In multipolar vertebrate neurons, action potentials (APs) initiate close to the soma, at the axonal initial segment. Invertebrate neurons are typically unipolar with dendrites integrating directly into the axon. Where APs are initiated in the axons of invertebrate neurons is unclear. Voltage-gated sodium (NaV) channels are a functional hallmark of the axonal initial segment in vertebrates.
Keywords: Drosophila, para, AIS, Distal Axonal Segment
Abstract
In multipolar vertebrate neurons, action potentials (APs) initiate close to the soma, at the axonal initial segment. Invertebrate neurons are typically unipolar with dendrites integrating directly into the axon. Where APs are initiated in the axons of invertebrate neurons is unclear. Voltage-gated sodium (NaV) channels are a functional hallmark of the axonal initial segment in vertebrates. We used an intronic Minos-Mediated Integration Cassette to determine the endogenous gene expression and subcellular localization of the sole NaV channel in both male and female Drosophila, para. Despite being the only NaV channel in the fly, we show that only 23 ± 1% of neurons in the embryonic and larval CNS express para, while in the adult CNS para is broadly expressed. We generated a single-cell transcriptomic atlas of the whole third instar larval brain to identify para expressing neurons and show that it positively correlates with markers of differentiated, actively firing neurons. Therefore, only 23 ± 1% of larval neurons may be capable of firing NaV-dependent APs. We then show that Para is enriched in an axonal segment, distal to the site of dendritic integration into the axon, which we named the distal axonal segment (DAS). The DAS is present in multiple neuron classes in both the third instar larval and adult CNS. Whole cell patch clamp electrophysiological recordings of adult CNS fly neurons are consistent with the interpretation that Nav-dependent APs originate in the DAS. Identification of the distal NaV localization in fly neurons will enable more accurate interpretation of electrophysiological recordings in invertebrates.
SIGNIFICANCE STATEMENT The site of action potential (AP) initiation in invertebrates is unknown. We tagged the sole voltage-gated sodium (NaV) channel in the fly, para, and identified that Para is enriched at a distal axonal segment. The distal axonal segment is located distal to where dendrites impinge on axons and is the likely site of AP initiation. Understanding where APs are initiated improves our ability to model neuronal activity and our interpretation of electrophysiological data. Additionally, para is only expressed in 23 ± 1% of third instar larval neurons but is broadly expressed in adults. Single-cell RNA sequencing of the third instar larval brain shows that para expression correlates with the expression of active, differentiated neuronal markers. Therefore, only 23 ± 1% of third instar larval neurons may be able to actively fire NaV-dependent APs.
Introduction
Action potentials (APs) are generated by the sequential opening of voltage-gated sodium (NaV) and potassium channels (KV) in the axons of neurons (Sherwood, 2008). Mammalian CNS neurons are typically multipolar; and APs initiate at the dense concentration of NaV channels in the axonal initial segment (AIS) close to the soma, and propagate along the axon via the nodes of Ranvier (Huxley and Stämpfli, 1949; Salzer, 2003; Palmer and Stuart, 2006; Shu et al., 2006; Kole et al., 2008; Rasband and Peles, 2016). In addition to AP initiation, the AIS forms a barrier between the soma and the axon, preventing the free diffusion of organelles, proteins, and lipids between the two compartments (Palay et al., 1968; Kobayashi et al., 1992; Winckler et al., 1999; Song et al., 2009). Invertebrate neurons are typically unipolar with the dendrites impinging on the axon distal to the cell body (Rolls, 2011). Whether an AIS is present in these neurons, and where it is located along the axon, is unresolved.
In order to determine the site of AP initiation, and if and where the AIS is in invertebrate neurons, we examined the location of the sole NaV channel gene in Drosophila melanogaster, paralytic (para). Unlike mammals, which have multiple NaV encoding genes (SCN1-5A, 8-11A) (Huang et al., 2017), the genome of D. melanogaster encodes only two genes predicted to encode NaV proteins para and Na channel protein 60E (NaCP60E) (D. T. Suzuki et al., 1971; Okamoto et al., 1987; Tseng-Crank et al., 1991; Hong and Ganetzky, 1994). para is the putative NaV channel as NaCP60E null animals are viable with no loss of inward sodium currents detected in neurons using patch clamp (Germeraad et al., 1992; Anholt et al., 1996; Kulkarni et al., 2002). In contrast, para null animals die as first instar larvae with no detectable inward sodium current in neurons using patch clamp (Loughney et al., 1989; O'Dowd et al., 1989; Hong and Ganetzky, 1994). Despite having one NaV gene, compared with nine in mammals, it is possible that a similar degree of channel protein diversity is achieved via alternate splicing. para has 60 predicted isoforms, some of which have different developmental expression (Lin et al., 2009; Baines et al., 2012). Very little is known about the expression pattern or subcellular localization of Para. ISH studies determined that para is expressed in the nervous system from embryos to adults (Amichot et al., 1993; Hong and Ganetzky, 1994). Whether para is expressed in all or just some cells in the nervous system, and where it is subcellularly localized, remains to be established.
To determine the expression pattern and protein localization of NaV channels in Drosophila neurons, we used previously established tools (Bateman et al., 2006; Venken et al., 2011) to develop two novel fly models: a model where the endogenous Para is tagged with GFP to determine Para subcellular localization and another with para replaced with GAL4 to determine para gene expression. Surprisingly, we find para present in a small fraction of CNS neurons in embryos and third instar larvae, while it is broadly expressed in neurons in the adult CNS. We also generated a single-cell transcriptomic atlas of the whole third instar larval brain to identify that para correlates with RNAs of active zone proteins and mature neuron markers; hence, para expression is restricted to active, differentiated neurons in larvae. Neurons that coexpress para and active zone protein RNAs are abundant in the adult CNS but only represent 23 ± 1% of neurons in third instar larvae. In neurons where para is expressed, Para protein is enriched at an AIS-like region in axons distal to where the dendritic tree connects to the axons in a distal axonal segment (DAS). Para localized far from the soma is functionally verified electrophysiologically. In longer neurons, Para is expressed throughout the axon, likely to maintain AP propagation to the synapses.
Materials and Methods
Reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by H.J.B. Flies generated in this study will be deposited to the Bloomington Drosophila Stock Center.
Model and subject details
For experiments using gene or protein-trapped para alleles, all stocks were kept at room temperature (22°C), all crosses were performed at 25°C, and both male and female flies were used for imaging experiments. For the single-cell RNA sequencing experiments on the larval brain, flies were raised on a yeast-based medium at 25°C on a 12 h/12 h day/night light cycle. All Drosophila lines used in the single-cell RNA-seq experiments are derived from the DGRP collection. One hybrid was created by crossing different DGRP lines, generating genetic diversity. Fly lines were obtained from Bloomington Drosophila Stock Center and the Kyoto Stock Center and are listed in Table 1.
Table 1.
Fly lines used in this study
| Name | Genotype | Stock # (BDSC unless noted) | Reference |
|---|---|---|---|
| paraMI08578 | y1 w* Mi{y[+mDint2]=MIC}para[MI08578] | #51087 | Venken et al., 2011 |
| para Df | Df(1)FDD-0230908, w1118/FM7c | #23296 | Venken et al., 2010 |
| para genomic rescue | w1118; Dp(1;3)DC134, PBac{DC134}VK00033 | #30274 | Venken et al., 2010 |
| paraA | y1 w* paraA P{neoFRT}19A/FM7c, P{GAL4-Kr.C}DC1, P{UAS-GFP.S65T}DC5, sn+ | #57108 | Yamamoto et al., 2014 |
| paraB | y1 w* paraB P{neoFRT}19A/FM7c, P{GAL4-Kr.C}DC1, P{UAS-GFP.S65T}DC5, sn+ | #57109 | Yamamoto et al., 2014 |
| UAS-mCD8::RFP | w*; P{y[+t7.7] w[+mC]=10×UAS-IVS-mCD8::RFP}attP40 | #32219 | Pfeiffer et al., 2010 |
| para-GFSTF | y[1] w[*] Mi{PT-GFSTF.0}para[MI08578-GFSTF.0] | Current study | |
| para-mCherry | y[1] w[*] Mi{PT-mCherry.0}para[MI08578-mCherry.0] | Current study | |
| para-RFP-3×HA | y[1] w[*] Mi{PT-RFP-3×HA.0}para[MI08578-RFP-3×HA.0] | Current study | |
| para-T2A-Gal4 | y[1] w[*] Mi{GT-TG4.0}para[MI08578-TG4.0]/Fm7c | Current Study | |
| FM7c Kr>eGFP | Df(1)JA27/FM7c, P{w[+mC]=GAL4-Kr.C}DC1, P{w[+mC]=UAS-GFP.S65T}DC5, sn+ | #5193 | Casso et al., 1999 |
| UAS-G-Trace | w*; P{w[+mC]=UAS-RedStinger}4, P{w[+mC]=UAS-FLP.D}JD1, P{w[+mC]=Ubi-p63E(FRT.STOP)Stinger}9F6/CyO | #28280 | Evans et al., 2009 |
| UAS-mCD8::GFP | w*; P{y[+t7.7] w[+mC]=10×UAS-mCD8::GFP}attP2 | #32184 | Pfeiffer et al., 2010 |
| UAS-DenMark | w1118; P{UAS-DenMark}2 | #33062 | Nicolaï et al., 2010 |
| 201Y-GAL4 | y1 w67c23; P{UAS-mCD8::GFP.L}LL5 P{GawB}Tab2201Y | #64296 | Yang et al., 1995 |
| OL0019B Split-Gal4 | w1118; P{y[+t7.7] w[+mC]=R35D04-p65.AD}attP40; P{y[+t7.7] w[+mC]=R22D06-GAL4.DBD}attP2 | #68336 | Wu et al., 2016; Dionne et al., 2018 |
| MB050B Split-Gal4 | w1118; P{y[+t7.7] w[+mC]=R65B09-p65.AD}attP40/CyO; P{y[+t7.7] w[+mC]=R11F03-GAL4.DBD}attP2 | #68365 | Aso et al., 2014; Dionne et al., 2018 |
| parats1 | parats1/C(1)DX, y1 f1 | Kyoto, #106393 | D. T. Suzuki et al., 1971 |
| elav-GAL4 | y,w; P[w+=elav-GAL4} | Luo et al., 1994 | |
| Pdf-GAL4 | P{w[+mC]=Pdf-GAL4.P2.4}X, y[1] w[*] | #6899 | Park et al., 2000 |
| D. melanogaster WT reference strains | DGRP-551, DGRP-45, DGRP-136, DGRP-360, DGRP-382, DGRP-409, DGRP-502, DGRP-639, DGRP-761, DGRP-907, DGRP-913 | #55026, #28128, #28142, #25186, #28189, #28278, #28204, #25199, #28227, #28262, #28265 | Huang et al., 2014 |
| D. melanogaster: hybrid (DGRP-551, DGRP-360, DGRP-907, DGRP-913) | Current study |
Methods
Generating fluorescently tagged para flies
Tagged alleles were generated as previously described (Venken et al., 2011). In brief, two separate plasmids, pBS-KS-attB1–2-PT-SA-SD-0-EGFP-FIAsH-StrepII-TEV-3xFlag (Drosophila Genomic Resource Center (DGRC) #1298) and another containing PhiC31 integrase mRNA, are injected into paraMI08578 embryos. These embryos are left to develop into adult flies where they are isolated after eclosion and crossed to w1118/FM7h flies. The loss of the yellow marker is screened for to detect successful recombination-mediated cassette exchange (RMCE). To check insertion orientation, DNA is isolated from yellow negative animals; and using primers on either side of the original attP sites, the orientation of the insertion is determined. Insertions in the same orientation as para transcription were kept. This process was also used for pBS-KS-attB1-2-PT-SA-SD-0-mCherry (DGRC#1299), pBS-KS-attB1-2-PT-SA-SD-0-TagRFP-T-3XHA (DGRC#1301), and pBS-KS-attB1-2-GT-SA-GAL4-Hsp70pA (DGRC#1412) constructs.
Confirmation of GFSTF incorporation into all para transcripts
To confirm incorporation of GFSTF into all para transcripts, we performed PCR across the GFSTF exon from para-GFSTF and para-Minos-Mediated Integration Cassette (MiMIC) cDNA. cDNA was generated using SuperScript IV First-Strand Synthesis System (Invitrogen) from RNA isolated from 10 adult fly heads. The fly heads were ground up, using a pestle, in 30 µl Trizol. The Trizol volume was increased to 400 µl and incubated at room temperature for 5 min after brief vortexing; 80 µl of chloroform is added, and the samples are incubated at room temperature after 15 s high speed vortexing. Samples were centrifuged at 14,000 rpm for 20 min at 4°C, the top layer was added to 200 µl isopropanol and incubated at room temperature for 10 min after mixing. The centrifugation step was repeated, and the supernatant removed. The pellet was washed with 500 µl 70% ethanol and the centrifugation step repeated once more. The pellet was then air dried and resuspended in DEPC H2O.
A forward primer in exon 1 (5′-CAGTTTGTTCCGTCCCTTTAC-3′) and a reverse primer in exon 4 (5′-TTCCTTTGCTTACAACTACGAATG-3′) were used to amplify the region. The bands were extracted using QIAquick (Invitrogen) and Sanger sequenced. To confirm the abundance of each tagged transcript a forward primer across the exon 1–2 boundary (5′-GAGAGGTGCCGCAATATGGTC-3′) and reverse primers in the GFP sequence (5′-AACAGCTCCTCGCCCTTG-3′) and across the exon 3–4 boundary (5′-ACAACTACGAATGTCAGTACATTGC-3′) were used for qPCR (SYBR Green, Bio-Rad) with primers in Actin 5C (forward, 5′-ACACACCAAATCTTACAAAATG-3′, reverse, 5′-CCACAATCGATGGGAAGAC-3′) used for control gene expression. qPCR was performed with three technical replicates for each of three biological replicates, with a negative control cDNA, which was generated without adding reverse transcriptase. Analysis was performed using Bio-Rad CFX manager 3.1 with the relative quantity (ΔCq) with reference to zero used for quantification, error bars show standard error of the mean.
Western blot confirmation of Para-GFSTF
For Western blot analysis, 20 adult fly heads of para-MiMIC and para-GFSTF animals were added to 60 μl Laemmli buffer with 5% β-mercaptoethanol and 1× GenDEPOT protease inhibitor (Thermo Fisher Scientific, #50-101-5485). Heads were ground using a pestle, then centrifuged for 10 min at 15,000 rpm at 4°C; 10 μl of sample supernatant was then loaded into a 4%-20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad, #4561096) as well as two ladders, a HiMark Pre-stained Protein Standard (Thermo Fisher Scientific, #LC5699) and a Precision Plus Protein Dual Color Standard (Bio-Rad, #1610374). Samples were run at a constant 75 mV for 10 min and then 200 mV until the 20 kDa band of the Precision Plus Protein Dual Color Standard reached the bottom of the gel and transferred to a nitrocellulose membrane at 400 mA for 90 min at 4°C. The membrane was then washed in TBS-Tween (TBST) (0.1%) and blocked for 1 h in 5% skimmed milk. The membrane was then incubated in rabbit anti-GFP (1:1000) (Thermo Fisher Scientific, #A-11122) overnight in a TBST (0.1%) 5% skim milk solution at 4°C. The antibody was removed, and blots washed in TBST (0.1%). HRP-conjugated goat anti-rabbit (1:10,000) (Jackson ImmunoResearch Laboratories) secondary antibodies were then incubated with the blot in a TBST (0.1%), 5% skim milk solution for 2 h at room temperature. Blots were washed again in TBST (0.1%) and then placed in a clear plastic film and 500 μl of SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific) added to the blot. The blot is then subsequently imaged using a ChemiDoc MP Imaging System (Bio-Rad). The blot is then stripped using a 15 min room temperature incubation with Restore Western Blot Stripping Buffer (Thermo Fisher Scientific), and the process is repeated from the initial wash and blocking steps using a mouse anti-Actin (1:5000) (EMD Millipore, #MAB1501) primary antibody and an HRP-conjugated goat anti-mouse (1:10 000) (Jackson ImmunoResearch Laboratories) secondary antibody.
Electroretinogram (ERG) assay
For ERG recordings, para-MiMIC and para-GFSTF flies were aged to 5 d at 25°C in 12 h light/12 h dark cycle or in 24 h darkness. ERG recordings were performed as previously described (Verstreken et al., 2003) using LabChart software (AD Instruments). At least 8 flies were examined for each genotype. Quantification was performed using Prism 8.0.
Temperature-sensitive paralysis assay
para-GFSTF, para-mCherry, para-RFP-3xHA, para-MiMIC, and parats1 males were collected at room temperature and aged for 4 d in vials of at most 10 flies. These flies were transferred to a fresh empty vial (2.5 cm diameter, 9.5 cm height) with a foam plug and given 5 min to acclimate to the new environment. These vials were then immersed in a water bath at 40°C for 1 min. Every 20 s, the number of flies that were unable to stand was recorded. After 1 min, the flies were removed from the water bath and placed on a bench top gently where they were monitored every 20 s, and the number of flies unable to stand was recorded until all flies were upright. At least 100 flies for each genotype were recorded. The results were plotted by accumulating all the data for each genotype and recording the paralysis as percentage of flies still standing. The error bars indicate SD.
Lethality screening of para-T2A-GAL4
para-T2A-GAL4 and two previously published para null alleles, paraA and paraB (Yamamoto et al., 2014), were balanced over FM7c, Kr>eGFP and placed in an embryo collection chamber on a grape juice plate with yeast to encourage egg laying. After 24 h, a fresh plate was placed in the chamber. Flies could lay on the plate for 24 h. After incubation, the plates were visualized using a fluorescence microscope to look for larvae devoid of the Kr>eGFP expression pattern and staged appropriately.
Immunofluorescent staining
Embryos
Parent flies are placed in an embryo collection chamber on a grape juice plate with yeast to encourage egg laying. After 24 h, acclimation to a fresh plate is added, and flies can lay eggs on the plate for 24 h at room temperature to enable collection of embryos at a variety of stages. Embryos are dislodged from the plate using a paintbrush and water and transferred to a cell strainer using a Pasteur pipette. Embryos are washed with water to remove the excess yeast and grape juice and are placed in a 50% bleach 50% water solution for 3 min to dechorinate the embryos. Embryos are then washed with water, blotted dry, and then transferred via paintbrush to a glass scintillation vial containing 4 ml N-heptane and 4 ml modified Steffani's fixative (4% formaldehyde, PA grade, methanol free, 15% saturated aqueous solution of picric acid, 75 mm disodium PIPES in PBS, pH 7.4) (Stefanini et al., 1967), which has been vigorously shaken and allowed to separate into layers. The embryos float between the heptane and fixative layers. The vial is then placed on a rotating platform for 25 min at room temperature. After 25 min, the lower fixative layer is removed and replaced with equal volume 100% methanol. The vial is shaken vigorously to remove the vitelline membrane, and embryos fall to the bottom of the vial. The lower methanol layer containing the embryos is then removed and placed in a separate vial where it is washed 2 more times with 100% methanol. Embryos are transferred to an Eppendorf tube and the methanol replaced with PBS-Tween (PBST) (0.2%). Embryos are washed 3 times with PBST (0.2%) before incubating in PBST (0.2%) with 10% normal goat serum (NGS) solution for 1 h on a rotating platform at room temperature. The blocking solution is then removed and a solution containing primary antibodies, 10% NGS and PBST (0.2%) is added, and embryos are incubated overnight at 4°C on a rotating platform. Primary antibodies are then removed, and embryos are washed in PBST (0.2%) 3 times for 10 min at room temperature before secondary antibodies are then added in a PBST (0.2%) with 10% NGS solution and incubated on a rotating platform overnight at 4°C. Secondary antibodies are then removed, and embryos are washed in PBST (0.2%) 3 times for 10 min at room temperature. PBST (0.2%) is removed and ProLong Gold mounting medium (Thermo Fisher Scientific, #P36930) added to the tube. Embryos are then transferred to a glass slide and sealed with a no. 1.5 coverslip and nail polish.
Primary antibodies used for imaging are mouse anti-Flag (1:200) (Sigma Millipore, #F1804) and rat anti-Elav (1:500) (DSHB, #7E8A10) (O'Neill et al., 1994). Corresponding goat secondary antibodies were used (1:500) (Jackson ImmunoResearch Laboratories) as well as Cy3-conjugated goat anti-HRP antibody (1:250) (Jackson ImmunoResearch Laboratories, #123-165-021).
Third instar larvae brain dissection
Wandering third instar larvae were collected from the side of vials and placed in cold PBS. The larval brains were dissected using fine forceps, and the brains were placed in cold 3.7% PFA in 1× PBS overnight at 4°C. The PFA solution is then aspirated off and washed twice with a 0.2% PBS-Triton-X (PBS-TX) solution and left to incubate at 4°C in 0.2% PBS-TX overnight. The 0.2% PBS-TX solution is aspirated and replaced with a primary antibody solution diluted in 0.2% PBS-TX solution with 5% NGS. Brains are left in the primary solution overnight at 4°C. The primary antibodies are then removed, and brains are washed quickly twice in 0.2% PBS-TX solution and incubated for 10 min at room temperature in 0.2% PBS-TX solution 3 times. The 0.2% PBS-TX solution is then removed and replaced with secondary antibodies diluted in 0.2% PBS-TX solution and 5% NGS and incubated at 4°C overnight. The secondary antibodies are then removed, and brains are washed quickly twice in 0.2% PBS-TX solution and incubated for 10 min at room temperature in 0.2% PBS-TX solution 3 times. The 0.2% PBS-TX solution is then removed and replaced with RapiClear 1.47 mounting medium (SUNJin Lab). The brains in the mounting medium are then placed inside a circular spacer on a glass slide. A no. 1.5 coverslip is then placed on top of the solution, and the coverslip is sealed with nail polish.
Primary antibodies used for imaging are rabbit anti-GFP (FITC-conjugated, 1:200; Thermo Fisher Scientific, #A-21311), rat anti-Elav (1:500) (DSHB, #7E8A10) (O'Neill et al., 1994), rabbit anti-mCherry (1:200; GeneTex, #GTX59788), and mouse anti-HA.11 (1:500; previously Covance, catalog #MMS-101R). Corresponding goat secondary antibodies were used (1:500; Jackson ImmunoResearch Laboratories) as well as Cy3-conjugated goat anti-HRP antibody (1:250; Jackson ImmunoResearch Laboratories, #123-165-021).
Adult brain dissection
For adult brain dissections, animals are anesthetized using CO2 and transferred using forceps to a Sylgard plate where Minutien pins are inserted, first into the abdomen and then the thorax of the fly, positioning the fly dorsal side up with the thorax at a 45° angle to the plate. The plates are rinsed with PBS and then briefly washed with 70% ethanol to remove the waxy layer on the cuticle. The flies are then rinsed 3 times with PBS and left submerged in PBS. Adult brains are dissected by making incisions in the cuticle using forceps from the posterior of the head and slowly peeling back the cuticle, leaving the brain untouched. The trachea is then peeled off the brain using fine forceps, and the brain is removed by pinching off at the posterior of the brain where it connects to the thorax. Brains are then placed in 3.7% PFA solution in PBS.
The staining protocol for adult brains is the same as the larval brain staining protocol above with the following exceptions: (1) 2% PBS-TX is used instead of 0.2%; (2) antibodies are incubated with the brains for 2 d at 4°C instead of one; and (3) before addition of primary antibodies, brains are placed under a vacuum 6 times for 10 min each to remove trapped air.
Primary antibodies used for imaging are the same as for the larval brain at the same concentrations.
Larvae neuromuscular junction (NMJ) dissection
Larvae are collected the same way as for the brain dissections. On the Sylgard plate, Minutien pins are placed in the posterior and anterior most regions of the larvae dorsal side up. Fine scissors are used to insert a horizontal incision at the posterior and then used to cut from posterior to anterior of the fly between the trachea. A second horizontal incision is then made at the anterior of the larvae. The internal organs and fat are all removed, leaving the brain attached. Pins are then used to secure each corner of the abdominal wall and spread the larva in a hexagonal shape. The following steps all occur with the larvae still pinned to the plate. The larvae are rinsed in PBS and fixed in 3.7% PFA for 20 min at room temperature. The larvae are rinsed with 0.2% PBS-TX quickly twice to remove residual PFA and then 3 times for 10 min each at room temperature. The larvae are then blocked in 5% NGS solution in 0.2% PBS-TX for 1 h at room temperature. The larvae are incubated with a primary antibody solution diluted in 0.2% PBS-TX solution with 5% NGS. Larvae are left in the primary solution overnight at 4°C, and the plate is placed in a black box with a moist towel to prevent evaporation. The primary antibodies are then removed, and larvae are washed quickly twice in 0.2% PBS-TX solution and incubated for 10 min at room temperature in 0.2% PBS-TX solution. The 0.2% PBS-TX solution is removed and replaced with secondary antibodies diluted in 0.2% PBS-TX solution and 5% NGS and incubated at 4°C overnight in the same box as before. The secondary antibody is then removed, and larvae are washed quickly twice in 0.2% PBS-TX solution and incubated for 10 min at room temperature in 0.2% PBS-TX solution. The larvae are then transferred to a slide and mounted in ProLong Gold mounting medium. A no. 1.5 coverslip is then placed on top of the solution and the coverslip is sealed with nail polish.
Primary antibodies used for imaging are rabbit anti-GFP (FITC-conjugated, 1:200; Thermo Fisher Scientific, #A-21311) and goat anti-HRP (Cy3-conjugated, 1:500; Jackson ImmunoResearch Laboratories, 123-165-021).
Confocal imaging
Most imaging was performed using Leica Microsystems SP8 and SP8X with Lightning deconvolution microscopes using a 20× or 63× oil immersion lens. NMJ imaging was performed on a Carl Zeiss Elyra 7 with Lattice SIM using a 63× immersion lens. Image analysis processing was performed using Imaris 9.3.1 (BitPlane).
Quantification of cells expressing para
To quantify the number of cells expressing para in the third instar larval and adult CNS, we used the Spots feature on Imaris 9.3.1 (BitPlane). For the third instar larval brain, we assigned spots as having an estimated diameter of 3.8 µm with a quality score >174. For the adult CNS, a spot had an estimated diameter of 2.5 µm and a quality score >56.1. We quantified the number of spots from 5 third instar larval brains and 5 1-2 day old adult animals with genotype para-T2A-GAL4/Fm7c; P{w[+mC]=UAS-RedStinger}4, P{w[+mC]=UAS-FLP.D}JD1, P{w[+mC]=Ubi-p63E(FRT.STOP)Stinger}9F6/CyO, and analyzed spots based on Elav and nls.RedStinger, to quantify all neurons and para-expressing neurons, respectively. Analysis was performed using Microsoft Excel, and error bars indicate SD.
Quantification of DAS length
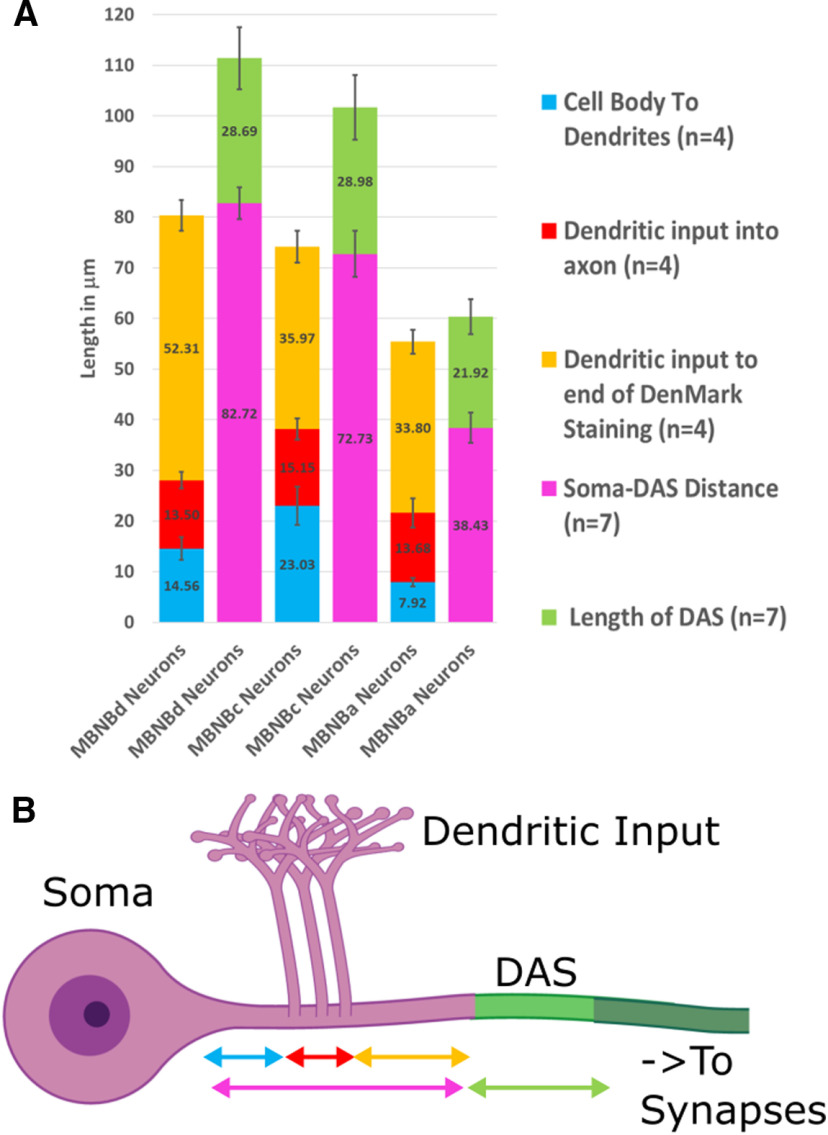
To detect the length of axonal compartments in adult mushroom body (MB) neurons, we used the measurement points feature in Imaris 9.3.1 (BitPlane) to manually trace MB neurons originating from MB neuroblast (MBNB) clusters a, c, and d (Kunz et al., 2012) from the soma to the anterior of the peduncle, generating intensity profiles of each fluorophore used. Tracing was performed on para-mCherry; 201Y-GAL4,UAS-mCD8::GFP (n = 7) and 201Y-GAL4-UAS-mCD8::GFP/UAS-DenMark (n = 4) 1-2 d old animals. Para-mCherry and DenMark fluorescence intensities were measured. The beginning of the DAS was defined by an increase to 25% of the maximum intensity of Para-mCherry signal and the end of the decrease in signal <25%. The increase to 25% of the maximum fluorescence intensity for DenMark staining was used to determine the start of the site of dendritic innervation, and the decrease <25% was used to define the end of the site of dendritic innervation. The boundary of the somatodendritic and axonal compartments is defined as the site where DenMark signal intensity was <5% of the maximum intensity. To get reliable measurements, three neurons were traced from each MBNB cluster in each animal; analysis was performed in Microsoft Excel, and error bars indicate SD.
Electrophysiology
For large ventral lateral clock neurons (lLNv), whole-cell patch-clamp recording, data acquisition, and analysis, methods were used as described previously in detail (Sheeba et al., 2008). Long-latency NaV currents were recorded with excellent access. In order to record such small cell bodies (3-4 μm), we fashioned 10 mΩ glass pipettes using a Narishige PP-83 two-step gravity puller. Cell-attached patch configuration was established by gentle negative pressure on the pipette holder. Subsequently, slightly stronger negative pressure was applied to achieve breakthrough of the membrane to the whole-cell configuration with giga-ohm seals. Whole-cell lLNv recordings are generally stable for ≥30 min. The following solution modifications were made to isolate TTX-sensitive NaV currents. A HEPES-buffered external solution was used to avoid precipitation of ion channel blockers that consisted of 110 mm NaCl, 2 mm CoCl2, 4 mm MgCl2, 5 mm glucose, and 10 mm HEPES. The internal patch electrode solution consisted of 102 mm D-gluconic acid, 102 mm CsOH, 0.085 mm CaCl2, 1.7 mm MgCl2, 17 mm NaCl, 0.94 mm EGTA, and 8.5 mm HEPES. Synaptic currents were blocked with 1 μm tubocurare, 10 μm picrotoxin, 5 μm CNQX, and 50 μm APV. K+ currents were blocked partially with 2 mm 4-AP and 10 mm tetra-ethyl ammonium (note the residual rapidly responding outward voltage-evoked K+ currents from KV channels expressed in or near neuronal cell bodies). Ca2+ currents were blocked with 2 mm CoCl2. Nav Para currents were confirmed by adding 100 nm TTX to the bath solution that abolished these currents.
Single-cell sequencing
Brain dissociation into single cells
Wandering third instar larvae were collected, and 30 brains were dissected and transferred to a tube containing 100 μl ice-cold Dulbecco's PBS (DPBS) solution. Next, the brains were centrifuged at 800 × g for 5 min, and the supernatant was replaced by 50 μl of dispase (3 mg/ml, Sigma Millipore, D4818-2 mg) and 75 μl collagenase I (100 mg/ml, Invitrogen, 17100-017). Brains were dissociated at 25°C with 500 rpm for 45-55 min. The enzymatic reaction was reinforced by pipette mixing every 15 min. Cells were washed with 1000 μl ice-cold DPBS solution and resuspended in 400 μl DPBS 0.04% BSA. Cell suspensions were passed through a 10 μm pluriStrainer (ImTec Diagnostics, 435001050), and cell viability and concentration were assessed by the LUNA-FL Dual Fluorescence Cell Counter. All genotypes were separately dissected and dissociated.
Methanol fixation
Fixation was performed after cell dissociation. The single-cell suspension was put in a thermoshaker at 4°C with 250 rpm, and ice-cold methanol was added dropwise. Next, the sample was incubated for 10 min at 4°C, followed by freezing and storage at −20°C.
10× genomics
Single-cell libraries were generated using the GemCode Single-Cell Instrument and Single-cell 3′ Library & Gel Bead Kit v2 and Chip Kit (10× Genomics) according to the manufacturer's protocol. Briefly, fly brain single cells were suspended in 0.04% BSA-PBS. For the fresh samples, the targeted cell recovery estimate was 5000 cells (using 8700 as input); while for the fixed genotype mix, the aim was to retrieve 9000 cells (using 15,800 cells as input). These cells were equally taken from each genotype separately in the genotype mix. After generation of nanoliter-scale Gel bead-in-EMulsions (GEMs), GEMs were reverse-transcribed in a C1000 Touch Thermal Cycler (Bio-Rad) programmed at 53°C for 45 min, 85°C for 5 min, and hold at 4°C. After reverse transcription, single-cell droplets were broken, and the single-strand cDNA was isolated and cleaned with Cleanup Mix containing DynaBeads (Thermo Fisher Scientific). cDNA was then amplified with a C1000 Touch Thermal Cycler programmed at 98°C for 3 min, 12 cycles of (98°C for 15 s, 67°C for 20 s, 72°C for 1 min), 72°C for 1 min, and held at 4°C twice. Subsequently, the amplified cDNA was fragmented, end-repaired, A-tailed, and index adaptor ligated, with SPRIselect Reagent Kit (Beckman Coulter) with cleanup in between steps. Postligation product was amplified with a C1000 Touch Thermal Cycler programmed at 98°C for 45 s, 14 cycles of (98°C for 20 s, 54°C for 30 s, 72°C for 20 s), 72°C for 1 min, and hold at 4°C. The sequencing-ready library was cleaned up with SPRIselect beads. CellRanger was used to detect cells from empty droplets.
Before sequencing, the fragment size of every library was analyzed on a Bioanalyzer high-sensitivity chip. The libraries were diluted to 2 nm and quantified by qPCR using primers against p5-p7 sequence. All 10× libraries were sequenced on NovaSeq6000 instruments (Illumina) with the following sequencing parameters: 28 bp read 1-8 bp index 1 (i7), 91 bp read 2. Sequencing information is available in Extended Data File 1.
Third instar larvae single-cell sequencing metrics, cluster composition and the gene list and GO terms used for analysis. Related to Figures 5-8. Download Extended Data File 1, XLSX file (50KB, xlsx)
Demuxlet
Demuxlet was used to demultiplex the different genotypes used in the methanol fixed DGRP-mix sample (Kang et al., 2018). This allows us to remove doublets consisting of cells with two different genetic backgrounds. The vcf file of the DGRP project (available at http://dgrp2.gnets.ncsu.edu/) was lifted over to dm6 genome. Next, we used bulk ATAC data to update the SNPs for DGRP-639 and to add SNP profiles for the hybrid. The vcf file was then filtered to only keep SNPs unique for one of the lines used. Demuxlet was run using this vcf file and on default parameters, leading to the identification of 970 doublets and 2937 singlets. Afterward, the datasets of the two runs were merged.
Scater
Scater was used to filter the merged dataset (McCarthy et al., 2017). First, cells were filtered using three quality characteristics: number of unique molecular identifiers (UMIs), number of genes, and percentage of mitochondrial genes. Cells that were >3 SDs away from the mean for number of UMIs and number of genes and cells that were >4 SDs away from the mean for percentage of mitochondrial genes were removed. Next, a principal component analysis was performed using quality characteristics (percentage of counts for the top 100 genes, number of genes, percentage of mitochondrial genes, number of mitochondrial genes, log10 of total counts, log10 of total mitochondrial counts), and outliers were removed. Finally, genes with an average expression < 0.01 were removed. Leading to a final dataset of 9853 genes by 5056 cells.
Seurat
Seurat v3 (Stuart et al., 2019) was used to integrate the data of the two different run conditions. Datasets were separately normalized using SCTransform (Hafemeister and Satija, 2019). Next, anchors were searched and integrated using 70 dimensions. Finally, the data were scaled; and principal component analysis, t-distributed stochastic neighbor embedding (tSNE), and Louvain clustering were performed using 70 dimensions. Louvain clustering was performed using resolution 2. To distinguish astrocytes from ensheathing glia, we used resolution 2.5, in which they split in two. We tested different resolutions in the Louvain clustering algorithm ranging from 0.2 to 4. The relationship between clusters across different resolution parameters was visualized using clustree (Zappia and Oshlack, 2018). Cluster composition is available in Extended Data File 1.
Trajectory inference
Trajectory inference was performed using Monocle3 (Cao et al., 2019). For Type 1 optic lobe (OL) neuroblasts, ordering genes for 7 principal components were used (hth, klu, ey, slp1, slp2, D, and tll). For the OL neuroepithelium, 50 components were used and for the whole dataset ordering; 5 principal components were used.
Loom
Data are available for visualization and downloading at http://scope.aertslab.org/#/Larval_Brain/*/welcome. Loom files of the data were generated using SCopeLoomR (https://github.com/aertslab/SCopeLoomR).
Single-cell data analysis
To identify the cells expressing para, we used the FeaturePlot and vlnPlot functions within the Seurat v3 package for the third instar larval CNS single-cell sequencing data generated in this study and the adult single-cell transcriptome atlas from Davie et al. (2018). We isolated the top 100 genes enriched in each of the 39 larval gene clusters using the FindAllMarkers function in Seurat. A Wilcoxon Rank Sum test was used to identify differentially expressed genes; the genes could not be expressed in >20% of all cells in each cluster to rule out ubiquitously expressed genes. The genes were ranked by log fold change in expression between the average expression in each cluster compared with expression in all other cells, and the top 100 in each dataset were selected. para was only present in the top 100 genes for clusters 3 and 33 (motor neurons) in the third instar larval brain single-cell sequencing data. The top 100 genes from both cluster 3 and 33 were submitted to http://pantherdb.org/ for gene ontology (GO) molecular function analysis (Ashburner et al., 2000; Gene Ontology Consortium, 2019; Mi et al., 2019). A PANTHER Overrepresentation test was performed using the top 100 genes in both cluster 3 and 33 separately with a reference list containing all genes in database for Drosophila. We annotated based on GO molecular function and identified a list of enriched terms using a Fisher's exact test with the Bonferroni correction for multiple testing (Tables 2, 3). A full list of genes used in each cluster and GO terms is available in Extended Data File 1.
Table 2.
para-positive cluster 3 of the single-cell transcriptomic atlas is enriched for genes required for neurotransmission and ion transporta
| GO molecular function complete | D. melanogaster - REFLIST (13781) | Cluster 3 (96) | Cluster 3 (expected) | Cluster 3 (over/under) | Cluster 3 (fold enrichment) | Cluster 3 (p) |
|---|---|---|---|---|---|---|
| Syntaxin binding (GO:0019905) | 26 | 5 | 0.18 | + | 27.61 | 2.87E-03 |
| Ion transmembrane transporter activity, phosphorylative mechanism (GO:0015662) | 21 | 4 | 0.15 | + | 27.34 | 3.32E-02 |
| Hormone activity (GO:0005179) | 49 | 8 | 0.34 | + | 23.44 | 6.67E-06 |
| Neuropeptide hormone activity (GO:0005184) | 33 | 5 | 0.23 | + | 21.75 | 8.16E-03 |
| SNARE binding (GO:0000149) | 48 | 6 | 0.33 | + | 17.94 | 2.53E-03 |
| ATPase-coupled cation transmembrane transporter activity (GO:0019829) | 50 | 6 | 0.35 | + | 17.23 | 3.14E-03 |
| ATPase-coupled ion transmembrane transporter activity (GO:0042625) | 54 | 6 | 0.38 | + | 15.95 | 4.74E-03 |
| Receptor ligand activity (GO:0048018) | 97 | 10 | 0.68 | + | 14.8 | 4.30E-06 |
| Neurotransmitter receptor activity (GO:0030594) | 70 | 7 | 0.49 | + | 14.36 | 1.35E-03 |
| Signaling receptor activator activity (GO:0030546) | 101 | 10 | 0.7 | + | 14.21 | 6.16E-06 |
| Receptor regulator activity (GO:0030545) | 104 | 10 | 0.72 | + | 13.8 | 8.00E-06 |
| Primary active transmembrane transporter activity (GO:0015399) | 121 | 7 | 0.84 | + | 8.3 | 3.97E-02 |
| Signaling receptor binding (GO:0005102) | 257 | 12 | 1.79 | + | 6.7 | 4.43E-04 |
| Monovalent inorganic cation transmembrane transporter activity (GO:0015077) | 240 | 10 | 1.67 | + | 5.98 | 1.19E-02 |
| Inorganic cation transmembrane transporter activity (GO:0022890) | 330 | 13 | 2.3 | + | 5.66 | 8.90E-04 |
| Cation transmembrane transporter activity (GO:0008324) | 346 | 13 | 2.41 | + | 5.39 | 1.49E-03 |
| Transmembrane signaling receptor activity (GO:0004888) | 342 | 11 | 2.38 | + | 4.62 | 4.31E-02 |
| Signaling receptor activity (GO:0038023) | 401 | 12 | 2.79 | + | 4.3 | 3.67E-02 |
| Molecular transducer activity (GO:0060089) | 402 | 12 | 2.8 | + | 4.29 | 3.76E-02 |
| Inorganic molecular entity transmembrane transporter activity (GO:0015318) | 511 | 15 | 3.56 | + | 4.21 | 4.13E-03 |
| Ion transmembrane transporter activity (GO:0015075) | 535 | 15 | 3.73 | + | 4.02 | 7.10E-03 |
| Molecular function regulator (GO:0098772) | 568 | 15 | 3.96 | + | 3.79 | 1.43E-02 |
| Transmembrane transporter activity (GO:0022857) | 731 | 17 | 5.09 | + | 3.34 | 1.76E-02 |
| Transporter activity (GO:0005215) | 791 | 17 | 5.51 | + | 3.09 | 4.75E-02 |
| Unclassified (UNCLASSIFIED) | 2872 | 10 | 20.01 | – | 0.5 | 0.00E + 00 |
aPANTHER overenrichment to determine enriched GO molecular functions in the top 100 enriched genes in cluster 3 of the single-cell third instar larval brain transcriptomic atlas shows that cluster 3 is enriched for genes involved in neuronal transmission, ATP synthesis, and ion transport. Process upregulated in actively firing neurons.
Table 3.
para-positive cluster 33 (motor neurons) of the single-cell transcriptomic atlas is enriched for genes required for neurotransmission and ion transporta
| GO molecular function complete | D. melanogaster - REFLIST (13781) | Cluster 33 (88) | Cluster 33 (expected) | Cluster 33 (over/under) | Cluster 33 (fold enrichment) | Cluster 33 (p) |
|---|---|---|---|---|---|---|
| Inositol monophosphate phosphatase activity (GO:0052834) | 7 | 3 | 0.04 | + | 67.12 | 3.92E-02 |
| Inositol monophosphate 1-phosphatase activity (GO:0008934) | 7 | 3 | 0.04 | + | 67.12 | 3.92E-02 |
| Proton-transporting ATPase activity, rotational mechanism (GO:0046961) | 35 | 6 | 0.22 | + | 26.85 | 2.82E-04 |
| ATPase activity, coupled to transmembrane movement of ions, rotational mechanism (GO:0044769) | 35 | 6 | 0.22 | + | 26.85 | 2.82E-04 |
| ATPase-coupled cation transmembrane transporter activity (GO:0019829) | 50 | 8 | 0.32 | + | 25.06 | 3.84E-06 |
| Pyrophosphate hydrolysis-driven proton transmembrane transporter activity (GO:0009678) | 39 | 6 | 0.25 | + | 24.09 | 5.01E-04 |
| ATPase-coupled ion transmembrane transporter activity (GO:0042625) | 54 | 8 | 0.34 | + | 23.2 | 6.64E-06 |
| ATPase-coupled transmembrane transporter activity (GO:0042626) | 101 | 8 | 0.64 | + | 12.4 | 5.86E-04 |
| Primary active transmembrane transporter activity (GO:0015399) | 121 | 8 | 0.77 | + | 10.35 | 2.12E-03 |
| Proton transmembrane transporter activity (GO:0015078) | 110 | 7 | 0.7 | + | 9.97 | 1.25E-02 |
| Active ion transmembrane transporter activity (GO:0022853) | 158 | 8 | 1.01 | + | 7.93 | 1.39E-02 |
| Monovalent inorganic cation transmembrane transporter activity (GO:0015077) | 240 | 12 | 1.53 | + | 7.83 | 8.23E-05 |
| Inorganic cation transmembrane transporter activity (GO:0022890) | 330 | 13 | 2.11 | + | 6.17 | 3.18E-04 |
| Cation transmembrane transporter activity (GO:0008324) | 346 | 13 | 2.21 | + | 5.88 | 5.36E-04 |
| Inorganic molecular entity transmembrane transporter activity (GO:0015318) | 511 | 15 | 3.26 | + | 4.6 | 1.33E-03 |
| Ion transmembrane transporter activity (GO:0015075) | 535 | 15 | 3.42 | + | 4.39 | 2.32E-03 |
| Transmembrane transporter activity (GO:0022857) | 731 | 16 | 4.67 | + | 3.43 | 2.21E-02 |
| Unclassified (UNCLASSIFIED) | 2872 | 11 | 18.34 | – | 0.6 | 0.00E + 00 |
aPANTHER overenrichment to determine enriched GO molecular functions in the top 100 enriched genes in cluster 33, which represents motorneurons, of the single-cell third instar larval brain transcriptomic atlas shows that cluster 33 is enriched for genes involved in ATP synthesis and ion transport. Both of these processes are upregulated in actively firing neurons.
To compare para expression with known neuronal activity genes, we collated a list of genes known to function in synaptic transmission using http://flybase.org (Thurmond et al., 2019) and identified activity-dependent genes from Chen et al. (2016), taking the 7 genes upregulated in 2 of 3 neuronal activity paradigms used in the study. To determine the correlation of para expression with the expression of active zone genes, activity-regulated genes (ARGs), and markers of neuronal differentiation, we averaged the log(CPM + 1) [CPM, counts per million] gene expression of all cells (including cells with zero expression) in each cluster for each gene. We then performed a Pearson's correlation analysis to compare the average expression of each gene across clusters in both the third instar larvae and adult single-cell sequencing datasets. To determine the significance of each correlation, we performed a two-tailed t test with the degrees of freedom equal to the number of clusters minus 2 (100 for the adult single-cell data and 39 for the larval single-cell dataset). We used Bonferroni correction to adjust our target p value of 0.05 for the 210 comparisons performed giving us an adjusted p value of 0.0002381 for both datasets. A t value of 4.0467 was used for the larval single-cell larvae correlations, and a t value of 3.8124 was used for the adult correlations.
Neuron diagrams
Diagrams of neurons in Figures 9D, 10B, 12D, 13B, and 14B were made using www.BioRender.com.
Figure 9.
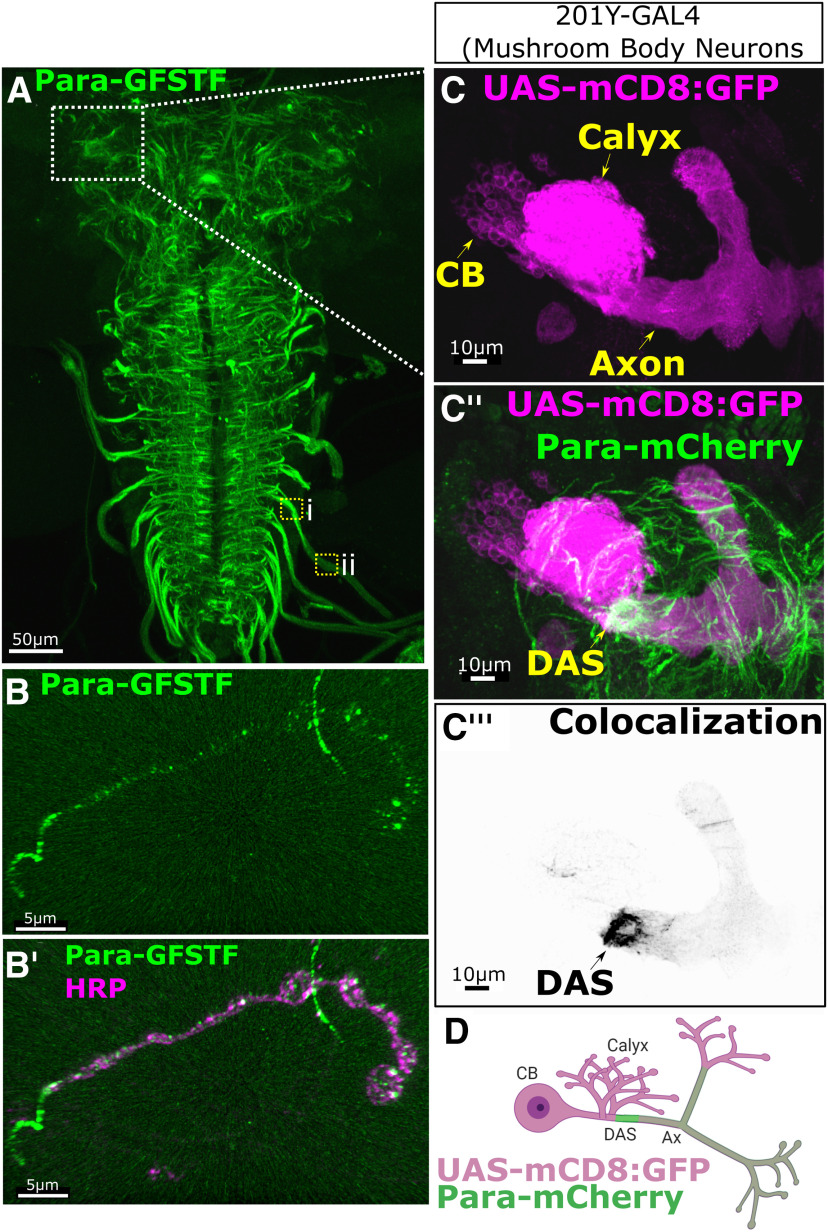
Para-GFSTF is localized to a Distal Axonal Segment (DAS) downstream of the site where dendrites impinge on axons in the third instar larval CNS. A, Para-GFSTF distribution in the larval brain is most abundantly expressed in thoracic and abdominal segments, the exiting axon tracks in the ventral nerve cord (VNC), and the median portion of the central brain (Movie 1). Clusters of Para-GFSTF localization can be seen in axon bundles leaving the VNC (inset box i). Para is less abundant in the more distal axons (inset, box ii). This pattern is also seen with other tagged Para isoforms (Extended Data Fig. 9-1A). B,B′, The distribution of Para-GFSTF extends to the neuromuscular junction where continual expression stops at the first bouton. Punctate expression of Para-GFSTF can be seen in each bouton. C,C′,C′′,C′′′, D, 201Y-GAL4, driving UAS-mCD8::GFP, labels the MB neurons in the third instar larvae (Extended Data Fig. 9-1B). 201YGAL4>UAS-mCD8::GFP labeling with Para-mCherry shows that Para is enriched in the MB neurons in the axon, after the site of dendritic innervation in the calyx, at a DAS. CB - Cell Body, Ax - Axon.
Figure 10.
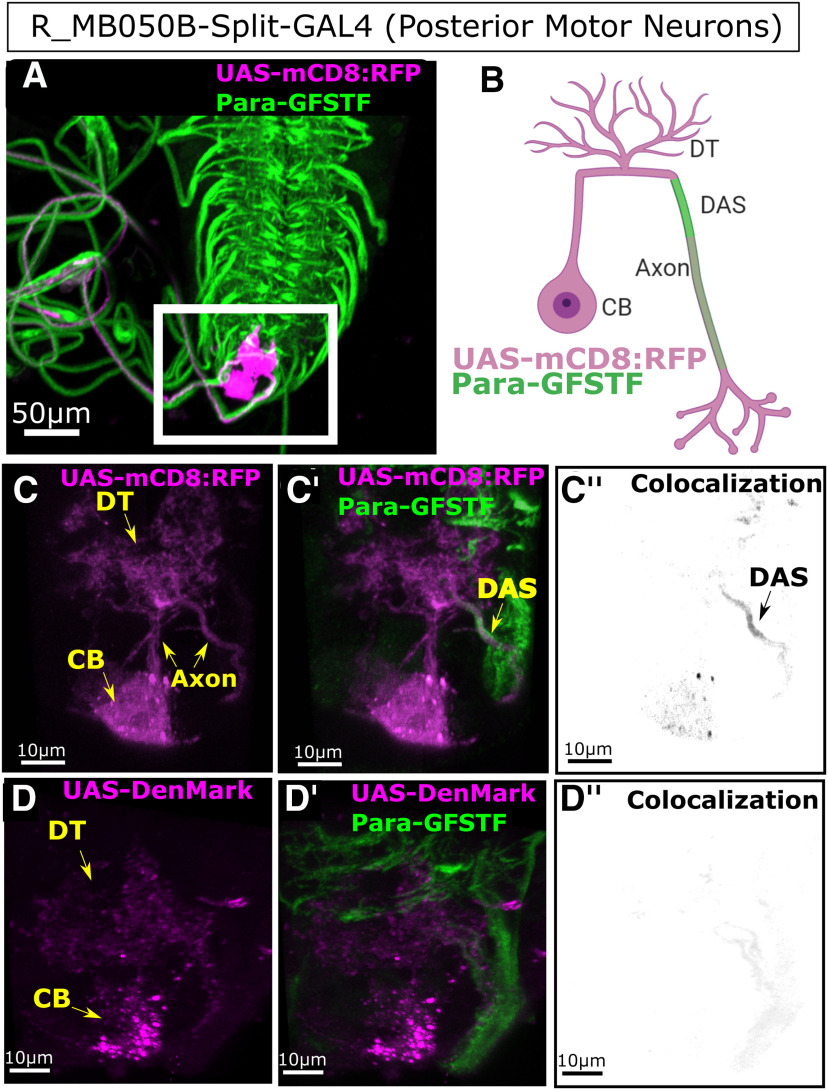
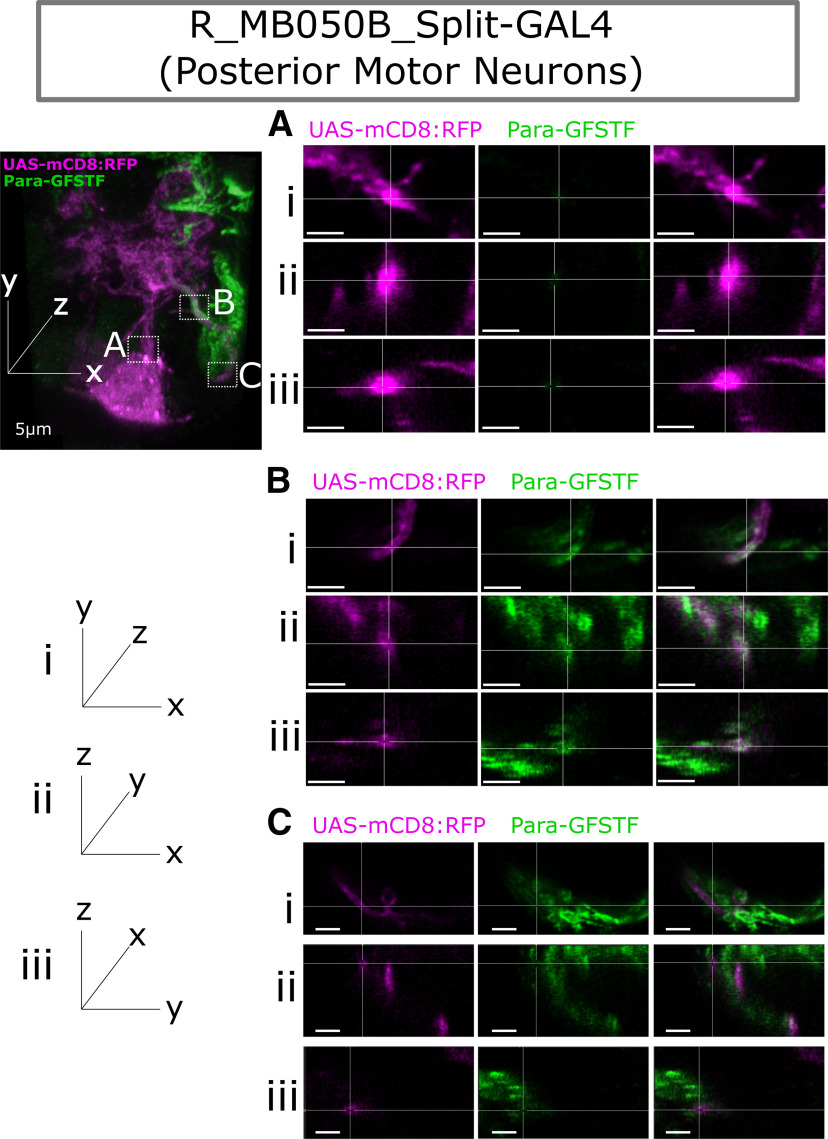
Para enrichment at the Distal Axonal Segment (DAS) is seen in posterior third instar larval motor neurons. A, Split-GAL4 line (R-MB050B) labels 2 motor neurons in the most posterior larval ventral nerve cord. B, C,C′,C′′, Using mCD8::RFP, we label the whole cell identifying the cell body (CB), dendritic tree (DT), and axon. Colocalization with Para-GFSTF is seen with mCD8::RFP at a DAS after the dendritic tree. D,D′,D′′, DenMark labels only the CB and the axon. No colocalization is seen between DenMark and Para-GFSTF, showing that the DAS is not in dendrites and is distal to the site of the dendritic tree.
Figure 12.
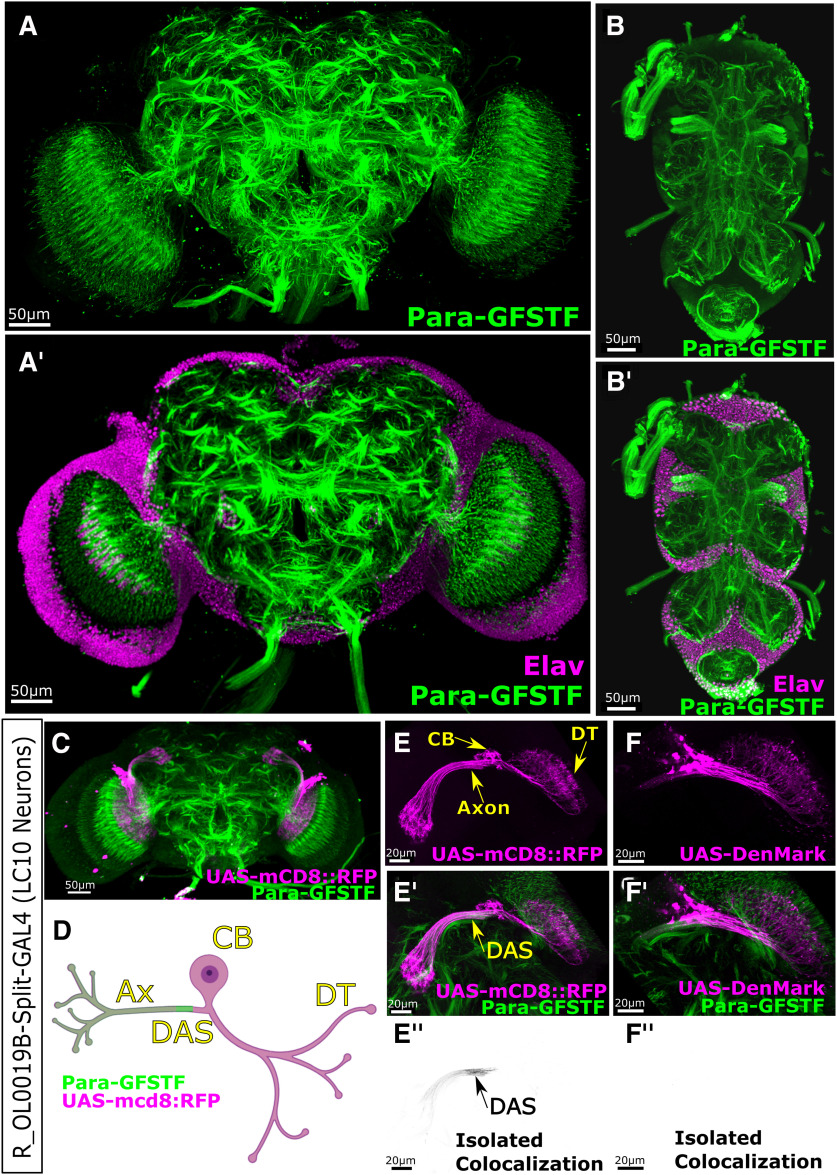
Para is localized to Distal Axonal Segments (DAS) in the adult central nervous system (CNS) neurons. Immunofluorescent imaging of Para-GFSTF subcellular localization of para in the adult CNS. A,A′, In the central brain, Para is enriched in axonal clusters in the neuropil (Movie 2). The Para clusters are not associated with the cell body of neurons (Elav) and are located distally to the cell body. B,B′, In the thoracic ganglion, the Para clusters can also be observed distally to the cell body. The additional labeling of axon tracts down the midline of the thoracic ganglion highlights the lower-intensity Para distribution in long axons. C, D, Split-GAL4 line R_OL0019B is specifically expressed in the LC10 (laminar columnar) neurons in the adult fly visual system. E,E′,E′′, Using UAS-mCD8::RFP, we can identify the cell body (CB), dendritic tree (DT), and axon (Ax). Para-GFSTF is localized to a DAS, in the axon, downstream of the dendritic tree. F,F′,F′′, Using UAS-DenMark to label the soma and dendritic tree of the LC10 neurons, we do not see any colocalization, with Para-GFSTF indicating that DAS is localized after the somatodendritic boundary.
Figure 13.
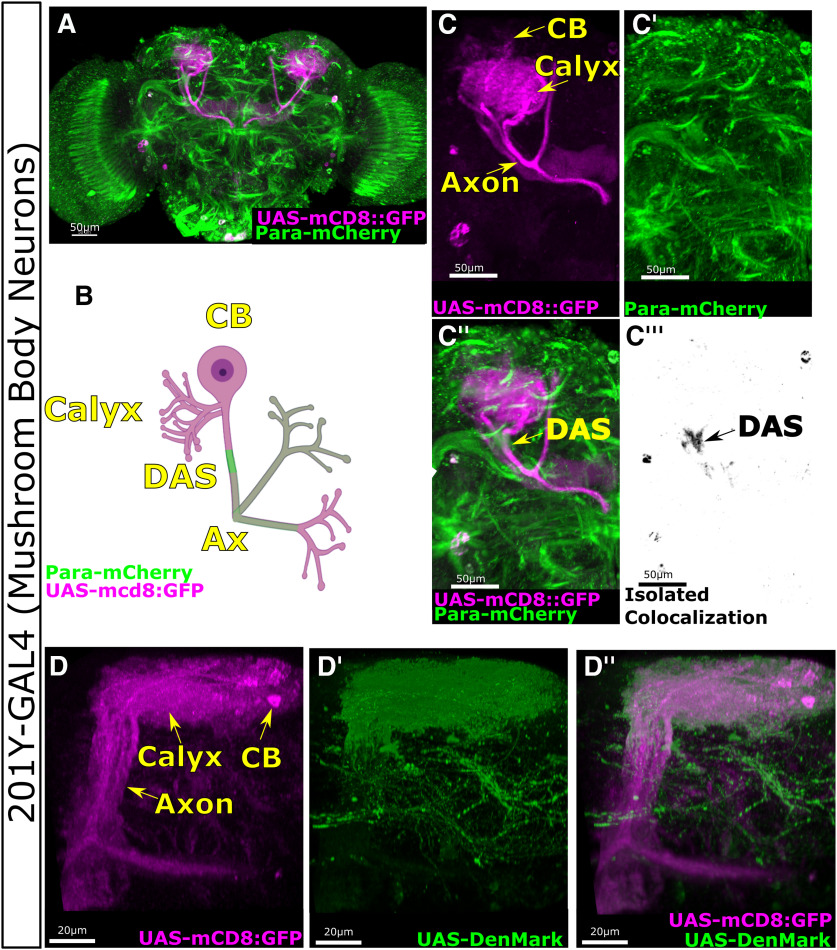
Para enrichment at the Distal Axonal Segment (DAS) is seen in the mushroom body (MB) neurons of the adult central nervous system (CNS). A, B, 201Y-GAL4 is expressed specifically in the neurons of the MB. C,C′,C′′,C′′′, Expression of mCD8:GFP in the MB clearly marks the cell body (CB), axon (Ax), and the Calyx where the neurons receive dendritic input. Para-mCherry is enriched at the DAS in the axon of the MB neurons, distal to the calyx. D,D′,D′′, UAS-DenMark expression in the MB neurons using 201Y-GAL4 colocalized with UAS-mCD8::GFP in the cell body and calyx region of the neurons but not in the peduncle beyond the DAS.
Figure 14.
The Distal Axonal Segment (DAS) is variable in size and location and forms the boundary between the somatodendritic and the distal region of the axon. A, Quantification of the sizes of axonal compartments (B) in neurons derived from mushroom body neuroblast lineages (MBNB) lineages a, c, and d. Cell body to dendrite, dendritic input, and dendritic input to end of DenMark staining measurements are derived from the intensity profile of UAS-DenMark using 201Y-GAL4 with UAS-mCD8::GFP (n = 4). Soma-DAS distance and DAS length are derived from the intensity profile of Para-mCherry with 201Y-GAL4 and UAS-mCD8::GFP. The DAS length ranges from 21.92 to 29.98 µm; neurons with shorter soma to DAS regions have a shorter DAS. DenMark does not, in any MBNB population, extend beyond the DAS indicating that the DAS forms a boundary between the somatodendritic proximal axon and the distal axon. Error bars indicate SD.
Data and code availability
Transcriptomic data are available for visualization and downloading at http://scope.aertslab.org/#/Larval_Brain/*/welcome and the code is released as open source code on Github. The scRNA-seq data has been deposited in GEO under accession code GEO: GSE157202. SCENIC for Drosophila is available at http://scenic.aertslab.org.
Results
Fluorescently tagged Para is a functional protein that is temperature-sensitive
To generate fluorescently tagged Para alleles, we used a MiMIC line: y1 w* Mi{y[+mDint2]=MIC}para[MI08578] constructed by the Gene Disruption Project (Nagarkar-Jaiswal et al., 2015). This line has two MiMICs inserted in the second coding intron of the para locus (Fig. 1A), which are incorporated into all 60 predicted isoforms of para. These MiMICs are inserted in the opposite orientation of the transcript, rendering them not mutagenic. We replaced each MiMIC through RMCE with a SA-EGFP-FlAsH-Strep-TEV-3xFlag-SD (Para-GFSTF) sequence to insert two artificial exons encoding several epitopes that allow visualization of the endogenous expression pattern (Fig. 1B) (Venken et al., 2011). As the cassette can integrate in either orientation, we confirmed the correct orientation for both with PCR. The same process was repeated for constructs containing SA-mCherry-SD and SA-RFP-3xHA-SD sequences. Sequencing of genomic DNA in the para-GFSTF animals revealed two GFSTF insertions 37 bp apart in the second coding intron indicating all animals contain 2 GFP insertions. Using an antibody against GFP, we performed Western blots of adult fly heads and detect a major broad band of ∼200-250 kDa in the Para-GFSTF sample (Fig. 1C), which is consistent with the expected molecular weight of 55 of the 60 Para isoforms. Because of the broadness of the band on the Western, we are unable to isolate bands corresponding to Para containing one or two copies of the GFSTF insert.
Figure 1.
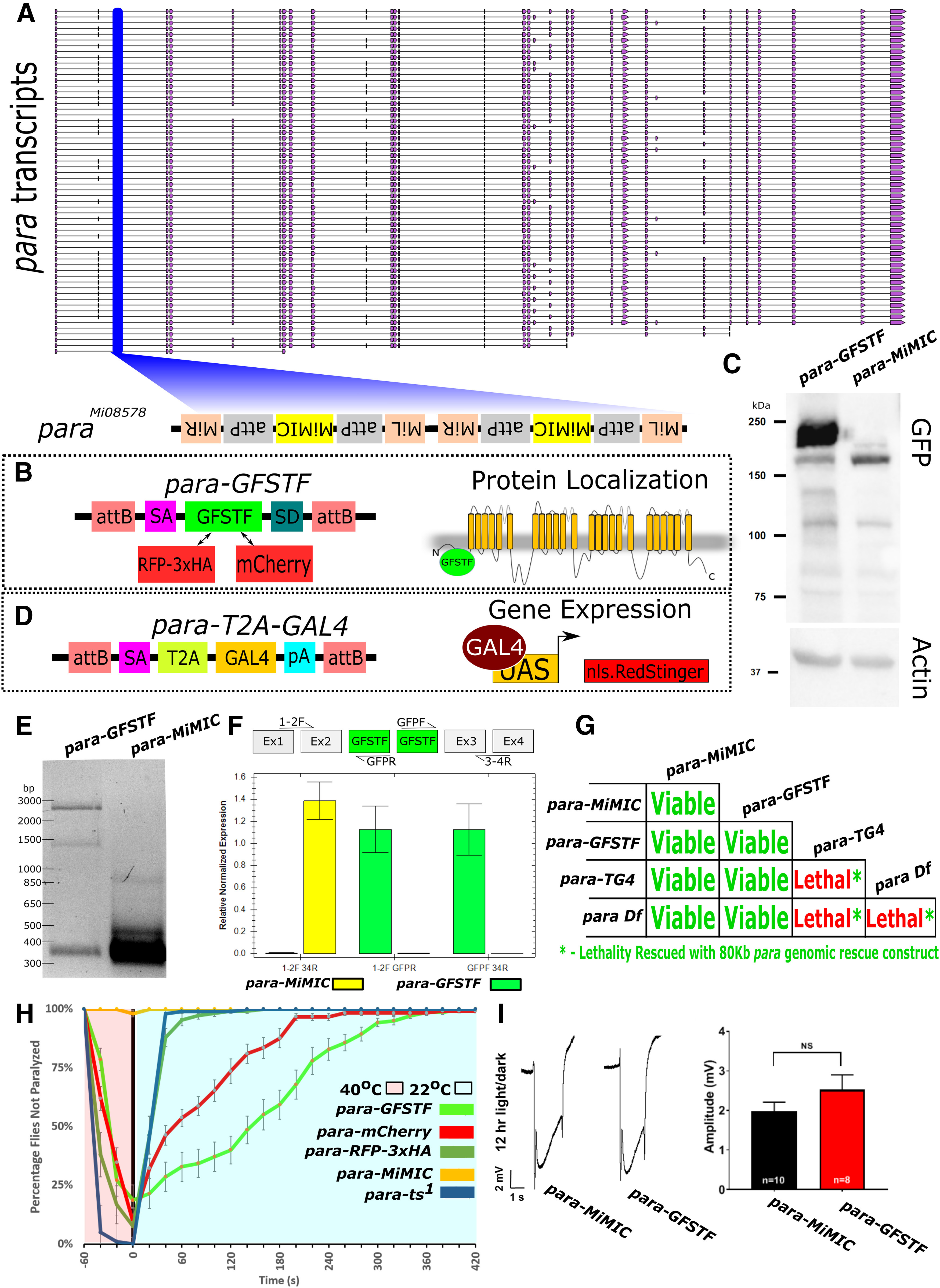
Fluorescent tagged Para and para-T2A-GAL4 alleles. A, paraMI08578 (para-MiMIC) contains two intronic MiMIC cassettes in the second coding intron of the para gene in the opposite orientation of transcription. B, Using RMCE, the MiMIC was replaced with two artificial exons that encode SA-EGFP-FlAsH-Strep-TEV-3xFlag-SD (GFSTF), SA-RFP-3xHA-SD (RFP-3xHA), or SA-mCherry-SD (mCherry) to detect protein expression. These artificial exons label all 60 isoforms based on the frames of the splice acceptor and splice donor sites. C, Western blot analysis of GFSTF tagged Para using an anti-GFP antibody confirms the presence of GFP-tagged Para proteins at ∼250 kDa, which corresponds to the expected molecular weight of 55/60 transcripts. An additional band can be seen at ∼130 kDa, which corresponds to the smaller isoforms of Para in A. D, A splice acceptor (SA)-T2A-GAL4-pA sequence was injected into para-MiMIC to generate a gene trap that expresses GAL4 in the same spatiotemporal pattern as para. The GAL4 produced by this artificial insertion/exon can activate the UAS-RedStinger.nls and allows the determination of the localization of the cell body of the para-expressing neurons. E, PCR amplification of para-GFSTF and para-MiMIC cDNA, using a forward primer in exon 1 and a reverse primer flanking exons 3 and 4, reveals one predominant band of ∼300 bp in the MiMIC control, which is the expected product sans any insertions, but three bands of ∼300, 1600, and 2400 bp, which correspond to 0, 1, and 2 artificial exons, respectively, are seen in the para-GFSTF sample. F, qPCR of Para-GFSTF to detect the relative proportion of para transcripts incorporating or skipping the GFSTF exon shows almost exclusive expression of GFSTF containing para transcripts. Amplicons measured span from the exons 1 and 2 boundary to the exons 3 and 4 boundary, from exons 1 and 2 to the GFSTF cassette, and from the GFSTF cassette to exons 3 and 4. Error bars indicate SEM. G, para-GFSTF animals are homozygous viable at room temperature and do not display any obvious defects. However, the gene trap para-T2A-GAL4-pA is homozygous lethal and fails to complement a deficiency (Df(1)FDD-0230908) that uncovers the para locus. The first instar lethal phase of the transheterozygous animals agrees with it being a null allele. para-T2A-GAL4 lethality can be rescued with a genomic rescue construct containing the para locus (P[acman Dp(1;3)DC134). H, Fluorescent-tagged Para animals display a temperature-sensitive paralysis phenotype. Most tagged animals are paralyzed after 60 s at 40°C. The rate of recovery is variable among the constructs with para-RFP-3xHA (n = 108) recovering within 60 s (similar rate to parats1, n = 102), whereas para-mCherry (n = 121) flies need 240 s to fully recover and para-GFSTF (n = 140) animals need 300 s. para-MiMIC (n = 101) was not paralyzed, suggesting that it does not affect the function of para. Error bars indicate SEM. I, ERGs show that the amplitude of off-transients of para-GFSTF and para-MiMIC are not significantly different from each other, indicating that the GFSTF tag does not impair channel function. Animals were raised in 12 h light/dark conditions. No significant differences were seen in 24 h dark conditions, or in the amplitude of on-transients (Extended Data Fig. 1-1). Unpaired, two-tailed t test was used. Error bars indicate SEM.
Para-GFSTF and para-MiMIC have comparable ERG recordings. ERGs were performed on para-GFSTF and para-MiMIC flies to measure neuronal activity in the visual system. (A) Representative trace of ERGs in 24 h dark conditions, with no significant difference seen in off transients between tagged and untagged animals. (B) No significant difference was seen in the amplitude of on transients in 12 h light/dark conditions or constant dark. The difference in phototransduction amplitude between the genotypes was not significant in 12 h light/dark conditions; in constant dark the amplitude of para-GFSTF flies was slightly larger than para-MiMIC flies. Unpaired, two-tailed t test. Error bars indicate SEM. Download Figure 1-1, TIF file (3.5MB, tif)
To validate that the GFSTF exons are spliced into all para isoforms, we PCR amplified the flanking regions of the MiMIC site with primers in exons 1 and 4. para-MiMIC cDNA had one band of ∼300 bp, which corresponds to the expected product without any GFSTF exon (Fig. 1E). The para-GFSTF sample contained three bands corresponding to ∼300, ∼1200, and ∼2000 bp (Fig. 1E). The GFSTF exon is 976 bp; therefore, each band corresponds to para transcripts with 0, 1, and 2 GFSTF exons incorporated, respectively, likely via splicing; this was confirmed by sequencing. The presence of duplicate MiMIC insertions was also seen in para-mCherry and para-RFP-3xHA animals. Saturated PCR conditions were used to detect the GFP inserts in the RNA product (Fig. 1E). To quantify the proportion of tagged versus untagged para transcripts in the para-GFSTF animals, we performed a qPCR using a forward primer in exons 1 and 2, and reverse primers in GFP and across exons 3 and 4 in para-GFSTF and para-MiMIC animals (Fig. 1F). The qPCR showed that only 1.61% (±0.39%) of all para transcripts in para-GFSTF animals do not incorporate a GFSTF exon. Therefore, the tagged form of para generated in this study represents >98% all the para transcripts produced in the fly.
To generate a gene reporter allele for para, we exchanged the MiMIC cassette in para-MiMIC with a SA-T2A-GAL4-pA (T2A-GAL4) sequence using RMCE (Fig. 1D). The splice acceptor is predicted to be spliced into all para transcripts and should generate a severe loss of function or null allele (P. T. Lee et al., 2018). para null alleles are lethal at the first instar larval stage (Loughney et al., 1989; O'Dowd et al., 1989; Hong and Ganetzky, 1994). para-T2A-GAL4 animals, in addition to two other reported para null alleles (paraA, paraB) (Yamamoto et al., 2014), are homozygous lethal at the same first instar larval stage, and fail to complement a deficiency that uncovers the para locus (Df(1)FDD-0230908) (Fig. 1G). Finally, an 80 kB genomic rescue construct (P[acman] Dp(1;3)DC134) (Venken et al., 2010) rescues para-T2A-GAL4 lethality and produces viable adults. Importantly, the above data show that there are no second site mutations on the chromosomes used in this study (Fig. 1G).
Partial loss-of-function para alleles (parats1) have been previously characterized using temperature-sensitive paralysis assays (D. T. Suzuki et al., 1971; Siddiqi and Benzer, 1976). To assess whether the para-GFSTF, para-mCherry, and para-RFP-3xHA alleles confer temperature sensitivity, we tested the effects of different temperatures on the animals. Homozygous animals that carry any of these three alleles are viable and appear healthy when raised at 25°C (Fig. 1G). At 29°C, the animals exhibit temperature-dependent paralysis and die after 24-36 h. However, they fully recover if returned to a lower temperature after 24 h. The temperature sensitivity of the tagged para alleles was compared with the classical parats1 allele using an assay at 40°C for 60 s (Fig. 1H). After 60 s, 80% of all para tagged flies are paralyzed, whereas 100% of parats1 animals are paralyzed. Interestingly, the different tagged alleles vary in the rate of paralysis and recovery. This implies that the different tagged insertions have varied effects on channel protein stability and that the refolding kinetics of each tagged allele likely are variable.
To test whether the para-GFSTF allele is functional at 25°C, we measured neuronal activity by recording ERGs (Fig. 1I; Extended Data Fig. 1-1). Loss-of-function alleles in para have been shown to have a loss of off-transients (Homyk and Pye, 1989), indicating neuronal dysfunction in the visual system downstream of photoreceptors (Joesch et al., 2010). No significant differences are found between the on- or off-transients in ERGs between the para-MiMIC and the para-GFSTF animals, suggesting that neuronal signaling is not affected by the presence of a GFSTF tag in para. We do find a slight increase in the amplitude in para-GFSTF animals compared with control para-MiMIC animals (Extended Data Fig. 1-1B). These data, in conjunction with mouse models of N-terminal tagged sodium channels that are physiologically comparable and in the same subcellular localization as nontagged channels (A. Lee and Goldin, 2009), argue that the GFSTF tag does not significantly impair channel function when animals are raised at 25°C. Therefore, the tagged para allele is representative of the endogenous expression and localization of untagged para.
Para localization is sparse in the embryonic nervous system
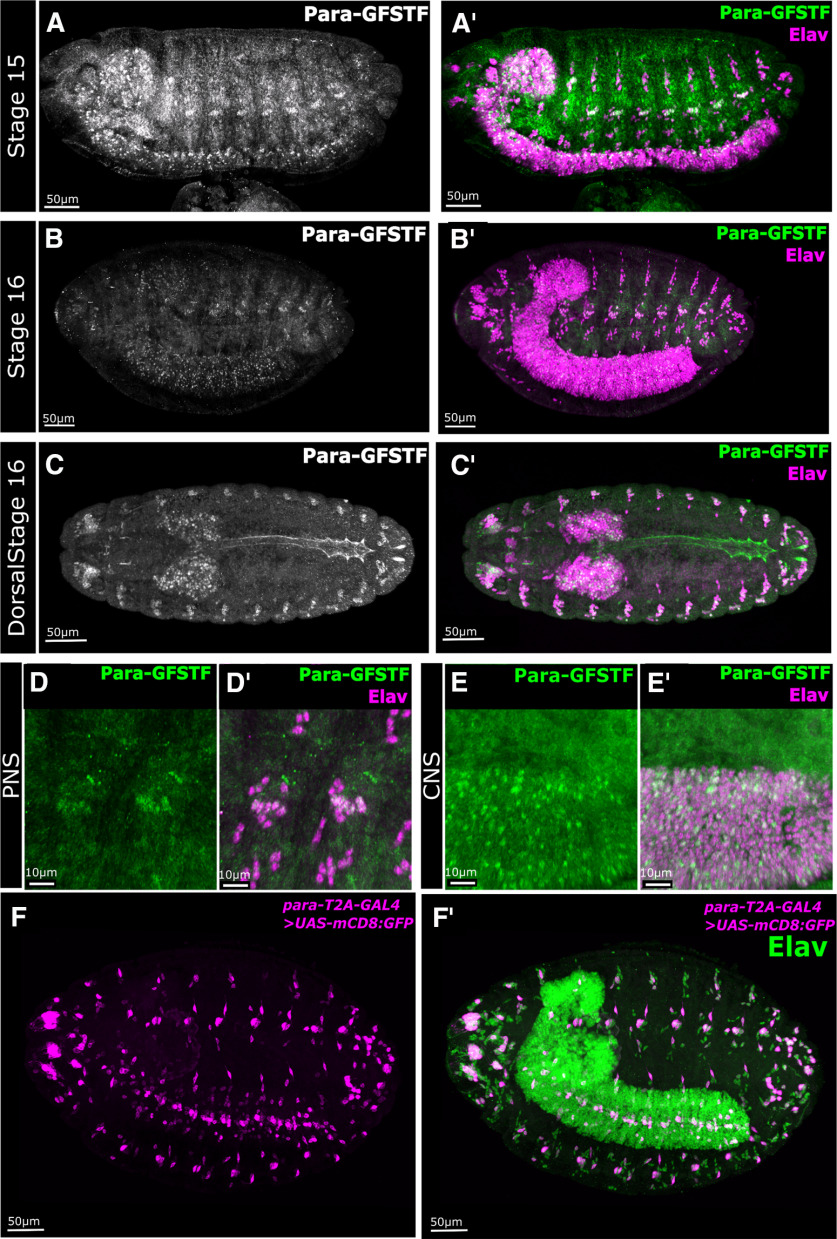
The subcellular localization of Para is unknown. ISH data have reported that para is broadly expressed in embryonic and adult neurons (Amichot et al., 1993; Hong and Ganetzky, 1994). However, the embryonic in situ data were presented at very low resolution (Hong and Ganetzky, 1994); and the only available Para antibody, though useful for Western blots (Xiao et al., 2017), does not work in fixed tissue in our hands. Using a brief fixation protocol, a pan-NaV antibody raised against vertebrate NaV channels has shown the presence of Para clusters in neuronal fascicles in the central brain of adult flies (Wang et al., 2020); however, the subcellular localization of Para remains unclear. To determine Para protein localization, we performed immunofluorescent imaging of para-GFSTF embryos using antibodies against the FLAG epitope. Para is not observed before Stage 15. In Stage 15/16 embryos, Para is localized to the cell bodies of both CNS and PNS neurons, but a large portion of CNS and PNS neurons remain unmarked (Fig. 2A,B). In the PNS, Para is most prominent in chordotonal neurons at the distal tip of the dendrites (Fig. 2D). Throughout the remainder of embryogenesis, Para localization in the CNS remains restricted to the cell bodies (Fig. 2E). In Stage 17 embryos, we also observe Para protein localization in cardiomyocytes in the dorsal tube (Fig. 2C). NaV channel expression in invertebrate cardiomyocytes has not previously been reported; however, mammalian cardiac muscle cells express a specific NaV channel (NaV 1.5), which is essential for AP propagation in the cardiac conduction system (Nuyens et al., 2001; Papadatos et al., 2002).
Figure 2.
Para-GFSTF is present in Central Nervous System (CNS) neurons, chordotonal neurons, and cardiomyocytes in embryos. A,A′, B,B′, Immunofluorescent staining of Para-GFSTF in Drosophila Stage 15 and 16 embryos, using an antibody against the Flag epitope, shows that Para localizes to the soma of some CNS and Peripheral Nervous System (PNS) neurons. C,C′, In Stage 16 embryos, Para expression is visible in the dorsal tube. D,D′, In the late stage, PNS Para-GFSTF is localized both to the cell body and to the dendrites of chordotonal neurons. E,E′, In the CNS, Para is restricted to the cell bodies of some CNS neurons. F,F′, Using membrane bound GFP (mCD8::GFP) driven by para-T2A-GAL4, we detect para gene expression in a restricted portion of CNS and PNS neurons in Stage 16 embryos.
To determine the cells that express para, we used para-T2A-GAL4 to drive UAS-mCD8::GFP. This typically reveals most or all cells as the GAL4 acts as an amplifier (P. T. Lee et al., 2018). Despite the mCD8::GFP having a membrane bound localization sequence, the marker only labels the soma. As shown in Figure 2F, para expression in the CNS and the PNS in Stage 16 embryos is restricted to a small number of neurons. In the PNS, labeling is mostly confined to the chordotonal neurons. The sparse expression of para and non-axonal localization, together with the data that para null animals hatch as first instar larvae imply that para is not essential to neuronal function in the embryo.
para expression is confined to less than one-quarter of the neuronal population in the third instar larval CNS but is broadly expressed in neurons in adults
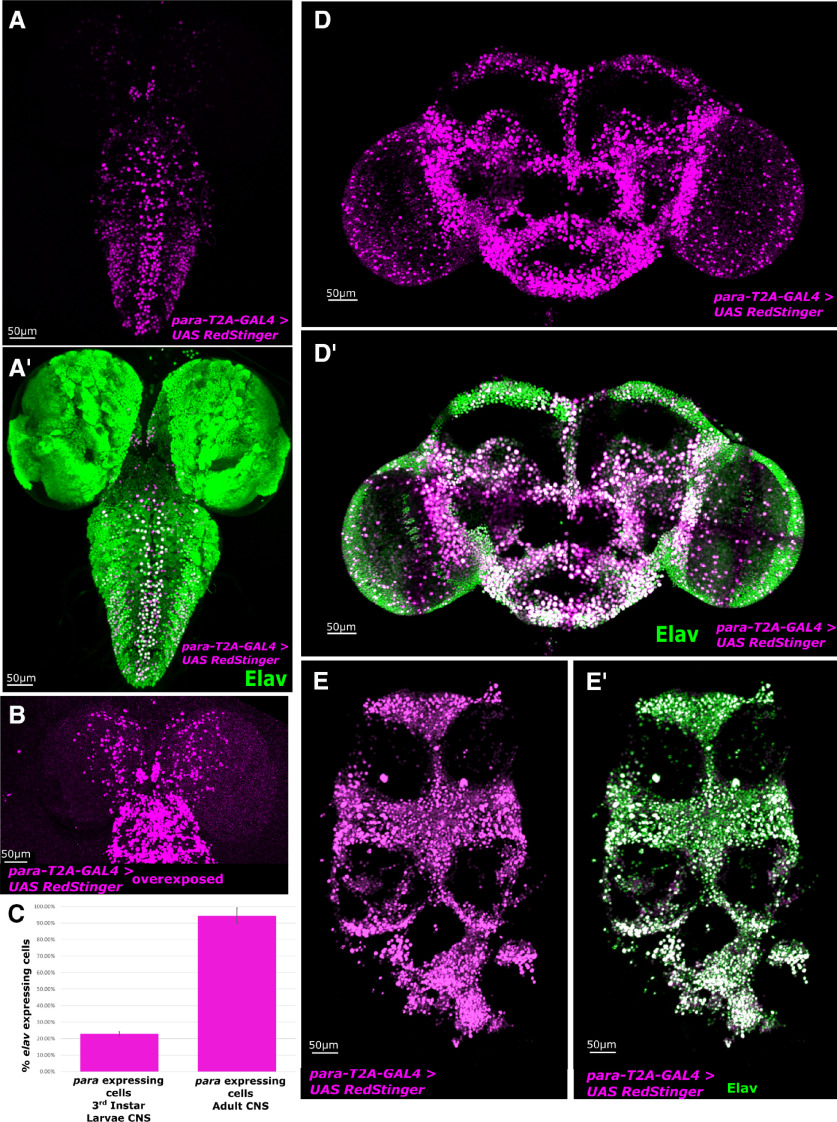
para expression is essential for larval viability (Loughney et al., 1989; O'Dowd et al., 1989; Hong and Ganetzky, 1996). To determine whether expression of para is more widespread in the third instar larval nervous system than in the embryonic nervous system, we used para-T2A-GAL4 to drive expression of a nuclear localized reporter UAS-RedStinger.nls (Fig. 3A). Surprisingly, para expression is limited to a small population of neurons in the ventral nerve cord (VNC) (23 ± 1%) compared with the pan-neuronal marker Elav (Fig. 3C). para expression is highest in the VNC, whereas cells in the central brain exhibit lower expression. By overexposing the signal, we identify additional para-positive cells in the central brain, whose expression levels are far lower than the VNC (Fig. 3B).
Figure 3.
para is expressed in only 23 ± 1% of neurons in the CNS of third instar larvae but most neurons in the adult CNS. Immunofluorescent staining of the CNS of third instar larvae and adult animals with para-T2A-GAL4 and UAS-RedStinger.nls compared with the broad neuronal marker Elav. A,A′, para-T2A-GAL4 driving nuclear localized UAS-RedStinger.nls is restricted to a limited number of CNS neurons in the larval brain with the strongest expression observed in the dorsal motor neurons in the VNC. B, para is expressed in a small amount of central brain neurons that can only be seen when the signal is overexposed. C, Quantification of neurons expressing para in the larvae and adult as a fraction of elav-expressing neurons (N = 4). Error bar indicates SD. D,D′, In the adult central brain, para-T2A-GAL4 driving nuclear localized RedStinger identifies a broad range of neurons that express para, with a strong overlap with Elav-expressing neurons. E,E′, The neurons in the thoracic ganglion also show almost 100% overlap with para-T2A-GAL4 driving nuclear localized RedStinger.
Using GAL4 as a reporter for gene expression provides a snapshot of a gene's current expression and does not provide information on a gene's historical expression (Evans et al., 2009; He et al., 2019). To determine the historical expression of para in the larval brain, we used UAS-G-Trace (Fig. 4) (Evans et al., 2009). The G-Trace construct contains two components: (1) UAS-RedStinger to label cells currently expressing the GAL4 with a nuclear localized mCherry protein; and (2) UAS-FLP, Ubi-FRT-STOP-FRT-nls.Stinger (GFP) sequence to label the nuclei of any cell that has ever expressed GAL4 with a nuclear GFP. GAL4 drives UAS-flippase that excises the STOP codon, revealing cells in which GAL4 was present at any time (Fig. 4A). A comparison of RedStinger.nls and G-Trace-induced GFP staining shows that many central brain and VNC neurons remain unlabeled, again indicating that many neurons in the larval CNS never express para (Fig. 4B,C). Importantly, the nuclear localized RedStinger nearly fully overlaps with the G-Trace staining which indicates that, once cells begin to express para, expression is maintained.
Figure 4.
Many third instar larval CNS neurons never express para. G-Trace signaling was used to determine whether CNS neurons of third instar larvae had historically expressed para. A, The G-Trace method labels present expression of para-T2A-GAL4 with UAS-RedStinger.nls and historical para-T2A-GAL4 expression with UAS-Flippase and Ubip63 >FRT-STOP-FRT Stinger.nls. B, Comparison of Stinger and RedStinger in the VNC shows almost a complete overlap indicating that, once cells activate para expression, it remains on. C, In the central brain, most of the historical and current staining overlaps; however, a small number of Stinger-positive, RedStinger-negative cells can be observed.
To determine para expression in the adult nervous system, we used para-T2A-GAL4 to drive UAS-RedStinger and the pan-neuronal nuclear marker Elav (Fig. 3D,E). para expression is far broader in the adult than in the third instar larvae, and almost all Elav-positive neurons in the central brain and in the thoracic ganglion colabel for para expression (94 ± 5%) (Fig. 3C). These data indicate that very few adult neurons, but over three-fourths of neurons in the third instar larval nervous system do not express NaV channels and may not be capable of generating NaV-dependent APs.
Single-cell sequencing of the third instar larval CNS identifies distinct neuronal, glial, and neuroblast populations
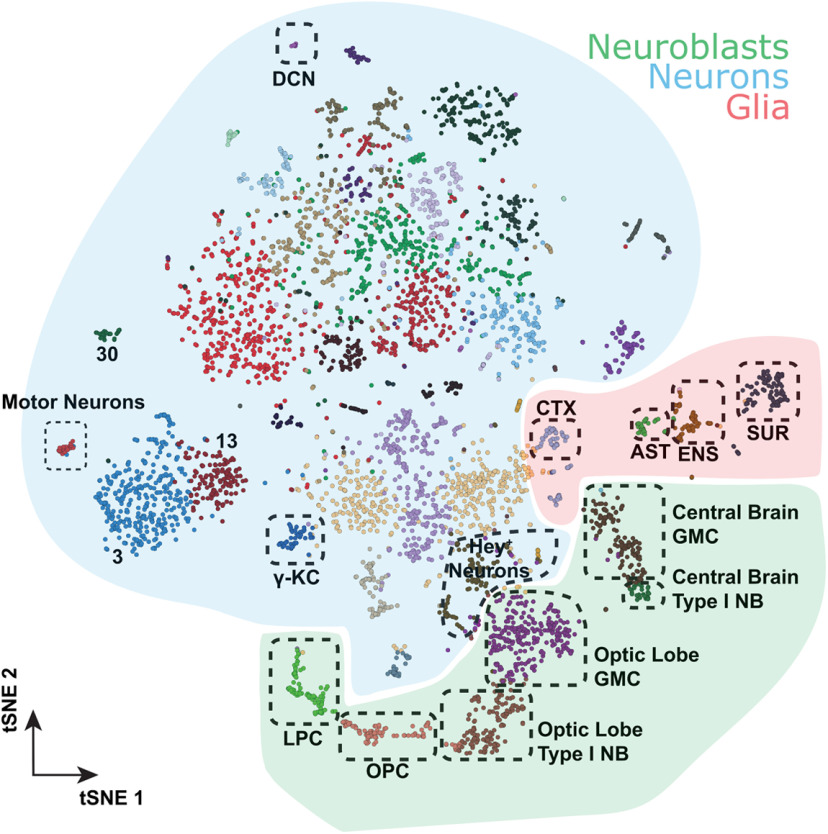
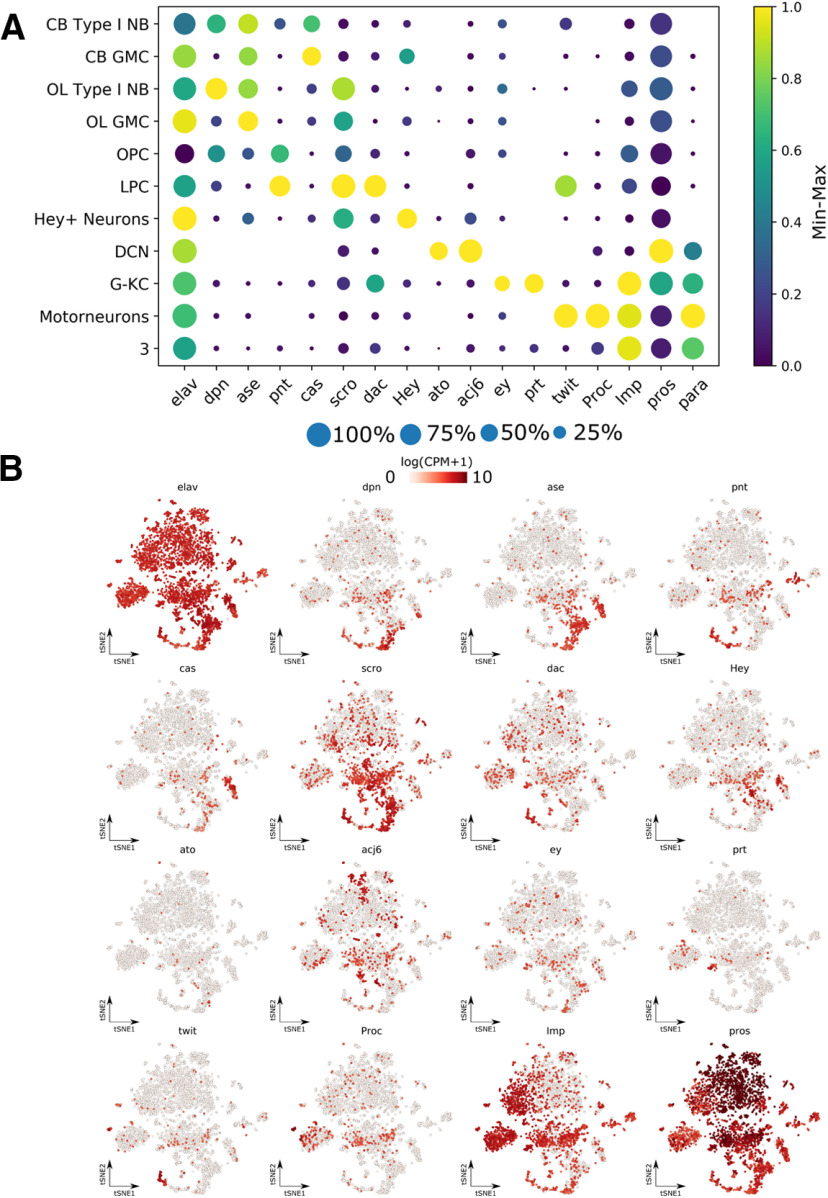
The restricted expression of para in the third instar larval nervous system to only 23 ± 1% of neurons, based on immunofluorescent data, indicates that the majority of neurons in the larval CNS may not fire NaV-based APs. To independently confirm this result, we isolated the CNS of whole third instar larval brains, containing both the brain lobes and VNC, and performed single-cell sequencing. We performed two sequencing runs (10× Chromium) for a total of ∼5000 cells from different genetic backgrounds. Seurat-integrated clustering of these cells identified 39 distinct cell clusters based on gene expression (Fig. 5), and these clusters show no bias for either sequencing run or fly genotypes (Extended Data Fig. 5-1A-C). Using markers for neurons (elav), glia (repo), or neuroblasts (dpn, ase), we were able to assign all cells to a major type (Fig. 5). Furthermore, we annotated all glial clusters and some neuronal clusters using known markers for specific cells based on published data (T. Lee et al., 1999; Hassan et al., 2000; Doherty et al., 2009; Spindler et al., 2009; DeSalvo et al., 2014; Davie et al., 2018) (Fig. 5).
Figure 5.
Building a single-cell transcriptome atlas of the whole third instar larval brain. Annotated cell types on Seurat tSNE of 5k cells from the central brain and ventral nerve chord of third instar larvae. AST, Astrocyte-like glia; CTX, cortex glia; DCN, dorsal cluster neurons; ENS; ensheathing glia; SUR, surface glia; LPC, lamina precursor cells; OPC, outer proliferation center; NB, neuroblasts; KC, Kenyon cells. The distribution of genotypes and/or fixation methods showed no bias in cluster annotation (Extended Data Fig. 5-1), and cluster assignment is stable (Extended Data Fig. 5-2). Sequencing metrics and cluster composition are listed in Extended Data File 1.
Composition of single-cell transcriptomic clusters. Distribution of (A) genotypes and (B) sample fixation type across annotated clusters. (C) Knee-plots showing distribution of valid barcodes and UMIs in fresh and fixed third instar larval brains. Download Figure 5-1, TIF file (1.9MB, tif)
Clusters are stable across multiple resolutions in the Louvain algorithm. Arrows linking clusters show overlap in percentage between clusters from different resolutions. Note that cluster 11 in resolution 0.2 becomes part of cluster 3 in resolution 0.6 but only accounts for 0.05% of cluster 3, making the arrow invisible. The squared box highlights the final chosen resolution. Download Figure 5-2, TIF file (5.7MB, tif)
para expression in neuronal progenitors has been implicated in cell proliferation (Piggott et al., 2019). Using dpn and ase, we can identify three major neuroblast clusters (Fig. 5). Two groups are dpn+ and ase+, and correspond to the Type I neuroblasts, while one group is ase-negative but shows slight dpn expression (Fig. 6A,B) (Bayraktar and Doe, 2013; Henson, 2017; Walsh and Doe, 2017). Furthermore, these cells express pnt, a marker for EGFR signaling, suggesting these are OL neuroepithelium (Fig. 6A,B) (Apitz and Salecker, 2014; Hakes et al., 2018). Two groups are dpn+ and ase+, and correspond to the Type I neuroblasts, while one group is only dpn+, the Type II neuroblasts (Fig. 6A,B) (Bayraktar and Doe, 2013; Henson, 2017; Walsh and Doe, 2017). Type I neuroblasts generate ganglion mother cells (GMCs), which only express ase and not dpn; and indeed, we can identify two clusters near each of the Type I neuroblasts that fulfill these requirements (Fig. 6A,B). Furthermore, one of these clusters is expressing cas, a transcription factor that plays a role during embryonic VNC neuroblast identity determination (Fig. 6A,B) (Cui and Doe, 1992; Mellerick et al., 1992), but has also been shown to be only expressed in the central brain GMCs in the third instar larval phase (Hitier et al., 2001). This suggests that these two clusters are derived from the central brain.
Figure 6.
Distribution of marker genes in the third instar larval CNS single-cell transcriptomic atlas. Marker genes are used to annotate cell clusters in the third instar larval brain. The selective expression of marker genes is visualized using dot plots (A) with a blue-yellow color scale representing min-max normalized log(CPM + 1) gene expression and a size scale representing the number of cells in each cluster expressing the gene. Individual tSNE plots (B) for each marker gene also show enrichments in specific clusters annotated in Figure 5. NB, Neuroblast; CB, central brain; DCN, dorsal cluster neurons; G-KC, γ Kenyon cells; LPC, lamina precursor cells; OPC, outer proliferation center. Sequencing metrics and cluster composition are listed in Extended Data File 1.
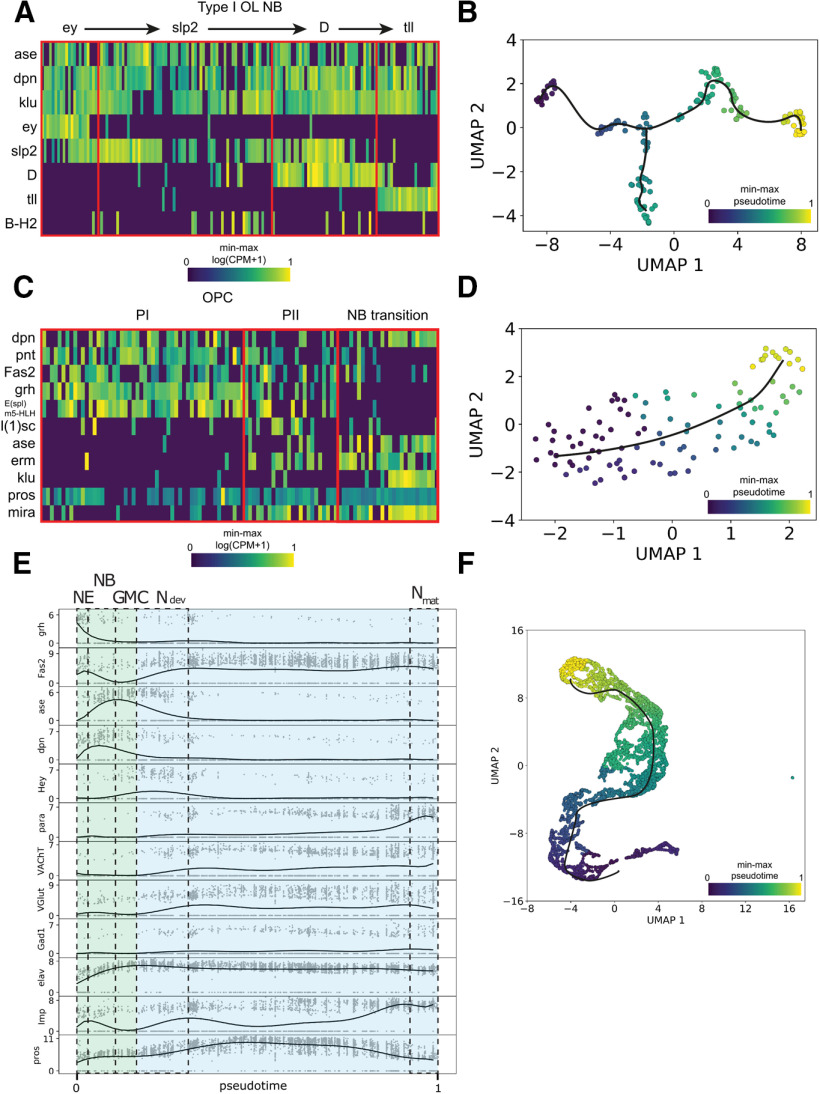
The other Type I cluster expresses many transcription factors involved in the temporal ordering of OL neuroblasts (Fig. 6A,B) (Li et al., 2013; T. Suzuki et al., 2013). Ordering the OL Type I neuroblast reveals that many transcription factors are involved in the temporal ordering of OL neuroblasts (Fig. 7A) (Li et al., 2013; T. Suzuki et al., 2013). Interestingly, we also notice a branch characterized by B-H2 expression. Next, we checked the cluster of the OL neuroepithelium. The OL neuroepithelium generates both the lamina precursor cells as the OL Type I neuroblasts previously described (Gold and Brand, 2014). The differentiation from neuroepithelial cell to neuroblast happens at the transition zone and is triggered by a proneural wave (Apitz and Salecker, 2014). Using trajectory inference, we can investigate the dynamics at this transition zone (Fig. 7B). Cells are ordered from a dpn low state with epithelial markers Fas2 and high Notch activity (Egger et al., 2010; Apitz and Salecker, 2014; Hakes et al., 2018), to a state with lethal of scute expression (Apitz and Salecker, 2014) and finally to neuroblast stage with earmuff expression (Hakes et al., 2018), fitting with previously described patterns. Finally, we note a cluster of Hey-positive neurons, a transcription factor found to be active in the Notch signaling pathway in newly born neurons (Fig. 6A,B) (Monastirioti et al., 2010).
Figure 7.
Lineage analysis of neuroblast differentiation from third instar larval single-cell transcriptomic data. A, Heatmap showing expression of OL temporal transcription factors in Type I neuroblast (NB) populations. Min-max normalized log(CPM + 1). B, Trajectory analysis for Type I OL NB. C, Heatmap showing effect of the neural wave in the neuroepithelium of the outer proliferation center (OPC), through Phase 1 (PI) and Phase 2 (PII) into NB. Min-max normalized log(CPM + 1). D, Trajectory analysis for OPC. E, Expression profiles of marker genes for neuroepithelial (NE) cells (grh), neuroblasts (NB) (dpn), GMC (ase), early neuronal development (Ndev) (Hey, pros), and mature and differentiated neurons (Nmat) (Fas2, VAChT, VGlut, Gad1, Imp), plotted against pseudotime showing dynamics through neuronal maturation. para expression increases greatly once neurons have matured and differentiated. Expression shown in log(CPM + 1), 10th degree polynomial fit through 15 cell moving average. NE, Neuroepithelium; NB, neuroblast; Ndev, developing neuron; Nmat, mature neuron. F, Trajectory analysis of neuronal maturation. Sequencing metrics and cluster composition are listed in Extended Data File 1.
To determine the stage of neuronal development in which para is expressed, we performed trajectory inference on the whole dataset (Fig. 7E,F). We identified a trajectory from neuroepithelium (grh+) to neuroblast (dpn+) to GMC (ase+) to developing neuron (Hey+) and then finally to mature neurons (VAChT+, VGLUT+, Gad1+) (Fig. 7E,F). In addition, a separation of imp+ and pros+ neurons has been demonstrated at various stages of Drosophila development (Etheredge, 2017; Davie et al., 2018; Allen et al., 2020). Higher imp expression occurs in embryonic derived, mature neurons in the larval VNC, while higher pros expression occurs in postembryonic, immature neurons in the larval VNC (Etheredge, 2017; Allen et al., 2020). This same separation can be observed in our trajectory (Fig. 7E). We do not detect para expression in any NB or GMC cell types identified (Figs. 6A, 7A,C). para expression follows both the expression of Imp and markers of neuronal differentiation into cholinergic (VAChT), GABAergic (Gad1) and glutaminergic (VGLUT) neurons; therefore, para is likely only expressed in mature, differentiated neurons. This contrasts with para's previously reported role in neuronal cell proliferation (Piggott et al., 2019).
para expression is correlated with active neuronal markers in the larval and adult CNS
Using the third instar larval brain transcriptome atlas, we can identify the cells expressing para and determine their identity. Plotting para expression on the larval tSNE (Fig. 8A), we identify that para is enriched in a small portion of the cells, with expression highest in neuronal clusters 3 and 33 (Fig. 8C), complementing the restricted population of neurons expressing para from the imaging results (Fig. 3A). To determine the identity of these clusters, we selected the top 100 genes that were enriched in each cluster compared with all other clusters and performed a PANTHER Overrepresentation Test to determine enriched GO molecular functions (Ashburner et al., 2000; Gene Ontology Consortium, 2019; Mi et al., 2019). Cluster 3 is enriched in genes that have been implicated in neurotransmission, ATP synthesis, and ion transport (Table 2), and cluster 33 is enriched in genes involved in ATP synthesis and ion transport (Table 3) and likely corresponds to motor neurons as these cells express target of wit (twit) and Proctolin (Proc), known markers for motor neurons (Fig. 6A,B) (Vuilleumier et al., 2019). Both ion transport and neurotransmission are necessary for neuronal activity, and ATPase activity is upregulated in active neurons (Johar et al., 2014). To determine whether para expression is representative of the active neuronal population, we calculated the Pearson's correlation between para expression and the expression of genes annotated in FlyBase as being present in the active zone (brp, cac, futsch, Nrx-1, nSyb, Rbp, RhoGAP100F, Syt1) as well as the coexpression of para and ARGs (Hr38, sr, CG14186, CG17778, CG8910, CG14024, CG13868) (Chen et al., 2016), in each cluster. We find that para expression is positively correlated with expression of active zone localized proteins and most ARG genes (Fig. 8D). These data indicate that para is expressed in a restricted population of neurons that are likely to be actively firing.
Figure 8.
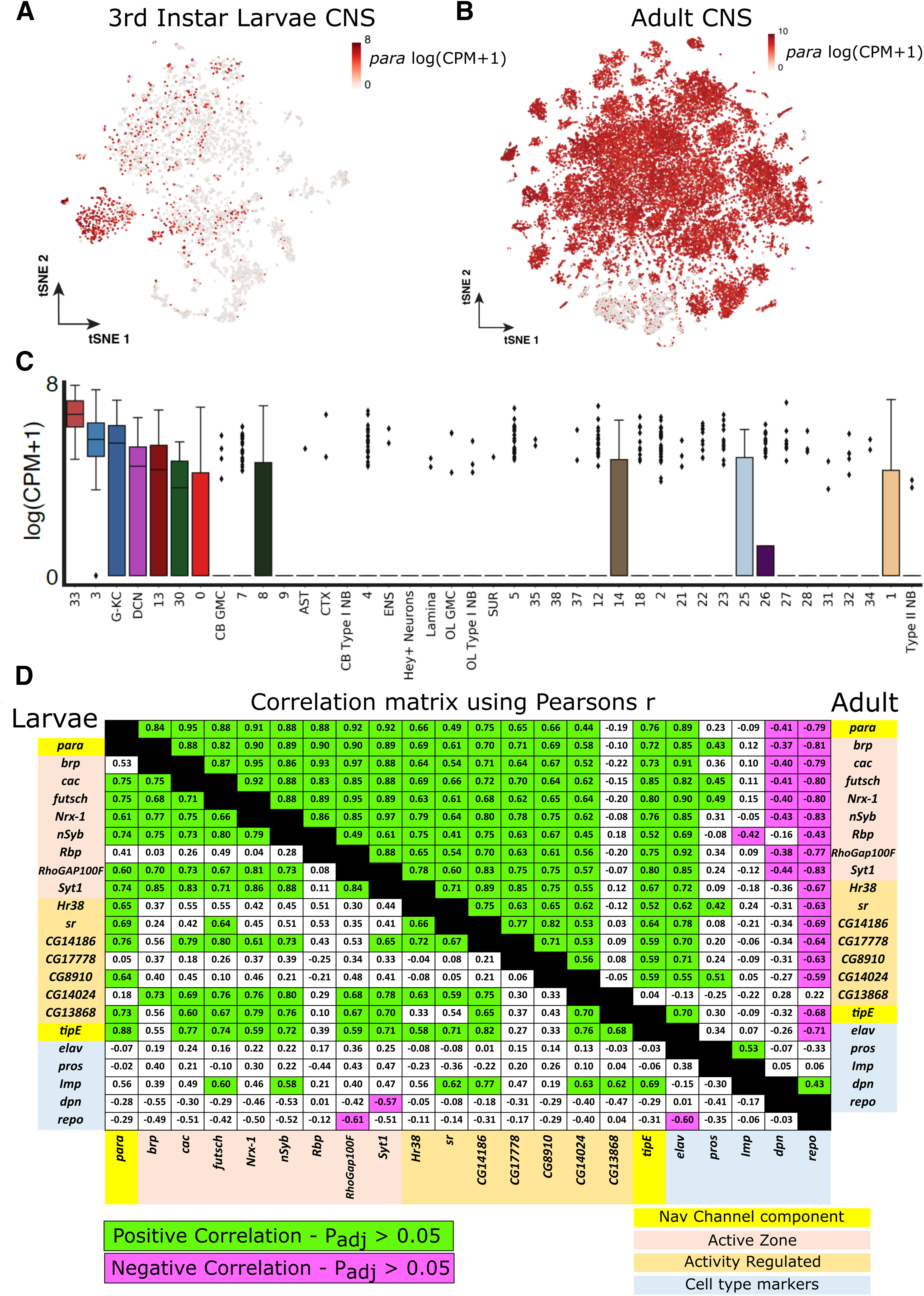
para is only expressed in neurons in both the third instar larval and adult CNS that are likely to be actively firing. A, Seurat tSNE plot of 5k cells from the whole third instar larval brain shows that para is only expressed in a small number of neurons. B, Seurat tSNE plot from the adult brain taken from Davie et al. (2018) (Extended Data Fig. 8-1) shows that para is widely expressed and is present in most neuronal cells. Pseudo-color refers to para CPM in each cell. C, Boxplots showing para expression for the different single-cell clusters from the third instar larval brain single-cell transcriptomic data; para is enriched in clusters 33 and 3 (a list of other enriched genes in these clusters and gene ontology analysis is in Extended Data File 1). D, Coexpression analysis of para with known active zone protein encoding genes, genes upregulated in response to neuronal activity in the third instar larvae (left hand side) and the adult (right hand side) CNS shows significant positive correlation (green highlight; two-tailed t test, padj <0.05). This suggests that neurons in which para is expressed are those that are actively firing. para expression positively correlates with the mature neuron marker Imp in the third instar larvae single-cell sequencing data and is negatively correlated with GMC (ase), immature neurons (pros), neuroblasts (dpn), and glial cells (repo) in both datasets; however, the correlation does not meet the threshold for significance in the larval single-cell sequencing data. This trend indicates that para is likely expressed only in mature neurons, which is evident by the significant positive correlation between para and elav in the adult single-cell sequencing data.
Distribution of para expressing cells in third instar larvae and adult transcriptomic data. (A) Annotated cell types on Seurat tSNE plot of cells from the adult brain as previously annotated by Davie et al. (2018). AST, astrocyte-like glia; CHI, chiasm glia; CTX, cortex glia; DCN, dorsal cluster neurons; DOP, dopaminergic neurons; ENS, ensheathing glia; HE, hemocytes; IPC, insulin-producing cells; KC, Kenyon cells; OCTY, octopaminergic-tyraminergic neurons; PEP, peptidergic neurons; PR, photoreceptors; PRN, perineurial glia; SER, serotonergic; SUB, subperineurial glia; SUR, surface glia; MBON, MB output neurons (B) Boxplots showing para expression for the different single-cell clusters from the adult brain single-cell transcriptomic data. Download Figure 8-1, TIF file (14.9MB, tif)
Analysis of the third instar larval transcriptomic atlas shows that many larval neurons express markers of immature neurons (Fig. 6A,B). We analyzed the correlation of para expression with known markers for neuronal differentiation (elav), neuroblast specific markers (dpn), or GMC markers (pros, ase) and found no or negative correlation coefficients (Fig. 8D). para does correlate with IGF-II mRNA-binding protein (Imp), an mRNA binding protein that is found to oppose prospero (pros) expression and is a marker of mature neurons (Davie et al., 2018). Davie et al. (2018) also found that Imp-positive clusters show higher expression of genes related to oxidative phosphorylation, hinting at an increase in neuronal activity. We did not detect para expression in any neuroblast lineages (Fig. 6A). The positive correlation of para only with mature neuronal markers and markers of active neuron firing suggests that a rather small fraction of neurons (23 ± 1%) in the larval CNS are mature neurons that can fire NaV-based APs.
We performed a similar analysis of single-cell RNA sequencing of the adult CNS (Davie et al., 2018). Seurat tSNE clustering separates the 57,000 cells into 102 unique clusters based on variable gene expression (Extended Data Fig. 8-1A). para expression is broadly distributed in the adult neurons with only a handful of clusters showing lack of para expression, including glial cells and photoreceptors (Fig. 8B; Extended Data Fig. 8-1B). In addition, in the adult CNS, para has a strong positive correlation with the pan neuronal marker elav, in agreement with the broad distribution of para expression seen in Figure 3D. To determine whether the increase in para expression correlates with an increase in the number of active neurons, we determined the Pearson's correlation coefficient of expression between para and several signatures of actively firing neurons (RNA for activity regulated and active zone proteins) (Fig. 8D). Similar to the larval data, para expression is positively correlated with all active zone genes and all ARGs. Hence, active neurons as defined by expression of active zone genes and ARGs in the adult express para. Together, these data suggest that the cells expressing para are active, differentiated neurons; and while this describes most neurons in the adult CNS, only 23 ± 1% of cells in larvae fit this criterion.
Para is enriched in a DAS in axons of third instar larval CNS neurons
As the only NaV channel in flies, the subcellular localization of Para shows where APs are likely initiated and propagated. In active, differentiated third instar larval neurons Para is confined to axons; however, Para is not uniformly distributed throughout the CNS (Fig. 9A; Extended Data Fig. 9-1A; Movie 1). To determine where along the axon Para is enriched, we looked at the third instar larval MB neurons. The anatomic structure of MB neurons in the larval CNS is well defined. The MB neurons have cell bodies, located in the ventral side of the larval central brain lobes, that project a single axon, which is intersected by dendrites in the calyx, and then continues along the α and β lobes where they synapse (Fig. 9C,D; Extended Data Fig. 9-1B). We used an MB-specific GAL4 (201Y-GAL4) (Yang et al., 1995) to drive expression of UAS-mCD8::GFP (to label the entire cell membranes) (T. Lee and Luo, 1999) in conjunction with Para-mCherry (Extended Data Fig. 9-1B) to determine where Para is localized. Para is enriched at a segment of the axon, after the site of dendritic integration at the calyx, after which it is distributed in a decreasing concentration toward the synapses (Fig. 9C,D). The same distribution of Para is also be observed in motor neuron axons exiting the VNC (Fig. 9A) with Para enriched as the axons leave the VNC (after dendritic innervation) (Fig. 9A, inset, box i) and then decreasing toward the NMJs (Fig. 9A, inset, box ii). The clustering of Para at the boundary of the somatodendritic and distal axonal compartments is reminiscent of the AIS location in vertebrates (Nelson and Jenkins, 2017). However, because of the unipolar organization of fly CNS neurons, this segment is distant from the soma. Conserved components of the vertebrate AIS have been shown to accumulate in fly axons, also at the distal boundary of dendrites and the remainder of the axon, at a region called an AIS-like compartment (Trunova et al., 2011; Smith-Trunova et al., 2015; Jegla et al., 2016). However, in CNS neurons, this segment is not at the initial part of the axon, but at a distal region; thus, we propose naming the segment the DAS to distinguish the invertebrate and vertebrate structures and avoid confusion.
Para-RFP-3xHA and Para-mCherry recapitulate the expression of Para-GFSTF in the third instar larval CNS. (A) Immunofluorescent imaging of Para-RFP-3xHA and Para-mCherry subcellular localization shows the same clustered distribution as Para-GFSTF. (B) 201Y-GAL4 labels the membrane of the MB neurons of third instar larvae, labeling these cells with Para-mCherry identifies the cluster of Para in at the DASs, yellow arrow. Download Figure 9-1, TIF file (19.8MB, tif)
Para-GFSTF expression in the third instar larval CNS.
To validate Para enrichment at the DAS across different larval CNS neuron types, we used split-GAL4 drivers (Luan et al., 2006; Pfeiffer et al., 2010) to assess Para distribution in an individual pair of motor neurons. Split-GAL4s are generated by separating the activation and DNA binding domains of GAL4, then expressing each domain under a separate promoter to restrict expression. The functional GAL4 protein is only present in cells that express both promoters. Split-GAL4 line R_MB050B is restricted to the two most posterior motor neurons in the larval VNC (Fig. 10A) (Aso et al., 2014). The cell bodies of each neuron are in the posterior of the VNC, with one dorsal cell body projecting an axon that exits the VNC on the right side, and a ventral cell body that projects an axon on the left side of the VNC with the dendritic tree for each cell in the midline of the VNC. Using pan-membrane labeling (UAS-mCD8::RFP), we observe localization of Para in the axons of each cell at the DAS, distal to the dendritic tree (Fig. 10B,C). We do not see the same colocalization when colabeling with the somatodendritic marker DenMark (Fig. 10D) (Nicolaï et al., 2010). Figure 11 shows cross sections through a posterior motor neuron, labeled with Split-GAL4 line R_MB050B driving UAS-mCD8::RFP, at the axon hillock (Fig. 11A) (where the AIS is located in vertebrates), in the axon distal to the dendritic tree where we predict the DAS to be located (Fig. 11B) and a section of the axon distal to the DAS (Fig. 11C). We clearly see Para localization distal to the dendritic tree at the DAS, whereas no Para is present at the axon hillock. Para is also enriched at the DAS (Fig. 11B) compared with a more distal segment of the axon (Fig. 11C). These images show that third instar larval neurons do contain segments of the axon where Nav channels are enriched like at the AIS in vertebrates; however, the subcellular localization of the segment is not shared. In addition, in motor neurons, the diffuse distribution of Para along the axon is maintained up to the first bouton of the NMJ (Fig. 9B). Interestingly, small clusters of Para can be observed near each bouton all along the length of the NMJ with super resolution microscopy. These data indicate that, in the neurons of third instar larvae, AP are likely initiated distally to the cell body, in contrast to the AIS initiation of AP in vertebrates. In addition, the low-level continual localization of Para between the DAS and synapses likely ensures AP are propagated to the synapses.
Figure 11.
Para is enriched at the Distal Axonal Segment (DAS) in third instar larval motor neurons. Cross sections through a posterior motor neuron labeled with Split-GAL4 line R-MB050B driving expression of UAS-mCD8::RFP. No axonal Para is detected at the axon hillock (A), where NaV channels cluster in vertebrates. B, Para is enriched at the DAS and is less abundant at a more distal axonal compartment (C). Scale bar, 5 μm. Axis orientation is displayed in i,ii and iii.
Para is enriched in a DAS in adult CNS neurons
para expression is greatly increased from the larval to the adult CNS. To determine whether the localization of Para to the DAS is conserved from the larval CNS to the adult CNS, we imaged the adult brain for Para-GFSTF. In the adult CNS, Para is localized to specific clusters of axons and not broadly distributed throughout the neuropil of the adult brain (Fig. 12A), and this distribution is comparable to staining's performed in adult fly brains with anti-NaV antibodies raised against vertebrate NaV channels (Wang et al., 2020). In addition, diffuse Para staining can be seen in the axons of the longest neurons of the CNS that project from the left to right brain hemisphere or vice-versa (Movie 2). The diffuse staining in long axons is clearly observed in axons of neurons in the thoracic ganglion where we see broad labeling of long axonal tracks (Fig. 12B). The enrichment of Para in the adult CNS is reminiscent of the DAS distribution observed in the larval CNS, not the AIS location as seen in mammals, as the clusters are not near the neuronal cell body marker Elav (Fig. 12A,B).
Para-GFSTF expression in the adult CNS.
The DAS in larval neurons is localized after the dendrites impinge on the axons. To determine where along the axon the Para-positive DAS clusters are localized in adult neurons, we labeled multiple classes of neurons using split-GAL4 line R_OL0019B-GAL4 (Fig. 12C–F) and 201Y-GAL4 (Fig. 13), labeling the laminar columnar neurons (LC10) and MB neurons, respectively. In both neuronal populations, Para is enriched in a DAS located in the axon after the last dendrite impinges on the axon (Figs. 12E, 13C). In LC10 neurons, Para enrichment in the DAS is seen when broadly labeling cells with mCD8::RFP but not when labeling the somatodendritic region of the neurons with DenMark (Fig. 12F). In addition to the DAS, we see lower-intensity Para distribution between the DAS and the synapses (Fig. 13E) for the remainder of axons. For the MB neurons in the adult, the DAS is present in the peduncle, beyond the localization of DenMark (Fig. 13D), as in the larval CNS (Figs. 10B,C, 13B,C). Hence, Para localization at the DAS, distal to the site where dendrites impinge on CNS neurons, and lower-intensity Para distribution beyond the DAS, is conserved from larvae to adults. These data show that Para is enriched in a DAS after dendritic innervation in multiple neuron classes throughout larval and adult development.
To determine the size of the DAS, we measured signal intensity of Para-mCherry along three classes of MB neurons that derive from MBNB clusters (MBNBa, c and d; Fig. 14A) (Kunz et al., 2012). In both MBNBc and MBNBd neurons, the DAS starts 82.72 ± 3.16 µm and 72.73 ± 5.54 µm from the soma and is 28.69 ± 6.12 µm and 28.69 ± 6.12 µm in length, respectively. The DAS in MBNBa neurons is both closer to the soma (38.43 ± 2.94 µm) and shorter in length (21.92 ± 3.44 µm) than the DAS in MBNBc and MBNBd neurons. The length of the DAS reported in this study is comparable with the length of low-frequency vertebrate AISs (Adachi et al., 2015), and the size of the AIS-like region previously reported in adult MB neurons (Trunova et al., 2011).
In vertebrate neurons, the length of the AIS is variable depending on the size and firing properties of the neuron (Adachi et al., 2015). To determine whether the size discrepancies between neuron population in the MB were as a result of differences in neuron size, we used UAS-DenMark (Fig. 14). DenMark labels the somatodendritic region of the neuron but is enriched in the dendritic tree (Fig. 13D). We can determine the length between the soma and the dendritic tree (weak DenMark signal to strong signal), the length of axon where dendrites impinge (strong DenMark signal to strong DenMark signal), and the distance from the dendritic tree to the boundary of the somatodendritic region (strong DenMark signal to no signal) using DenMark signal intensity (Fig. 14). We show that, in all three MBNB populations measured (a, c, and d), the length of the region of the axon where dendrites impinge is similar (13.68 ± 2.88, 15.15 ± 2.12, and 13.50 ± 1.68 µm, respectively) (Fig. 14A). The distance between the soma and the boundary of the somatodendritic compartment of the cell is variable. MBNBc and MBNBd cells have a soma to somatodendritic boundary length of 74.15 ± 8.98 and 80.38 ± 6.94 µm, respectively, which is comparable to the soma to DAS distance in each neuronal population. In MBNBa cells, the soma to somatodendritic boundary was shorter (55.40 ± 6.01 µm), which corresponds to the shorter soma to DAS distance seen in MBNBa neurons. This indicates that the DAS length is shorter in neurons with a shorter soma to DAS distance, and that the DAS does act as a boundary between the somatodendritic region of the axon and the distal axon as defined by absence of DenMark staining.
Voltage-gated Para currents are evoked distal from the soma
Para's distal localization far from the soma suggests that NaV-based APs in Drosophila neurons initiate far from the soma. NaV currents in mammalian neurons are readily evoked by voltage steps. These voltage-evoked NaV currents show little or no temporal delay (1-2 ms) when recorded from the soma because of the close proximity of the AIS (Magistretti et al., 2006) (see Fig. 1). In contrast, evoked Para currents recorded at the soma should show delays following voltage steps and exhibit features of poor space clamp because of their distance from the soma. To determine this, we measured Para NaV voltage-evoked currents by whole-cell voltage clamp in lLNvs, pharmacologically isolated by blockers of voltage-gated calcium channels (CaV) and voltage-gated KV channels and synaptic currents. Depolarizing voltage steps evoke rapidly inactivating inward Para currents as shown by downward-deflecting currents (Fig. 15A). Para currents are abolished by the NaV-specific blocker TTX (Fig. 15A′) and show peak maximum voltage activation at −35 mV (Fig. 15A′′). The short-latency upward-deflecting outward currents seen in Figure 15A, A′ may be somatic or near-somatic voltage-gated KV currents that are not completely blocked by 4-AP and tetra-ethyl ammonium. Voltage-evoked Para currents decrease as voltage steps approach the Na+ reversal potential (Fig. 15A′′). Consistent with localization originating in the DAS, TTX-sensitive inward Para currents show long delays measured at the soma following voltage steps and features of poor space clamp and poor voltage control (Fig. 15A,A′′). To determine Para localization in the lLNv neurons, we used Pdf-GAL4 to drive expression of UAS-mCD8::RFP and UAS-DenMark, to label the neuronal membrane and dendrites, respectively (Fig. 15B,C), in I-LNv and s-LNv neurons. Para is enriched in the axons of I-LNv and s-LNv neurons after the integration of the dendritic tree in a DAS. Para is not localized to the soma, proximal axon or the dendritic projections. This is consistent with the delay in evoked Para currents and indicates that the DAS is the likely site of these currents. Therefore, NaV-evoked AP are likely generated at the DAS.
Figure 15.
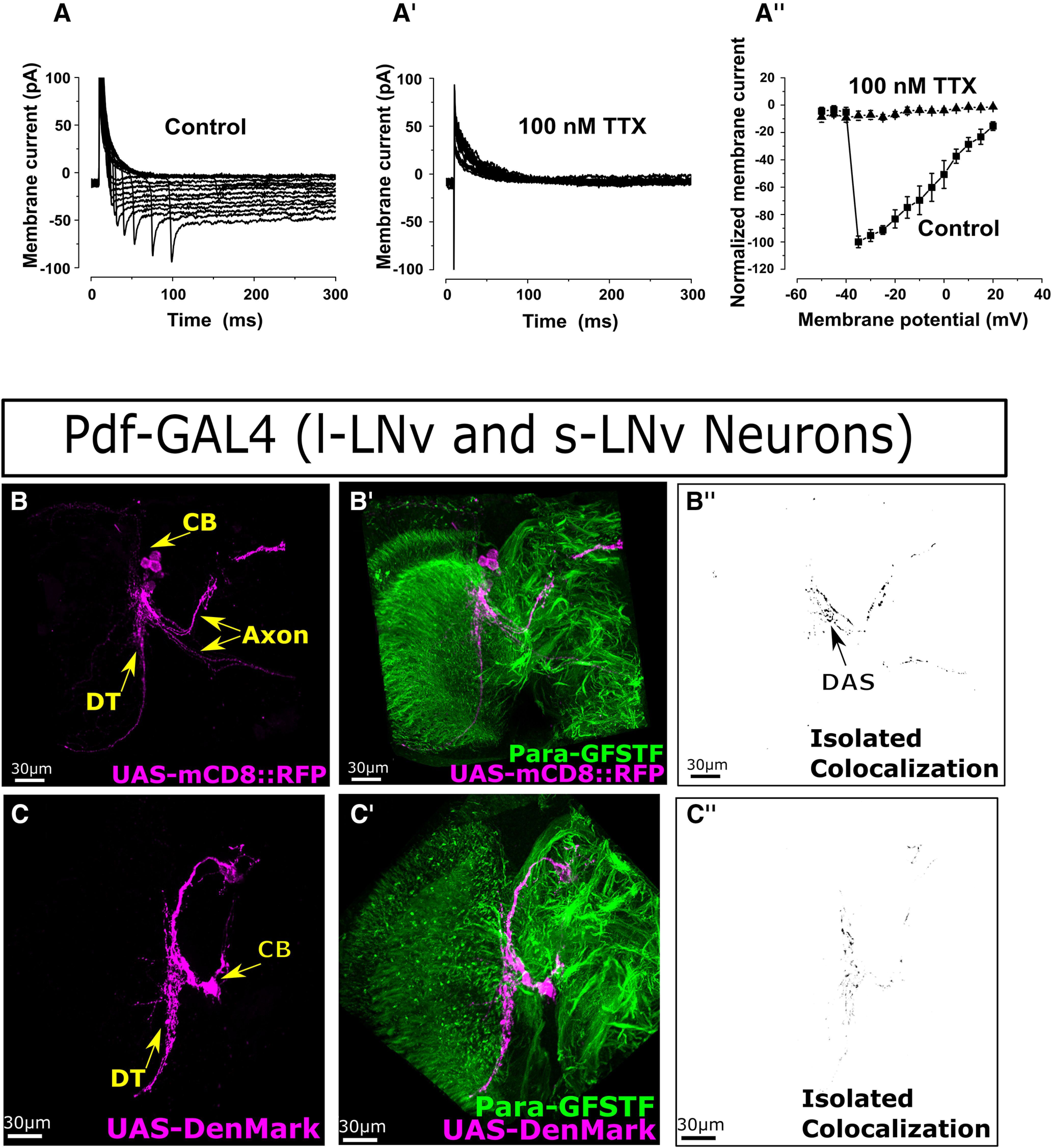
Inward activating NaV currents are generated distal to the soma in I-LNv neurons in the adult CNS. A, Depolarizing steps in whole-cell patch recordings in voltage-clamp mode in the presence of blockers of synaptic currents, KV and CaV channels reveal the functional expression of voltage-gated Para sodium channels in lLNv processes. These inward, rapidly activating currents show an average peak value of 91 ± 3 mV, at the maximum activation voltage of –35 mV (n = 5). The long delays in inward current initiation relative to voltage steps show poor space clamp and poor voltage control, indicating that Para channels are located distally in neuronal processes rather than in or close to the neuronal cell body. Fourth power exponential time constant for the activation was 2.1 ± 0.7 ms, whereas the single exponential fit inactivation time constant was 5.9 ± 2.6 (n = 5). A′, Currents were completely blocked by perfusing with 100 nm TTX. A′′, Voltage-current relationship for the inward sodium currents (squares) is shown, along with the effect of TTX channel block (triangles). B,B′,B′′, Pdf-GAL4 is specifically expressed in the small and large lateral ventral neurons (s-LNv, l-LNv) in the adult CNS. Using Pdf-GAL4 to drive expression of UAS-mCD8::RFP, we can identify the cell body of the l-LNv neurons (CB), the dendritic tree (DT), and axons (Ax) for both s-LNv (dorsal) and l-LNv neurons. Colabeling of these neurons with Para-GFSTF shows that Para is localized only to DASs in both neuron types with no Para seen in the soma, proximal axon, or dendritic tree. C,C′,C′′, UAS-DenMark is localized to the soma and dendritic projections. In the I-LNv and s-LNv neurons, the dendritic projections extend across the accessory medulla. Colabeling of Pdf-GAL4 driving UAS-DenMark and Para-GFSTF does not show any Para localization in the dendritic tree, the soma, or the proximal axon.
Discussion
The site of AP initiation in invertebrate neurons is unclear. In vertebrates, APs are generated at the AIS. The major functional hallmark of vertebrate AIS is clustering of NaV channels. Therefore, we characterized the gene expression and subcellular localization of the sole NaV channel in Drosophila, (para). Despite being the only NaV channel in flies, we find that para is only expressed in 23 ± 1% of neurons in the third instar larval CNS. In contrast, in the adult, para expression is far broader. By generating a single-cell transcriptomic atlas of the third instar larval brain, we determined that para expression correlates with the expression of ARGs, genes expressed in active zones, and markers of mature neurons. This implies that cells expressing para, while proportionally small, likely represent the active population of neurons in the third instar larval CNS. This correlation occurs also in the adult CNS, indicating that the number of active neurons is likely much higher in the adult than in the third instar larvae. In the neurons expressing para, we find that Para distributed into axonal segments that are far removed from the soma, distal to where dendrites integrate into the axon. This structure shares some features of the mammalian AIS, and whole-cell patch-clamp electrophysiological recordings are consistent with the interpretation that Nav-dependent APs initiate there; however, because of its distal location to the cell soma, we name this region the DAS.
The AIS identifies the origin of AP propagation (Westenbroek et al., 1989; Whitaker et al., 2000) through the high density of NaV channels (Mainen et al., 1995). The clustering of NaV channels at the DAS suggests that this is the site of AP initiation in fly neurons. Our identification that TTX-sensitive, inward sodium currents that occur after long delays in response to somatic depolarization are also suggestive of a distal site of AP initiation. In vertebrate neurons, TTX-sensitive inward sodium currents are measured immediately after somatic depolarization as the AIS is so close to the soma (Kuo and Bean, 1994; Magistretti et al., 2006; Lewis and Raman, 2014). Single-cell electrophysiological recordings performed on fly neurons in the CNS are a challenge because of their small size. Recordings performed in the neurons of larger invertebrates, Aplysia, crab, and leeches also exhibit delayed activation and poor space clamp, indicating that APs are initiated at a site distal relative to the cell body (Tauc, 1962; Tauc and Hughes, 1963; Meyrand et al., 1992; Tobin et al., 2006). This suggests that AP initiation at the DAS is conserved across invertebrate species. Because of their small size, patch-clamp recordings are made on the soma (Wilson et al., 2004; Gouwens and Wilson, 2009). Therefore, the soma receives back-propagating APs from the DAS, and passive fluctuations in membrane potential are readily measured in the soma as well in Drosophila CNS neurons (Sheeba et al., 2008). While APs appear to initiate from the DAS, the sources and ionic mechanisms of passive membrane potentials are not clear; however, fluctuations in membrane potential clearly influence the pattern of AP firing in Drosophila neurons. For example, large fluctuations in membrane potential drive burst firing in lLNv, whereas stable membrane potential is associated with regular tonic AP firing (Sheeba et al., 2008). Further, transition between burst and tonic firing AP firing pattern is CaV-dependent as it is modulated by cobalt block of CaV channels. Surprisingly, either TTX or cobalt abolishes lLNv APs altogether, suggesting complex interactions between Para and CaVs in Drosophila neurons (Sheeba et al., 2008). Therefore, the electrical activity recorded at the soma may not be representative of the neuron's actual firing activity, as in some cases the DAS is far away from the soma and these cell bodies may not undergo any depolarization. This emphasizes the need to establish voltage-sensing reporters to simultaneously enable accurate reporting of APs and calcium events at high spatiotemporal resolution (Simpson and Looger, 2018). While genetically encoded calcium indicators are often used for a proxy for inferring APs, intracellular calcium sources are highly diverse (CaVs, TRP channels, nonselective cation channels, multiple intracellular stores, etc.).
As an alternative to electrophysiological recordings, computational models of compartmentalized fly neurons have been used to model their electrophysiological properties. Models of different neurons in the fly CNS predict that the site of AP propagation occurs, not proximal to the cell body as in mammals, but distally in the axon after the last dendrite innervates the axon (Gouwens and Wilson, 2009; Günay et al., 2015). The mapping of the DAS and the ability to visualize it in any fly neuron using Para-GFSTF endorse the accuracy of these models and enable the generation of models with greater precision to better predict and analyze neuron activity and dynamics.
Some studies have indicated that an AIS-like region may be present in some Drosophila neurons, based on GAL4-mediated overexpression of Drosophila homologs of vertebrate AIS proteins, such as Ank1 and Shal in MB neurons (Trunova et al., 2011; Smith-Trunova et al., 2015). The overexpressed proteins were shown to cluster in axons downstream of the dendritic tree, in the MB of the third instar larval and the adult CNS. Using a similar approach, it has been shown that proximal axons of ddaE neurons in the PNS also may contain AIS-like regions (Jegla et al., 2016). However, the overexpression of proteins may lead to their restriction in cellular compartments as Katsuki et al. (2009) showed that the mobility of transmembrane molecules is restricted to areas before and after dendritic branches, suggesting that there are barriers in axons that restrict protein localization (Katsuki et al., 2009). The DAS reported in this study is of comparable size and location to the AIS-like region previously reported in adult MB neurons (Trunova et al., 2011). AnkG is key for NaV channel clustering at the vertebrate AIS (Jenkins and Bennett, 2001). The similarity in the reported AIS-like localization of Ank1 localization from Trunova et al. (2011) and Nav enrichment at the DAS as defined in this study indicate that the role of ankyrins in NaV clustering may be conserved in invertebrates, despite differences in ankyrin structure across species (Jegla et al., 2016). Interestingly, we found that the distance between the soma and the length of the DAS is correlated, with a longer DAS present in neurons where the DAS is more distal. This is in contrast to what is observed in vertebrates where the more distal the AIS, the shorter the length of the AIS (Adachi et al., 2015).
We observe low levels of Para distribution along long axons, suggesting that AP propagation is maintained by Para across the length of the axon. APs are all-or-nothing signals that rapidly propagate along axons; however, if no NaV channel is present, the signal should decay rapidly. We argue that the low levels of Para distributed along the length of long axons enable the APs to reach the synapse without signal depletion. We see this clearly in neurons, such as motor neurons in larval and adult CNS, and neurons in the adult brain that cross from one hemisphere to the other. Neurons with smaller axons may also have Para continually distributed along axons at undetectable levels or may not require Para at all. However, given the expression data discussed below, the latter is unlikely.
The sparse number of CNS neurons in the third instar larvae that express para is an unanticipated observation. Only 23 ± 1% of elav-positive neurons in the larval CNS express para, and historical gene expression tracing shows that those neurons that are para-negative never express para. These data are also in agreement with the single-cell sequencing data shown in Figure 8A, which document a highly restricted expression pattern for para in the third instar larval CNS. Neurons expressing para also express genes that produce neurotransmitters as well as proteins that have been shown to be upregulated in many Drosophila models of enhanced neuronal activity (Harris and Littleton, 2015; Chen et al., 2016), both in the third instar larval and adult brain single-cell RNA sequencing datasets. This provides evidence that this restricted cell population is likely capable of firing APs, that para expression is a potential marker for active AP firing neurons, and that only 23 ± 1% of cells in the third instar larvae fit this criterion. Whether the para-negative cells still have electrical activity in lieu of NaV channels, either through passive signaling or using CaV, is unknown. para expression has been demonstrated to positively correlate with neural progenitor proliferation in third instar larvae (Piggott et al., 2019). The single-cell transcriptomic atlas of the whole third instar larval CNS generated in this study identifies neuronal progenitors at immature, intermediate, and mature maturation steps, and we failed to detect para expression in any neuroblasts lineage at any stage of maturation (Figs. 6, 7).
In this study, we have identified the DAS, a compartment of Drosophila axons analogous to the AIS in vertebrates. We report the characterization of NaV channels throughout development in D. melanogaster, uncovering the presence of the DAS in most axons after the dendritic tree, where AP is likely initiated, and continual low-level Para distribution in long axons to maintain AP. Additionally, we performed comprehensive single-cell RNA sequencing of the third instar larval CNS. This single-cell atlas in conjunction with para-T2A-GAL4 revealed that only 23 ± 1% of the cells in the third instar larval brain express para and other genes expressed in AP firing neurons. These data are consistent with the notion that most neurons are developing in third instar larvae, especially in the brain lobes (Hartenstein et al., 2008). In contrast, para expression is broad in the adult CNS. The models generated herein should allow the mapping of DAS in any neuron and provide a GAL4 driver to target differentiated, AP firing neurons.
Footnotes
This work was supported in part by National Institutes of Health R01GM067858 and the Huffington Foundation. T.A.R. was supported by the Cullen Foundation. P.C.M. was supported by Canadian Institutes of Health Research MFE-164712. J.J. was supported by The Research Foundation–Flanders PhD fellowship FWO 1199518N. T.C.H. was supported by the National Institutes of Health/National Institute of General Medical Sciences R35GM127102. H.J.B. is an Investigator of the Howard Hughes Medical Institute. Confocal microscopy was performed in the Neurovisualization core of the BCM IDDRC (supported by National Institute of Child Health and Human Development U54HD083092). We thank the BDSC (National Institutes of Health P40OD018537), DGRC, BDGP, and the Developmental Studies Hybridoma Bank for flies, molecular biology reagents, and antibodies; Margaret Ho for the Para antibody; Kartik Venkatachalam, Herman Dierick, Matthew Rasband, Shinya Yamamoto, Joshua Shulman, Mingshan Xue, Hyunglok Chung, Amir Fayyazuddin, Lindsey Goodman, and Karen Schulze for helpful discussions and comments; Dinghui Yu for assistance with microscopes; and Yuchun He for injections to create transgenic flies.
The authors declare no competing financial interests.
References
- Adachi R, Yamada R, Kuba H (2015) Plasticity of the axonal trigger zone. Neuroscientist 21:255–265. 10.1177/1073858414535986 [DOI] [PubMed] [Google Scholar]
- Allen AM, Neville MC, Birtles S, Croset V, Treiber CD, Waddell S, Goodwin SF (2020) A single-cell transcriptomic atlas of the adult Drosophila ventral nerve cord. eLife 9:e54074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amichot M, Castella C, Bergé JB, Pauron D (1993) Transcription analysis of the para gene by in situ hybridization and immunological characterization of its expression product in wild-type and mutant strains of Drosophila. Insect Biochem Mol Biol 23:381–390. 10.1016/0965-1748(93)90022-k [DOI] [PubMed] [Google Scholar]
- Anholt RR, Lyman RF, Mackay TF (1996) Effects of single P-Element insertions on olfactory behavior in Drosophila melanogaster. Genetics 143:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apitz H, Salecker I (2014) A challenge of numbers and diversity: neurogenesis in the Drosophila optic lobe. J Neurogenet 28:233–249. 10.3109/01677063.2014.922558 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, Rubin GM (2014) The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3:e04577. 10.7554/eLife.04577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines RA, Lin WH, Günay C, Marley R, Prinz AA (2012) Activity-dependent alternative splicing increases persistent sodium current and promotes seizure. J Neurosci 32:7267–7277. 10.1523/JNEUROSCI.6042-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT (2006) Site-specific transformation of Drosophila via φC31 integrase-mediated cassette exchange. Genetics 173:769–777. 10.1534/genetics.106.056945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar OA, Doe CQ (2013) Combinatorial temporal patterning in progenitors expands neural diversity. Nature 498:449–455. 10.1038/nature12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J (2019) The single-cell transcriptional landscape of mammalian organogenesis. Nature 566:496–502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casso D, Ramírez-Weber FA, Kornberg TB (1999) GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech Dev 88:229–232. [DOI] [PubMed] [Google Scholar]
- Chen X, Rahman R, Guo F, Rosbash M (2016) Genome-wide identification of neuronal activity-regulated genes in Drosophila. eLife 5:e19942 10.7554/eLife.19942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Doe CQ (1992) Ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development 116:943–952. [DOI] [PubMed] [Google Scholar]
- Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft Ł, Aibar S, Makhzami S, Christiaens V, Bravo González-Blas C, Poovathingal S, Hulselmans G, Spanier KI, Moerman T, Vanspauwen B, Geurs S, Voet T, Lammertyn J, Thienpont B, Liu S, et al. (2018) A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174:982–998.e20. 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalvo MK, Hindle SJ, Rusan ZM, Orng S, Eddison M, Halliwill K, Bainton RJ (2014) The Drosophila surface glia transcriptome: evolutionary conserved blood-brain barrier processes. Front Neurosci 8:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne H, Hibbard KL, Cavallaro A, Kao JC, Rubin GM (2018) Genetic reagents for making split-GAL4 lines in Drosophila. Genetics 209:31–35. 10.1534/genetics.118.300682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Tasdemir-Yilmaz OE, Freeman MR (2009) Ensheathing Glia Function as Phagocytes in the Adult Drosophila Brain. J Neurosci 929:4768–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH (2010) Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development 137:2981–2987. 10.1242/dev.051250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheredge J. (2017) Transcriptional Profiling of Drosophila Larval Ventral Nervous System Hemilineages and Neuroscience. (Doctoral thesis) University of Cambridge Development Physiology. [Google Scholar]
- Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, Patananan AN, Sitz D, Tran PT, Do MT, Yackle K, Cespedes A, Hartenstein V, Call GB, Banerjee U (2009) G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods 6:603–605. 10.1038/nmeth.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2019) The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germeraad S, O'Dowd D, Aldrich RW (1992) Functional assay of a putative Drosophila sodium channel gene in homozygous deficiency neurons. J Neurogenet 8:1–16. 10.3109/01677069209167268 [DOI] [PubMed] [Google Scholar]
- Gold KS, Brand AH (2014) Optix defines a neuroepithelial compartment in the optic lobe of the Drosophila brain. Neural Dev 9:18. 10.1186/1749-8104-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouwens NW, Wilson RI (2009) Signal propagation in Drosophila central neurons. J Neurosci 29:6239–6249. 10.1523/JNEUROSCI.0764-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günay C, Sieling FH, Dharmar L, Lin WH, Wolfram V, Marley R, Baines RA, Prinz AA (2015) Distal spike initiation zone location estimation by morphological simulation of ionic current filtering demonstrated in a novel model of an identified Drosophila motoneuron. PLoS Comput Biol 11:e1004189 10.1371/journal.pcbi.1004189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, Satija R (2019) Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakes AE, Otsuki L, Brand AH (2018) A newly discovered neural stem cell population is generated by the optic lobe neuroepithelium during embryogenesis in Drosophila melanogaster. Development 145:dev166207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KP, Littleton JT (2015) Transmission, development, and plasticity of synapses. Genetics 201:345–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Spindler S, Pereanu W, Fung S (2008) The development of the Drosophila larval brain. Adv Exp Med Biol 628:1–31. 10.1007/978-0-387-78261-4_1 [DOI] [PubMed] [Google Scholar]
- Hassan BA, Bermingham NA, He Y, Sun Y, Jan YN, Zoghbi HY, Bellen HJ (2000) Atonal regulates neurite arborization but does not act as a proneural gene in the Drosophila brain. Neuron 25:549–561. 10.1016/S0896-6273(00)81059-4 [DOI] [PubMed] [Google Scholar]
- He L, Binari R, Huang J, Falo-Sanjuan J, Perrimon N (2019) In vivo study of gene expression with an enhanced dual-color fluorescent transcriptional timer. eLife 8:e46181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson PM. (2017) Annual review of cell and developmental biology cell removal: efferocytosis. Annu Rev Cell Dev Biol 33:127–171. 10.1146/annurev-cellbio-111315-125315 [DOI] [PubMed] [Google Scholar]
- Hitier R, Chaminade M, Préat T (2001) The Drosophila castor gene is involved in postembryonic brain development. Mech Dev 103:3–11. 10.1016/s0925-4773(01)00312-4 [DOI] [PubMed] [Google Scholar]
- Homyk T, Pye Q (1989) Some mutations affecting neural or muscular tissues alter the physiological components of the electroretinogram in Drosophila. J Neurogenet 5:37–48. 10.3109/01677068909167263 [DOI] [PubMed] [Google Scholar]
- Hong CS, Ganetzky B (1994) Spatial and temporal expression patterns of two sodium channel genes in Drosophila. J Neurosci 14:5160–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Ganetzky B (1996) Molecular characterization of neurally expressing genes in the para sodium channel gene cluster of Drosophila. Genetics 142:879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, Peiffer J, Ràmia M, Tarone AM, Turlapati L, Zichner T, Zhu D, Lyman RF, Magwire MM, Blankenburg K, Carbone MA, Chang K, Ellis LL, Fernandez S, Han Y, Highnam G, Hjelmen CE, Jack JR, et al. (2014) Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res 24:1193–1208. 10.1101/gr.171546.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Liu M, Yan SF, Yan N (2017) Structure-based assessment of disease-related mutations in human voltage-gated sodium channels. Protein Cell 8:401–438. 10.1007/s13238-017-0372-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley AF, Stämpfli R (1949) Evidence for saltatory conduction in peripheral myelinated nerve fibres. J Physiol 108:315–339. 10.1113/jphysiol.1949.sp004335 [DOI] [PubMed] [Google Scholar]
- Jegla T, Nguyen MM, Feng C, Goetschius DJ, Luna E, van Rossum DB, Kamel B, Pisupati A, Milner ES, Rolls MM (2016) Bilaterian giant ankyrins have a common evolutionary origin and play a conserved role in patterning the axon initial segment. PLoS Genet 12:e1006457 10.1371/journal.pgen.1006457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins SM, Bennett V (2001) Ankyrin-G coordinates assembly of the spectrin-based membrane skeleton, voltage-gated sodium channels, and L1 CAMs at Purkinje neuron initial segments. J Cell Biol 155:739–745. 10.1083/jcb.200109026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joesch M, Schnell B, Raghu SV, Reiff DF, Borst A (2010) ON and off pathways in Drosophila motion vision. Nature 468:300–304. 10.1038/nature09545 [DOI] [PubMed] [Google Scholar]
- Johar K, Priya A, Wong-Riley MT (2014) Regulation of Na+/K+-ATPase by neuron-specific transcription factor Sp4: implication in the tight coupling of energy production, neuronal activity and energy consumption in neurons. Eur J Neurosci 39:566–578. 10.1111/ejn.12415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA, Ye CJ (2018) Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 36:89–94. 10.1038/nbt.4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuki T, Ailani D, Hiramoto M, Hiromi Y (2009) Intra-axonal patterning: intrinsic compartmentalization of the axonal membrane in Drosophila neurons. Neuron 64:188–199. 10.1016/j.neuron.2009.08.019 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Storrie B, Simons K, Dotti CG (1992) A functional barrier to movement of lipids in polarized neurons. Nature 359:647–650. 10.1038/359647a0 [DOI] [PubMed] [Google Scholar]
- Kole MH, Ilschner SU, Kampa BM, Williams SR, Ruben PC, Stuart GJ (2008) Action potential generation requires a high sodium channel density in the axon initial segment. Nat Neurosci 11:178–186. 10.1038/nn2040 [DOI] [PubMed] [Google Scholar]
- Kulkarni NH, Yamamoto AH, Robinson KO, Mackay TFC, Anholt RR (2002) The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics 161:1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz T, Kraft KF, Technau GM, Urbach R (2012) Origin of Drosophila mushroom body neuroblasts and generation of divergent embryonic lineages. Development 139:2510–2522. 10.1242/dev.077883 [DOI] [PubMed] [Google Scholar]
- Kuo CC, Bean BP (1994) Na+ channels must deactivate to recover from inactivation. Neuron 12:819–829. [DOI] [PubMed] [Google Scholar]
- Lee A, Goldin AL (2009) Role of the terminal domains in sodium channel localization. Channels (Austin) 3:171–180. 10.4161/chan.3.3.8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PT, Zirin J, Kanca O, Lin WW, Schulze KL, Li-Kroeger D, Tao R, Devereaux C, Hu Y, Chung V, Fang Y, He Y, Pan H, Ge M, Zuo Z, Housden BE, Mohr SE, Yamamoto S, Levis RW, Spradling AC, et al. (2018) A gene-specific T2A-GAL4 library for Drosophila. eLife 7:e35574 10.7554/eLife.35574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L (1999) Mosaic analysis with a repressible neurotechnique cell marker for studies of gene function in neuronal morphogenesis. Neuron 22:451–461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L (1999) Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126:4065–4076. [DOI] [PubMed] [Google Scholar]
- Lewis AH, Raman IM (2014) Resurgent current of voltage-gated Na+ channels. J Physiol 592:4825–4838. 10.1113/jphysiol.2014.277582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen Z, Desplan C (2013) Temporal patterning of neural progenitors in Drosophila. Curr Top Dev Biol 105:69–96. 10.1016/B978-0-12-396968-2.00003-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WH, Wright DE, Muraro NI, Baines RA (2009) Alternative splicing in the voltage-gated sodium channel DmNav regulates activation, inactivation, and persistent current. J Neurophysiol 102:1994–2006. 10.1152/jn.00613.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K, Kreber R, Ganetzky B (1989) Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell 58:1143–1154. 10.1016/0092-8674(89)90512-6 [DOI] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH (2006) Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52:425–436. 10.1016/j.neuron.2006.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN (1994) Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev 8:1787–1802. 10.1101/gad.8.15.1787 [DOI] [PubMed] [Google Scholar]
- Magistretti J, Castelli L, Forti L, D'Angelo E (2006) Kinetic and functional analysis of transient, persistent and resurgent sodium currents in rat cerebellar granule cells in situ: an electrophysiological and modelling study. J Physiol 573:83–106. 10.1113/jphysiol.2006.106682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainen ZF, Joerges J, Huguenard JR, Sejnowski TJ (1995) A model of spike initiation in neocortical pyramidal neurons. Neuron 15:1427–1439. 10.1016/0896-6273(95)90020-9 [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun AT, Wills QF (2017) Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerick DM, Kassis JA, Zhang SD, Odenwald WF (1992) castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron 9:789–803. 10.1016/0896-6273(92)90234-5 [DOI] [PubMed] [Google Scholar]
- Meyrand P, Weimann JM, Marder E (1992) Multiple axonal spike initiation zones in a motor neuron: serotonin activation. J Neurosci 12:2803–2812. 10.1523/JNEUROSCI.12-07-02803.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47:D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastirioti M, Giagtzoglou N, Koumbanakis KA, Zacharioudaki E, Deligiannaki M, Wech I, Almeida M, Preiss A, Bray S, Delidakis C (2010) Drosophila Hey is a target of Notch in asymmetric divisions during embryonic and larval neurogenesis. Development 137:191–201. 10.1242/dev.043604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal S, Deluca SZ, Lee PT, Lin WW, Pan H, Zuo Z, Lv J, Spradling AC, Bellen HJ (2015) A genetic toolkit for tagging intronic MiMIC containing genes. eLife 4:2–9. 10.7554/eLife.08469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AD, Jenkins PM (2017) Axonal membranes and their domains: assembly and function of the axon initial segment and node of Ranvier. Front Cell Neurosci 11:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaï LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA (2010) Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci USA 107:20553–20558. 10.1073/pnas.1010198107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuyens D, Stengl M, Dugarmaa S, Rossenbacker T, Compernolle V, Rudy Y, Smits JF, Flameng W, Clancy CE, Moons L, Vos MA, Dewerchin M, Benndorf K, Collen D, Carmeliet E, Carmeliet P (2001) Abrupt rate accelerations or premature beats cause life-threatening arrhythmias in mice with long-QT3 syndrome. Nat Med 7:1021–1027. 10.1038/nm0901-1021 [DOI] [PubMed] [Google Scholar]
- O'Dowd DK, Germeraad SE, Aldrich RW (1989) Alterations in the expression and gating of Drosophila sodium channels by mutations in the para gene. Neuron 2:1301–1311. 10.1016/0896-6273(89)90068-8 [DOI] [PubMed] [Google Scholar]
- Okamoto H, Sakai K, Goto S, Takasu-Ishikawa E, Hotta Y (1987) Isolation of Drosophila genomic clones homologous to the eel sodium channel gene. Proc Jpn Acad B Phys Biol Sci 63:284–288. 10.2183/pjab.63.284 [DOI] [Google Scholar]
- O'Neill EM, Rebay I, Tjian R, Rubin GM (1994) The activities of two Ets-related transcription factors required for drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78:137–147. 10.1016/0092-8674(94)90580-0 [DOI] [PubMed] [Google Scholar]
- Palay SL, Sotelo C, Peters A, Orkand PM (1968) The axon hillock and the initial segment. J Cell Biol 38:193–201. 10.1083/jcb.38.1.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LM, Stuart GJ (2006) Site of action potential initiation in layer 5 pyramidal neurons. J Neurosci 26:1854–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadatos GA, Wallerstein PM, Head CE, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AE, Huang CL, Vandenberg JI, Colledge WH, Grace AA (2002) Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci USA 99:6210–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA 97:3608–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM (2010) Refinement of tools for targeted gene expression in Drosophila. Genetics 186:735–755. 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggott BJ, Peters CJ, He Y, Huang X, Younger S, Jan LY, Jan YN (2019) Paralytic, the Drosophila voltage-gated sodium channel, regulates proliferation of neural progenitors. Genes Dev 33:1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband MN, Peles E (2016) The nodes of Ranvier: molecular assembly and maintenance. Cold Spring Harb Perspect Biol 8:a020495 10.1101/cshperspect.a020495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM. (2011) Neuronal polarity in Drosophila: sorting out axons and dendrites. Dev Neurobiol 71:419–429. 10.1002/dneu.20836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer JL. (2003) Polarized domains of myelinated axons. Neuron 40:297–318. [DOI] [PubMed] [Google Scholar]
- Sheeba V, Gu H, Sharma VK, O'Dowd DK, Holmes TC (2008) Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol 99:976–988. 10.1152/jn.00930.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood L. (2008) Human physiology: from cells to systems. Boston, MA: Cengage Learning. [Google Scholar]
- Shu Y, Hasenstaub A, Duque A, Yu Y, McCormick DA (2006) Modulation of intracortical synaptic potentials by presynaptic somatic membrane potential. Nature 441:761–765. 10.1038/nature04720 [DOI] [PubMed] [Google Scholar]
- Siddiqi O, Benzer S (1976) Neurophysiological defects in temperature sensitive paralytic mutants of Drosophila melanogaster. Proc Natl Acad Sci USA 73:3253–3257. 10.1073/pnas.73.9.3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JH, Looger LL (2018) Functional imaging and optogenetics in Drosophila. Genetics 208:1291–1309. 10.1534/genetics.117.300228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Trunova S, Prithviraj R, Spurrier J, Kuzina I, Gu Q, Giniger E (2015) Cdk5 regulates developmental remodeling of mushroom body neurons in Drosophila. Dev Dyn 244:1550–1563. 10.1002/dvdy.24350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song AH, Wang D, Chen G, Li Y, Luo J, Duan S, Poo MM (2009) A selective filter for cytoplasmic transport at the axon initial segment. Cell 136:1148–1160. 10.1016/j.cell.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Spindler SR, Ortiz I, Fung S, Takashima S, Hartenstein V (2009) Drosophila cortex and neuropile glia influence secondary axon tract growth, pathfinding, and fasciculation in the developing larval brain. Dev Biol 334:355–368. 10.1016/j.ydbio.2009.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini M, Martino CD, Zamboni L (1967) Fixation of ejaculated spermatozoa for electron microscopy. Nature 216:173–174. 10.1038/216173a0 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R (2019) Comprehensive integration of single-cell data. Cell 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki DT, Grigliatti T, Williamson R (1971) Temperature-sensitive mutations in Drosophila melanogaster: VII. A mutation (para-ts) causing reversible adult paralysis. Proc Natl Acad Sci USA 68:890–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kaido M, Takayama R, Sato M (2013) A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev Biol 380:12–24. 10.1016/j.ydbio.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Tauc L. (1962) Site of origin and propagation in spike in the giant neuron of Aplysia. J Gen Physiol 45:1077–1097. 10.1085/jgp.45.6.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauc L, Hughes GM (1963) Modes of initiation and propagation of spikes in the branching axons of molluscan central neurons. J Gen Physiol 46:533–549. 10.1085/jgp.46.3.533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, Kaufman TC, Calvi BR; FlyBase Consortium (2019) FlyBase 2.0: the next generation. Nucleic Acids Res 47:D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin AE, Van Hooser SD, Calabrese RL (2006) Creation and reduction of a morphologically detailed model of a leech heart interneuron. J Neurophysiol 96:2107–2120. 10.1152/jn.00026.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunova S, Baek B, Giniger E (2011) Cdk5 regulates the size of an axon initial segment-like compartment in mushroom body neurons of the Drosophila central brain. J Neurosci 31:10451–10462. 10.1523/JNEUROSCI.0117-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng-Crank J, Pollock JA, Hayashi I, Tanouye MA (1991) Expression of ion channel genes in Drosophila. J Neurogenet 7:229–239. 10.3109/01677069109167435 [DOI] [PubMed] [Google Scholar]
- Venken KJ, Popodi E, Holtzman SL, Schulze KL, Park S, Carlson JW, Hoskins RA, Bellen HJ, Kaufman TC (2010) A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics 186:1111–1125. 10.1534/genetics.110.121285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, Bellen HJ (2011) MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods 8:737–747. 10.1038/nmeth.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Koh TW, Schulze KL, Zhai RG, Hiesinger PR, Zhou Y, Mehta SQ, Cao Y, Roos J, Bellen HJ (2003) Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron 40:733–748. 10.1016/S0896-6273(03)00644-5 [DOI] [PubMed] [Google Scholar]
- Vuilleumier R, Lian T, Flibotte S, Khan ZN, Fuchs A, Pyrowolakis G, Allan DW (2019) Retrograde BMP signaling activates neuronal gene expression through widespread deployment of a conserved BMP-responsive cis-regulatory activation element. Nucleic Acids Res 47:679–699. 10.1093/nar/gky1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KT, Doe CQ (2017) Drosophila embryonic Type II neuroblasts: origin, temporal patterning, and contribution to the adult central complex. Development 144:4552–4562. 10.1242/dev.157826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Foquet B, Dewell RB, Song H, Dierick HA, Gabbiani F (2020) Molecular characterization and distribution of the voltage-gated sodium channel, Para, in the brain of the grasshopper and vinegar fly. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 206:289–307. 10.1007/s00359-019-01396-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenbroek RE, Merrick DK, Catterall WA (1989) Differential subcellular localization of the RI and RII Na+ channel subtypes in central neurons. Neuron 3:695–704. 10.1016/0896-6273(89)90238-9 [DOI] [PubMed] [Google Scholar]
- Whitaker WR, Clare JJ, Powell AJ, Chen YH, Faull RL, Emson PC (2000) Distribution of voltage-gated sodium channel alpha-subunit and beta-subunit mRNAs in human hippocampal formation, cortex, and cerebellum. J Comp Neurol 422:123–139. [DOI] [PubMed] [Google Scholar]
- Wilson RI, Turner GC, Laurent G (2004) Transformation of olfactory representations in the Drosophila antennal lobe. Science 303:366–370. 10.1126/science.1090782 [DOI] [PubMed] [Google Scholar]
- Winckler B, Forscher P, Mellman I (1999) A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature 397:698–701. 10.1038/17806 [DOI] [PubMed] [Google Scholar]
- Wu M, Nern A, Williamson WR, Morimoto MM, Reiser MB, Card GM, Rubin GM (2016) Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. Elife 5:e21022 10.7554/eLife.21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Chen C, Yu TM, Ou J, Rui M, Zhai Y, He Y, Xue L, Ho MS (2017) Molecular chaperone calnexin regulates the function of Drosophila sodium channel paralytic. Front Mol Neurosci 10:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wisniewski W, Sandoval H, Haelterman NA, Xiong B, Zhang K, Bayat V, David G, Li T, Chen K, Gala U, Harel T, Pehlivan D, Penney S, Vissers LE, et al. (2014) A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159:200–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K (1995) Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron 15:45–54. 10.1016/0896-6273(95)90063-2 [DOI] [PubMed] [Google Scholar]
- Zappia L, Oshlack A (2018) Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience 7:giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Third instar larvae single-cell sequencing metrics, cluster composition and the gene list and GO terms used for analysis. Related to Figures 5-8. Download Extended Data File 1, XLSX file (50KB, xlsx)
Para-GFSTF and para-MiMIC have comparable ERG recordings. ERGs were performed on para-GFSTF and para-MiMIC flies to measure neuronal activity in the visual system. (A) Representative trace of ERGs in 24 h dark conditions, with no significant difference seen in off transients between tagged and untagged animals. (B) No significant difference was seen in the amplitude of on transients in 12 h light/dark conditions or constant dark. The difference in phototransduction amplitude between the genotypes was not significant in 12 h light/dark conditions; in constant dark the amplitude of para-GFSTF flies was slightly larger than para-MiMIC flies. Unpaired, two-tailed t test. Error bars indicate SEM. Download Figure 1-1, TIF file (3.5MB, tif)
Composition of single-cell transcriptomic clusters. Distribution of (A) genotypes and (B) sample fixation type across annotated clusters. (C) Knee-plots showing distribution of valid barcodes and UMIs in fresh and fixed third instar larval brains. Download Figure 5-1, TIF file (1.9MB, tif)
Clusters are stable across multiple resolutions in the Louvain algorithm. Arrows linking clusters show overlap in percentage between clusters from different resolutions. Note that cluster 11 in resolution 0.2 becomes part of cluster 3 in resolution 0.6 but only accounts for 0.05% of cluster 3, making the arrow invisible. The squared box highlights the final chosen resolution. Download Figure 5-2, TIF file (5.7MB, tif)
Distribution of para expressing cells in third instar larvae and adult transcriptomic data. (A) Annotated cell types on Seurat tSNE plot of cells from the adult brain as previously annotated by Davie et al. (2018). AST, astrocyte-like glia; CHI, chiasm glia; CTX, cortex glia; DCN, dorsal cluster neurons; DOP, dopaminergic neurons; ENS, ensheathing glia; HE, hemocytes; IPC, insulin-producing cells; KC, Kenyon cells; OCTY, octopaminergic-tyraminergic neurons; PEP, peptidergic neurons; PR, photoreceptors; PRN, perineurial glia; SER, serotonergic; SUB, subperineurial glia; SUR, surface glia; MBON, MB output neurons (B) Boxplots showing para expression for the different single-cell clusters from the adult brain single-cell transcriptomic data. Download Figure 8-1, TIF file (14.9MB, tif)
Para-RFP-3xHA and Para-mCherry recapitulate the expression of Para-GFSTF in the third instar larval CNS. (A) Immunofluorescent imaging of Para-RFP-3xHA and Para-mCherry subcellular localization shows the same clustered distribution as Para-GFSTF. (B) 201Y-GAL4 labels the membrane of the MB neurons of third instar larvae, labeling these cells with Para-mCherry identifies the cluster of Para in at the DASs, yellow arrow. Download Figure 9-1, TIF file (19.8MB, tif)
Para-GFSTF expression in the third instar larval CNS.
Para-GFSTF expression in the adult CNS.
Data Availability Statement
Transcriptomic data are available for visualization and downloading at http://scope.aertslab.org/#/Larval_Brain/*/welcome and the code is released as open source code on Github. The scRNA-seq data has been deposited in GEO under accession code GEO: GSE157202. SCENIC for Drosophila is available at http://scenic.aertslab.org.