replicates (n) are reported in the figure legends.
Supplementary Material
3D confocal composite stitch image reconstruction of the infarcted LV wall (7 days post-MI) from a cleared Gata6H2B-venus bone marrow chimera mouse heart. Box dimensions-1000 μm (h) x 950 μm (w) x 150 μm (d).
Highlights.
Pericardial cavity in mouse and human serves as a reservoir for GPCMs
GPCMs are transcriptionally distinct from neighboring cardiac macrophages
GPCMs relocate rapidly to the ischemic heart and undergo phenotypic alterations
Absence of GPCMs in Lyz2cre;Gata6fl/fl mice promotes post-injury cardiac fibrosis
Acknowledgments
We thank Trecia Nussbaumer for the breeding of mice. We thank Dr. Karen Poon at the Nicole Perkins Microbial Communities Core Labs for assistance with flow cytometry. Next Generation Sequencing was performed by the NIAMS Sequencing Core This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov)
Funding: P.K. is supported by the Heart and Stroke Foundation of Canada, Canadian Institutes of Health Research, and the Canada Research Chairs Program. P.W.M.F. is supported by Heart and Stroke Foundation of Canada, Canadian Institutes of Health Research, and National Institutes of Health. J.F.D. is supported by a fellowship from Canadian Institutes of Health Research. S.K.J. is supported by a fellowship from the DFG (JO1497/2–1). C.D. is supported by a fellowship from the DFG (DE2654/1–1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
Authors declare no competing interests.
Data and Software Availability
The raw RNA sequencing data, normalized data and differentially expressed analysis has been deposited to GEO with accession number GEO: GSE131724.
References
- Beltrami C, Besnier M, Shantikumar S, Shearn AI, Rajakaruna C, Laftah A, Sessa F, Spinetti G, Petretto E, Angelini GD, and Emanueli C (2017). Human Pericardial Fluid Contains Exosomes Enriched with Cardiovascular-Expressed MicroRNAs and Promotes Therapeutic Angiogenesis. Mol Ther 25, 679–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butts B, Goeddel LA, George DJ, Steele C, Davies JE, Wei CC, Varagic J, George JF, Ferrario CM, Melby SJ, and Dell’Italia LJ (2017). Increased Inflammation in Pericardial Fluid Persists 48 Hours After Cardiac Surgery. Circulation 136, 2284–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh S, Aronoff L, et al. (2019). Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol 20, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, et al. (2014). Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer L, Schroter C, Saiz N, Schrode N, Nowotschin S, Martinez-Arias A, and Hadjantonakis AK (2015). A loss-of-function and H2B-Venus transcriptional reporter allele for Gata6 in mice. BMC Dev Biol 15, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Ikemoto M, Kishishita M, Otani H, Nohara R, Tanaka T, Tamaki S, Yamazato A, and Sasayama S (1996). Elevated basic fibroblast growth factor in pericardial fluid of patients with unstable angina. Circulation 94, 610–613. [DOI] [PubMed] [Google Scholar]
- Fujita M, Ikemoto M, Tanaka T, Tamaki S, Yamazato A, Sawamura T, Hasegawa K, Kihara Y, Nohara R, and Sasayama S (1998). Marked elevation of vascular endothelial growth factor and basic fibroblast growth factor in pericardial fluid of patients with angina pectoris. Angiogenesis 2, 105–108. [DOI] [PubMed] [Google Scholar]
- Gautier EL, Ivanov S, Williams JW, Huang SC, Marcelin G, Fairfax K, Wang PL, Francis JS, Leone P, Wilson DB, et al. (2014). Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J Exp Med 211, 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidt T, Courties G, Dutta P, Sager HB, Sebas M, Iwamoto Y, Sun Y, Da Silva N, Panizzi P, van der Laan AM, et al. (2014). Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ Res 115, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf I, Gerhardt LM, Tan TC, Winter C, Holderried TA, Chousterman BG, Iwamoto Y, Liao R, Zirlik A, Scherer-Crosbie M, et al. (2014). Ly-6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ Res 114, 1611–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horckmans M, Bianchini M, Santovito D, Megens RTA, Springael JY, Negri I, Vacca M, Di Eusanio M, Moschetta A, Weber C, et al. (2018). Pericardial Adipose Tissue Regulates Granulopoiesis, Fibrosis, and Cardiac Function After Myocardial Infarction. Circulation 137, 948–960. [DOI] [PubMed] [Google Scholar]
- Horckmans M, Ring L, Duchene J, Santovito D, Schloss MJ, Drechsler M, Weber C, Soehnlein O, and Steffens S (2017). Neutrophils orchestrate post-myocardial infarction healing by polarizing macrophages towards a reparative phenotype. Eur Heart J 38, 187–197. [DOI] [PubMed] [Google Scholar]
- Jellis C, Martin J, Narula J, and Marwick TH (2010). Assessment of nonischemic myocardial fibrosis. J Am Coll Cardiol 56, 89–97. [DOI] [PubMed] [Google Scholar]
- Klingberg A, Hasenberg A, Ludwig-Portugall I, Medyukhina A, Mann L, Brenzel A, Engel DR, Figge MT, Kurts C, and Gunzer M (2017). Fully Automated Evaluation of Total Glomerular Number and Capillary Tuft Size in Nephritic Kidneys Using Lightsheet Microscopy. J Am Soc Nephrol 28, 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewhort HEM, Svystonyuk DA, Turnbull JD, Teng G, Belke DD, Guzzardi DG, Park DS, Kang S, Hollenberg MD, and Fedak PWM (2017). Bioactive Extracellular Matrix Scaffold Promotes Adaptive Cardiac Remodeling and Repair. JACC Basic Transl Sci 2, 450–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, and Pittet MJ (2007). The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med 204, 3037–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H, Moran TP, Nakano K, Gerrish KE, Bortner CD, and Cook DN (2015). Complement receptor C5aR1/CD88 and dipeptidyl peptidase-4/CD26 define distinct hematopoietic lineages of dendritic cells. J Immunol 194, 3808–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani T, Shinohara H, Fukuo Y, Morisawa S, and Matsuda T (1988). Pericardium of rodents: pores connect the pericardial and pleural cavities. Anat Rec 220, 132–137. [DOI] [PubMed] [Google Scholar]
- Okabe Y, and Medzhitov R (2014). Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas M, Davies LC, Giles PJ, Liao CT, Kharfan B, Stone TC, O’Donnell VB, Fraser DJ, Jones SA, and Taylor PR (2014). The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science 344, 645–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup N, Cardona AE, Croft K, Mizutani M, Cotleur AC, Tsou CL, Ransohoff RM, and Charo IF (2010). Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS One 5, e13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager HB, Hulsmans M, Lavine KJ, Moreira MB, Heidt T, Courties G, Sun Y, Iwamoto Y, Tricot B, Khan OF, et al. (2016). Proliferation and Recruitment Contribute to Myocardial Macrophage Expansion in Chronic Heart Failure. Circ Res 119, 853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, Figueiredo JL, Kohler RH, Chudnovskiy A, Waterman P, et al. (2009). Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 325, 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, and Kubes P (2016). A Reservoir of Mature Cavity Macrophages that Can Rapidly Invade Visceral Organs to Affect Tissue Repair. Cell 165, 668–678. [DOI] [PubMed] [Google Scholar]
- Weber GF (2015). Immune targeting of the pleural space by intercostal approach. BMC Pulm Med 15, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Anzai A, Katsumata Y, Matsuhashi T, Ito K, Endo J, Yamamoto T, Takeshima A, Shinmura K, Shen W, et al. (2013). Temporal dynamics of cardiac immune cell accumulation following acute myocardial infarction. J Mol Cell Cardiol 62, 24–35. [DOI] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, et al. (2011). Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest 121, 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D confocal composite stitch image reconstruction of the infarcted LV wall (7 days post-MI) from a cleared Gata6H2B-venus bone marrow chimera mouse heart. Box dimensions-1000 μm (h) x 950 μm (w) x 150 μm (d).


